Abstract
Mammalian evolution has occurred in the presence of mutualistic, commensal, and pathogenic micro- and macro-organisms for millennia. The presence of these organisms during mammalian evolution has allowed for intimate crosstalk between these colonizing species and the host immune system. In this review, we introduce the concept of the ‘multibiome’ to holistically refer to the biodiverse collection of bacteria, viruses, fungi and multicellular helminthic worms colonizing the mammalian intestine. Furthermore, we discuss new insights into multibiome-host interactions in the context of host-protective immunity and immune-mediated diseases, including inflammatory bowel disease and multiple sclerosis. Finally, we provide reasons to account for the multibiome in experimental design, analysis and in therapeutic applications.
Abbreviations: CID, chronic inflammatory disease; DSS, dextran sodium sulfate; EAE, experimental autoimmune encephalomyelitis; EBV, Epstein-Barr virus; EE, environmental enteropathy; GF, germ-free; HuNoV, Human Norovirus; IBD, inflammatory bowel disease; MNV, murine norovirus; MS, multiple sclerosis; SFB, Segmented Filamentous Bacteria; TSO, Trichuris suis ova
Keywords: Microbiome, Mucosal immunology, Autoimmune disease, Inflammatory bowel disease, Helminth immunotherapy
Highlights
-
•
The intestinal multibiome is composed of bacteria, viruses, fungi, and eukaryotes.
-
•
Mammals evolved alongside a complex and biodiverse multibiome.
-
•
Cross-talk between the multibiome and the host regulates immunity and inflammation.
1. Introduction
“Everything is alive, everything is interconnected.”
–Cicero 106 BCE
Complex and coevolved interdependence is commonly observed between organisms that occupy the same ecological niches. However, considering the ecosystem of the mammalian intestine as complex and important is relatively novel, with the earliest microbiome studies occurring in the last century (Rosebury et al., 1954). Since then, studies have demonstrated a largely mutualistic relationship between the host and its bacterial microbiota, which promotes intestinal vascularization and epithelial cell function, supports nutrient absorption, and limits pathogen invasion (Brestoff and Artis, 2013, Stappenbeck et al., 2002, Hooper et al., 2012). Moreover, crosstalk between intestinal bacterial communities and the host fundamentally influences immune homeostasis, host-protective immunity and disease-associated inflammation (Brestoff and Artis, 2013, Belkaid and Hand, 2014). However, our understanding of how the other biological entities (archaea, viruses, fungi, and mammalian parasites) that colonize the intestine communicate with one another and the host is incomplete. ‘Transkingdom’ interactions between these entities and the host can influence intestinal ecosystem dynamics and immune homeostasis (Pfeiffer and Virgin, 2016). Here, we introduce the term ‘multibiome’ to encompass the biodiverse collection of microscopic (bacteria, archaea, fungi) and macroscopic (multicellular worms) organisms, as well as viruses (not represented in phylogenetic kingdoms) that colonize mammals (Fig. 1). In this review, we will discuss how each group individually influences the host immune system, how intestinal colonization or infection is impacted by other members of the multibiome, and how crosstalk between members of the multibiome and the host influences local and systemic immune homeostasis, health, and disease.
Fig. 1.
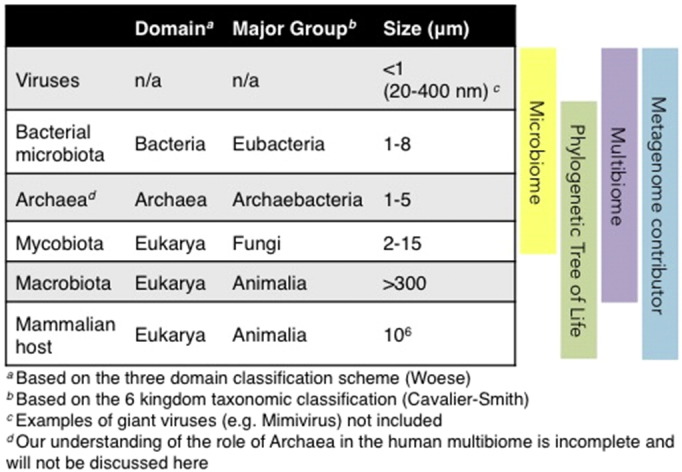
The multibiome.
Due to the shortcomings of the terms microbiome and trans-kingdom, we propose the introduction of the term multibiome. This all-encompassing term accounts for viruses (that are not part of any taxonomic kingdom) and for macrobionts such as helminth parasites. Each multibiome member (when present, in the case of helminths) and the mammalian host, together, contribute their genetic information to the holistic metagenome.
2. The Multibiome: Importance of Individual Groups
Mammalian physiology has undergone evolutionary optimization in collaboration with the bacterial microbiome, as illustrated by the physiologic defects that have been described in germ-free (GF) animals, including impaired intestinal function, immune and metabolic homeostasis. Bacterial microbiome reconstitution can restore the majority of these defects (Brestoff and Artis, 2013, Belkaid and Hand, 2014, El Aidy et al., 2012). However, the recent demonstration that persistent intestinal virus colonization can restore a subset of intestinal morphology and immune defects of germ-free mice indicates that non-bacterial members of the multibiome can also provide critical developmental cues to the host (Kernbauer et al., 2014).
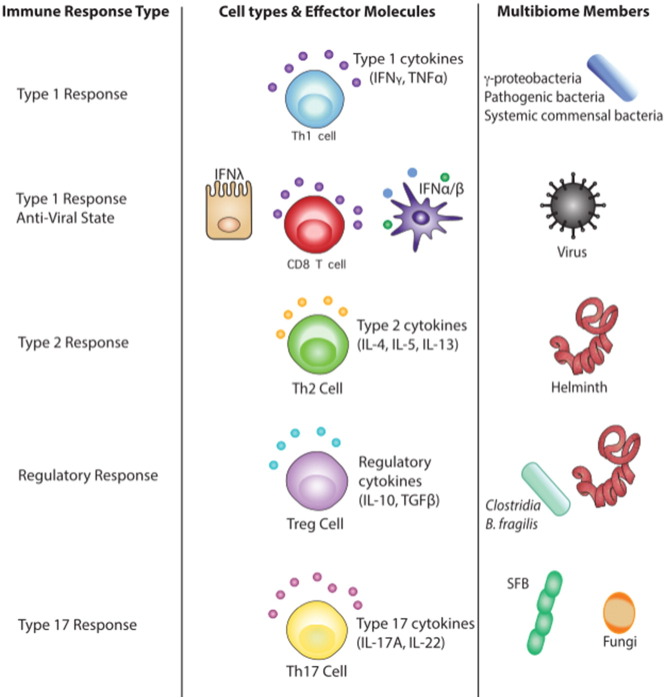
The mammalian immune system has evolved multiple immune pathways characterized by T helper (Th) cell subsets and their associated cytokines that are tailored toward distinct types of pathogens. Intracellular bacteria and viruses typically induce pro-inflammatory type 1 responses (classically activated macrophage, cytotoxic T cells and Th1 cells), multicellular helminths can induce wound healing type 2 responses (alternatively activated macrophage and Th2 cells) and/or tolerogenic responses (regulatory T cells (Treg)), and certain extracellular bacteria and most fungi elicit type 17 responses (Th17 cells) (Fig. 2). In this section, we will discuss how distinct members of the multibiome can elicit these pathways to influence immunity and inflammation.
Fig. 2.
The immune response associated with each constituent of the multibiome.
Type 1 responses are initiated by detecting microbe-associated molecular patterns. Type 1 pro-inflammatory cytokines and cells target intracellular microbes, including bacterial pathogens, opportunistic commensal bacterial infection and viruses. The innate type 1 and 3 IFN pathways are critical components of the antiviral immune response. Type 1 responses are pro-inflammatory and can cause collateral damage to the host if the immune response is not balanced and regulated.
Type 2 immune responses are triggered by helminth infections. A type 2 responses coordinates worm expulsion and rapid wound healing; however, it can lead to fibrosis if the immune response is not balanced and regulated.
Regulatory responses are promoted by certain bacterial species (some Clostridia species, B. fragilis), and many helminth infections. Their broad immunosuppressive actions contribute to self-tolerance and commensal multibiome-tolerance. They are also initiated during the resolution phase of type 1, 2, and 3-promoting infections.
Type 17 responses are triggered by extracellular bacterial pathogens, fungi and some epithelial cell binding commensal bacterial species. Type 17-associated cytokines promote mucosal barrier function, but are also widely implicated in autoimmune and CIDs.
2.1. The Bacterial Microbiome
The intestinal bacterial microbiome is comprised of trillions of individual bacteria from approximately 1000 different species (Brestoff and Artis, 2013). Within this vast diversity, examples of specific microbes and collections of bacterial consortium have been shown to elicit immune polarization (Fig. 2) through direct interaction with host intestinal epithelial or dendritic cells as well as indirect mechanisms that rely on bacterial metabolism. Multiple chronic inflammatory disorders (CIDs) including inflammatory bowel diseases (IBD) like Crohn's and ulcerative colitis and extra-intestinal autoimmune disorders (multiple sclerosis and rheumatoid arthritis) have been shown to be associated with intestinal dysbiosis, which suggests that the bacterial microbiome may contribute to the development or progression of inflammatory diseases (Belkaid and Hand, 2014). However, it is important to note that the dialog between the bacterial microbiome and the host is highly contextual and commensal-induced responses can be influenced by genetics and environmental factors including infection history, nutrient availability, and age, which have been reviewed elsewhere (Nicholson et al., 2012).
2.2. The Virome
The virome consists of all bacteriophage, mammalian viruses and the endogenous retroviruses that have integrated into the host's genome (Pfeiffer and Virgin, 2016). Despite the enormity of the intestinal virome (estimated ten-fold more particles than bacterial microbes), understanding its impact on health and disease is in its infancy. However, the intestinal virome is likely an important regulator of immune homeostasis; colonizing GF mice with a single persistent viral strain was sufficient to correct a subset of immune defects, including Type 1-associated interferon responses (Kernbauer et al., 2014) (Fig. 2). Supporting the hypothesis that the virome and the mammalian immune system engage in ongoing cross-talk, the intestinal virome is expanded in the context of Human Immunodeficiency Virus (HIV) and Simian Immunodeficiency Virus (SIV)-induced acquired immunodeficiency (Monaco et al., 2016, Handley et al., 2012) and is dysregulated in IBD patients (Norman et al., 2015). Additionally, a number of viral infections have been associated with autoimmunity (enterovirus with Type 1 Diabetes and Epstein Barr virus with MS) (Richardson and Horwitz, 2014, Casiraghi et al., 2012). Thus, our current understanding indicates that in certain contexts the virome can contribute to the development or progression of various CIDs.
Curating and characterizing the human virome is challenging due to the absence of a conserved gene region (e.g. bacterial 16S) and incomplete viral genome libraries (Pfeiffer and Virgin, 2016). However, with the advent of metagenomic shotgun sequencing it is anticipated that we will gain a new appreciation of the role of the virome in maintaining intestinal and immune homeostasis, and how dysregulation of host-virome interactions can contribute to disease.
2.3. The Mycobiome
The mycobiome (fungal constituent of the multibiome) is less diverse and abundant than the bacterial microbiome (Underhill and Iliev, 2014). Recent shotgun sequencing approaches suggest that fungi account for approximately 0.1% of the intestinal microbiome, although this is likely an underestimate of their true representation (Underhill and Iliev, 2014). Fungi activate the type 17 axis of the immune system (Fig. 2) and can contribute to local (gastric ulcers, food allergy sensitization and colitis) and systemic (allergic airway) diseases (Mason et al., 2012, Wheeler et al., 2016, Yamaguchi et al., 2006). Recent advances in the sequencing and phylogenetic assignment of the mycobiome may reveal further impacts on mammalian health and immunity (Underhill and Iliev, 2014).
2.4. The Macrobiome
The macrobiome consists of intestinal multicellular parasitic worms, most commonly referred to as helminths (from the Greek word for worm) (Hotez et al., 2008). Unlike bacteria, fungi, and viruses, helminths are only present in about one-third of the global population (Maizels, 2016). Many helminths complete part of their life cycle in the host's intestine by securing themselves into the intestinal epithelium, during which they disrupt the intestinal ecosystem and damage the epithelium (Allen and Maizels, 2011). In response, mammalian hosts activate type 2 responses that promote rapid intestinal epithelial cell turnover, mucus production and increased gut motility to encourage helminth expulsion (Allen and Maizels, 2011) (Fig. 2). This can be paired with an expanded Treg population and production of wound healing molecules to limit inflammation and promote intestinal repair. The coevolution of helminths and the host type 2 and regulatory responses has resulted in an immune-mediated truce where worms are tolerated and host tissue damage is minimized (Allen and Maizels, 2011). Although helminths are commonly thought of as parasites, the ‘Old Friends’ hypothesis suggests that lack of exposure to these organisms could have detrimental effects (Rook, 2010). The impact of helminth-induced immune modulation in the context of co-infection, vaccination efficacy, and CID development is discussed more thoroughly in a later section.
2.5. Communication Between Multibiome Members and the Host Regulates Immune Homeostasis
Collectively, these findings indicate that constituents of the multibiome can individually influence the host immune system. In a diverse intestinal ecosystem composed of all members of the multibiome, each corresponding arm of the immune system can receive basal stimulation (Fig. 3). Type 1 and type 2 immune axes reciprocally inhibit each other, and regulatory T cells can dampen Th1, Th2 and Th17-mediated inflammation (Fig. 3). Thus, a rich and diverse multibiome may promote immune homeostasis, while the absence of helminths or a low abundance of immunomodulatory bacteria may alter the response to other viral, bacterial and fungal stimuli. This imbalance may play a role in the development or progression of various chronic inflammatory or autoimmune diseases.
Fig. 3.
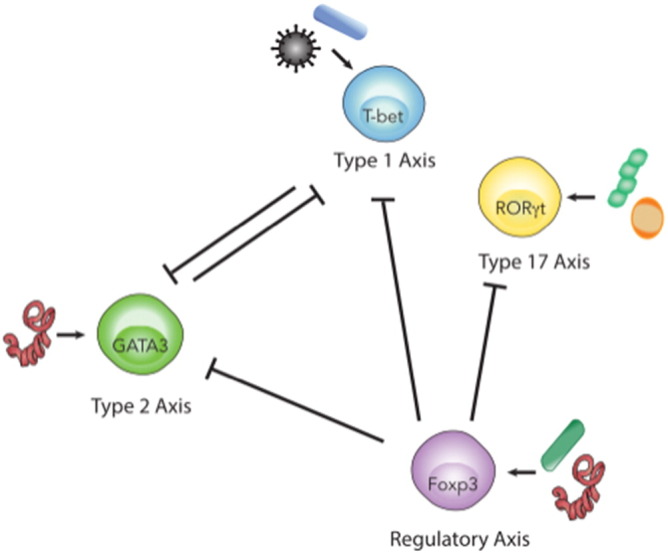
Multibiome-mediated regulation of immune polarization and homeostasis.
This illustration simplifies the many interactions and pathways involved in multibiome-host cross-talk down to the core T helper (Th) cell subsets they promote. It also demonstrates how each Th subset is able to regulate, inhibit, or promote the activation or function of another subset. During a proper immune response, balancing these subsets is crucial to avoid pathology and generate a good memory response. Immune cross-regulation is mediated through a variety of mechanisms including cytokines, chemokines, receptor interactions, and transcription factor expression. Certain members of the multibiome promote the expansion of different subsets of Th cells, which in turn inhibit, promote, and/or balance the other subsets. T-bet expressing Th1 (activated in response to bacteria and viruses) and GATA3 expressing Th2 cells (helminth-induced), antagonize each other. Some bacterial species and helminths promote Foxp3-expressing Tregs, which in turn limit the activation of Th1, Th2 and Th17 cells.
3. Multibiome Members Interact in a Dynamic Ecosystem
The intestinal lumen harbors a complex and interactive ecosystem, where colonization or infection can be influenced by inter-multibiome cross-talk or by host responses. In this section, we will focus on inter-multibiome interactions that influence colonization across taxonomic boundaries.
3.1. Interactions Between Members of the Multibiome
Similar to other ecosystems, the intestinal community is dynamic, responsive, and regulated by interactions between distinct biological entities. For example, bacteriophage can shape the bacterial microbiome by lysing commensal or pathogenic bacteria and driving bacterial evolution (Duerkop and Hooper, 2013). The bacterial microbiome can inhibit colonization of other commensal bacteria and/or pathogens by occupying ecological niches, competing for resources, producing metabolites and stimulating immune responses (Lawley and Walker, 2013, Caballero and Pamer, 2015, Belkaid and Hand, 2014). In addition, bacterial-derived fatty-acid metabolites can impair Candida albicans colonization of intestinal tissue, and a complex bacterial microbiome inhibits translocation of pathogenic C. albicans across the intestinal barrier (Kennedy and Volz, 1985). Consistent with resource competition models, fungi often overgrow post-antibiotic treatment and are associated with antibiotic-induced diarrhea (Sullivan et al., 2001, Krause et al., 2001, Mason et al., 2012). Conversely, introduction of fungi or helminths can alter the composition of local, downstream, and upstream bacterial microbiota (Kreisinger et al., 2015). Mechanisms of microbiome remodeling are largely unknown, but in the context of helminth infection it has been proposed that helminth-induced alterations to nutrient access, antimicrobial peptide secretion, mucus production and immune modulation may produce a ripple effect that can alter multibiome composition at distant sites (Kreisinger et al., 2015). Together, these data demonstrate that interactions between constituents of the multibiome shape the overall structure of the intestinal community (Fig. 4). The resulting disruptions in ecological homeostasis may have far-reaching implications on host physiology, immune homeostasis, and CID development, as discussed below.
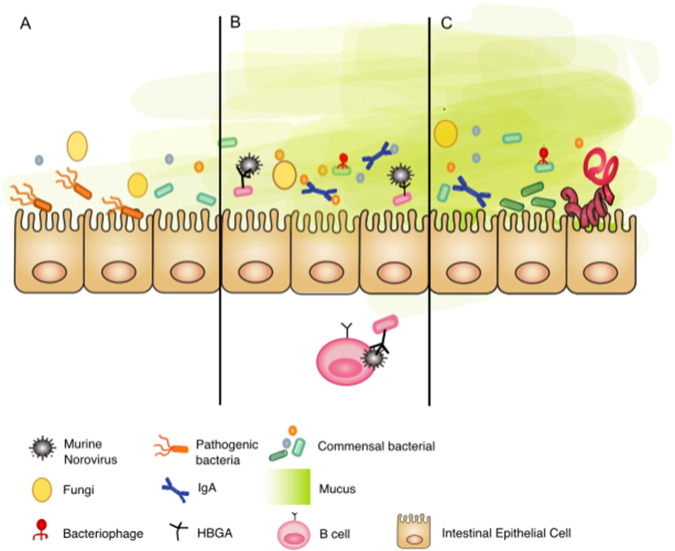
Fig. 4.
Interactions between multibiome members regulate the intestinal community.
In addition to immune-mediated regulation, composition of the intestinal ecosystem is dynamically regulated by interactions across taxonomic boundaries.
A) A diverse commensal bacterial community limits outgrowth of opportunistic or pathogenic bacteria and Candida albicans by niche competition for space and nutrients, production of metabolites and immune-mediated mechanisms.
B) Bacteriophage diversity can dictate bacterial community composition and drive evolution, through a predator-prey relationship. Some enteric pathogens have evolved to take advantage of the ubiquitous presence of intestinal commensal bacteria. Bacteria expressing histo-blood group antigen (HBGA) promote human and murine Norovirus can promote infection of B cells.
C) Helminths have evolved to utilize cues from commensals, to ensure the life cycle is completed in the appropriate location and host. Helminth-induced type 2 responses promote a thickening of the mucus layer, which can alter the colonizing microbiota species.
3.2. Coevolution Between Intestinal Pathogens and the Multibiome: The Bacterial Microbiome Cues ‘Home’
The gastrointestinal tract is colonized with commensal species along its entire length. Although bacterial populations differ depending on physical location (mouth, stomach, small and large intestine), a number of pathogens have evolved to utilize conserved microbial features such as biofilm formation and microbe-associated molecular patterns (MAMPs) to their advantage. Bacterial biofilms provide a platform for fungal colonization (Shirtliff et al., 2009, Iliev et al., 2012) and intestinal helminths have evolved to use commensal bacteria as cues that they have reached the appropriate destination to develop (Hayes et al., 2010, Reynolds et al., 2014). Once established, some helminths support expansion of immunoregulatory bacterial populations, which promote Treg differentiation as an additional and indirect means of creating a tolerant immune environment that favors helminth persistence (Zaiss et al., 2015). Moreover, multiple enteric viruses (huNoV, poliovirus, reovirus and mouse mammary tumor virus) have evolved to utilize bacteria to improve infectivity and transmission by either stabilizing virion structure, enhancing host receptor-binding, or inducing a tolerant environment (Karst, 2016). These findings indicate that intestinal pathogens have evolved to utilize the bacterial microbiome to enhance their fitness and infectivity (Fig. 4).
4. Host-multibiome Interactions Influence Infection and Immunity
4.1. Nature vs. Nurture: Environmental Contributions to Immune Homeostasis
The crosstalk between environmental factors (including the intestinal multibiome) and the mammalian immune system significantly shapes immune homeostasis, accounting for up to 75% of an individual's cellular immune profile, whereas genetic factors account for ~ 25–50% (Orru et al., 2013, Brodin et al., 2015). Socio-economic status, diet, infection history, pet ownership, and exercise are all associated with inter-individual differences in immune homeostasis and intestinal multibiome composition (Carr et al., 2016, Song et al., 2013). Intimate sharing of an environment while co-habiting and co-parenting is linked to immune profile convergence between unrelated individuals (Carr et al., 2016). Co-habitation is also coupled to bacterial microbiome convergence between unrelated individuals and even between their pets (Carr et al., 2016, Song et al., 2013). Although these studies focused on the intestinal bacterial microbiome, environmental factors likely influence other members of the multibiome as well. Indeed, the intestinal virome is highly similar between infant co-twins and as the twins are exposed to different environmental conditions, similarity of the DNA virome diminishes by adulthood (Reyes et al., 2010). Together, these findings indicate that the intestinal multibiome is sensitive to environmental exposures and contributes to immune homeostasis. This concept adds a layer of complexity to the future of personalized medicine and suggests that the human metagenome (an individual's genome plus the genomes of their unique multibiome) may influence not only the development of disease, but also responsiveness to therapeutic intervention.
4.2. The Multibiome Influences the Immune Response to Pathogens
The multibiome's influence on immune cells can affect the response to infectious agents and vaccinations. Depletion of the murine bacterial microbiome with antibiotics resulted in enhanced susceptibility to severe systemic Lymphocytic Choriomeningitis Virus (LCMV) and Influenza A Virus lung infections (Abt et al., 2012, Ichinohe et al., 2011) that was linked to commensal bacteria-dependent basal stimulation of antiviral signaling pathways in innate immune cells. Alternatively, enhancing innate immune signaling pathways with intestinal viral infection or exposure to viral ligands was sufficient to diminish colonization of vancomycin-resistant Enterococcus faecium, an opportunistic bacteria (Abt et al., 2016). These data indicate that innate immune cells integrate signals from commensal bacteria or viruses to remain poised to respond to pathogen infection across taxonomic boundaries.
Past infection history can also influence the outcome of future infections. Measles virus infection induces a long-term immunosuppressive state and disables immune memory formation, increasing the risk of secondary bacterial or viral infection in children for 2–3 years after the initial infection (Mina et al., 2015). Helminth-induced immunomodulation can also impair antiviral immunity to newly acquired intestinal viral infection (murine Norovirus, MNV) and enhance reactivation of latent herpesvirus infection in a type 2 cytokine-dependent manner (Osborne et al., 2014, Reese et al., 2014). It has been hypothesized that helminth-induced alterations in the bacterial microbiome can indirectly influence immune homeostasis and responses to pathogens. However, in the case of helminth and MNV co-infection, the helminth-induced impairment of antiviral immunity was independent of alterations in the bacterial microbiome, indicating that coevolution of helminths with the mammalian host has enabled helminths to directly manipulate immune pathways (Osborne et al., 2014).
Many orally-administered vaccines (e.g. Rotarix, for Rotavirus and the Oral Polio Vaccine) developed and trialed in industrialized countries are less effective in developing regions (Jiang et al., 2010, Naylor et al., 2015). The impact of environmental exposures such as helminth infection, malnutrition and access to clean water all likely contribute to diminished vaccine efficacy. Emerging human data indicates that the helminth-induced promotion of immunoregulatory CD4+ Treg cells contribute to suppressed vaccine-induced immunity (Maizels, 2016), and anti-helminthic therapies have been demonstrated to improve vaccine responses (Labeaud et al., 2009). Independent studies indicate that unsanitary living conditions, bacterial dysbiosis and subclinical but insidious environmental enteropathy (EE) are also associated with diminished efficacy of orally administered vaccines (Naylor et al., 2015). However, most studies do not test for helminth co-infections in children with EE, thus the interplay between various environmental infections and their impact on the immune response remains unknown. Recently, it was demonstrated that multibiome co-infections in mice alter immune homeostasis and impact vaccine responses (Reese et al., 2016). A sequential co-infection model could be useful to interrogate the interplay between malnourishment and host-multibiome interactions in the context of infection and immunity. This line of study could lead to new understanding of how immune dysregulation can be overcome in these settings to provide better protection from infectious disease.
5. Multibiome-host Interactions Influence Immune-mediated Disease in the Gut and Beyond
Independent epidemiological studies have revealed a robust correlation between bacterial microbiota dysbiosis and autoimmune and CIDs. Given the comprehensive discussion of the importance of the bacterial microbiome in these diseases (Belkaid and Hand, 2014), we will focus on the influence of other multibiome members in regulating intestinal inflammation.
5.1. Multibiome-host Interactions Influence Intestinal Inflammation and Inflammatory Bowel Disease
Emerging data indicate that interactions between the virome, intestinal bacteria and host genetics can all contribute to IBD. Polymorphisms in the autophagy gene Atg16L1 are associated with IBD (Cadwell et al., 2010). Mice with a mutation in this gene infected with a persistent strain of MNV and given dextran sodium sulfate (DSS - a chemical that induces intestinal inflammation) developed enhanced colitis compared to littermate controls. Notably, inflammation was dependent on the presence of commensal bacteria, providing support for a multi-hit model of IBD, in which viral infection, the bacterial microbiome and genetic susceptibility collaborate in the development of IBD (Cadwell et al., 2010). Similar reports of MNV-microbiome-host interactions involved in IBD development have been observed in mice lacking a key regulatory cytokine (IL-10) that mediates immune tolerance to the bacterial microbiota and resolves inflammation after infection (Basic et al., 2014). Providing clinical relevance for these murine studies, epidemiological data suggests that huNoV infections are associated with post-infectious chronic irritable bowel syndrome and enteropathy development (Zanini et al., 2012, Woodward et al., 2015) although the genetics of susceptible populations remain unknown.
In other settings, the intestinal virome may mitigate the severity of intestinal inflammation (Yang et al., 2016). IBD patients with polymorphisms in the genes encoding viral recognition proteins Toll-like receptor (TLR) 3 and TLR7 have an elevated risk of hospitalization and TLR3/7-deficient mice develop exacerbated DSS-induced colitis compared to wildtype animals, collectively suggesting that TLR3/7-mediated recognition of the virome limits intestinal inflammation (Yang et al., 2016). Further, depleting the murine virome with broad-spectrum (DNA and RNA) antiviral treatment exacerbated DSS-induced colitis in wildtype mice. Although the virome and bacterial microbiome were both altered by antiviral treatment, exogenous treatment with TLR3 and TLR7 agonists reduced colitis severity independently of the microbiome, indicating that a virome-TLR3/7 signaling pathway may provide tonic anti-inflammatory signals to the host directly (Yang et al., 2016). While broad-spectrum antivirals are not commonly used, it is worth noting that disrupting the commensal enteric virome could influence multibiome composition and/or alter immune homeostasis.
Cross-talk between host genetics and the mycobiota also contributes to intestinal immune homeostasis. Polymorphisms in the fungal pattern recognition receptor Dectin-1 have been detected in a subset of IBD patients, and Dectin-1 deficient mice developed severe DSS-induced colitis that was associated with fungal dysbiosis (Iliev et al., 2012). In the absence of this genetic susceptibility, long-term antifungal treatment enhanced disease severity in two models of IBD (Wheeler et al., 2016). Notably, bacterial dysbiosis is also associated with antifungal treatment, including decreased representation of immunoregulatory Lactobacilli, which may alter the tolerant intestinal immune environment (Wheeler et al., 2016). The recent identification of a fungal-bacterial biofilm that is associated with IBD (Hoarau et al., 2016) supports the hypothesis that fungi can influence IBD progression through inter-multibiome as well as mycobiome-host interactions.
Together, these studies suggest common themes in virome and mycobiome interactions with the bacterial microbiome and host genetics. In otherwise healthy individuals, both the virome and mycobiome provide tonic signals that contribute to immune and microbiome homeostasis. However, in the context of a genetic susceptibility that impairs recognition or response to these multibiome members, the bacterial microbiome is implicated in intestinal inflammation.
5.2. Multibiome Influences on Systemic Chronic Inflammatory Diseases
Multiple sclerosis (MS) is a progressive autoimmune disease of the central nervous system (Sospedra and Martin, 2016). Similar to other CIDs, genetic susceptibility cannot fully explain the increased incidence and prevalence of MS in industrialized regions and the impact of the virome, bacterial microbiome and mycobiome on disease susceptibility and progression are being investigated (Ascherio and Munger, 2016). Epidemiologic data indicate an association between Epstein-Barr Virus (EBV) infection history and development of MS, especially if it manifested as severe infectious mononucleosis (Ascherio and Munger, 2016). Supporting this correlation, using a pre-clinical mouse model of MS (experimental autoimmune encephalomyelitis (EAE)), mice that harbored latent murine gammaherpesvirus-68 (the murine homolog of EBV) infection demonstrated more severe neuroinflammation and an immune profile that more closely resembled MS than the standard EAE model (Casiraghi et al., 2012).
Emerging clinical data now suggest that the intestinal microbiome is also disturbed in MS patients compared to healthy control populations, although it remains unknown whether these alterations in bacterial community populations are a cause or consequence of disease (Jangi et al., 2016, Miyake et al., 2015). Notably, successful therapeutic intervention is accompanied by restoration of a ‘normal’ bacterial microbiota (Jangi et al., 2016). Pre-clinical animal models support the hypothesis that the intestinal multibiome composition can impact neuronal immunopathology. EAE symptoms do not develop in GF mice, but colonizing GF mice with the Th17 promoting bacteria Segmented Filamentous Bacteria (SFB) restores EAE susceptibility (Lee et al., 2011). Similarly, the Th17 promoting fungus Candida albicans has been associated with exacerbated EAE and has been detected in the cerebrospinal fluid of MS patients (Fraga-Silva et al., 2015). Conversely, probiotic mixtures of regulatory T cell-promoting bacteria can dampen neuroinflammation and EAE symptoms (Kwon et al., 2013). Collectively, these findings indicate that the microbiome can tune the immune response to impart protective or pathogenic effects during MS or EAE.
To our knowledge, studies specifically examining interactions between viruses, bacteria and the mycobiota in the context of MS have not been reported. Although not discussed here, similar themes have emerged in the context of other autoimmune diseases, including rheumatoid arthritis and Type 1 Diabetes, suggesting that multibiome interactions with the host immune system could impact the onset, progression or severity of numerous autoimmune diseases. Our understanding of how these findings relate to human disease is still in its infancy, but microbial interventions may one day be incorporated into treatment plans for multiple autoimmune disorders.
6. Helminth Immunomodulation and Chronic Inflammatory Disease
The rising incidence and prevalence of autoimmune and CIDs in industrialized global regions is especially pronounced in comparison to the low, yet stable prevalence of these diseases in lower income regions (Maizels, 2016). The increased incidence of CIDs far exceeds the rate of population evolution, implicating environmental changes associated with industrialization as drivers of CIDs in the genetically predisposed. The near abolishment of helminth infections in industrialized areas is one recent, and potentially biologically important, environmental change (Maizels, 2016). As an example, treatment of Gabonese children with an anti-helminthic regimen resulted in a higher incidence of allergic reactions compared to untreated controls (van den Biggelaar et al., 2004). Parasitic infections have been major drivers of human evolution; populations from parasite-endemic regions have elevated frequencies of polymorphisms in genes associated with immune function (Fumagalli et al., 2011). These observations have lead to the hypothesis that ancestral heritage from parasite-endemic regions imparts an elevated risk of developing CIDs when paired with Western environmental exposures and the absence of helminth-mediated immune regulation (Fig. 5).
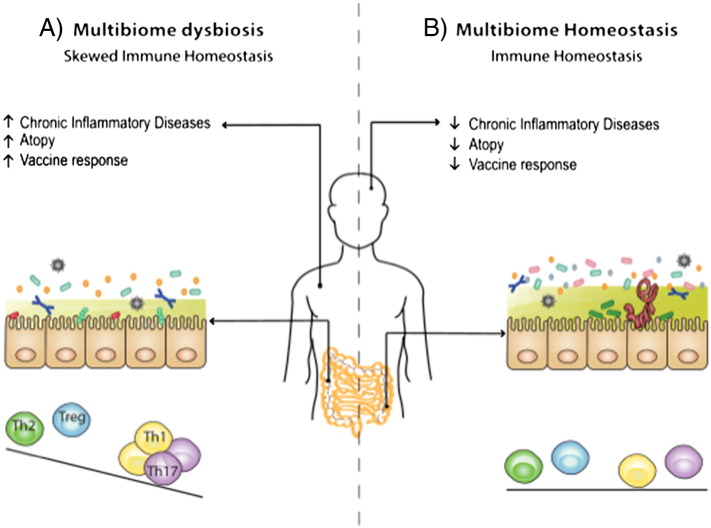
Fig. 5.
The multibiome as a critical regulator of immune responses.
Differences between industrialized and non-industrialized regions (infrastructure, sanitation, diet, medical interventions, and multibiome exposures) are factors associated with the disparity in the prevalence of CIDs and the efficacy of vaccines between different human populations.
A) An intestinal ecosystem with limited diversity is associated with antibiotic usage, decreased exposures to the multibiome and/or a ‘Westernized’ diet (high fat and sugar, low fiber). In industrialized nations, this is associated with good vaccine efficacy and decreased childhood mortality to infectious disease, but increased rates of autoimmune and CIDs.
B) An intestinal ecosystem rich in diversity may drive a balanced immune system. Non-industrialization is associated with endemic helminth infections, malnutrition, childhood morbidity and mortality, and diminished vaccine efficacy. Nevertheless, these regions have less autoimmune and CID incidence, indicating that factors that promote a complex multibiome might support a balanced immune system. Further research could help determine how to establish optimal multibiome diversity to limit CIDs without compromising host-protective immunity.
The immunomodulatory effects of helminth colonization have been proposed to limit intestinal inflammation and this is supported in both murine and non-human primate models of IBD (Hang et al., 2010, Broadhurst et al., 2012). As an example of bacteria-dependent helminth immunomodulation, we highlight studies that examined intestinal inflammation in genetically susceptible Nod2–/– mice (another risk allele for human IBD development). A commensal bacterial species, Bacteroides vulgatus, is responsible for the enhanced IBD severity noted in Nod2–/– mice (Ramanan et al., 2014). Infection with the helminth Heligmosomoides polygyrus prior to inducing intestinal inflammation was sufficient to reduce B. vulgatus colonization and diminish IBD severity in a type 2-dependent manner (Ramanan et al., 2016). Although Nod2 polymorphisms and/or dysregulated B. vulgatus colonization are not uniform features of IBD, these studies highlight the importance of host-multibiome interactions in the development and treatment of etiologically heterogeneous CIDs like IBD. However, despite anecdotal successes, clinical trials of helminth immunotherapy have been relatively unsuccessful (Maizels, 2016). These trials have primarily used the eggs of a pig whipworm (Trichuris suis ova, TSO). Further trials with other helminths, helminth-derived molecules or study design that accounts for host-multibiome interactions may yield more promising results.
Helminth immunotherapy is also being evaluated as a therapy for MS, with varying success. So far, clinical trials with TSO in the context of MS have demonstrated modest to no clinical improvement (Wolff et al., 2012). However, in a prospective study where MS patients were enrolled during remission, a subset of patients adventitiously acquired a helminth infection and remained in remission while the helminth-free cohort experienced periods of relapse and/or exacerbation (Correale and Farez, 2007). Additionally, patients that received anti-helminthic therapy relapsed and had reduced levels of circulating regulatory cytokines (Correale and Farez, 2007). This data supports the hypothesis that helminth-inspired therapies may aid MS patients, but much remains unknown about the underlying immunologic and/or multibiome-mediated mechanisms.
It is also important to recognize the inherent risks associated with helminth infections. These organisms are a significant cause of morbidity and mortality in developing regions and can impair nutrient absorption, vaccine efficacy and pathogen-specific immunity to viral, bacterial, and malarial co-infections (McKay, 2015). Helminth-derived products and type 2 cytokine therapy, instead of live-egg or worm infection, are being actively investigated as a treatment for multiple CIDs, which will hopefully provide similar benefits while negating the risks of helminth infections (Maizels, 2016, Ramanan et al., 2016).
7. Outstanding Questions in Multibiome-host Research and Translation
We are at the forefront of understanding the complex intestinal ecosystem and the impact of host-multibiome interactions on immune homeostasis, health, and disease. Relatively novel kingdom agnostic metagenomics may help account for the possible influences of all multibiome constituents on a specific research question (Norman et al., 2014). Increasingly available cell-type specific genetic and gnotobiotic animal models can be used to experimentally determine how diverse members of the multibiome influence each other and the host. These approaches, along with standardized experimental conditions and reporting (Stappenbeck and Virgin, 2016) will advance our understanding of multibiome-host interactions and the development of effective treatment strategies for multiple human diseases.
The American baseball manager Casey Stengel once said, “Finding good players is easy. Getting them to play as a team is another story”. In order to harness our expanding knowledge of how individual multibiome members influence immune homeostasis and inflammatory diseases, we must embrace the holistic nature of the intestinal ecosystem and aim to understand how these biodiverse entities interact with each other and their host.
8. Search Strategy and Selection Criteria
Data for this review were identified by searches of PubMed and references from relevant articles using the search terms “microbiome”, “co-infection”, “helminth immunotherapy”, “mycobiome” and “virome”. Only articles published in English between 1954 and 2016 were included; however, preference was given to articles published between 2013 and 2016.
Conflict of Interest Statement
The authors have no conflicts of interest to report.
Acknowledgments
The authors would like to thank Dr Cara Haney and Blair Hardman for their thoughtful comments on the manuscript and Juliane Poschinski for help with figures. We apologize to colleagues whose work could not be directly quoted due to space constraints. The Osborne lab is supported by the Natural Sciences and Engineering Research Council of Canada (NSERC) and the Canadian Institutes of Health Research (CIHR). HAF holds a CIHR Canada Graduate Scholarship Master's award and LCO is supported by the Canada Research Chairs program.
References
- Abt M.C., Osborne L.C., Monticelli L.A., Doering T.A., Alenghat T., Sonnenberg G.F., Paley M.A., Antenus M., Williams K.L., Erikson J., Wherry E.J., Artis D. Commensal bacteria calibrate the activation threshold of innate antiviral immunity. Immunity. 2012;37:158–170. doi: 10.1016/j.immuni.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abt M.C., Buffie C.G., Susac B., Becattini S., Carter R.A., Leiner I., Keith J.W., Artis D., Osborne L.C., Pamer E.G. TLR-7 activation enhances IL-22-mediated colonization resistance against vancomycin-resistant enterococcus. Sci. Transl. Med. 2016;8:327ra25. doi: 10.1126/scitranslmed.aad6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen J.E., Maizels R.M. Diversity and dialogue in immunity to helminths. Nat. Rev. Immunol. 2011;11:375–388. doi: 10.1038/nri2992. [DOI] [PubMed] [Google Scholar]
- Ascherio A., Munger K.L. Epidemiology of multiple sclerosis: from risk factors to prevention-an update. Semin. Neurol. 2016;36:103–114. doi: 10.1055/s-0036-1579693. [DOI] [PubMed] [Google Scholar]
- Basic M., Keubler L.M., Buettner M., Achard M., Breves G., Schroder B., Smoczek A., Jorns A., Wedekind D., Zschemisch N.H., Gunther C., Neumann D., Lienenklaus S., Weiss S., Hornef M.W., Mahler M., Bleich A. Norovirus triggered microbiota-driven mucosal inflammation in interleukin 10-deficient mice. Inflamm. Bowel Dis. 2014;20:431–443. doi: 10.1097/01.MIB.0000441346.86827.ed. [DOI] [PubMed] [Google Scholar]
- Belkaid Y., Hand T.W. Role of the microbiota in immunity and inflammation. Cell. 2014;157:121–141. doi: 10.1016/j.cell.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brestoff J.R., Artis D. Commensal bacteria at the interface of host metabolism and the immune system. Nat. Immunol. 2013;14:676–684. doi: 10.1038/ni.2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broadhurst M.J., Ardeshir A., Kanwar B., Mirpuri J., Gundra U.M., Leung J.M., Wiens K.E., Vujkovic-Cvijin I., Kim C.C., Yarovinsky F., Lerche N.W., Mccune J.M., Loke P. Therapeutic helminth infection of macaques with idiopathic chronic diarrhea alters the inflammatory signature and mucosal microbiota of the colon. PLoS Pathog. 2012;8 doi: 10.1371/journal.ppat.1003000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodin P., Jojic V., Gao T., Bhattacharya S., Angel C.J., Furman D., Shen-Orr S., Dekker C.L., Swan G.E., Butte A.J., Maecker H.T., Davis M.M. Variation in the human immune system is largely driven by non-heritable influences. Cell. 2015;160:37–47. doi: 10.1016/j.cell.2014.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caballero S., Pamer E.G. Microbiota-mediated inflammation and antimicrobial defense in the intestine. Annu. Rev. Immunol. 2015;33:227–256. doi: 10.1146/annurev-immunol-032713-120238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadwell K., Patel K.K., Maloney N.S., Liu T.C., Ng A.C., Storer C.E., Head R.D., Xavier R., Stappenbeck T.S., Virgin H.W. Virus-plus-susceptibility gene interaction determines Crohn's disease gene Atg16L1 phenotypes in intestine. Cell. 2010;141:1135–1145. doi: 10.1016/j.cell.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr E.J., Dooley J., Garcia-Perez J.E., Lagou V., Lee J.C., Wouters C., Meyts I., Goris A., Boeckxstaens G., Linterman M.A., Liston A. The cellular composition of the human immune system is shaped by age and cohabitation. Nat. Immunol. 2016;17:461–468. doi: 10.1038/ni.3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casiraghi C., Shanina I., Cho S., Freeman M.L., Blackman M.A., Horwitz M.S. Gammaherpesvirus latency accentuates EAE pathogenesis: relevance to Epstein-Barr virus and multiple sclerosis. PLoS Pathog. 2012;8 doi: 10.1371/journal.ppat.1002715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correale J., Farez M. Association between parasite infection and immune responses in multiple sclerosis. Ann. Neurol. 2007;61:97–108. doi: 10.1002/ana.21067. [DOI] [PubMed] [Google Scholar]
- Duerkop B.A., Hooper L.V. Resident viruses and their interactions with the immune system. Nat. Immunol. 2013;14:654–659. doi: 10.1038/ni.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Aidy S., Van Baarlen P., Derrien M., Lindenbergh-Kortleve D.J., Hooiveld G., Levenez F., Dore J., Dekker J., Samsom J.N., Nieuwenhuis E.E., Kleerebezem M. Temporal and spatial interplay of microbiota and intestinal mucosa drive establishment of immune homeostasis in conventionalized mice. Mucosal Immunol. 2012;5:567–579. doi: 10.1038/mi.2012.32. [DOI] [PubMed] [Google Scholar]
- Fraga-Silva T.F., Mimura L.A., Marchetti C.M., Chiuso-Minicucci F., Franca T.G., Zorzella-Pezavento S.F., Venturini J., Arruda M.S., Sartori A. Experimental autoimmune encephalomyelitis development is aggravated by Candida albicans infection. J. Immunol. Res. 2015;2015:635052. doi: 10.1155/2015/635052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumagalli M., Sironi M., Pozzoli U., Ferrer-Admetlla A., Pattini L., Nielsen R. Signatures of environmental genetic adaptation pinpoint pathogens as the main selective pressure through human evolution. PLoS Genet. 2011;7 doi: 10.1371/journal.pgen.1002355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handley S.A., Thackray L.B., Zhao G., Presti R., Miller A.D., Droit L., Abbink P., Maxfield L.F., Kambal A., Duan E., Stanley K., Kramer J., Macri S.C., Permar S.R., Schmitz J.E., Mansfield K., Brenchley J.M., Veazey R.S., Stappenbeck T.S., Wang D., Barouch D.H., Virgin H.W. Pathogenic simian immunodeficiency virus infection is associated with expansion of the enteric virome. Cell. 2012;151:253–266. doi: 10.1016/j.cell.2012.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hang L., Setiawan T., Blum A.M., Urban J., Stoyanoff K., Arihiro S., Reinecker H.C., Weinstock J.V. Heligmosomoides polygyrus infection can inhibit colitis through direct interaction with innate immunity. J. Immunol. 2010;185:3184–3189. doi: 10.4049/jimmunol.1000941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes K.S., Bancroft A.J., Goldrick M., Portsmouth C., Roberts I.S., Grencis R.K. Exploitation of the intestinal microflora by the parasitic nematode Trichuris muris. Science. 2010;328:1391–1394. doi: 10.1126/science.1187703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoarau G., Mukherjee P.K., Gower-Rousseau C., Hager C., Chandra J., Retuerto M.A., Neut C., Vermeire S., Clemente J., Colombel J.F., Fujioka H., Poulain D., Sendid B., Ghannoum M.A. Bacteriome and mycobiome interactions underscore microbial dysbiosis in familial Crohn's disease. MBio. 2016;7 doi: 10.1128/mBio.01250-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper L.V., Littman D.R., Macpherson A.J. Interactions between the microbiota and the immune system. Science. 2012;336:1268–1273. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotez P.J., Brindley P.J., Bethony J.M., King C.H., Pearce E.J., Jacobson J. Helminth infections: the great neglected tropical diseases. J. Clin. Invest. 2008;118:1311–1321. doi: 10.1172/JCI34261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichinohe T., Pang I.K., Kumamoto Y., Peaper D.R., Ho J.H., Murray T.S., Iwasaki A. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc. Natl. Acad. Sci. U. S. A. 2011;108:5354–5359. doi: 10.1073/pnas.1019378108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliev I.D., Funari V.A., Taylor K.D., Nguyen Q., Reyes C.N., Strom S.P., Brown J., Becker C.A., Fleshner P.R., Dubinsky M., Rotter J.I., Wang H.L., Mcgovern D.P., Brown G.D., Underhill D.M. Interactions between commensal fungi and the C-type lectin receptor Dectin-1 influence colitis. Science. 2012;336:1314–1317. doi: 10.1126/science.1221789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jangi S., Gandhi R., Cox L.M., Li N., Von Glehn F., Yan R., Patel B., Mazzola M.A., Liu S., Glanz B.L., Cook S., Tankou S., Stuart F., Melo K., Nejad P., Smith K., Topcuolu B.D., Holden J., Kivisakk P., Chitnis T., De Jager P.L., Quintana F.J., Gerber G.K., Bry L., Weiner H.L. Alterations of the human gut microbiome in multiple sclerosis. Nat. Commun. 2016;7:12015. doi: 10.1038/ncomms12015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang V., Jiang B., Tate J., Parashar U.D., Patel M.M. Performance of rotavirus vaccines in developed and developing countries. Hum. Vaccin. 2010;6:532–542. doi: 10.4161/hv.6.7.11278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karst S.M. The influence of commensal bacteria on infection with enteric viruses. Nat. Rev. Microbiol. 2016;14:197–204. doi: 10.1038/nrmicro.2015.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy M.J., Volz P.A. Ecology of Candida albicans gut colonization: inhibition of Candida adhesion, colonization, and dissemination from the gastrointestinal tract by bacterial antagonism. Infect. Immun. 1985;49:654–663. doi: 10.1128/iai.49.3.654-663.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kernbauer E., Ding Y., Cadwell K. An enteric virus can replace the beneficial function of commensal bacteria. Nature. 2014;516:94–98. doi: 10.1038/nature13960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause R., Schwab E., Bachhiesl D., Daxbock F., Wenisch C., Krejs G.J., Reisinger E.C. Role of Candida in antibiotic-associated diarrhea. J. Infect. Dis. 2001;184:1065–1069. doi: 10.1086/323550. [DOI] [PubMed] [Google Scholar]
- Kreisinger J., Bastien G., Hauffe H.C., Marchesi J., Perkins S.E. Interactions between multiple helminths and the gut microbiota in wild rodents. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 2015;370 doi: 10.1098/rstb.2014.0295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon H.K., Kim G.C., Kim Y., Hwang W., Jash A., Sahoo A., Kim J.E., Nam J.H., Im S.H. Amelioration of experimental autoimmune encephalomyelitis by probiotic mixture is mediated by a shift in T helper cell immune response. Clin. Immunol. 2013;146:217–227. doi: 10.1016/j.clim.2013.01.001. [DOI] [PubMed] [Google Scholar]
- Labeaud A.D., Malhotra I., King M.J., King C.L., King C.H. Do antenatal parasite infections devalue childhood vaccination? PLoS Negl. Trop. Dis. 2009;3 doi: 10.1371/journal.pntd.0000442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawley T.D., Walker A.W. Intestinal colonization resistance. Immunology. 2013;138:1–11. doi: 10.1111/j.1365-2567.2012.03616.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.K., Menezes J.S., Umesaki Y., Mazmanian S.K. Proinflammatory T-cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. Proc. Natl. Acad. Sci. U. S. A. 2011;108(Suppl 1):4615–4622. doi: 10.1073/pnas.1000082107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maizels R.M. Parasitic helminth infections and the control of human allergic and autoimmune disorders. Clin. Microbiol. Infect. 2016;22:481–486. doi: 10.1016/j.cmi.2016.04.024. [DOI] [PubMed] [Google Scholar]
- Mason K.L., Erb Downward J.R., Falkowski N.R., Young V.B., Kao J.Y., Huffnagle G.B. Interplay between the gastric bacterial microbiota and Candida albicans during postantibiotic recolonization and gastritis. Infect. Immun. 2012;80:150–158. doi: 10.1128/IAI.05162-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mckay D.M. Not all parasites are protective. Parasite Immunol. 2015;37:324–332. doi: 10.1111/pim.12160. [DOI] [PubMed] [Google Scholar]
- Mina M.J., Metcalf C.J., De Swart R.L., Osterhaus A.D., Grenfell B.T. Long-term measles-induced immunomodulation increases overall childhood infectious disease mortality. Science. 2015;348:694–699. doi: 10.1126/science.aaa3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyake S., Kim S., Suda W., Oshima K., Nakamura M., Matsuoka T., Chihara N., Tomita A., Sato W., Kim S.W., Morita H., Hattori M., Yamamura T. Dysbiosis in the gut microbiota of patients with multiple sclerosis, with a striking depletion of species belonging to Clostridia XIVa and IV clusters. PLoS One. 2015;10 doi: 10.1371/journal.pone.0137429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaco C.L., Gootenberg D.B., Zhao G., Handley S.A., Ghebremichael M.S., Lim E.S., Lankowski A., Baldridge M.T., Wilen C.B., Flagg M., Norman J.M., Keller B.C., Luevano J.M., Wang D., Boum Y., Martin J.N., Hunt P.W., Bangsberg D.R., Siedner M.J., Kwon D.S., Virgin H.W. Altered virome and bacterial microbiome in human immunodeficiency virus-associated acquired immunodeficiency syndrome. Cell Host Microbe. 2016;19:311–322. doi: 10.1016/j.chom.2016.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naylor C., Lu M., Haque R., Mondal D., Buonomo E., Nayak U., Mychaleckyj J.C., Kirkpatrick B., Colgate R., Carmolli M., Dickson D., Van Der Klis F., Weldon W., Steven Oberste M., Teams P.S., Ma J.Z., Petri W.A., JR. Environmental enteropathy, oral vaccine failure and growth faltering in infants in Bangladesh. EBioMedicine. 2015;2:1759–1766. doi: 10.1016/j.ebiom.2015.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson J.K., Holmes E., Kinross J., Burcelin R., Gibson G., Jia W., Pettersson S. Host-gut microbiota metabolic interactions. Science. 2012;336:1262–1267. doi: 10.1126/science.1223813. [DOI] [PubMed] [Google Scholar]
- Norman J.M., Handley S.A., Virgin H.W. Kingdom-agnostic metagenomics and the importance of complete characterization of enteric microbial communities. Gastroenterology. 2014;146:1459–1469. doi: 10.1053/j.gastro.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman J.M., Handley S.A., Baldridge M.T., Droit L., Liu C.Y., Keller B.C., Kambal A., Monaco C.L., Zhao G., Fleshner P., Stappenbeck T.S., Mcgovern D.P., Keshavarzian A., Mutlu E.A., Sauk J., Gevers D., Xavier R.J., Wang D., Parkes M., Virgin H.W. Disease-specific alterations in the enteric virome in inflammatory bowel disease. Cell. 2015;160:447–460. doi: 10.1016/j.cell.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orru V., Steri M., Sole G., Sidore C., Virdis F., Dei M., Lai S., Zoledziewska M., Busonero F., Mulas A., Floris M., Mentzen W.I., Urru S.A., Olla S., Marongiu M., Piras M.G., Lobina M., Maschio A., Pitzalis M., Urru M.F., Marcelli M., Cusano R., Deidda F., Serra V., Oppo M., Pilu R., Reinier F., Berutti R., Pireddu L., Zara I., Porcu E., Kwong A., Brennan C., Tarrier B., Lyons R., Kang H.M., Uzzau S., Atzeni R., Valentini M., Firinu D., Leoni L., Rotta G., Naitza S., Angius A., Congia M., Whalen M.B., Jones C.M., Schlessinger D., Abecasis G.R., Fiorillo E., Sanna S., Cucca F. Genetic variants regulating immune cell levels in health and disease. Cell. 2013;155:242–256. doi: 10.1016/j.cell.2013.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne L.C., Monticelli L.A., Nice T.J., Sutherland T.E., Siracusa M.C., Hepworth M.R., Tomov V.T., Kobuley D., Tran S.V., Bittinger K., Bailey A.G., Laughlin A.L., Boucher J.L., Wherry E.J., Bushman F.D., Allen J.E., Virgin H.W., Artis D. Coinfection. Virus-helminth coinfection reveals a microbiota-independent mechanism of immunomodulation. Science. 2014;345:578–582. doi: 10.1126/science.1256942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer J.K., Virgin H.W. Viral immunity. Transkingdom control of viral infection and immunity in the mammalian intestine. Science. 2016;351 doi: 10.1126/science.aad5872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanan D., Tang M.S., Bowcutt R., Loke P., Cadwell K. Bacterial sensor Nod2 prevents inflammation of the small intestine by restricting the expansion of the commensal Bacteroides vulgatus. Immunity. 2014;41:311–324. doi: 10.1016/j.immuni.2014.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanan D., Bowcutt R., Lee S.C., Tang M.S., Kurtz Z.D., Ding Y., Honda K., Gause W.C., Blaser M.J., Bonneau R.A., Lim Y.A., Loke P., Cadwell K. Helminth infection promotes colonization resistance via type 2 immunity. Science. 2016;352:608–612. doi: 10.1126/science.aaf3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese T.A., Wakeman B.S., Choi H.S., Hufford M.M., Huang S.C., Zhang X., Buck M.D., Jezewski A., Kambal A., Liu C.Y., Goel G., Murray P.J., Xavier R.J., Kaplan M.H., Renne R., Speck S.H., Artyomov M.N., Pearce E.J., Virgin H.W. Helminth infection reactivates latent gamma-herpesvirus via cytokine competition at a viral promoter. Science. 2014;345:573–577. doi: 10.1126/science.1254517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese T.A., BI K., Kambal A., Filali-Mouhim A., Beura L.K., Burger M.C., Pulendran B., Sekaly R.P., Jameson S.C., Masopust D., Haining W.N., Virgin H.W. Sequential infection with common pathogens promotes human-like immune gene expression and altered vaccine response. Cell Host Microbe. 2016;19:713–719. doi: 10.1016/j.chom.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes A., Haynes M., Hanson N., Angly F.E., Heath A.C., Rohwer F., Gordon J.I. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature. 2010;466:334–338. doi: 10.1038/nature09199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds L.A., Smith K.A., Filbey K.J., Harcus Y., Hewitson J.P., Redpath S.A., Valdez Y., Yebra M.J., Finlay B.B., Maizels R.M. Commensal-pathogen interactions in the intestinal tract: Lactobacilli promote infection with, and are promoted by, helminth parasites. Gut Microbes. 2014;5:522–532. doi: 10.4161/gmic.32155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson S.J., Horwitz M.S. Is type 1 diabetes “going viral”? Diabetes. 2014;63:2203–2205. doi: 10.2337/db14-0510. [DOI] [PubMed] [Google Scholar]
- Rook G.A. 99th Dahlem conference on infection, inflammation and chronic inflammatory disorders: Darwinian medicine and the ‘hygiene’ or ‘old friends’ hypothesis. Clin. Exp. Immunol. 2010;160:70–79. doi: 10.1111/j.1365-2249.2010.04133.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosebury T., Gale D., Taylor D.F. An approach to the study of interactive phenomena among microorganisms indigenous to man. J. Bacteriol. 1954;67:135–152. doi: 10.1128/jb.67.2.135-152.1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirtliff M.E., Peters B.M., Jabra-Rizk M.A. Cross-kingdom interactions: Candida albicans and bacteria. FEMS Microbiol. Lett. 2009;299:1–8. doi: 10.1111/j.1574-6968.2009.01668.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S.J., Lauber C., Costello E.K., Lozupone C.A., Humphrey G., Berg-Lyons D., Caporaso J.G., Knights D., Clemente J.C., Nakielny S., Gordon J.I., Fierer N., Knight R. Cohabiting family members share microbiota with one another and with their dogs. Elife. 2013;2 doi: 10.7554/eLife.00458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sospedra M., Martin R. Immunology of multiple sclerosis. Semin. Neurol. 2016;36:115–127. doi: 10.1055/s-0036-1579739. [DOI] [PubMed] [Google Scholar]
- Stappenbeck T.S., Virgin H.W. Accounting for reciprocal host-microbiome interactions in experimental science. Nature. 2016;534:191–199. doi: 10.1038/nature18285. [DOI] [PubMed] [Google Scholar]
- Stappenbeck T.S., Hooper L.V., Gordon J.I. Developmental regulation of intestinal angiogenesis by indigenous microbes via Paneth cells. Proc. Natl. Acad. Sci. U. S. A. 2002;99:15451–15455. doi: 10.1073/pnas.202604299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan A., Edlund C., Nord C.E. Effect of antimicrobial agents on the ecological balance of human microflora. Lancet Infect. Dis. 2001;1:101–114. doi: 10.1016/S1473-3099(01)00066-4. [DOI] [PubMed] [Google Scholar]
- Underhill D.M., Iliev I.D. The mycobiota: interactions between commensal fungi and the host immune system. Nat. Rev. Immunol. 2014;14:405–416. doi: 10.1038/nri3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Biggelaar A.H., Rodrigues L.C., Van Ree R., Van Der Zee J.S., Hoeksma-Kruize Y.C., Souverijn J.H., Missinou M.A., Borrmann S., Kremsner P.G., Yazdanbakhsh M. Long-term treatment of intestinal helminths increases mite skin-test reactivity in Gabonese schoolchildren. J. Infect. Dis. 2004;189:892–900. doi: 10.1086/381767. [DOI] [PubMed] [Google Scholar]
- Wheeler M.L., Limon J.J., Bar A.S., Leal C.A., Gargus M., Tang J., Brown J., Funari V.A., Wang H.L., Crother T.R., Arditi M., Underhill D.M., Iliev I.D. Immunological consequences of intestinal fungal dysbiosis. Cell Host Microbe. 2016;19:865–873. doi: 10.1016/j.chom.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff M.J., Broadhurst M.J., Loke P. Helminthic therapy: improving mucosal barrier function. Trends Parasitol. 2012;28:187–194. doi: 10.1016/j.pt.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward J.M., Gkrania-Klotsas E., Cordero-Ng A.Y., Aravinthan A., Bandoh B.N., Liu H., Davies S., Zhang H., Stevenson P., Curran M.D., Kumararatne D. The role of chronic norovirus infection in the enteropathy associated with common variable immunodeficiency. Am. J. Gastroenterol. 2015;110:320–327. doi: 10.1038/ajg.2014.432. [DOI] [PubMed] [Google Scholar]
- Yamaguchi N., Sugita R., Miki A., Takemura N., Kawabata J., Watanabe J., Sonoyama K. Gastrointestinal candida colonisation promotes sensitisation against food antigens by affecting the mucosal barrier in mice. Gut. 2006;55:954–960. doi: 10.1136/gut.2005.084954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J.Y., Kim M.S., Kim E., Cheon J.H., Lee Y.S., Kim Y., Lee S.H., Seo S.U., Shin S.H., Choi S.S., Kim B., Chang S.Y., Ko H.J., Bae J.W., Kweon M.N. Enteric viruses ameliorate gut inflammation via toll-like receptor 3 and toll-like receptor 7-mediated interferon-beta production. Immunity. 2016;44:889–900. doi: 10.1016/j.immuni.2016.03.009. [DOI] [PubMed] [Google Scholar]
- Zaiss M.M., Rapin A., Lebon L., Dubey L.K., Mosconi I., Sarter K., Piersigilli A., Menin L., Walker A.W., Rougemont J., Paerewijck O., Geldhof P., Mccoy K.D., Macpherson A.J., Croese J., Giacomin P.R., Loukas A., Junt T., Marsland B.J., Harris N.L. The intestinal microbiota contributes to the ability of helminths to modulate allergic inflammation. Immunity. 2015;43:998–1010. doi: 10.1016/j.immuni.2015.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanini B., Ricci C., Bandera F., Caselani F., Magni A., Laronga A.M., Lanzini A., San Felice del Benaco Study, I. Incidence of post-infectious irritable bowel syndrome and functional intestinal disorders following a water-borne viral gastroenteritis outbreak. Am. J. Gastroenterol. 2012;107:891–899. doi: 10.1038/ajg.2012.102. [DOI] [PubMed] [Google Scholar]