Abstract
Introduction
Long-term natural history cohorts of HIV-1 in the absence of treatment provide the best measure of virulence by different viral subtypes.
Methods
Newly HIV infected Ugandan and Zimbabwean women (N = 303) were recruited and monitored for clinical, social, behavioral, immunological and viral parameters for 3 to 9.5 years.
Results
Ugandan and Zimbabwean women infected with HIV-1 subtype C had 2.5-fold slower rates of CD4 T-cell declines and higher frequencies of long-term non-progression than those infected with subtype A or D (GEE model, P < 0.001), a difference not associated with any other clinical parameters. Relative replicative fitness and entry efficiency of HIV-1 variants directly correlated with virulence in the patients, subtype D > A > C (P < 0.001, ANOVA).
Discussion
HIV-1 subtype C was less virulent than either A or D in humans; the latter being the most virulent. Longer periods of asymptomatic HIV-1 subtype C could explain the continued expansion and dominance of subtype C in the global epidemic.
Keywords: HIV-1 diversity, Pathogenesis, Disease progression, Africa, Subtypes
Highlights
-
•
Subtype C is the dominant HIV-1 strain in the epidemic infecting > 15 million.
-
•
Women infected with HIV-1 subtype C viruses have slower disease than those infected with A or D viruses.
-
•
Long term non-progression is more common in subtype C as compared to A or D infected women.
-
•
In CD4 + T cells HIV-1 subtype C viruses are less fit than A or D viruses.
-
•
Subtype C virus envelopes function with reduced kinetic rate in both cell-free and cell-cell assays.
HIV-1 subtype C has expanded in the human population to dominate the global epidemic in just 10–20 years. Despite more 15 million people infected with subtype C, our 15 year study on the natural history of HIV infection clearly shows a slower rate of disease in women infected with subtype C than the other two prevalent subtypes, A and D in Africa. In vitro studies show that subtype C HIV-1 was less fit in human T cell cultures than subtype A and D HIV-1 derived from acute/early infection in these women.
1. Introduction
Human immunodeficiency virus type 1 (HIV-1) emerged in the human population shortly after the turn of the 19th century. Distribution of HIV-1 across the globe over the past 30–35 years can be traced to founder events with primordial HIV strains from sub-Saharan Africa (Sharp and Hahn, 2011, Arien et al., 2007, Arien et al., 2007, Tebit and Arts, 2011), a region which is still the HIV epicenter with 30 million historic and 28 million current infections (Sharp and Hahn, 2011, Arien et al., 2007, Tebit and Arts, 2011). Even in light of the burden of HIV in Africa, our knowledge of HIV-1 disease is still largely limited to subtype B HIV-1, a strain responsible for 3 million infections in North America and Europe as compared to the 33 million that are infected with HIV-1 subtypes A, C, D, and circulating and unique recombinant forms (CRFs and URFs, Fig. 1). Over the past 25 years (1990 to 2015), the expansion of 27.2 million new infections is largely due to the predominant spread of HIV-1 subtype C (accounting for 14.5 million new infections) over all other HIV-1 subtypes and recombinant forms. As documented in many reports, different HIV subtypes pre-dated the introduction of subtype C in southern Africa, South Asia, and Brazil but the rapid expansion of these epidemics in the past 25 years appears to coincide with the founding of HIV-1 subtype C in the heterosexual population.
Fig. 1.
The number of people living with HIV-1 between 1980 and 2015 in the four UNAIDS Regions with the greatest number of infections as well as globally. The distribution of subtypes in 1990 and 2015 are shown in the inset pie graphs, scaled by the number of infections. Relative amounts of subtype C infections per UNAIDS Region are reported on the pie graphs and via red shading. Data compiled from the World Health Organization, UNAIDS and published literature.
The extreme genetic diversity between HIV-1 subtypes has phenotypic consequences in vitro including differential HIV-1 mRNA transcriptional control (Montano et al., 2000, van et al., 2004), protease activity (Velazquez-Campoy et al., 2001), integration site selection (Demeulemeester et al., 2014), MHC class I downregulation (Mann et al., 2014), and entry efficiency (Marozsan et al., 2005). Overall, subtype C HIV-1 isolates have lower replicative fitness in tissue culture relative to HIV-1 isolates of any other subtype (Abraha et al., 2009, Ball et al., 2003, Arien et al., 2005). Relevance of these in vitro HIV studies to virulence would require comprehensive analyses on the natural history of infections with different HIV-1 subtypes, which could then in turn guide treatment and prevention strategies in the majority of people currently living with HIV-1. Longitudinal and cross-sectional studies in sub-Saharan Africa have suggested different disease courses and treatment outcomes in individuals infected with subtype A, B, C, D and URFs/CRFs (Amornkul et al., 2013, Rainwater et al., 2005, Baeten et al., 2007, Kaleebu et al., 2002, Palm et al., 2014, Kanki et al., 1999, Kiwanuka et al., 2009), the most significant being the faster disease progression and higher rates of treatment failures in subtype D versus subtype A infections (Baeten et al., 2007, Kaleebu et al., 2002, Kyeyune et al., 2013).
From 1999 to 2003, over 4400 HIV-negative women in Uganda and Zimbabwe were enrolled in the Hormonal Contraception and the Risk of HIV Acquisition (HC-HIV) Study (Morrison et al., 2007), during which 303 women were identified with incident HIV infection and participated in the Hormonal Contraception and HIV-1 Genital Shedding and Disease Progression among Women with Primary HIV Infection (GS) Study (Morrison et al., 2010, Morrison et al., 2011, Lemonovich et al., 2015). The 76 subtype A, 177 subtype C, 31 subtype D and two URF HIV infected women (286 of 303 women) were followed for an average of 5 years, currently the largest natural history cohort of non-subtype B infections. Consistent with WHO recommendations at the time of the study, treatment was provided when HIV-infected participants reached CD4 cell counts below 200/ml on two consecutive tests or were diagnosed with stage IV or advanced III disease (WHO Classification). It should be noted that anti-retroviral therapy (ART) was not yet routinely available in these countries at the start of this study period. This study was designed to monitor immune and viral parameters and to compare these biomarkers of disease progression to country of origin, contraceptive use, infecting subtype, and phenotypic properties of the virus.
2. Materials and Methods
2.1. Participants and Clinical Tests
A complete description of the Ugandan and Zimbabwean participants is previously described (Morrison et al., 2011, Lemonovich et al., 2015). Women who became HIV infected while participating in the Hormonal Contraception and Risk of HIV Acquisition Study in Uganda and Zimbabwe (Morrison et al., 2007) were enrolled upon primary HIV-1 infection into a subsequent study, the Hormonal Contraception and HIV-1 Genital Shedding and Disease Progression among Women with Primary HIV Infection (GS) Study (Morrison et al., 2010, Morrison et al., 2011, Lemonovich et al., 2015). Ethical approval was obtained from the Institutional Review boards (IRBs) from the Joint Clinical Research Centre and UNST in Uganda, from University of Zimbabwe, from the University Hospitals of Cleveland, and recently, from Western University. Protocol numbers and documentation of these approvals/renewals are available upon request. 71% (N = 203 of 286) were enrolled within 18 weeks of the infection date. Blood and cervical samples were collected every month for the first six months, then every three months for the first two years, and then every six months up to 9.5 years. Women who had CD4 lymphocyte counts of 200 cells/ml and/or who developed severe symptoms of HIV infection (WHO clinical stage IV or advanced stage III disease) were offered combination ART (cART) and trimethoprim-sulfamethoxazole (for prophylaxis against bacterial infections and Pneumocystis jeroveci pneumonia). In addition, information was collected at every visit related to changes in sociodemographic information, sexual behavior, sexually transmitted and reproductive tract infections, opportunistic infections, and diet.
Following primary infection, 112 Uganda and 174 Zimbabwean women were followed for an average 1826 and 1899 days (respectively) for a maximum of 3453 days (9.5 years) (Suppl. Fig.1a). Antiretroviral treatment was initiated in 23% of the women in both countries but 100 days sooner on average in the Ugandan versus Zimbabwean women (1458 versus 1558 days) (Suppl. Fig.1b). As described (Morrison et al., 2010), cervical and plasma viral loads were determined with the Roche Amplicor HIV-1 Monitor Test, version 1.5 using cryopreserved samples from every three month visits. CD4 T-lymphocyte counts were determined by standard flow cytometry using FACSCalibur (Becton Dickinson, Sparks, MD, USA).
2.2. Models to Determine Viral Load Increases and CD4 Cell Declines
A marginal model with the generalized estimating equation (GEE) approach including autoregressive error structures (accounting for repeated measurements on the same individual) was used to determine the plasma viral load changes and compare rates of CD4 cell decline (Morrison et al., 2010). Multivariable analyses included demographic, sexual, contraceptive and medical history data and underwent stepwise model simplification with a significance threshold of p ≤ 0.05. All analyses used Stata Version 14 (StataCorp LP, College Station, TX).
2.3. DNA Sequencing and Subtyping
DNA was extracted from patient PBMC samples obtained at the seroconversion visit and every 3 months post HIV infection. Primers specific to the protease (PR) and polymerase regions of reverse transcriptase (RT), and C2-V3 envelope (env) coding regions were used to PCR 300, 800, and 450 nt regions as described (Abraha et al., 2009) in a Virology Quality Assurance (VQA) certified laboratory in Kampala, Uganda (Morrison et al., 2010). The HIV-1 sequences were edited using BioEdit v7.0.4 and PR-RT coding regions were uploaded into the Stanford University HIV Drug resistance database (http://hivdb.stanford.edu) to obtain drug resistance profiles. Phylogenetic alignments were performed using maximum likelihood methods (MUSCLE (Edgar, 2004)) and trees constructed using SEAVIEW 4 (Gouy et al., 2010) and FigTree 1.4 (http://tree.bio.ed.ac.uk/software/figtree/). Each participant PR, RT, and env sequence was aligned to curated set of subtype A, B, C, D, G, and CRFs HIV-1 reference sequences (hiv.lanl.gov).
2.4. Virus Cloning and Propagation
The entire gp120 coding region and the extracellular domain of gp41 (referred to as env DNA) was PCR amplified with the ENV REC primer set from PBMC DNA derived from the seroconversion visit. This Env PCR product was cloned into pREC_nflNL4–3 ∆ env by yeast recombination/gap repair as described previously. pREC_nfl (containing the patient derived env gene) was transfected to produce virus-like particles for entry and cell fusion assays. Co-transfections with pCMV_cplt in 293T cells produces replication-competent virus for replicative fitness (Dudley et al., 2009).
2.5. Replicative Fitness
47 primary HIV-1 isolates of subtypes A (Arien et al., 2005), C (Dudley et al., 2009), and D (Mann et al., 2014) derived from patients from around the world were tittered on PHA-activated, IL2-treated PBMCs of HIV-negative donors using the standard Reed-Munch TCID50 determination (Reed and Muench, 1938) then competed against 25 subtype B primary HIV-1 in the same PBMCs. The primary isolates of HIV-1 subtype A, B, C and D were all obtained from chronic infection, many are part of the WHO collection, whereas others were isolated from our laboratory in Uganda. A complete summary of all the viruses is available upon request. All of the viruses have been previously screened for co-receptor usage. Any CXCR4-using or dual tropic viruses were excluded from this analysis. We also competed chimeric env viruses derived from the acute/early infection samples of 5 subtype A, 3 subtype D, and 5 subtype C infections of this cohort against two reference chimeric env viruses derived from subtype B HIV-1 samples, B-Q0 and B-K44. The reference B-Q0 and B-K44 were derived from untreated patient Q at early infection (month 0) and from untreated patient K at 44 months post infection (Troyer et al., 2005, King et al., 2013). The chimeric env viruses had the env gene from the AHI sample or the chronic reference sample cloned into the HIV-1 NL4-3 backbone using our yeast-based cloning technique (Dudley et al., 2009). The reference chimeric env virus with chronic subtype B HIV-1 has been employed in multiple published and in press studies and serves as a reference competitor virus to standardize relative fitness. The relative production of two viruses in each competition was measured by a radiolabeled heteroduplex tracking assays (HTA) or using next generation sequencing (NGS) (Abraha et al., 2009, Ball et al., 2003, Arien et al., 2005, Rubio et al., 2014). It is important to note that same relative fitness values (< 5% variance) was obtained using the HTA and NGS, a finding that will be reported in submitted methods paper. Assay sensitivity is 100-fold differences in replicative fitness expressed as percentages.
2.6. Cell-to-Cell Infections
Veritrop involved the transfection (Fugene 6, Promega) of the pRECnfl constructs containing patient envs into 6.5 × 104 HEK293T cells (Weber et al., 2013) and cocultured with target U87 cells. Detailed methods are published previously. Fully inhibitory concentrations of Enfuvirtide (3 μM) were added at discrete time points (t = 4 to 7 h) and luciferase production was quantified with the Bright-Glo Luciferase Assay System (Promega) to measure relative cell fusion. Drug treated cells were compared to untreated controls to calculate percent maximal fusion. Time required to reach 50% maximal fusion by different env in pREC-NFL constructs subtype were statistically compared using ANOVA (STATA Version 14).
2.7. Virus Entry Into Host Cells
Cell-free viral entry utilized the pMM310 plasmid (NIH AIDS Repository, cat #11,444) to generate patient env viruses containing the HIV-1 accessory protein Vpr fused to E. coli β-lactamase (BlaM). Cells were placed on a plate reader and virus entry was quantified every 15 min by comparing changes in emission spectra (460 nm cleaved dye relative to 530 nm uncleaved dye), then normalized relative to cell-free and virus-free controls to determine time required to reach 75% maximal viral entry (ANOVA).
3. Results
3.1. Monitoring Disease Progression in the Absence of Treatment
Newly infected women in Uganda (N = 112) and Zimbabwe (N = 174) were followed after acute/early HIV infection (AHI) for an average of 5.1 years up to 9.5 years in the absence of combination antiretroviral therapy (cART), initiated when CD4 cell counts dropped below 200/mm3 (Suppl. Fig. 1). Complete sociodemographic, clinical blood chemistries, and other clinical data has been reported (Morrison et al., 2010, Morrison et al., 2011, Lemonovich et al., 2015, Lovvorn et al., 2010).
Lower CD4 cell levels were reported in Zimbabwean as compared to Ugandan women (Lovvorn et al., 2010), both uninfected and in women with AHI. Despite this difference between countries, declines in CD4 T-cells was 2-fold slower in the Zimbabwean than in the Ugandan women in the absence of treatment, translating to 2 to 5 years of longer asymptomatic disease (Fig. 2a, Suppl. Fig. 2) (GEE model; P < 0.001). Virus levels at both the set point (Morrison et al., 2010) and all subsequent time points in the plasma and endocervix did not significantly differ by country (Suppl. Fig. 3a & b). Approximately 8.1% (n = 9) and 7.4% (n = 13) were classified as rapid progressors in Uganda and Zimbabwe (CD4 decline to below 200 cells/ml within 2 years of infection) but slow progressors/controllers (stable CD4 cell counts above 350 and viral loads < 2000 copies/ml in plasma for > 3 years post-infection) were observed in < 3% (n = 3) of Ugandan women versus 10% (n = 18) of Zimbabwean women (Fig. 2b). It is important to note that when rapid progressors and controllers were removed from the analyses, the CD4 declines were still significantly slower in Zimbabwean versus Ugandan women (P < 0.001).
Fig. 2.
CD4 + cell declines in HIV-1 infected Ugandan (n = 112) and Zimbabwean women (n = 174) during the natural course of disease. (a) The mean loss of CD4 cells per mm3 per week of infection in Ugandan and Zimbabwean women and (c) in those women infected with subtypes A, C, and D. (b) The percentage of rapid progressors and slow progressors/controllers in Ugandan and Zimbabwean women and (d) in those women infected with subtypes A, C, and D. Rapid progressors are defined as HIV-1 infected women with a drop to 200 CD4 cell counts/mm3 within 2 years of infection and sustained viral RNA loads > 2000 copies/ml. Slow progressors/controllers had viral RNA loads stable CD4 cell counts above 350 and viral loads < 2000 copies/ml in plasma for > 3 years.
3.2. Possible Factors Contributing to Country-Specific Difference in Disease Progression
To explain these differences in CD4 T-cell decline in HIV-1 infected women between countries, we analyzed numerous factors previously described as correlates of disease progression. Appearance of various opportunistic infections and secondary infections (malaria, TB, various STIs) were not significantly different in the cohorts and each infection was treated accordingly upon diagnosis. Other factors such as diet, recurrent sexually transmitted diseases (chlamydia, trichomonas, HSV-2, bacterial vaginosis), smoking, sexual behavior, depot-medroxyprogesterone acetate (DMPA) use, and combined oral contraceptives (COC) use did not show significant differences in HIV-1 disease progression between countries (Morrison et al., 2010, Morrison et al., 2011, Lemonovich et al., 2015, Lovvorn et al., 2010, Bark et al., 2010).
There may be a perceived difference in human genetics between the two countries. Our informed consent and IRB approvals did not permit genetic testing in these patients. These women self-reported being members of the Shona tribe of Zimbabwe and Buganda tribe of Uganda. Both are of Bantu origin which has the highest HLA diversity among humans as well as low frequencies of “HIV protective alleles”, HLA B57 (3–5%) and B27 (< 1%) (Cao et al., 2004, Paximadis et al., 2012, Kijak et al., 2009) when compare to Caucasian populations (7.0 and 9.2%, respectively) (www.allelefrequencies.net/). There are also several reports of the Bantu (of both Uganda and Zimbabwe) and African populations in general having low frequencies of CCR5Δ32 deletions. Genome-wide association studies in African ancestry populations did not reveal a single genetic polymorphism associated with slow disease progression and protection from HIV acquisition whereas several polymorphisms in the HLA region of chromosome 6 (e.g. HLA B5701) did appear at sufficient frequency in in European ancestry population to impact HIV-1 disease (Limou and Zagury, 2013). Thus, genetic polymorphisms are unlikely to be the only explanation for the striking differences in CD4 decline in the HIV-infected Ugandan and Zimbabwean women.
3.3. Infecting HIV-1 Subtype Correlates to Disease Progression
Based on the pol and env coding regions, the Ugandan women, all residing in Kampala, were predominantly infected with HIV-1 subtype A (68%, n = 76) followed by subtype D (28%, n = 31), 3 cases of subtype C, and 2 intersubtype A/D recombinants (Suppl. Figs. 5 & 6). Previous studies in Kampala, Uganda involving over 1500 HIV-infected patients showed a similar frequency of HIV-1 subtype and less than a 5% frequency of intersubtype HIV-1 recombinants (within or between pol and env genes, roughly three quarters of the genome) (Kyeyune et al., 2013). With full genome sequencing, intersubtype recombinants are generally detected at a 5–10% frequency in Kampala, Uganda. Most HIV-1 recombinants in East Africa are unique recombinant forms that have recently emerged or have very limited transmission chains with minimal evidence of extensive spread as observed with circulating recombinant forms (CRFs, specifically 02 and 06) found in the historic origin/epicenter, i.e. the Congo basin and its watershed regions in West Africa.
All 174 Zimbabwean women were infected with HIV-1 subtype C. Based on the infecting subtype, women with subtype C HIV-1 infections had CD4 T-cell declines (− 0.489 cells/week, GEE model) 2.5-fold and 1.6-fold slower than with subtype D and A infections, respectively (Fig. 2c). The faster CD4 T-cell declines in subtype D infections also resulted in more patients (45%, n = 14) requiring cART than in women infected with subtype A and C (28% and 32%) (Suppl. Fig. 1b). Slow/non-progression (defined as controllers) was not observed among the subtype D infected women as compared to the 4% (n = 3) and 10% in the subtype A and C infected women, respectively. In contrast, subtype A, D, and C infected women had similar frequencies of rapid progressors (8.5, 11, and 7.5%, respectively) (Fig. 2d). Again, viral levels at set point (Suppl. Fig. 3c) in plasma or endocervical mucosa or in plasma during disease (Suppl. Fig. 3 d, e & f) did not differ by subtype (statistical analyses in Suppl. Fig. 3g).
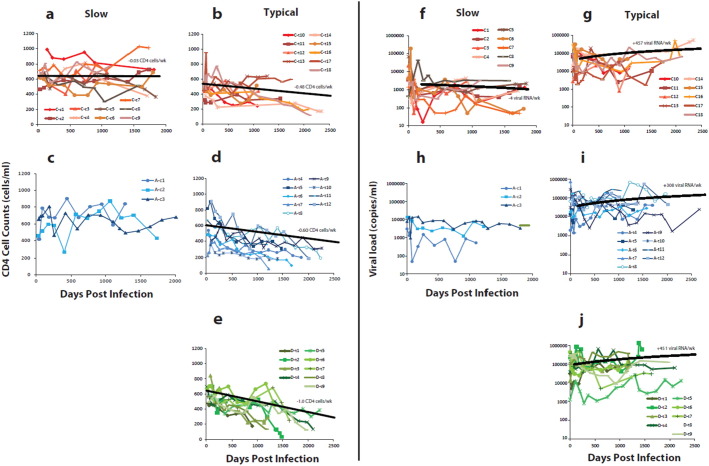
To illustrate the subtype-specific differences, CD4 T-cell and viral RNA levels were plotted for subtype C and A controllers (Fig. 3a & c respectively) and subtype C, A, and D typical progressors (Fig. 3b, d & e respectively) (nine patients for each). The controller status in subtype C infected patients is quite evident with < 0.03 CD4 cells lost per week (based on all 13 controllers) (Fig. 3a) as compared to the 0.48 lost per week (linear regression or GEE model) in the 148 typical subtype C progressors (Fig. 3b). This rate of CD4 T-cell decline in subtype C is slower than that observed in the typical subtype D (Fig. 2e) and subtype A (Fig. 3d) progressors (− 1.0 and − 0.60 CD4 cell loss/week, respectively, linear regression and GEE). Following the set point, viral load increases were similar in all the subtype A, C, and D progressors (Fig. 3g, i, & j) but viral loads remained the lowest in the subtype C controllers with an actual decrease over time (− 4 HIV-1 RNA copies/ml/week; Fig. 3f).
Fig. 3.
Comparing disease progression in a subset of 56 women in Uganda and Zimbabwe infected by different subtypes. (a)(b)(c)(d)(e) CD4 cell counts/ml were plotted over time in subtype C infected women defined as slow progressors/controllers (a), as typical progressors (b), in subtype A infected women defined as slow progressors/controllers (c), as typical progressors (d), and in subtype D infected women defined as typical progressors (e). (f)(g)(h)(i)(j) Viral RNA loads in plasma of subtype C slow progressors, C typical progressors, A slow progressors, A typical progressors and D typical progressors, respectively.
3.4. Phenotypic Differences between HIV-1 Subtypes
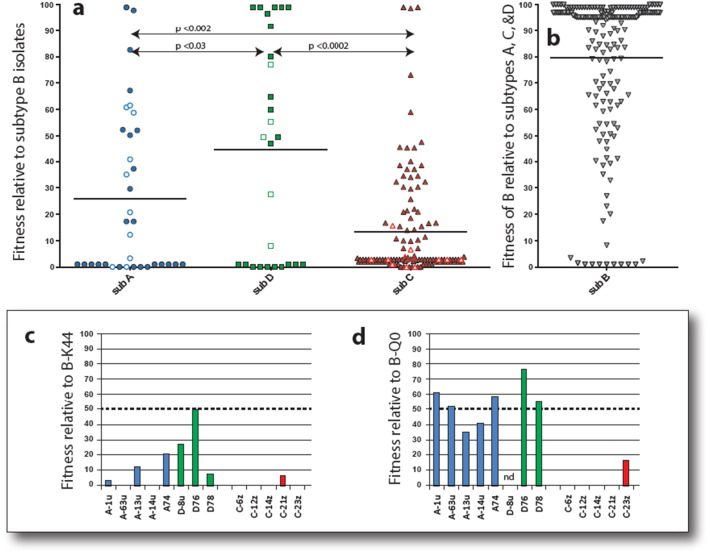
For this study, we analyzed 185 dual HIV-1 competitions in HIV-negative PBMCs involving 47 subtypes A, D, and C HIV-1 isolates against 25 subtype B isolates (Fig. 4a & b). In these dual infection/competitions, absolute virus production can vary 100-fold in PBMCs of different human donors but the relative production (i.e. fitness) of one virus versus the other typically varies < 10% (Abraha et al., 2009, Ball et al., 2003, Arien et al., 2005). Subtype C HIV-1 isolates are clearly less fit in PBMCs than subtype A and D HIV-1 isolates (P < 0.002) when competing against subtype B HIV-1 (Fig. 4a). Subtype C viruses were not detected (i.e. 100-fold less fit) in 60% (n = 74/123) of the competitions despite the fact that these same subtype C viruses were readily detected in mono-infections or competitions against other subtype C isolates (Abraha et al., 2009, Ball et al., 2003, Arien et al., 2005). These competitions suggest a fitness order of HIV-1 subtype B = D > A > C in human PBMC or primary CD4 T-cells (Fig. 4) (Abraha et al., 2009, Ball et al., 2003, Arien et al., 2005). We have previously reported that the replicative fitness of primary HIV-1 isolates is dominated by the env gene and by the rate of cell entry, i.e. the rate limiting step in HIV-1 replication (Marozsan et al., 2005, Ball et al., 2003). Five subtype A, 5 C, and 3 D HIV-1 env genes (from 13 patients) derived from AHI were used to produce chimeric virus (subtype B NL4-3 backbone) which were then competed against the B-QO and B-K44 subtype B reference viruses (Fig. 4c & d, respectively). The AHI subtype D Env chimeric viruses were able to compete with both the B-Q0 and B-K44 reference viruses whereas the AHI subtype A Env chimeric viruses were less fit (Fig. 4c & d). In contrast, AHI subtype C Env chimeric viruses had extremely low replicative fitness or were completely outcompeted by the B-K44 and B-Q0 reference viruses. To demonstrate that these differences are due to subtype rather than within patient fitness increase, the relative fitness of viral isolates from chronic infections were compared to isolates from AHI for both individual subtypes and all isolates (p > 0.05, Suppl. Fig. 4). Thus, lower “pathogenic” fitness of subtype C HIV-1 is also observed at initial infection and may be predictive of slower disease progression.
Fig. 4.
Relative replicative fitness of HIV-1 subtype A, B, C, and D in human peripheral blood mononuclear cells. (a) 12 subtype A, 8 subtype D, and 27 subtype C HIV-1 primary isolates were competed against 25 subtype B HIV-1 isolates in PHA-activated, IL2-treated PBMCs of HIV-negative donors in series of published and unpublished studies. All competitions in PBMCs involved the use of 0.001 infectious units of virus to 1 cell. The relative production of two viruses in each competition was measured by a radiolabeled heteroduplex tracking assays or using next generation sequencing (Abraha et al., 2009, Ball et al., 2003, Arien et al., 2005). Production of individual HIV-1 isolates in a dual infection (f0) divided by its initial proportion in the inoculum (i0) is referred to as the relative fitness (w = f0/i0). In this study, w is expressed as percent replication of the subtype A, C, or D HIV-1 isolates relative to subtype B HIV-1. Assay sensitivity is 100-fold differences in replicative fitness. (b) The relative replicative fitness of the subtype B HIV-1 isolates in competition with the subtype A, C, and D HIV-1 (the reciprocal of a). Symbols with thick outlines represent replicative fitness of the classified virus at > 100-fold or < 0.01-fold to competitor virus. The shaded symbols represent the replicative fitness values of primary HIV-1 isolates. The open/white symbols represent the replicative fitness values of replication-competent chimeric viruses containing the env gene of a subtype A, C, or D virus within the NL4-3 subtype B backbone. (c) and (d) Replicative fitness of chimeric viruses derived from env gene of 5 subtype A, 3 subtype D, and 5 subtype C AHI of this cohort. These 13 AHI chimeric viruses were competed against the reference subtype B HIV-1 isolates, B-QO (c) and B-K44 (triplicate).
HIV-1 infection can be mediated by cell-to-cell virus transfer or by direct virus infection. We examined cell-to-cell infection using the more physiological Veritrop system (Weber et al., 2013) where transfection of the pREC-nfl HIV-1 vector into 293T effector cells can produce HIV-1 particles (non-infectious) and can mediate fusion with the U87.CD4.CCR5 target cell. Initial assays without Enfuvirtide were used to validate CCR5 tropism in all patient envelopes (Fig. 5a) In addition, we only detected Env-mediated cell fusion with U87.CD4.CCR5 target cells and not with U87.CD4.CXCR4 cells. Absolute level of cell-to-cell HIV transmission was similar with 34 AHI env genes in pREC-nfl over a 12 h incubation (Fig. 5a). Cell-to-cell fusion kinetics was then measured using a time-of-drug-addition experiment where Enfuvirtide, an inhibitor of HIV-1 entry, was added at different times post co-incubation of effector + target cells (Fig. 5b). AHI subtype C HIV-1 Env glycoproteins mediated significantly slower rates of cell-to-cell HIV-1 infection than did AHI subtype A (P < 0.001) or subtype D (P < 0.001) Env glycoproteins based on the T1/2 of Enfuvirtide inhibition (Fig. 5c).
Fig. 5.
Function, cell-to-cell transmission efficiency and viral fusion of the Env glycoprotein derived from acute subtype A, C, and D HIV-1 infections. The HIV-1 env gene of acute/early infections in Ugandan and Zimbabwean women was cloned into the pREC-nfl HIV-1 genomic vector by yeast-based recombination/gap repair as described. The function and co-receptor usage of the Env glycoprotein from these 14 subtype A, 4 subtype D, and 16 subtype C acute/early infections was accessed using the Veritrop cell-to-cell fusion assay. None of the 34 Env produced in the context of virus-like particles (VLPs) could mediate cell fusion via the CXCR4 co-receptor within U87.CD4.CXCR4 cells (below negative control and 1000 RLUs) (data below range of graph in (a)). (a) Cell-to-cell fusion between the U87.CD4.CCR5 target cells expressing Firefly Luciferase upon Tat/Rev-mediate expression from pDM1.1 and the 293T effector cells expressing the 34 Env glycoproteins in context with HIV-1 VLPs. The NL4-3 nfl VLPs are morphologically identical to wild type HIV but is incapable of reverse transcription and cannot induce luciferase expression in the target cells. (b) Schematic of the time-of-drug-addition experiment using the Veritrop assay. 2 μM of Enfuvirtide was added at 30 min to 10 h post incubation of the effector and target cells. (c) Box plot of the time to 50% inhibition by Enfuvirtide of cell-to-cell fusion mediated by 34 Env glycoproteins expressed in context with the HIV-1 VLPs. (c) Schematic of the viral fusion assay which can be monitored by HIV-1 carrying a BlaM-Vpr fusion protein that cleaves the CCF2 dye in target cells and changes the fluorescent spectrum. (d) Relative entry over time (0–600 min) into U87.CD4.CCR5 cells by the chimeric HIV nfl carrying the 14 subtype A, 4 subtype D, and 16 subtype C Env glycoproteins from acute/early infections. (e) Spectral shift curves and box plot of the time required for maximal virus entry into U87.CD4.CCR5 cells carrying the CCF2 dye.
Finally, the rate of free virus entry into host cells (U87.CD4.CCR5) was measured using these same 34 AHI HIV-1. In this Miller et al. system (Lineberger et al., 2002), pREC-nfl was co-transfected with pMM310 BlaM-Vpr to produce HIV-1 particles harboring a Vpr-BlaM fusion protein (Fig. 5d). Upon de novo infection, the HIV_nfl is fully capable of host cell entry which releases the Vpr-Beta-lactamase to cleave the fluorescent CCF2 dye and change fluorescent spectra (Fig. 5d) (Lineberger et al., 2002). The rate of virus entry into host cells was the slowest with the 16 HIV_nfl harboring the AHI subtype C envelopes as compared to those carrying the AHI subtype A (P < 0.05) and subtype D envelopes (P < 0.001) (Fig. 5e & f).
4. Discussion
By the 1980s, all the HIV-1 subtypes had emerged, were circulating and recombining in the Congo basin as well as spreading to neighboring countries/regions (Tebit and Arts, 2011). Various HIV-1 subtypes were introduced in multiple geographic regions (South Africa, Zimbabwe, Brazil, India, etc) (Arien et al., 2007) by the late 1990s and yet, subtype C HIV-1 rose in prevalence faster than any other subtype in the heterosexual population (< 10% in early 1990s to > 50% today) (Arien et al., 2007) (Fig. 1). We have previously reported that HIV-1 subtype C isolates had lower replicative fitness in human T-cells and macrophages than other group M subtypes (Abraha et al., 2009, Ball et al., 2003, Arien et al., 2005) but could only speculate that HIV-1 subtype C may cause slower disease progression (Arien et al., 2007). Aside from regions in Brazil, Tanzania, and Kenya, HIV-1 subtype C typically dominates in regional pandemics and does not co-circulate at high frequencies with other HIV-1 subtypes in human populations (Arien et al., 2007). As a consequence, comparing disease progression related to different HIV-1 subtype infections in a single country or region is very difficult. Thus, we screened for and recruited 300 AHI in Ugandan and Zimbabwean women and then followed the natural history of these HIV infections for 5–9 years in the absence of treatment. With this cohort in Uganda and Zimbabwe, we have a population of only women, all of Bantu origin, all recruited within AHI (following heterosexual transmission), and finally, representing the subtype A, C and D HIV cohorts for sufficient statistical power to measure differences in disease progression. Independent statisticians at two different institutions (FHI 360 and Case Western Reserve University) analyzed the cohort data presented herein. The 2-fold difference in disease progression between these two countries was not attributable to diet, secondary/opportunistic infections, sexual habits, or age (Lemonovich et al., 2015). Of course, we could not screen for presence or absence of all pathogens, sociodemographic or tribal/population differences but the questionnaires administered by medical officers at each patient visit was extensive as was the tests for various clinical chemistries and other health indicators. Examples of the questionnaire and acquired laboratory data is available upon request. We are currently interest in screening the microbiota in the vaginal tract of these women to assess possible difference which may be associated with disease progression but again, we do anticipate differences that segregate by country given the similar diet and diverse human genetics. Of all the parameters tested, only HIV-1 subtypes could clearly delineate differences in the rate of CD4 cell decline in blood. Infection with subtype C HIV-1 resulted in the slowest declines in CD4 T-cell counts (0.489 cells/week) as compared to subtype A (0.781 cells/week) and then subtype D HIV-1 infection with the most rapid declines in CD4 T-cell counts (1.231 cells/week).
Like other natural history cohorts, rapid disease progression was observed at a 7–9% frequency regardless of HIV-1 subtype but slow progressors/controllers appeared only in subtype A and C infections. The differential rates of CD4 T-cell declines (D > A > C) was still significant when rapid progressors and controllers were removed from the analyses. We have proposed that a combination of “good” host genetics (e.g. HLA B27 or B57) and infection with HIV of low replicative fitness may result in “elite” HIV control (Lobritz et al., 2011), i.e. a rare condition in Africa but closely approximated by the 13 subtype C infections with no declines in CD4 cell counts and < 103 copies/ml of virus for > 5 years of infection (Fig. 3a & f).
Based on previous human genetic studies across Africa (Cao et al., 2004, Paximadis et al., 2012, Kijak et al., 2009, McLaren and Fellay, 2015, McLaren and Carrington, 2015) (www.allelefrequencies.net/), we now know that the Bantu population has the greatest genetic diversity of Homo sapiens. Low prevalence of “HIV protective” polymorphisms/alleles in the Shona and Buganda tribes (both Bantu) (e.g. < 5% of CCR5Δ32, CCR2a 64I, HLA-B57 and B27) could not explain the dramatic differences in disease progression. Interestingly, there were no significant differences between countries or HIV-1 subtypes in the viral RNA levels at set point or over the course of infection. In our previous report using 188 patients for this cohort and with minimal analyses of follow up (< 3 years), we observed a slightly higher viral loads at set point in subtype C and D versus A infections (p < 0.04, ANOVA) that has not held significance when expanding to the 286 patients in this study (Morrison et al., 2010). Instead, expansion of the cohort size now showed a trend for lower viral load set points in subtype C infections versus A or D (Suppl Fig. 3c). Regardless, it is again important to stress that we did not observe any significant differences in viral load during disease based on infection by specific HIV-1 subtype.
Most studies examining disease progression in natural history cohorts are now impossible and unethical based on WHO guidelines for the initiation of cART in all HIV positive patients regardless of CD4 T-cells counts (http://www.who.int/hiv/pub/guidelines/earlyrelease-arv/en/). With the start of this study in 2000, we followed WHO/UNAIDS guidelines to treat with CD4 cell counts ≤ 200/ml which was controversial because few charities, governments, and international organizations (e.g. WHO, PEPFAR) had rolled out their treatment programs in Africa. During this ten year cohort study, 33% of the participants received treatment at an average of 1500 days post-infection. Subtype D infected women were ~ 1.7-fold more likely to receive treatment than subtype A or C infected women. By modelling the rates of CD4 declines in this natural history cohort, the projected time to reach ≤ 200/ml or AIDS in these women was 1.3 fold longer with subtype C (estimated mean of 12.3 years) than A (9.5 years) and 2.0 fold longer with C than subtype D (6.2 years). Historical data of subtype B infections in North America and Europe suggest 6–8 years as an approximate time to reach AIDS (CD4 T-cells < 200/ml) (Munoz et al., 1995, Mellors et al., 1996) but there are no natural history cohorts from diagnosis (prior to treatment) to establish accurate estimates. Interestingly, despite (i) different geographical regions, (ii) different human populations, and (iii) higher rates of parasitic and other co-morbities in Uganda, infections with subtype B in North America and D in Uganda may have similar rates of CD4 T-cell declines and time to AIDS. Subtype B and D HIV-1 share the most sequence homology of all subtypes and several studies suggest that subtype B was a sub-branch and “member” of subtype D super-cluster. Earlier reports have also described faster disease progression and reduced response to treatment in East Africans infected with HIV-1 subtype D than subtype A (Baeten et al., 2007, Kaleebu et al., 2002, Kyeyune et al., 2013).
HIV-1 subtype C isolates are less fit than subtype A which are both less fit than subtype B and D HIV-1 isolates, a finding established by over 2000 direct head-to-head dual virus competitions in primary CD4 + T-cells and macrophages from HIV-negative donors of different races/ethnicities (Abraha et al., 2009, Ball et al., 2003, Arien et al., 2005). These findings suggest a direct association between the replicative fitness of HIV-1 and subsequent disease progression. In addition, the HIV-1 env genes derived from 47 women with AHI also showed that subtype C as compared to subtype A and D were slower in host cell entry by free virus and slower in cell-to-cell transfer of virus. Previous reports have also demonstrated that subtype C viruses rarely switch to a CXCR4 using or dual co-receptor using phenotype whereas subtype D HIV-1 shows higher percentage in switching to CXCR4 co-receptor usage during disease as compare to patients infected with subtype A HIV-1 (or HIV-1C). More rapid progression in subtype D infected patients may be associated with the more common switch to CXCR4 tropic virus but again, this appears to be an inherent attribute of the viral subtype and not due to host genetics. HIV-1 subtype B found in North America and Europe is closely related to subtype D found in East Africa and both B and D have similar propensities to switch to a CXCR4 using phenotype late in disease progression. Overall, these observations suggest that infecting HIV-1 subtype may actually set the course of subsequent disease progression.
In summary, this largest natural history cohort of non-subtype B infected women revealed a significant difference in HIV-1 subtype virulence with ramifications for HIV-1 spread and treatment in the global epidemic. Women infected with HIV-1 subtype C progressed to AIDS at least 1.5-fold slower than women infected with subtype A or D. Longer asymptomatic periods with subtype C infections (between 3 and 5 years longer) will lead to greater probability of transmission, especially among those discordant couples with unknown infection status (http://www.who.int/hiv/pub/progressreports/update2014/en/). Using human cervical and penile explant tissues, transmission “fitness” appears to be similar among most HIV-1 subtypes (Abraha et al., 2009, King et al., 2013). Thus, the expansion of HIV-1 subtype C infections in the global epidemic may be related to the low virulence of subtype C, resulting in long periods of asymptomatic periods, and leading to increased opportunity for heterosexual transmission over other HIV-1 subtypes. Ultimately, HIV-1 subtype C may be more “fit” in the human population based on low subtype virulence and high transmission efficiency.
Funding
This clinical study was funded by the Eunice Kennedy Shriver National Institute of Child Health and Human Development (N01-HD-0-3310) to CM as the principal investigator. F.K. was funded by a scholarship from the NIH Fogarty International Center grants D43-TW000011 and D43-TW009780. E.J.A. was funded by NIAID/NIH AI49170 and the CWRU/UH Center for AIDS Research (P30 AI036219). E.J.A. currently holds the Canadian Research Chair in HIV-1 Pathogenesis and Viral Control.
Competing Interests
E.J.A. holds the patent Grant US8586295B2 entitled “Method for screening HIV drug sensitivity” which describes the yeast-based cloning technology employed herein. BVDP reports receiving research support, honoraria or consulting fees from the following: Atlas Genetics; BD Diagnostics; Beckman Coulter; Hologic; Rheonix: and, Roche Molecular.
Author Contributions
TC, JB, PM, RAS, CSM, SR, EJA, and CK designed the clinical study, directed the recruitment of patients, and maintained the human ethics approvals. EJA directed and supervised all the analyses presented herein especially those related to replicative fitness. CSM was the overall PI of the GS study with TS as site PI in Zimbabwe, JB, PM, and RAS as site PIs in Uganda, and BVdP as the laboratory consultant for both sites. KD, IN, FK, and KD coordinated all of the clinical assays in Uganda and collaborated with MM who did the same in Zimbabwe. PLC and CMV performed all the statistical analyses with CK maintaining the clinical databases. IN, FK, and KD processed all the clinical samples on site, preformed subtyping, and prepared the clinical HIV isolates. Finally, CMV performed all the replicative fitness assays, entry assays, and analyzed the clinical data. EJA and CMV wrote the manuscript with editing by all the other authors.
Acknowledgements
We thank all of participants of the study entitled Hormonal Contraception and Risk of HIV acquisition (HC) and the follow up study entitled Hormonal Contraception and HIV-1 Genital Shedding and Disease Progression among Women with Primary HIV Infection (GS). We also thank all of the nurses, counselors, research assistants, medical officers, data managers, study coordinators, and home visitors involved in this 10 year study.
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.ebiom.2016.10.014.
Appendix A. Supplementary Data
Supplementary Figures 1–6
References
- Abraha A., Nankya I.L., Gibson R., Demers K., Tebit D.M., Johnston E. J. Virol. 2009 March 18 doi: 10.1128/JVI.02051-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amornkul P.N., Karita E., Kamali A., Rida W.N., Sanders E.J., Lakhi S. Disease progression by infecting HIV-1 subtype in a seroconverter cohort in sub-Saharan Africa. AIDS. 2013 November 13;27(17):2775–2786. doi: 10.1097/QAD.0000000000000012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arien K.K., Abraha A., Quinones-Mateu M.E., Kestens L., Vanham G., Arts E.J. The replicative fitness of primary human immunodeficiency virus type 1 (HIV-1) group M, HIV-1 group O, and HIV-2 isolates. J. Virol. 2005 July;79(14):8979–8990. doi: 10.1128/JVI.79.14.8979-8990.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arien K.K., Vanham G., Arts E.J. Is HIV-1 evolving to a less virulent form in humans? Nat. Rev. Microbiol. 2007 February;5(2):141–151. doi: 10.1038/nrmicro1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baeten J.M., Chohan B., Lavreys L., Chohan V., McClelland R.S., Certain L. HIV-1 subtype D infection is associated with faster disease progression than subtype A in spite of similar plasma HIV-1 loads. J Infect Dis. 2007 April 15;195(8):1177–1180. doi: 10.1086/512682. [DOI] [PubMed] [Google Scholar]
- Ball S.C., Abraha A., Collins K.R., Marozsan A.J., Baird H., Quinones-Mateu M.E. Comparing the ex vivo fitness of CCR5-tropic human immunodeficiency virus type 1 isolates of subtypes B and C 1. J. Virol. 2003 January 15;77(2):1021–1038. doi: 10.1128/JVI.77.2.1021-1038.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bark C.M., Morrison C.S., Salata R.A., Byamugisha J.K., Katalemwa N.H., Mugerwa R.D. Acceptability of treatment of latent tuberculosis infection in newly HIV-infected young women in Uganda. Int J Tuberc Lung Dis. 2010 December;14(12):1647–1649. [PubMed] [Google Scholar]
- Cao K., Moormann A.M., Lyke K.E., Masaberg C., Sumba O.P., Doumbo O.K. Differentiation between African populations is evidenced by the diversity of alleles and haplotypes of HLA class I loci. Tissue Antigens. 2004 April;63(4):293–325. doi: 10.1111/j.0001-2815.2004.00192.x. [DOI] [PubMed] [Google Scholar]
- Demeulemeester J., Vets S., Schrijvers R., Madlala P., De M.M., De R.J. HIV-1 integrase variants retarget viral integration and are associated with disease progression in a chronic infection cohort. Cell Host Microbe. 2014 November 12;16(5):651–662. doi: 10.1016/j.chom.2014.09.016. [DOI] [PubMed] [Google Scholar]
- Dudley D.M., Gao Y., Nelson K.N., Henry K.R., Nankya I., Gibson R.M. A novel yeast-based recombination method to clone and propagate diverse HIV-1 isolates. BioTechniques. 2009 May;46(6):458–467. doi: 10.2144/000113119. [DOI] [PubMed] [Google Scholar]
- Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouy M., Guindon S., Gascuel O. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 2010 February;27(2):221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- Kaleebu P., French N., Mahe C., Yirrell D., Watera C., Lyagoba F. Effect of human immunodeficiency virus (HIV) type 1 envelope subtypes A and D on disease progression in a large cohort of HIV-1-positive persons in Uganda. J Infect Dis. 2002 May 1;185(9):1244–1250. doi: 10.1086/340130. [DOI] [PubMed] [Google Scholar]
- Kanki P.J., Hamel D.J., Sankale J.L., Hsieh C., Thior I., Barin F. Human immunodeficiency virus type 1 subtypes differ in disease progression. J Infect Dis. 1999 January;179(1):68–73. doi: 10.1086/314557. [DOI] [PubMed] [Google Scholar]
- Kijak G.H., Walsh A.M., Koehler R.N., Moqueet N., Eller L.A., Eller M. HLA class I allele and haplotype diversity in Ugandans supports the presence of a major east African genetic cluster. Tissue Antigens. 2009 March;73(3):262–269. doi: 10.1111/j.1399-0039.2008.01192.x. [DOI] [PubMed] [Google Scholar]
- King D.F., Siddiqui A.A., Buffa V., Fischetti L., Gao Y., Stieh D. Mucosal tissue tropism and dissemination of HIV-1 subtype B acute envelope-expressing chimeric virus. J. Virol. 2013 January;87(2):890–899. doi: 10.1128/JVI.02216-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiwanuka N., Robb M., Laeyendecker O., Kigozi G., Wabwire-Mangen F., Makumbi F.E. HIV-1 viral subtype differences in the rate of CD4 + T-cell decline among HIV seroincident antiretroviral naive persons in Rakai District, Uganda. J. Acquir. Immune Defic. Syndr. 2009 December;11 doi: 10.1097/QAI.0b013e3181c98fc0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyeyune F., Nankya I., Metha S., Akao J., Ndashimye E., Tebit D.M. Treatment failure and drug resistance is more frequent in HIV-1 subtype D versus A infected Ugandans: a ten year history in Kampala, Uganda. AIDS. 2013 March 18 doi: 10.1097/QAD.0b013e3283610ec7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemonovich T.L., Watkins R.R., Morrison C.S., Kwok C., Chipato T., Musoke R. Differences in clinical manifestations of acute and early HIV-1 infection between HIV-1 subtypes in African women. J Int Assoc Provid AIDS Care. 2015 September;14(5):415–422. doi: 10.1177/2325957413504827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limou S., Zagury J.F. Immunogenetics: genome-wide association of non-progressive HIV and viral load control: HLA genes and beyond. Front. Immunol. 2013;4:118. doi: 10.3389/fimmu.2013.00118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lineberger J.E., Danzeisen R., Hazuda D.J., Simon A.J., Miller M.D. Altering expression levels of human immunodeficiency virus type 1 gp120-gp41 affects efficiency but not kinetics of cell-cell fusion. J. Virol. 2002 April;76(7):3522–3533. doi: 10.1128/JVI.76.7.3522-3533.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobritz M.A., Lassen K.G., Arts E.J. HIV-1 replicative fitness in elite controllers. Curr. Opin. HIV AIDS. 2011 May;6(3):214–220. doi: 10.1097/COH.0b013e3283454cf5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovvorn A.E., Patnaik P., Walker C.J., Kwok C., van der Pol B., Chipato T. Variations in CD4 cell counts among HIV-uninfected and infected women in Uganda and Zimbabwe. Int. J. STD AIDS. 2010 May;21(5):342–345. doi: 10.1258/ijsa.2009.009020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann J.K., Chopera D., Omarjee S., Kuang X.T., Le A.Q., Anmole G. Nef-mediated down-regulation of CD4 and HLA class I in HIV-1 subtype C infection: association with disease progression and influence of immune pressure. Virology. 2014 November;468–470:214–225. doi: 10.1016/j.virol.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marozsan A.J., Moore D.M., Lobritz M.A., Fraundorf E., Abraha A., Reeves J.D. Differences in the fitness of two diverse wild-type human immunodeficiency virus type 1 isolates are related to the efficiency of cell binding and entry. J. Virol. 2005 June;79(11):7121–7134. doi: 10.1128/JVI.79.11.7121-7134.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren P.J., Carrington M. The impact of host genetic variation on infection with HIV-1. Nat. Immunol. 2015 June;16(6):577–583. doi: 10.1038/ni.3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren P.J., Fellay J. Human genetic variation in HIV disease: beyond genome-wide association studies. Curr. Opin. HIV AIDS. 2015 March;10(2):110–115. doi: 10.1097/COH.0000000000000133. [DOI] [PubMed] [Google Scholar]
- Mellors J.W., Rinaldo C.R., Jr., Gupta P., White R.M., Todd J.A., Kingsley L.A. Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science. 1996 May 24;272(5265):1167–1170. doi: 10.1126/science.272.5265.1167. [DOI] [PubMed] [Google Scholar]
- Montano M.A., Nixon C.P., Ndung'u T., Bussmann H., Novitsky V.A., Dickman D. Elevated tumor necrosis factor-alpha activation of human immunodeficiency virus type 1 subtype C in Southern Africa is associated with an NF-kappaB enhancer gain-of-function. J Infect Dis. 2000 January;181(1):76–81. doi: 10.1086/315185. [DOI] [PubMed] [Google Scholar]
- Morrison C.S., Richardson B.A., Mmiro F., Chipato T., Celentano D.D., Luoto J. Hormonal contraception and the risk of HIV acquisition. AIDS. 2007 January 2;21(1):85–95. doi: 10.1097/QAD.0b013e3280117c8b. [DOI] [PubMed] [Google Scholar]
- Morrison C.S., Demers K., Kwok C., Bulime S., Rinaldi A., Munjoma M. Plasma and cervical viral loads among Ugandan and Zimbabwean women during acute and early HIV-1 infection. AIDS. 2010 February 20;24(4):573–582. doi: 10.1097/QAD.0b013e32833433df. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison C.S., Chen P.L., Nankya I., Rinaldi A., van der Pol B., Ma Y.R. Hormonal contraceptive use and HIV disease progression among women in Uganda and Zimbabwe. J. Acquir. Immune Defic. Syndr. 2011 February;24 doi: 10.1097/QAI.0b013e318214ba4a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz A., Kirby A.J., He Y.D., Margolick J.B., Visscher B.R., Rinaldo C.R. Long-term survivors with HIV-1 infection: incubation period and longitudinal patterns of CD4 + lymphocytes. J Acquir Immune Defic Syndr Hum Retrovirol. 1995 April 15;8(5):496–505. doi: 10.1097/00042560-199504120-00010. [DOI] [PubMed] [Google Scholar]
- Palm A.A., Esbjornsson J., Mansson F., Kvist A., Isberg P.E., Biague A. Faster progression to AIDS and AIDS-related death among seroincident individuals infected with recombinant HIV-1 A3/CRF02_AG compared with sub-subtype A3. J Infect Dis. 2014 March 1;209(5):721–728. doi: 10.1093/infdis/jit416. [DOI] [PubMed] [Google Scholar]
- Paximadis M., Mathebula T.Y., Gentle N.L., Vardas E., Colvin M., Gray C.M. Human leukocyte antigen class I (A, B, C) and II (DRB1) diversity in the black and Caucasian South African population. Hum. Immunol. 2012 January;73(1):80–92. doi: 10.1016/j.humimm.2011.10.013. [DOI] [PubMed] [Google Scholar]
- Rainwater S., Devange S., Sagar M., Ndinya-Achola J., Mandaliya K., Kreiss J.K. No evidence for rapid subtype C spread within an epidemic in which multiple subtypes and intersubtype recombinants circulate. AIDS Res. Hum. Retrovir. 2005 December;21(12):1060–1065. doi: 10.1089/aid.2005.21.1060. [DOI] [PubMed] [Google Scholar]
- Reed L.J., Muench H. A simple method of estimating fifty percent endpoints. Am. J. Hyg. 1938;27:493–497. Ref Type: Journal (Full) [Google Scholar]
- Rubio A.E., Abraha A., Carpenter C.A., Troyer R.M., Reyes-Rodriguez A.L., Salomon H. Similar replicative fitness is shared by the subtype B and unique BF recombinant HIV-1 isolates that dominate the epidemic in Argentina. PLoS One. 2014;9(4) doi: 10.1371/journal.pone.0092084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P.M., Hahn B.H. Origins of HIV and the AIDS pandemic. Cold Spring Harb Perspect Med. 2011 September;1(1):a006841. doi: 10.1101/cshperspect.a006841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tebit D.M., Arts E.J. Tracking a century of global expansion and evolution of HIV to drive understanding and to combat disease. Lancet Infect. Dis. 2011 January;11(1):45–56. doi: 10.1016/S1473-3099(10)70186-9. [DOI] [PubMed] [Google Scholar]
- Troyer R.M., Collins K.R., Abraha A., Fraundorf E., Moore D.M., Krizan R.W. Changes in human immunodeficiency virus type 1 fitness and genetic diversity during disease progression. J. Virol. 2005 July;79(14):9006–9018. doi: 10.1128/JVI.79.14.9006-9018.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van O.T., Jeeninga R.E., Boerlijst M.C., Pollakis G.P., Zetterberg V., Salminen M. Human immunodeficiency virus type 1 subtypes have a distinct long terminal repeat that determines the replication rate in a host-cell-specific manner. J. Virol. 2004 April;78(7):3675–3683. doi: 10.1128/JVI.78.7.3675-3683.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velazquez-Campoy A., Todd M.J., Vega S., Freire E. Catalytic efficiency and vitality of HIV-1 proteases from African viral subtypes. Proc. Natl. Acad. Sci. U. S. A. 2001 May 22;98(11):6062–6067. doi: 10.1073/pnas.111152698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber J., Vazquez A.C., Winner D., Gibson R.M., Rhea A.M., Rose J.D. Sensitive cell-based assay for determination of human immunodeficiency virus type 1 coreceptor tropism. J. Clin. Microbiol. 2013 May;51(5):1517–1527. doi: 10.1128/JCM.00092-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures 1–6