Abstract
The human gut microbiome is a complex ecosystem of fundamental importance to human health. Our increased understanding of gut microbial composition and functional interactions in health and disease states has spurred research efforts examining the gut microbiome as a valuable target for therapeutic intervention. This review provides updated insight into the state of the gut microbiome in recurrent Clostridium difficile infection (CDI), ulcerative colitis (UC), and obesity while addressing the rationale for the modulation of the gut microbiome using fecal microbiota transplant (FMT)-based therapies. Current microbiome-based therapeutics in pre-clinical or clinical development are discussed. We end by putting this within the context of the current regulatory framework surrounding FMT and related therapies.
Keywords: Gut, Microbiome, Dysbiosis, Therapeutic, Fecal, Transplant
Highlights
-
•
There is an increased understanding of gut microbial composition and functional interactions in health and disease states.
-
•
FMT is effective for rCDI and has led to the development of fecal microbiota-based therapeutics for other indications.
-
•
For stool-substitute therapies to enter mainstream medicine, further mechanistic studies, RCTs and regulations are needed.
1. Introduction
Our microbial inhabitants that form the human gut microbiome influence many aspects of our physiology that were previously unrecognized. As the gut microbiome has recently become acknowledged as a virtual ‘organ’ in human biology, many studies have associated problems of this ecosystem with a growing number of both gastrointestinal (GI) and non-GI disease states. Because gut microbial composition tends to remain relatively temporally stable within a given individual, there is potential for this ecosystem to be exploited both as a predictor of health-status and a valuable target for therapeutic intervention (Faith et al., 2013). In this review, we will examine how our increased understanding of gut microbiota has led to a surge in microbiome-based therapeutics for human medicine.
2. The Human Gut Microbiome
The human gut microbiome is the collection of microorganisms, their gene products and corresponding physiological functions found in the human GI tract (Gill et al., 2006). Much of our current understanding of the bacterial composition of the human gut microbiome has come from ‘omics-based’ survey initiatives such as the Human Microbiome Project (Human Microbiome Project Consortium, 2012). These studies have provided great insight into gut microbial diversity, however, understanding of the function of the microbiome has lagged behind. To appreciate how the gut microbiome influences health, we must consider not only the composition of the microbiome but also microbe-microbe and host-microbe interactions.
2.1. How Is a ‘Healthy’ Microbiome Defined?
Our gut microbial inhabitants collectively provide a myriad of beneficial physiological functions; for example, production of vitamins and nutrients, regulation of metabolism, exclusion of pathogens, and maintenance of the immune system (Lozupone et al., 2012). The average healthy adult colon contains a dense microbial community of approximately 160 bacterial species (Qin et al., 2010). Phylogenomic studies of stool samples, primarily from Western populations, have shown that although gut microbial composition varies greatly between individuals there are general trends among healthy adults. Bacteroidetes and Firmicutes are the most prevalent bacterial phyla, while Actinobacteria and Proteobacteria are also prominent members (Human Microbiome Project Consortium, 2012). The large inter-individual variation in the ratios of these representative phyla can be attributed to numerous influences; a recent study found 69 factors that significantly correlated with overall microbiota compositional variation including medication/drug use, health-status, age, sex, lifestyle, host genetics, diet, and animal exposure (Falony et al., 2016).
The functional profile of the gut microbiome is more conserved than the taxonomic composition (Qin et al., 2010). Multiple studies have shown that the overall metabonomic (complex system associated metabolomic profile) (Marchesi and Ravel, 2015) and genetic capabilities of gut microbial ecosystems appear to be somewhat consistent across healthy adults (Human Microbiome Project (HMP) Consortium, 2012, Qin et al., 2010, Turnbaugh et al., 2009). For example, short-chain fatty acids (SCFAs), produced by many gut microbial species through fermentation, provide 10% of our daily energy requirements, regulate host energy demands, intestinal epithelial cell homeostasis, and immune function, and also support the growth of the gut microbiota (Marchesi et al., 2015). Overall, a diverse microbial consortium displaying functional redundancy will preserve and augment essential functions associated with gut homeostasis (Rampelli et al., 2015).
2.2. Gut Microbial ‘Dysbiosis’?
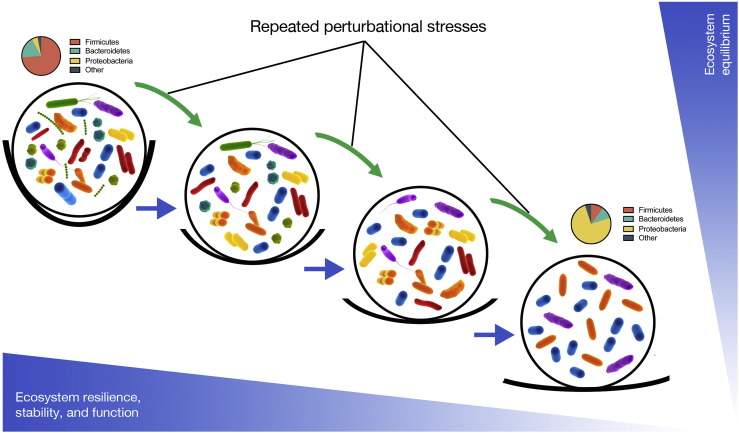
Gut dysbiosis can be defined as an imbalance in a microbial ecosystem characterized by a shift in the composition or function of microbes, which can result in pathogenesis (Fig. 1). Antibiotics, toxic compounds, poor diet, medical interventions, and disease can cause disturbances to, and potentially loss of diversity within the gut microbiota ecosystem (Hell et al., 2013). Diet is a major modulator of the gut microbiota; in the absence of dietary complex carbohydrates (the main substrates for SCFA production), microbial metabolism will shift towards proteolytic fermentation, which results in the generation of potentially toxic, pro-inflammatory compounds such as amines, ammonia, phenols and sulfides (Vipperla and O'Keefe, 2016). Such compounds have been implicated in gut dysbiosis and the development of colorectal cancer and inflammatory bowel disease (IBD) (Vipperla and O'Keefe, 2016). However, defining gut microbial dysbiosis is challenging because of the compositional variability of the microbial ecosystem across different individuals. Although currently there is no biomarker for dysbiosis, recent work suggests that certain correlations may become diagnostically useful. For example, low fecal chromogranin A (CgA) concentrations, alongside higher microbial diversity and functional richness has been found to associate with higher high-density lipoprotein (HDL) concentrations (Zhernakova et al., 2016).
Fig. 1.
Alternative stable states (ball and cup model) of human gut microbial ecosystems. Here we conceptualize the ecological stability of the human gut microbiota as inferred through its species richness. The gut microbial ecosystem is represented as a ball that exists within an equilibrium represented as a cup. The depth of each cup is symbolic of the resilience of a given ecosystem and is related to species richness, where lower diversity leads to lower resilience and the greater the likelihood that the “ball” will roll out of the “cup” into a new state of equilibrium that may be less stable that the first. Perturbational stresses caused by numerous factors such as poor diet, disease state, drug use (including antimicrobials), immunosenescence as examples can impact bacterial diversity and force the ecosystem into a less-stable equilibrium state. Repeated stresses can cause a situation where function may be reduced to the point that dysbiosis and the development of disease ensue.
Adapted from (Relman, 2012) and (Folke et al., 2004).
3. Gut Dysbiosis in Clostridium difficile Infection (CDI)
C. difficile is an anaerobic, sporulating, bacterial pathogen that is the etiological agent of CDI (Abt et al., 2016). CDI is a leading cause of nosocomial, antibiotic-associated diarrhea and the disease pathology is mediated by bacterial secreted toxins (Abt et al., 2016). Under normal circumstances, the resident gut microbiota are thought to facilitate colonization resistance against C. difficile suppressing its pathogenic activity in the colon (Theriot et al., 2014). However, broad-spectrum antibiotic use disrupts host-microbiota homeostasis by decreasing gut microbiota abundance, diversity, community structure, and altering metabonomic functional profiles (Dethlefsen et al., 2008, Perez-Cobas et al., 2013a, Perez-Cobas et al., 2013b, Theriot et al., 2016, Theriot et al., 2014). Many studies have described microbial taxonomic alterations in CDI patients (Table 1). In general, the gut microbiota of CDI patients have reduced overall bacterial diversity, characterized by increases in Proteobacteria with decreases in Firmicutes and Bacteroidetes phyla in comparison to healthy subjects (Antharam et al., 2013, Chang et al., 2008, Fuentes et al., 2014, Rea et al., 2012, Schubert et al., 2014, Shahinas et al., 2012). However, taxonomic changes do not appear to be consistent between distinct patient cohorts likely because of the large inter-individual variation in human gut microbiota (Girotra et al., 2016, Schubert et al., 2014).
Table 1.
Microbial taxa positively correlated to select gut conditions in human subjects.
| Condition | Study size/details | Microbial taxonomic changes associated with conditiona | Reference |
|---|---|---|---|
| CDI | Fecal samples from n = 14 recurrent CDI patients (4 within this group given FMT) | ↑ Proteobacteria, ↓ Bacteroidetes, ↓ Firmicutes |
Weingarden et al., 2015 |
| Fecal samples from n = 94 CDI, and n = 89 non-CDI inpatients | ↑ Proteobacteria, ↓ Firmicutes (except Enterococcus and Erysipelotrichia and some Lachnospiraceae) | Schubert et al., 2014 | |
| Fecal samples from n = 9 recurrent CDI patients given FMT in van Nood et al. (2013) study | ↑ Proteobacteria, ↑ Bacilli, ↓ Bacteroidetes, ↓ Firmicutes, ↓ overall microbial diversity compared to post-FMT and healthy donor samples |
Fuentes et al., 2014 | |
| Fecal samples from n = 22 C. difficile culture-positive elderly subjects, and n = 252 healthy elderly subjects | ↑ Proteobacteria, Firmicutes (↓ Enterococcaceae and ↑ Lactobacillaceae), ↓ overall microbial diversity | Rea et al., 2012 | |
| Fecal samples from n = 39 CDI patients, n = 36 C. difficile-negative nosocomial diarrhea patients, and n = 40 healthy control subjects | ↓ Firmicutes, ↑ Proteobacteria, ↓ Microbial diversity in both diarrheal groups compared to healthy controls |
Antharam et al., 2013 | |
| Stool samples from n = 6 CDI patients given FMT | ↑ Proteobacteria, ↓ Bacteroidetes, ↓ Firmicutes (specifically Lachnospiraceae, ↓ overall microbial diversity |
Shahinas et al., 2012 | |
| UC | Fecal samples from n = 15 UC patients and n = 15 control subjects | ↓ Bacteroidetes, ↓ Firmicutes, ↓ overall microbial diversity |
Rajilic-Stojanovic et al., 2013 |
| RCT on FMT for UC (58 fecal samples total from n = 34 UC patients, and n = 24 healthy donors) | ↓ Firmicutes, ↑ Proteobacteria ↑ Bacteroidetes, ↑ Bacilli, no change in bacterial diversity |
Rossen et al., 2015 | |
| Sequencing completed on inflamed and non-inflamed mucosal biopsies from n = 6 UC patients, and n = 5 healthy controls | ↑ Bacteroidetes, ↓ Firmicutes | Walker et al., 2011 | |
| Fecal samples from n = 36 active UC patients treated with FMT, and n = 34 active UC patients given placebo | Reduced bacterial diversity, no information on phyla, but: ↓ Lachnospiraceae, ↓ Ruminococcus |
Moayyedi et al., 2015 | |
| Sigmoid colon biopsies from n = 8 monozygotic twin pairs discordant for UC, and n = 10 healthy individuals | ↑ Actinobacteria, ↑ Proteobacteria (↓ Bacteroidetes), no change in Firmicutes, ↓ Overall bacterial diversity |
Lepage et al., 2011 | |
| Obesity | Gut microbial communities profiled from n = 31 monozygotic twin pairs, and n = 23 dizygotic twin pairs generally concordant for obesity or leanness | ↓ Bacteroidetes, ↑ Actinobacteria, no change in Firmicutes | Turnbaugh et al., 2009 |
| Stool samples from n = 2 lean, and n = 12 obese individuals | ↓ Bacteroidetes, ↑ Firmicutes | Ley et al., 2006 | |
| Stool samples from n = 1 normal-weight adolescent, and n = 1 obese adolescent | ↓ Bacteroidetes, ↑ Firmicutes | Ferrer et al., 2013 | |
| Collected fecal samples from n = 52 lean, and n = 42 overweight or obese individuals | No change in Firmicutes to Bacteroidetes (F:B) ratio between lean and obese groups | Fernandes et al., 2014 | |
| Stool samples from n = 30 normal range, n = 35 overweight, n = 33 obese (defined by BMI) | ↑ Bacteroidetes, no change in Firmicutes | Schwiertz et al., 2010 | |
| Stool samples from n = 9 metabolic syndrome patients undergoing allogenic gut microbiota (n = 9) from healthy lean individuals or autologous FMT (n = 9) | ↓ Overall bacterial diversity, and ↑ Bacteroidetes, ↓ Lachnospiraceae in obese subjects compared to lean prior to FMT | Vrieze et al., 2012 |
Abbreviations: Body mass index (BMI); fecal microbiota transplantation (FMT); Clostridium difficile infection (CDI); randomized controlled clinical trial (RCT); ulcerative colitis (UC).
Phylum level taxonomic changes are listed unless otherwise described.
Metabonomic alterations as a result of antibiotic use are important in the context of CDI (Theriot et al., 2014). As an example, primary and secondary bile acids have markedly different effects on C. difficile; primary bile acids promote growth of this pathogen while secondary bile acids are inhibitory. Thus, an antibiotic-induced perturbation that reduces secondary bile acid production can promote CDI (Theriot et al., 2016). In addition, mucosal carbohydrate concentrations, which can increase when antibiotics are present, can promote C. difficile expansion (Ng et al., 2013).
The standard treatment for CDI is metronidazole or oral vancomycin, which kill C. difficile, providing symptomatic relief and allowing restoration of colonization resistance (Surawicz et al., 2013). However, in approximately 25–30% of cases, persistent C. difficile (as endospores) or introduction of a new strain can lead to disease recurrence, which is more likely when there is incomplete recovery of the gut microbiota (Cornely et al., 2012). Management of recurrent CDI (rCDI) is a major clinical challenge; patients that are non-responsive to antimicrobial therapy require alternative treatment options (Lapointe-Shaw et al., 2016).
4. Gut Dysbiosis in Ulcerative Colitis (UC)
UC, a form of IBD, is a chronic, relapsing, idiopathic, inflammatory disorder of the colon and rectum (Bouma and Strober, 2003). In the last decade, there have been increases in UC incidence and prevalence worldwide, making it an important emerging global disease (Molodecky et al., 2012). Although the pathogenesis of UC is complex, multifactorial, and not fully understood, aberrant host immune responses, a dysfunctional intestinal barrier, and gut microbiota dysbiosis have been associated with this condition (Lepage et al., 2011, Michail et al., 2012).
Various studies have demonstrated a reduction of gut microbial diversity and taxonomic compositional differences in UC patients compared to healthy individuals, although less stereotypical trends are observed in taxonomic variation in comparison with CDI patients (Table 1). Notably, UC has been associated with decreases in species within the Lachnospiraceae family, and Faecalibacterium prausnitzii (Lepage et al., 2011, Rossen et al., 2015). As known butyrate producers, their reduction may account for the depletion of this SCFA observed in some UC patients (Lepage et al., 2011). Additionally, Akkermansia muciniphila, a commensal gut microbe that contributes to the homeostatic balance of the intestinal mucus layer was found to be reduced in UC patients (Png et al., 2010) potentially resulting in decreased mucosal thickness corroborating histologic findings in rectal biopsies from active UC patients (Pullan et al., 1994). Driven by observations of gut microbial dysbiosis in UC, there is interest in the development and application of microbiome-based therapeutics for this indication.
5. Gut Dysbiosis in Obesity
Obesity is a severe worldwide public health issue that has reached epidemic status in many industrialized countries (Villanueva-Millan et al., 2015), and is defined as the accumulation of excess adipose tissue to the detriment of health (Boulange et al., 2016) defined as a body mass index (BMI) > 30 in adults. Dietary intake is a principal contributor to the pathophysiology of obesity, however, gut microbes can modulate nutrient uptake and energy regulation providing an important, albeit underexplored environmental factor in metabolic syndrome and obesity-related disorders (Boulange et al., 2016). The connection between obesity and the gut microbiome is complicated by heterogeneity of patient diet, genetics, age, lifestyle, hormones, and disease as well as the complexity of clinical presentation of metabolic disorders (Boulange et al., 2016).
The gut microbiome has been implicated as an important player in obesity following the seminal discovery that the ratio of Firmicutes to Bacteroidetes (F:B), and energy harvest capacity differs in obese versus lean animal and human subjects (Ley et al., 2006, Turnbaugh et al., 2009). However, other studies of this link have provided conflicting results (Duncan et al., 2008, Fernandes et al., 2014, Schwiertz et al., 2010), thus, currently, there is no consensus of the importance of the F:B ratio as an indicator of obesity. In spite of this, many groups have found that both obese animal and human subjects have an altered gut microbiota compared to their lean counterparts characterized by reduced bacterial diversity, and altered colonic fermentation potential (Fernandes et al., 2014, Le Chatelier et al., 2013, Schwiertz et al., 2010, Teixeira et al., 2013, Vrieze et al., 2012) (Table 1). Metagenomically, the gut microbiome can be classified into two groups: high gene count (HGC) or low gene count (LGC). The HGC group, most often seen in lean individuals, was associated with increased F. prausnitzii and butyrate (SCFA) levels, while the LGC group, most often seen in obese individuals, was associated with lower butyrate production, but higher levels of Bacteroides spp. and Ruminococcus gnavus and more genes putatively associated with pro-carcinogenic metabolite production and oxidative stress (Le Chatelier et al., 2013). Diet-induced weight loss interventions in the LGC group could partially restore these metabolic changes, illustrating the plasticity of the gut microbiome and the significant impact of diet in obesity-related diseases (Cotillard et al., 2013).
6. Modulation of the Gut Microbiome Using Stool-based Therapeutics
In Table 2, we list representative examples of live microbial preparations and microbial-based products that are currently being developed with therapeutic manipulation of the gut microbiome or its associated host interactions in mind. Fecal-microbiota derived products, probiotic, prebiotic and targeted molecule therapeutics have been developed for a variety of indications. In the following section, we specifically discuss the use of complex stool-based therapeutics as the most commonly used treatment modalities to date.
Table 2.
Select gut microbiome-based therapies in clinical or pre-clinical testing.
| Active agent | Presumed mechanism of action | Indication | Research status | Company/innovator | Reference/clinical trial identifiera |
|---|---|---|---|---|---|
| Stool | Restoration of colonization resistance through FMT | CDI | Non-profit stool bank, screening, and FMT product development | OpenBiome | Kazerouni et al., 2015 |
| Stool bank and screening | Advancing Bio | AdvancingBio, 2016 | |||
| RBX2660 | FMT-derived bacteria to restore gut microbiota | CDI, IBD | Phase II RCT complete for CDI | Rebiotix Inc. | NCT01925417 |
| MET-1 | 33 strain defined bacterial ecosystem that restores colonization resistance | Recurrent CDI | Proof-of-principle clinical study | NuBiyota | Petrof et al., 2013 |
| SER-109 | Purified bacterial spore-based ecological formulation to restore gut microbiota after antibiotic treatment | Recurrent CDI | Missed primary endpoint in phase II RCT | Seres Therapeutics Inc. | Khanna et al., 2016; NCT02437487 |
| SER-287 | Mild-to-moderate UC | Phase I RCT | NCT02618187 | ||
| VE-202 | Oral formulation of live Clostridial bacteria that restore immune system homeostasis (increase regulatory T cells) | IBD | Pre-clinical | Vedanta | Atarashi et al., 2013, Furusawa et al., 2013 |
| CBM588 | Clostridium butyricum bacterial probiotic | Pediatric AAD | Phase II RCT | Osel Inc. | Seki et al., 2003 |
| UC with pouchitis | Phase II RCT | Yasueda et al., 2016 | |||
| Lactin-V | Lactobacillus crispatus bacterial probiotic | Bacterial vaginosis | Phase II RCT complete | NCT00635622; Hemmerling et al., 2010 | |
| Recurrent UTIs | Phase II RCT complete | NCT00305227; Stapleton et al., 2011 | |||
| Lactobacillus plantarum | Probiotic to modulate the gut microbiome | CVD and obesity | Phase II RCT complete | OptiBiotix Health PLC | Patent #: WO2015067948A1 |
| CNDO-201 | Trichuris suis ova, nonpathogenic helminth probiotic to modulate immune system | Crohn's disease | Missed primary endpoints in two phase II RCTs | Fortress Biotech | NCT01576471; NCT01279577 |
| Blautix | Proprietary live bacterial therapeutic | IBS | Phase I RCT complete | 4D Pharma Research Ltd. | RNS Number: 4931E |
| Thetanix | Anti-inflammatory protein derived from Bacteroides thetaiotaomicron | Pediatrics Crohn's disease | Phase I RCT | NCT02704728; Eudract 2014-005666-29 | |
| SYN-004 | Targeted molecule that degrades IV β-lactam antibiotics specifically in gut to protect microbiome | CDI, antibiotic-resistant infections and AAD | Phase II RCT | Synthetic Biologics Inc. | NCT02563106 |
| Avidocins | Genetically modified R-type bacteriocins that have specific antibacterial activity | CDI, foodborne pathogens | Pre-clinical | AvidBiotics | Gebhart et al., 2015, Scholl et al., 2012, Scholl et al., 2009 |
| AG014 | Genetically modified Lactococcus lactis probiotic strain that secretes anti-inflammatory factors | IBD | Phase I RCT complete | Intrexon | Vandenbroucke et al., 2010 |
| SHP-01 | Narrow-spectrum lysin (antimicrobial enzyme) specifically targeting C. difficile | Acute and recurrent CDI | Pre-clinical | Symbiotic Health | Hirsch et al., 2015, Wang et al., 2015 |
| VT301 | Genetically-engineered lactic acid bacterial probiotic that produce elafin to heal gut epithelial lining | IBD | Pre-clinical | ViThera Pharmaceuticals | Motta et al., 2011 |
| SGM-1019 | Small molecule inhibitor that modulates host-microbiota interactions | IBD | Phase I RCT complete | Second Genome | Ratner, 2015 |
| NM504/505 | Modulating the gut microbiome using prebiotics to improve glucose tolerance and other metabolic parameters | Metformin-intolerant type 2 diabetes | Phase 0 RCT | MicroBiome Therapeutics | NCT01866462; Burton et al., 2015 |
Abbreviations: Antibiotic-associated diarrhea (AAD); Clostridium difficile infection (CDI); cardiovascular disease (CVD); fecal microbiota transplant (FMT); inflammatory bowel disease (IBD); irritable bowel syndrome (IBS); randomized controlled clinical trial (RCT); ulcerative colitis (UC); urinary tract infection (UTI).
Applicable clinical trial registry numbers or primary literature references are included where publicly available.
6.1. Fecal Microbiota Transplantation (FMT)
FMT aims to restore gut microbiota diversity by transferring feces from a healthy donor to a sick patient. The therapy has been extensively used in the treatment of rCDI with good success, likely because the donated gut microbial ecosystem can replace the microbiota lost through antibiotic use and thus suppress C. difficile overgrowth, promoting patient recovery (Seekatz et al., 2014). Recent studies have demonstrated effectiveness of FMT for clinical cure of rCDI around 90% (Agrawal et al., 2016, Kelly et al., 2016, van Nood et al., 2013, Youngster et al., 2016, Youngster et al., 2014). Failure of a first FMT is strongly associated with previous history of CDI-related hospitalization events, preexisting IBD, being an inpatient while receiving FMT, or severe/complicated rCDI status (Fischer et al., 2016, Khoruts et al., 2016).
Donor strains introduced into the GI tract via FMT durably colonize and establish themselves alongside or in place of the pre-existing microbiota (Li et al., 2016, Seekatz et al., 2014). In general, decreases in Proteobacteria, and increases in Firmicutes and Bacteroidetes have been widely observed in CDI patients following FMT (Fuentes et al., 2014, Hamilton et al., 2013, Kelly et al., 2016, Khoruts et al., 2010, Seekatz et al., 2014, Shahinas et al., 2012, van Nood et al., 2013, Weingarden et al., 2015) (see Table 1). Butyrate-producing bacteria, mainly from Ruminococcaceae and Lachnospiraceae families, are depleted in CDI (Antharam et al., 2013), and the observation that FMT can restore SCFA production by the gut microbiota (Fuentes et al., 2014) has encouraged the investigation of metabonomic changes in the gut microbiome post-FMT. In one study it was shown that post-FMT patient metabolome profiles shifted from their pre-treatment state to closely resemble their donor metabolome counterparts (Weingarden et al., 2014). Increased amounts of secondary bile acids were found to be the main FMT-induced metabolic change, suggesting that restoring bile acid composition with FMT may play an important role in treating rCDI (Weingarden et al., 2014).
Given the success of treating rCDI with FMT, there has been great interest in extending this treatment modality to other gut diseases, notably UC (Table 2). However, while CDI clearly results from a simple breakdown in gut microbiota diversity, the underlying cause of UC is as yet unknown but likely to be more complex in etiology (Petrof and Khoruts, 2014). Data from individual case studies have suggested that FMT may be of benefit for UC, however there are mixed results from RCTs. To date, FMT has been assessed as a novel therapeutic for UC in two RCTs (Moayyedi et al., 2015, Rossen et al., 2015). The first study found that 24% of patients who received FMT and 5% who received placebo entered remission after 7 weeks, and identified that FMT patients had increased microbial diversity in comparison to patients given placebo. An interesting trend was noted in that the majority of patients who went into remission after FMT received stool from a single healthy donor, suggesting particular benefits of this donor's microbiota above others (Moayyedi et al., 2015). Although the study did not reach its primary efficacy endpoint, it attained significance for the end point of remission. Interestingly, a greater success in inducing remission was seen in patients who had been recently diagnosed with UC (within the year previous to treatment) compared to patients with a longer history of disease (75% vs. 18%), suggesting a temporal window when UC patients may be more receptive to FMT (Moayyedi et al., 2015).
In contrast, Rossen et al. showed no statistically significant difference in clinical remission between UC patients who received FMT sourced from healthy donors or autologous FMT (their own fecal microbiota), and only 41% of patients who received donor FMT achieved clinical and endoscopic remission. The authors of this study carried out a detailed assessment of the fecal microbiota taxonomic composition pre- and post-FMT and demonstrated that the microbial ecosystems of patients who responded to FMT from a healthy donor increased in the numbers of bacterial species from Clostridium clusters IV, XIVa, XVIII while the amount of Bacteroidetes species was reduced (Rossen et al., 2015).
UC has a complex etiology and it is not yet understood whether gut microbial dysbiosis is a cause or effect of disease; this may underlie the apparent range of outcomes after FMT. When treatment is successful in inducing remission, the heterogeneity of patient, donor, and mode of delivery complicate the understanding of which factors contribute to optimal clinical resolution.
Overall, there have been a limited number of studies evaluating the effectiveness of FMT for the treatment of obesity. When microbiota contents were transferred from obese mice into lean germ-free mice, the recipient mice showed an increase in adiposity compared to control animals (Turnbaugh et al., 2006). Similarly, the transfer of stool from monozygotic or dizygotic twins discordant for obesity into ‘humanized’ germ-free mice resulted in increased adiposity and the transmission of obesity-associated metabolic phenotypes (Ridaura et al., 2013). Because the gut microbiota imparts such clear phenotypic changes to distinct recipient mammalian hosts, this holds promise for the therapeutic manipulation of the microbiome to treat obesity-related disorders. There is currently only one published report investigating the effects FMT has on metabolic syndrome in human subjects. In this small RCT, FMT from healthy lean donors to patients with metabolic syndrome resulted in increased insulin sensitivity 6 weeks after transfer (Vrieze et al., 2012). This improvement was associated with increased gut microbial diversity (including butyrate-producing bacteria, particularly Roseburia intestinalis) (Vrieze et al., 2012). Butyrate is known to play an important role in promoting insulin sensitivity in mice (Lin et al., 2012), thus it is possible that an increase in butyrate production may be responsible for the observed therapeutic effect of FMT (Vrieze et al., 2012). Further studies investigating global compositional and metabolic changes resulting from FMT are ongoing and will be necessary to bring targeted-microbiome therapeutics into clinical practice.
6.2. Stool-substitute Therapies
Stool-substitute therapies aim to combine the high-effectiveness of FMT with the excellent safety-profile of probiotics to mitigate many of the concerns of each respective therapeutic option. To date, this approach has been used only for the treatment of rCDI. Stool-substitute therapies use defined, standardized preparations of stool-derived products, while retaining the compositional, metabolic and transcriptomic properties of fecal communities. Tvede and Rask-Madsen were among the first to use a stool-derived microbial consortium of 10 bacterial strains to cure six patients with CDI (Tvede and Rask-Madsen, 1989). However, at the time of the study, CDI was considered an uncommon complication of antibiotic use and no real progress was made in the field for almost three decades.
In a small proof-of-principle trial we demonstrated a successful cure of two rCDI patients, using a stool-substitute preparation of 33 purified bacterial strains (MET-1) which had been derived from a single, healthy donor (Petrof et al., 2013). As for FMT, the microbiota profiles of the treated patients indicated incorporation of the MET-1 composition strains after a single administration of the therapeutic. Signatures identifying with MET-1 strains were present in the patients 6 months post-treatment, suggesting that a subset was able to colonize the recipients; this sets MET-1 apart from conventional probiotics assessed to date, and highlights the therapeutic potential representative of a multi-species microbial ecosystem (Petrof et al., 2013). Seres Therapeutics Inc. has also developed stool-substitute therapies for rCDI with a varied approach by using a spore-based microbial ecosystem formulation. Results from their first RCT with product SER-109 were promising, however interim results from their phase II RCT have been largely disappointing (Seres Therapeutics Inc., 2016). In general, as with FMT, the utility of stool-substitute therapies for treatment of rCDI requires further research to understand, and capitalize upon, their mechanisms of action.
7. Regulation of Stool-based Therapeutics
There is persuasive evidence for the use of FMT in rCDI, and perhaps also for a number of different diseases where the gut microbiota may play a pivotal role. FMT is cost effective and now recommended for treating rCDI nonresponsive to conventional antibiotic therapy (Health Quality Ontario (HQO), 2016, Lapointe-Shaw et al., 2016, Merlo et al., 2016). As such, two registered stool banks have emerged in the USA that screen and provide FMT products for use in the treatment of rCDI (Kazerouni et al., 2015, Advancing Bio, 2016) (Table 2). These organizations aim to facilitate and expand FMT access, nonetheless, the FMT regulatory landscape poses one of the biggest obstacles to efficient delivery of FMT to rCDI patients, as well as to the exploration of potential uses of FMT for other diseases.
In many countries that recommend FMT for rCDI (e.g. England, Austria, Australia), FMT is not considered a drug by regulatory bodies (The Gastroenterological Society of Australia (GESA), 2015, Kump et al., 2014, The National Institute for Health and Care Excellence (NICE). In contrast, Health Canada, the Federal Drug Administration, (FDA, USA), and the Agence nationale de sécurité du médicament, (ANSM, France) all classify FMT as a drug (L'Agence nationale de sécurité du médicament et des produits, Federal Drug Administration (FDA), 2013, Health Canada, 2015). Because fecal material is extremely complex containing living microbes, microbial metabolites, host cells, and food particles; it is not inert and thus cannot be standardized as a drug in terms of pharmacokinetics, pharmacodynamics, or consistency of chemical composition. Labeled as a drug, storage of FMT falls under the jurisdiction of the pharmacist, which has caused problems for implementing FMT in North America but perhaps more so in France, where the preparation and documentation of FMT material is the responsibility of the pharmacist (Kump et al., 2014). Pharmacies not equipped for preparation of FMT material must coordinate with a microbiology laboratory, adding an additional layer of complexity. In addition, for tracking purposes donor fecal samples must be stored for a minimum of two years. Across regulatory bodies, there is also no clear consensus on donor screening tests. The ANSM is the most specific, requiring a two-step screening process and indicates which types of culture media and PCR tests should be employed (ANSM, 2014). Health Canada provides less specific recommendations, suggesting a list of potential pathogens for testing at the discretion of the treating physician (Health Canada, 2015).
Currently the FDA will allow FMT for the treatment of rCDI without the need for an Investigational New Drug (IND) application, provided the donor and donor stool are adequately screened, and the health care provider obtains adequate consent from the patient after discussion of risks (FDA, 2013). An IND is required if the above conditions are not met, and for FMT for any indication other than rCDI. Health Canada has released similar restrictions (Health Canada, 2015). In March 2016, the FDA proposed an additional “enforcement discretion” clause that requires the donor be “known” to the treating physician, i.e. that the FMT product used is not obtained from a stool bank (IDSA, 2016). These restrictions, while aimed at improving safety, create delays, as healthy donors need to be sourced and screened. The FDA currently continues to exercise “enforcement discretion”, however, the Infectious Disease Society of America (IDSA) has recently requested that IND regulations for using FMT from stool banks to treat rCDI be waived, as they may cause potentially fatal delays in providing treatment for rCDI (IDSA, 2016). IDSA also pointed out that stool banks very comprehensively screen their donors. Indeed, the development of centralized, registered stool banks, with rigorous donor screening and FMT processing protocols in place, would be more efficient overall.
Another issue raised by the IDSA and others is the urgent need to establish a national FMT registry in order to monitor safety and long-term adverse events of FMT (Hecht et al., 2014, Vyas et al., 2015). Earlier this year, the American Gastroenterological Association (AGA) received funding from the National Institutes of Health (NIH) for creation of the first national registry for patients receiving FMT (AGA, 2016). The registry, designed to assess efficacy, patient outcomes and safety, aims to collect data from over 4000 patients over a 10-year period and will begin prospectively collecting clinical data starting in 2017. Such registries have been established for other human tissue products (e.g. blood, semen, breastmilk and organs). The FDA regulates each of these products differently, and the development of a new category of “tissue” for FMT, as advocated by many (Vyas et al., 2015) would address most of the issues surrounding standardization of the practice of FMT. Though regulatory agencies have yet to adopt this stance for FMT, new knowledge obtained from such a registry may provide valuable information in this regard.
This leaves us with the emerging approach of stool-substitute therapies. Several groups are pursuing this avenue of research (Table 2) in the hope of expediting the commercial availability of an effective FMT-like treatment for rCDI that lacks many of its drawbacks.
Regulatory bodies acknowledge that their requirements are not perfect; their goal is to provide optimal safety conditions for the FMT recipient based on the current state of knowledge on FMT, and they recognize that more clinical trials and long-term outcome data are needed. However, regulation must be carefully balanced with the fact that stool is widely available, and overly restrictive policies will drive patients to seek DIY approaches, easily found on the web, which can be dangerous without appropriate medical oversight. As regulatory bodies continue to grapple with the most appropriate way to regulate FMT, either there will need to be a change in regulations to better reflect the complex nature of stool, or the field will be forced to move more towards developing defined mixtures and away from FMT.
8. Outstanding Questions
Given the potential of microbiome-based products for rCDI, there is interest in examining whether stool-derived therapeutics will prove beneficial for other indications including inflammatory intestinal diseases. As we continue to unravel the mechanisms behind the effectiveness of stool-derived therapies for rCDI, many findings emphasize the importance of a robust gut microbiota to host health. Investigating whether such mechanisms hold true for other GI and non-GI indications are important future research directions to pursue. As FMT has been adopted into clinical practice in many countries, the proper standardization of FMT across regulatory bodies, for example through the use of standardized preparations, guidelines surrounding indication, and a better definition of what is considered to be a healthy microbiota will be necessary for efficient, consistent and safe use of this treatment modality (Abt et al., 2016).
9. Search Strategies and Selection Criteria
Data for this review were identified by searches of PubMed, and references from relevant articles using the search terms “gut microbiome”, “gut microbiota”, “antibiotics”, “probiotics”, “fecal microbiota transplant”, “clostridium difficile”, “ulcerative colitis”, “obesity”. Unpublished reports were included in the FMT regulatory section. An exhaustive web search was conducted to compile the microbiome-based therapeutics in pre-clinical or clinical testing described in Table 2.
Contributor Information
Christian Carlucci, Email: ccarlucc@uoguelph.ca.
Elaine O. Petrof, Email: eop@queensu.ca.
Emma Allen-Vercoe, Email: eav@uoguelph.ca.
References
- Abt M.C., McKenney P.T., Pamer E.G. Clostridium difficile colitis: pathogenesis and host defence. Nat. Rev. Microbiol. 2016;10:609–620. doi: 10.1038/nrmicro.2016.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Advancing Bio 2016. https://http://www.advancingbio.org
- Agrawal M., Aroniadis O.C., Brandt L.J., Kelly C., Freeman S., Surawicz C., Broussard E., Stollman N., Giovanelli A., Smith B., Yen E., Trivedi A., Hubble L., Kao D., Borody T., Finlayson S., Ray A., Smith R. The long-term efficacy and safety of fecal microbiota transplant for recurrent, severe, and complicated Clostridium difficile infection in 146 elderly individuals. J. Clin. Gastroenterol. 2016;5:403–407. doi: 10.1097/MCG.0000000000000410. [DOI] [PubMed] [Google Scholar]
- American Gastroenterological Association (AGA) AGA establishes NIH-funded registry to track fecal microbiota transplants. 2016. http://www.gastro.org/press_releases/aga-establishes-nih-funded-registry-to-track-fecal-microbiota-transplants
- Antharam V.C., Li E.C., Ishmael A., Sharma A., Mai V., Rand K.H., Wang G.P. Intestinal dysbiosis and depletion of butyrogenic bacteria in Clostridium difficile infection and nosocomial diarrhea. J. Clin. Microbiol. 2013;9:2884–2892. doi: 10.1128/JCM.00845-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atarashi K., Tanoue T., Oshima K., Suda W., Nagano Y., Nishikawa H., Fukuda S., Saito T., Narushima S., Hase K., Kim S., Fritz J.V., Wilmes P., Ueha S., Matsushima K., Ohno H., Olle B., Sakaguchi S., Taniguchi T., Morita H., Hattori M., Honda K. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;7461:232–236. doi: 10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- Boulange C.L., Neves A.L., Chilloux J., Nicholson J.K., Dumas M.E. Impact of the gut microbiota on inflammation, obesity, and metabolic disease. Genome. Med. 2016;1:42. doi: 10.1186/s13073-016-0303-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouma G., Strober W. The immunological and genetic basis of inflammatory bowel disease. Nat. Rev. Immunol. 2003;7:521–533. doi: 10.1038/nri1132. [DOI] [PubMed] [Google Scholar]
- Burton J.H., Johnson M., Johnson J., Hsia D.S., Greenway F.L., Heiman M.L. Addition of a gastrointestinal microbiome modulator to metformin improves metformin tolerance and fasting glucose levels. J. Diabetes Sci. Technol. 2015;4:808–814. doi: 10.1177/1932296815577425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J.Y., Antonopoulos D.A., Kalra A., Tonelli A., Khalife W.T., Schmidt T.M., Young V.B. Decreased diversity of the fecal microbiome in recurrent Clostridium difficile-associated diarrhea. J. Infect. Dis. 2008;3:435–438. doi: 10.1086/525047. [DOI] [PubMed] [Google Scholar]
- Cornely O.A., Miller M.A., Louie T.J., Crook D.W., Gorbach S.L. Treatment of first recurrence of Clostridium difficile infection: fidaxomicin versus vancomycin. Clin. Infect. Dis. 2012;S154-61 doi: 10.1093/cid/cis462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotillard A., Kennedy S.P., Kong L.C., Prifti E., Pons N., Le Chatelier E., Almeida M., Quinquis B., Levenez F., Galleron N., Gougis S., Rizkalla S., Batto J.M., Renault P., Dore J., Zucker J.D., Clement K., Ehrlich S.D. Dietary intervention impact on gut microbial gene richness. Nature. 2013;7464:585–588. doi: 10.1038/nature12480. [DOI] [PubMed] [Google Scholar]
- Dethlefsen L., Huse S., Sogin M.L., Relman D.A. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008;11:e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan S.H., Lobley G.E., Holtrop G., Ince J., Johnstone A.M., Louis P., Flint H.J. Human colonic microbiota associated with diet, obesity and weight loss. Int. J. Obes. 2008;11:1720–1724. doi: 10.1038/ijo.2008.155. [DOI] [PubMed] [Google Scholar]
- Faith J.J., Guruge J.L., Charbonneau M., Subramanian S., Seedorf H., Goodman A.L., Clemente J.C., Knight R., Heath A.C., Leibel R.L., Rosenbaum M., Gordon J.I. The long-term stability of the human gut microbiota. Science. 2013;6141:1237439. doi: 10.1126/science.1237439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falony G., Joossens M., Vieira-Silva S., Wang J., Darzi Y., Faust K., Kurilshikov A., Bonder M.J., Valles-Colomer M., Vandeputte D., Tito R.Y., Chaffron S., Rymenans L., Verspecht C., de Sutter L., Lima-Mendez G., D'Hoe K., Jonckheere K., Homola D., Garcia R., Tigchelaar E.F., Eeckhaudt L., Fu J., Henckaerts L., Zhernakova A., Wijmenga C., Raes J. Population-level analysis of gut microbiome variation. Science. 2016;6285:560–564. doi: 10.1126/science.aad3503. [DOI] [PubMed] [Google Scholar]
- Federal Drug Administration (FDA) Enforcement policy regarding investigational new drug requirements for use of fecal microbiota for transplantation to treat Clostridium difficile infection not responsive to standard therapies. 2013. http://www.fda.gov/downloads/BiologicsBloodVaccines/GuidanceComplianceRegulatoryInformation/Guidances/Vaccines/UCM361393.pdf
- Fernandes J., Su W., Rahat-Rozenbloom S., Wolever T.M., Comelli E.M. Adiposity, gut microbiota and faecal short chain fatty acids are linked in adult humans. Nutr. Diabetes. 2014 doi: 10.1038/nutd.2014.23. (e121-e121) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrer M., Ruiz A., Lanza F., Haange S.B., Oberbach A., Till H., Bargiela R., Campoy C., Segura M.T., Richter M., von Bergen M., Seifert J., Suarez A. Microbiota from the distal guts of lean and obese adolescents exhibit partial functional redundancy besides clear differences in community structure. Environ. Microbiol. 2013;1:211–226. doi: 10.1111/j.1462-2920.2012.02845.x. [DOI] [PubMed] [Google Scholar]
- Fischer M., Kao D., Mehta S.R., Martin T., Dimitry J., Keshteli A.H., Cook G.K., Phelps E., Sipe B.W., Xu H., Kelly C.R. Predictors of early failure after fecal microbiota transplantation for the therapy of Clostridium difficile infection: a multicenter study. Am. J. Gastroenterol. 2016;7:1024–1031. doi: 10.1038/ajg.2016.180. [DOI] [PubMed] [Google Scholar]
- Folke C., Carpenter S., Walker B., Scheffer M., Elmqvist T., Gunderson L., Holling C.S. Regime shifts, resilience, and biodiversity in ecosystem management. Annu. Rev. Ecol. Evol. Syst. 2004;1:557–581. [Google Scholar]
- Fuentes S., van Nood E., Tims S., Heikamp-de Jong I., ter Braak C.J., Keller J.J., Zoetendal E.G., de Vos W.M. Reset of a critically disturbed microbial ecosystem: faecal transplant in recurrent Clostridium difficile infection. ISME J. 2014;8:1621–1633. doi: 10.1038/ismej.2014.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furusawa Y., Obata Y., Fukuda S., Endo T.A., Nakato G., Takahashi D., Nakanishi Y., Uetake C., Kato K., Kato T., Takahashi M., Fukuda N.N., Murakami S., Miyauchi E., Hino S., Atarashi K., Onawa S., Fujimura Y., Lockett T., Clarke J.M., Topping D.L., Tomita M., Hori S., Ohara O., Morita T., Koseki H., Kikuchi J., Honda K., Hase K., Ohno H. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;7480:446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- Gebhart D., Lok S., Clare S., Tomas M., Stares M., Scholl D., Donskey C.J., Lawley T.D., Govoni G.R. A modified R-type bacteriocin specifically targeting Clostridium difficile prevents colonization of mice without affecting gut microbiota diversity. MBio. 2015;2 doi: 10.1128/mBio.02368-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill S.R., Pop M., Deboy R.T., Eckburg P.B., Turnbaugh P.J., Samuel B.S., Gordon J.I., Relman D.A., Fraser-Liggett C.M., Nelson K.E. Metagenomic analysis of the human distal gut microbiome. Science. 2006;5778:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girotra M., Garg S., Anand R., Song Y., Dutta S.K. Fecal microbiota transplantation for recurrent Clostridium difficile infection in the elderly: long-term outcomes and microbiota changes. Dig. Dis. Sci. 2016;10:3007–3015. doi: 10.1007/s10620-016-4229-8. [DOI] [PubMed] [Google Scholar]
- Hamilton M.J., Weingarden A.R., Unno T., Khoruts A., Sadowsky M.J. High-throughput DNA sequence analysis reveals stable engraftment of gut microbiota following transplantation of previously frozen fecal bacteria. Gut Microbes. 2013;2:125–135. doi: 10.4161/gmic.23571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Health Canada Guidance document: fecal microbiota therapy used in the treatment of Clostridium difficile infection not responsive to conventional therapies. 2015. http://www.hc-sc.gc.ca/dhp-mps/consultation/biolog/fecal_microbiota-bacterio_fecale-eng.php
- Health Quality Ontario (HQO) Fecal microbiota therapy for Clostridium difficile infection: OHTAC recommendation. 2016. http://www.hqontario.ca/evidence-to-improve-care/recommendations-and-reports/OHTAC/fecal-microbiota-therapy [PMC free article] [PubMed]
- Hecht G.A., Blaser M.J., Gordon J., Kaplan L.M., Knight R., Laine L., Peek R., Sanders M.E., Sartor B., Wu G.D., Yang V.W. What is the value of a food and drug administration investigational new drug application for fecal microbiota transplantation to treat Clostridium difficile infection? Clin. Gastroenterol. Hepatol. 2014;2:289–291. doi: 10.1016/j.cgh.2013.10.009. [DOI] [PubMed] [Google Scholar]
- Hell M., Bernhofer C., Stalzer P., Kern J.M., Claassen E. Probiotics in Clostridium difficile infection: reviewing the need for a multistrain probiotic. Benefic. Microbes. 2013;1:39–51. doi: 10.3920/BM2012.0049. [DOI] [PubMed] [Google Scholar]
- Hemmerling A., Harrison W., Schroeder A., Park J., Korn A., Shiboski S., Foster-Rosales A., Cohen C.R. Phase 2a study assessing colonization efficiency, safety, and acceptability of Lactobacillus crispatus CTV-05 in women with bacterial vaginosis. Sex. Transm. Dis. 2010;12:745–750. doi: 10.1097/OLQ.0b013e3181e50026. [DOI] [PubMed] [Google Scholar]
- Hirsch B.E., Saraiya N., Poeth K., Schwartz R.M., Epstein M.E., Honig G. Effectiveness of fecal-derived microbiota transfer using orally administered capsules for recurrent Clostridium difficile infection. BMC Infect. Dis. 2015;191-191 doi: 10.1186/s12879-015-0930-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Human Microbiome Project (HMP) Consortium Structure, function and diversity of the healthy human microbiome. Nature. 2012;7402:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazerouni A., Burgess J., Burns L.J., Wein L.M. Optimal screening and donor management in a public stool bank. Microbiome. 2015;75 doi: 10.1186/s40168-015-0140-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly C.R., Khoruts A., Staley C., Sadowsky M.J., Abd M., Alani M., Bakow B., Curran P., McKenney J., Tisch A., Reinert S.E., Machan J.T., Brandt L.J. Effect of fecal microbiota transplantation on recurrence in multiply recurrent Clostridium difficile infection: a randomized trial. Ann. Intern. Med. 2016 doi: 10.7326/M16-0271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna S., Pardi D.S., Kelly C.R., Kraft C.S., Dhere T., Henn M.R., Lombardo M.J., Vulic M., Ohsumi T., Winkler J., Pindar C., McGovern B.H., Pomerantz R.J., Aunins J.G., Cook D.N., Hohmann E.L. A novel microbiome therapeutic increases gut microbial diversity and prevents recurrent Clostridium difficile infection. J Infect Dis. 2016;2:173–181. doi: 10.1093/infdis/jiv766. [DOI] [PubMed] [Google Scholar]
- Khoruts A., Dicksved J., Jansson J.K., Sadowsky M.J. Changes in the composition of the human fecal microbiome after bacteriotherapy for recurrent Clostridium difficile-associated diarrhea. J. Clin. Gastroenterol. 2010;5:354–360. doi: 10.1097/MCG.0b013e3181c87e02. [DOI] [PubMed] [Google Scholar]
- Khoruts A., Rank K.M., Newman K.M., Viskocil K., Vaughn B.P., Hamilton M.J., Sadowsky M.J. Inflammatory bowel disease affects the outcome of fecal microbiota transplantation for recurrent Clostridium difficile infection. Clin Gastroenterol H. 2016;14:1433–1438. doi: 10.1016/j.cgh.2016.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kump P.K., Krause R., Allerberger F., Hogenauer C. Faecal microbiota transplantation—the Austrian approach. Clin. Microbiol. Infect. 2014;11:1106–1111. doi: 10.1111/1469-0691.12801. [DOI] [PubMed] [Google Scholar]
- L'Agence nationale de sécurité du médicament et des produits de santé (ANSM) La transplantation de microbiote fecal et son encadrement dans les essais cliniques. 2014. http://ansm.sante.fr/var/ansm_site/storage/original/application/5e5e01018303790194275ded0e02353c.pdf
- Lapointe-Shaw L., Tran K.L., Coyte P.C., Hancock-Howard R.L., Powis J., Poutanen S.M., Hota S. Cost-effectiveness analysis of six strategies to treat recurrent Clostridium difficile infection. PLoS One. 2016;2 doi: 10.1371/journal.pone.0149521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Chatelier E., Nielsen T., Qin J., Prifti E., Hildebrand F., Falony G., Almeida M., Arumugam M., Batto J.M., Kennedy S., Leonard P., Li J., Burgdorf K., Grarup N., Jorgensen T., Brandslund I., Nielsen H.B., Juncker A.S., Bertalan M., Levenez F., Pons N., Rasmussen S., Sunagawa S., Tap J., Tims S., Zoetendal E.G., Brunak S., Clement K., Dore J., Kleerebezem M., Kristiansen K., Renault P., Sicheritz-Ponten T., de Vos W.M., Zucker J.D., Raes J., Hansen T., Bork P., Wang J., Ehrlich S.D., Pedersen O. Richness of human gut microbiome correlates with metabolic markers. Nature. 2013;7464:541–546. doi: 10.1038/nature12506. [DOI] [PubMed] [Google Scholar]
- Lepage P., Hasler R., Spehlmann M.E., Rehman A., Zvirbliene A., Begun A., Ott S., Kupcinskas L., Dore J., Raedler A., Schreiber S. Twin study indicates loss of interaction between microbiota and mucosa of patients with ulcerative colitis. Gastroenterology. 2011;1:227–236. doi: 10.1053/j.gastro.2011.04.011. [DOI] [PubMed] [Google Scholar]
- Ley R.E., Turnbaugh P.J., Klein S., Gordon J.I. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;7122:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- Li S.S., Zhu A., Benes V., Costea P.I., Hercog R., Hildebrand F., Huerta-Cepas J., Nieuwdorp M., Salojarvi J., Voigt A.Y., Zeller G., Sunagawa S., de Vos W.M., Bork P. Durable coexistence of donor and recipient strains after fecal microbiota transplantation. Science. 2016;6285:586–589. doi: 10.1126/science.aad8852. [DOI] [PubMed] [Google Scholar]
- Lin H.V., Frassetto A., Kowalik E.J., Jr., Nawrocki A.R., Lu M.M., Kosinski J.R., Hubert J.A., Szeto D., Yao X., Forrest G., Marsh D.J. Butyrate and propionate protect against diet-induced obesity and regulate gut hormones via free fatty acid receptor 3-independent mechanisms. PLoS One. 2012;4:e35240. doi: 10.1371/journal.pone.0035240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C.A., Stombaugh J.I., Gordon J.I., Jansson J.K., Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;7415:220–230. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchesi J.R., Ravel J. The vocabulary of microbiome research: a proposal. Microbiome. 2015;1:1–3. doi: 10.1186/s40168-015-0094-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchesi J.R., Adams D.H., Fava F., Hermes G.D.a., Hirschfield G.M., Hold G., Quraishi M.N., Kinross J., Smidt H., Tuohy K.M., Thomas L.V., Zoetendal E.G., Hart A. The gut microbiota and host health: a new clinical frontier. Gut. 2015:1–10. doi: 10.1136/gutjnl-2015-309990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlo G., Graves N., Brain D., Connelly L. Economic evaluation of fecal microbiota transplantation for the treatment of recurrent Clostridium difficile infection in Australia. J. Gastroenterol. Hepatol. 2016 doi: 10.1111/jgh.13402. [DOI] [PubMed] [Google Scholar]
- Michail S., Durbin M., Turner D., Griffiths A.M., Mack D.R., Hyams J., Leleiko N., Kenche H., Stolfi A., Wine E. Alterations in the gut microbiome of children with severe ulcerative colitis. Inflamm. Bowel Dis. 2012;10:1799–1808. doi: 10.1002/ibd.22860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moayyedi P., Surette M.G., Kim P.T., Libertucci J., Wolfe M., Onischi C., Armstrong D., Marshall J.K., Kassam Z., Reinisch W., Lee C.H. Fecal microbiota transplantation induces remission in patients with active ulcerative colitis in a randomized controlled trial. Gastroenterology. 2015;1:102–109. doi: 10.1053/j.gastro.2015.04.001. (e6) [DOI] [PubMed] [Google Scholar]
- Molodecky N.A., Soon I.S., Rabi D.M., Ghali W.A., Ferris M., Chernoff G., Benchimol E.I., Panaccione R., Ghosh S., Barkema H.W., Kaplan G.G. Increasing incidence and prevalence of the inflammatory bowel diseases with time, based on systematic review. Gastroenterology. 2012;1:46–54. doi: 10.1053/j.gastro.2011.10.001. (quiz e30) [DOI] [PubMed] [Google Scholar]
- Motta J.P., Magne L., Descamps D., Rolland C., Squarzoni-Dale C., Rousset P., Martin L., Cenac N., Balloy V., Huerre M., Frohlich L.F., Jenne D., Wartelle J., Belaaouaj A., Mas E., Vinel J.P., Alric L., Chignard M., Vergnolle N., Sallenave J.M. Modifying the protease, antiprotease pattern by elafin overexpression protects mice from colitis. Gastroenterology. 2011;4:1272–1282. doi: 10.1053/j.gastro.2010.12.050. [DOI] [PubMed] [Google Scholar]
- Ng K.M., Ferreyra J.A., Higginbottom S.K., Lynch J.B., Kashyap P.C., Gopinath S., Naidu N., Choudhury B., Weimer B.C., Monack D.M., Sonnenburg J.L. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature. 2013;7469:96–99. doi: 10.1038/nature12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Cobas A.E., Artacho A., Knecht H., Ferrus M.L., Friedrichs A., Ott S.J., Moya A., Latorre A., Gosalbes M.J. Differential effects of antibiotic therapy on the structure and function of human gut microbiota. PLoS One. 2013;11:e80201. doi: 10.1371/journal.pone.0080201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Cobas A.E., Gosalbes M.J., Friedrichs A., Knecht H., Artacho A., Eismann K., Otto W., Rojo D., Bargiela R., von Bergen M., Neulinger S.C., Daumer C., Heinsen F.A., Latorre A., Barbas C., Seifert J., dos Santos V.M., Ott S.J., Ferrer M., Moya A. Gut microbiota disturbance during antibiotic therapy: a multi-omic approach. Gut. 2013;11:1591–1601. doi: 10.1136/gutjnl-2012-303184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrof E.O., Khoruts A. From stool transplants to next-generation microbiota therapeutics. Gastroenterology. 2014;6:1573–1582. doi: 10.1053/j.gastro.2014.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrof E.O., Gloor G.B., Vanner S.J., Weese S.J., Carter D., Daigneault M.C., Brown E.M., Schroeter K., Allen-Vercoe E. Stool substitute transplant therapy for the eradication of Clostridium difficile infection: ‘RePOOPulating’ the gut. Microbiome. 2013;1:3. doi: 10.1186/2049-2618-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Png C.W., Linden S.K., Gilshenan K.S., E.G Z. Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Am. J. Gastroenterol. 2010;11:2420–2428. doi: 10.1038/ajg.2010.281. [DOI] [PubMed] [Google Scholar]
- Pullan R.D., Thomas G.A., Rhodes M., Newcombe R.G., Williams G.T., Allen A., Rhodes J. Thickness of adherent mucus gel on colonic mucosa in humans and its relevance to colitis. Gut. 1994;3:353–359. doi: 10.1136/gut.35.3.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J., Li R., Raes J., Arumugam M., Burgdorf K.S., Manichanh C., Nielsen T., Pons N., Levenez F., Yamada T., Mende D.R., Li J., Xu J., Li S., Li D., Cao J., Wang B., Liang H., Zheng H., Xie Y., Tap J., Lepage P., Bertalan M., Batto J.M., Hansen T., Le Paslier D., Linneberg A., Nielsen H.B., Pelletier E., Renault P., Sicheritz-Ponten T., Turner K., Zhu H., Yu C., Li S., Jian M., Zhou Y., Li Y., Zhang X., Li S., Qin N., Yang H., Wang J., Brunak S., Doré J., Guarner F., Kristiansen K., Pedersen O., Parkhill J., Weissenbach J., Bork P., Ehrlich S.D., Wang J. A human gut microbial gene catalog established by metagenomic sequencing. Nature. 2010;7285:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajilic-Stojanovic M., Shanahan F., Guarner F., de Vos W.M. Phylogenetic analysis of dysbiosis in ulcerative colitis during remission. Inflamm. Bowel Dis. 2013;3:481–488. doi: 10.1097/MIB.0b013e31827fec6d. [DOI] [PubMed] [Google Scholar]
- Rampelli S., Schnorr S.L., Consolandi C., Turroni S., Severgnini M., Peano C., Brigidi P., Crittenden A.N., Henry A.G., Candela M. Metagenome sequencing of the Hadza hunter-gatherer gut microbiota. Curr. Biol. 2015;13:1682–1693. doi: 10.1016/j.cub.2015.04.055. [DOI] [PubMed] [Google Scholar]
- Ratner M. Microbial cocktails join fecal transplants in IBD treatment trials. Nat. Biotechnol. 2015;8:787–788. doi: 10.1038/nbt0815-787. [DOI] [PubMed] [Google Scholar]
- Rea M.C., O'Sullivan O., Shanahan F., O'Toole P.W., Stanton C., Ross R.P., Hill C. Clostridium difficile carriage in elderly subjects and associated changes in the intestinal microbiota. J. Clin. Microbiol. 2012;3:867–875. doi: 10.1128/JCM.05176-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relman D.A. The human microbiome: ecosystem resilience and health. Nutr. Rev. 2012;S2-9 doi: 10.1111/j.1753-4887.2012.00489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridaura V.K., Faith J.J., Rey F.E., Cheng J., Duncan A.E., Kau A.L., Griffin N.W., Lombard V., Henrissat B., Bain J.R., Muehlbauer M.J., Ilkayeva O., Semenkovich C.F., Funai K., Hayashi D.K., Lyle B.J., Martini M.C., Ursell L.K., Clemente J.C., van Treuren W., Walters W.A., Knight R., Newgard C.B., Heath A.C., Gordon J.I. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science. 2013;6150:1241214. doi: 10.1126/science.1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossen N.G., Fuentes S., van der Spek M.J., Tijssen J.G., Hartman J.H., Duflou A., Lowenberg M., van den Brink G.R., Mathus-Vliegen E.M., de Vos W.M., Zoetendal E.G., D'Haens G.R., Ponsioen C.Y. Findings from a randomized controlled trial of fecal transplantation for patients with ulcerative colitis. Gastroenterology. 2015;1:110–118. doi: 10.1053/j.gastro.2015.03.045. (e4) [DOI] [PubMed] [Google Scholar]
- Scholl D., Cooley M., Williams S.R., Gebhart D., Martin D., Bates A., Mandrell R. An engineered R-type pyocin is a highly specific and sensitive bactericidal agent for the food-borne pathogen Escherichia coli O157:H7. Antimicrob. Agents Chemother. 2009;7:3074–3080. doi: 10.1128/AAC.01660-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholl D., Gebhart D., Williams S.R., Bates A., Mandrell R. Genome sequence of E. coli O104:H4 leads to rapid development of a targeted antimicrobial agent against this emerging pathogen. PLoS One. 2012;3:e33637. doi: 10.1371/journal.pone.0033637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert A.M., Rogers M.A., Ring C., Mogle J., Petrosino J.P., Young V.B., Aronoff D.M., Schloss P.D. Microbiome data distinguish patients with Clostridium difficile infection and non-C. difficile-associated diarrhea from healthy controls. MBio. 2014;3:e01021–01014. doi: 10.1128/mBio.01021-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwiertz A., Taras D., Schafer K., Beijer S., Bos N.A., Donus C., Hardt P.D. Microbiota and SCFA in lean and overweight healthy subjects. Obesity. 2010;1:190–195. doi: 10.1038/oby.2009.167. [DOI] [PubMed] [Google Scholar]
- Seekatz A.M., Aas J., Gessert C.E., Rubin T.A., Saman D.M., Bakken J.S., Young V.B. Recovery of the gut microbiome following fecal microbiota transplantation. MBio. 2014;3 doi: 10.1128/mBio.00893-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki H., Shiohara M., Matsumura T., Miyagawa N., Tanaka M., Komiyama A., Kurata S. Prevention of antibiotic-associated diarrhea in children by Clostridium butyricum MIYAIRI. Pediatr. Int. 2003;1:86–90. doi: 10.1046/j.1442-200x.2003.01671.x. [DOI] [PubMed] [Google Scholar]
- Seres Therapeutics Inc. Seres Therapeutics announces interim results from SER-109 phase 2 ECOSPOR Study in multiply recurrent Clostridium difficile infection. 2016. http://www.businesswire.com/news/home/20160729005385/en
- Shahinas D., Silverman M., Sittler T., Chiu C., Kim P., Allen-Vercoe E., Weese S., Wong A., Low D.E., Pillai D.R. Toward an understanding of changes in diversity associated with fecal microbiome transplantation based on 16S rRNA gene deep sequencing. MBio. 2012;5 doi: 10.1128/mBio.00338-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapleton A.E., Au-Yeung M., Hooton T.M., Fredricks D.N., Roberts P.L., Czaja C.A., Yarova-Yarovaya Y., Fiedler T., Cox M., Stamm W.E. Randomized, placebo-controlled phase 2 trial of a Lactobacillus crispatus probiotic given intravaginally for prevention of recurrent urinary tract infection. Clin. Infect. Dis. 2011;10:1212–1217. doi: 10.1093/cid/cir183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surawicz C.M., Brandt L.J., Binion D.G., Ananthakrishnan A.N., Curry S.R., Gilligan P.H., McFarland L.V., Mellow M., Zuckerbraun B.S. Guidelines for diagnosis, treatment, and prevention of Clostridium difficile infections. Am. J. Gastroenterol. 2013;4:478–498. doi: 10.1038/ajg.2013.4. (quiz 499) [DOI] [PubMed] [Google Scholar]
- Teixeira T.F., Grzeskowiak L., Franceschini S.C., Bressan J., Ferreira C.L., Peluzio M.C. Higher level of faecal SCFA in women correlates with metabolic syndrome risk factors. Br. J. Nutr. 2013;5:914–919. doi: 10.1017/S0007114512002723. [DOI] [PubMed] [Google Scholar]
- The Gastroenterological Society of Australia (GESA) Position statement of faecal microbiota transplant (FMT) 2015. http://www.gesa.org.au/news.asp?id=541
- The Infectious Diseases Society of America (IDSA) Fecal microbiota transplantation: incestigational new drug protocol. 2016. http://www.idsociety.org/FMT
- The National Institute for Health and Care Excellence (NICE) Faecal microbiota transplant for recurrent Clostridium difficile infection. 2014. http://www.nice.org.uk/guidance/ipg485
- Theriot C.M., Koenigsknecht M.J., Carlson P.E., Jr., Hatton G.E., Nelson A.M., Li B., Huffnagle G.B., Z Li J., Young V.B. Antibiotic-induced shifts in the mouse gut microbiome and metabolome increase susceptibility to Clostridium difficile infection. Nat. Commun. 2014:3114. doi: 10.1038/ncomms4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theriot C.M., Bowman A.A., Young V.B. Antibiotic-induced alterations of the gut microbiota alter secondary bile acid production and allow for Clostridium difficile spore germination and outgrowth in the large intestine. mSphere. 2016;1 doi: 10.1128/mSphere.00045-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh P.J., Ley R.E., Mahowald M.A., Magrini V., Mardis E.R., Gordon J.I. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;7122:1027–1131. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- Turnbaugh P.J., Hamady M., Yatsunenko T., Cantarel B.L., Duncan A., Ley R.E., Sogin M.L., Jones W.J., Roe B.A., Affourtit J.P., Egholm M., Henrissat B., Heath A.C., Knight R., Gordon J.I. A core gut microbiome in obese and lean twins. Nature. 2009;7228:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tvede M., Rask-Madsen J. Bacteriotherapy for chronic relapsing Clostridium difficile diarrhoea in six patients. Lancet. 1989;8648:1156–1160. doi: 10.1016/s0140-6736(89)92749-9. [DOI] [PubMed] [Google Scholar]
- van Nood E., Vrieze A., Nieuwdorp M., Fuentes S., Zoetendal E.G., de Vos W.M., Visser C.E., Kuijper E.J., Bartelsman J.F., Tijssen J.G., Speelman P., Dijkgraaf M.G., Keller J.J. Duodenal infusion of donor feces for recurrent Clostridium difficile. N. Engl. J. Med. 2013;5:407–415. doi: 10.1056/NEJMoa1205037. [DOI] [PubMed] [Google Scholar]
- Vandenbroucke K., de Haard H., Beirnaert E., Dreier T., Lauwereys M., Huyck L., van Huysse J., Demetter P., Steidler L., Remaut E., Cuvelier C., Rottiers P. Orally administered L. lactis secreting an anti-TNF Nanobody demonstrate efficacy in chronic colitis. Mucosal Immunol. 2010;1:49–56. doi: 10.1038/mi.2009.116. [DOI] [PubMed] [Google Scholar]
- Villanueva-Millan M.J., Perez-Matute P., Oteo J.A. Gut microbiota: a key player in health and disease. A review focused on obesity. J. Physiol. Biochem. 2015;3:509–525. doi: 10.1007/s13105-015-0390-3. [DOI] [PubMed] [Google Scholar]
- Vipperla K., O'Keefe S.J. Diet, microbiota, and dysbiosis: a ‘recipe’ for colorectal cancer. Food Funct. 2016;4:1731–1740. doi: 10.1039/c5fo01276g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrieze A., Van Nood E., Holleman F., Salojarvi J., Kootte R.S., Bartelsman J.F., Dallinga-Thie G.M., Ackermans M.T., Serlie M.J., Oozeer R., Derrien M., Druesne A., van Hylckama Vlieg J.E., Bloks V.W., Groen A.K., Heilig H.G., Zoetendal E.G., Stroes E.S., de Vos W.M., Hoekstra J.B., Nieuwdorp M. Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology. 2012;4:913–916. doi: 10.1053/j.gastro.2012.06.031. (e7) [DOI] [PubMed] [Google Scholar]
- Vyas D., Aekka A., Vyas A. Fecal transplant policy and legislation. World J. Gastroenterol. 2015;1:6–11. doi: 10.3748/wjg.v21.i1.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker A.W., Sanderson J.D., Churcher C., Parkes G.C., Hudspith B.N., Rayment N., Brostoff J., Parkhill J., Dougan G., Petrovska L. High-throughput clone library analysis of the mucosa-associated microbiota reveals dysbiosis and differences between inflamed and non-inflamed regions of the intestine in inflammatory bowel disease. BMC Microbiol. 2011;7-7 doi: 10.1186/1471-2180-11-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Euler C.W., Delaune A., Fischetti V.A. Using a novel lysin to help control Clostridium difficile infections. Antimicrob Agents Ch. 2015;12:7447–7457. doi: 10.1128/AAC.01357-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weingarden A.R., Chen C., Bobr A., Yao D., Lu Y., Nelson V.M., Sadowsky M.J., Khoruts A. Microbiota transplantation restores normal fecal bile acid composition in recurrent Clostridium difficile infection. Am. J. Phys. Gastrointest. Liver. 2014;4:G310–G319. doi: 10.1152/ajpgi.00282.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weingarden A., Gonzalez A., Vazquez-Baeza Y., Weiss S., Humphry G., Berg-Lyons D., Knights D., Unno T., Bobr A., Kang J., Khoruts A., Knight R., Sadowsky M.J. Dynamic changes in short- and long-term bacterial composition following fecal microbiota transplantation for recurrent Clostridium difficile infection. Microbiome. 2015;10 doi: 10.1186/s40168-015-0070-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasueda A., Mizushima T., Nezu R., Sumi R., Tanaka M., Nishimura J., Kai Y., Hirota M., Osawa H., Nakajima K., Mori M., Ito T. The effect of Clostridium butyricum MIYAIRI on the prevention of pouchitis and alteration of the microbiota profile in patients with ulcerative colitis. Surg. Today. 2016;8:939–949. doi: 10.1007/s00595-015-1261-9. [DOI] [PubMed] [Google Scholar]
- Youngster I., Sauk J., Pindar C., Wilson R.G., Kaplan J.L., Smith M.B., Alm E.J., Gevers D., Russell G.H., Hohmann E.L. Fecal microbiota transplant for relapsing Clostridium difficile infection using a frozen inoculum from unrelated donors: a randomized, open-label, controlled pilot study. Clin. Infect. Dis. 2014;11:1515–1522. doi: 10.1093/cid/ciu135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngster I., Mahabamunuge J., Systrom H.K., Sauk J., Khalili H., Levin J., Kaplan J.L., Hohmann E.L. Oral, frozen fecal microbiota transplant (FMT) capsules for recurrent Clostridium difficile infection. BMC Med. 2016;1:134. doi: 10.1186/s12916-016-0680-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhernakova A., Kurilshikov A., Bonder M.J., Tigchelaar E.F., Schirmer M., Vatanen T., Mujagic Z., Vila A.V., Falony G., Vieira-Silva S., Wang J., Imhann F., Brandsma E., Jankipersadsing S.A., Joossens M., Cenit M.C., Deelen P., Swertz M.A., Weersma R.K., Feskens E.J., Netea M.G., Gevers D., Jonkers D., Franke L., Aulchenko Y.S., Huttenhower C., Raes J., Hofker M.H., Xavier R.J., Wijmenga C., Fu J. Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science. 2016;6285:565–569. doi: 10.1126/science.aad3369. [DOI] [PMC free article] [PubMed] [Google Scholar]