Abstract
Background
Increasing studies showed that abnormal changes in single nucleotide polymorphisms (SNPs) of DNMTs (DNMT1, DNMT3A and DNMT3B) were associated with occurrence or decrease of various tumors. However, the associations between DNMTs variations and gastric cancer (GC) risk were still conflicting. We aimed to assess the effect of DNMTs polymorphisms on the susceptibility to GC.
Methods
Firstly, we did a meta-analysis for 7 SNPs (rs16999593, rs2228611, rs8101866 in DNMT1, rs1550117, rs13420827 in DNMT3A, rs1569686, rs2424913 in DNMT3B). Four genetic models (homozygote, heterozygote, dominant and recessive model) were used. Moreover, a meta-sensitivity and subgroup analysis was performed to clarify heterogeneity source. Lastly, 17 SNPs that couldn't be meta-analyzed were presented in a systematic review.
Findings
20 studies were included, 13 studies could be meta-analyzed and 7 ones could not. Firstly, a meta-analysis on 13 studies (3959 GC cases and 5992 controls) for 7 SNPs showed that GC risk increased in rs16999593 (heterozygote model: OR 1.36, 95%CI 1.14–1.61; dominant model: OR 1.36, 95%CI 1.15–1.60) and rs1550117 (homozygote model: OR 2.03, 95%CI 1.38–3.00; dominant model: OR 1.20, 95%CI 1.01–1.42; recessive model: OR 1.96, 95%CI 1.33–2.89) but decreased in rs1569686 (dominant model: OR 0.74, 95%CI 0.61–0.90). The remaining SNPs were not found associated with GC risk. Furthermore, the subgroup analysis indicated that for rs1550117 and rs1569686, the significant associations were particularly found in people from Chinese Jiangsu province (rs1550117, OR 1.77, 95%CI 1.25–2.51; rs1569686, OR 0.48, 95%CI 0.36–0.64) and that PCR-RFLP was a sensitive method to discover significant associations (rs1550117, OR 1.77, 95%CI 1.25–2.51; rs1569686, OR 0.49, 95%CI 0.37–0.65). Lastly, a systematic review on 7 studies for 17 SNPs suggested that rs36012910, rs7560488 and rs6087990 might have a potential effect on GC initiation.
Conclusion
This meta-analysis demonstrated that rs16999593 and rs1550117 could contribute to GC risk and that rs1569686 might be a protective factor against gastric carcinogenesis. By using these SNPs as biomarkers, it is feasible to estimate the risk of acquiring GC and thus formulate timely preventive strategy.
Keywords: DNA methyltransferase, Single nucleotide polymorphism, Gastric cancer, Meta-analysis, Systematic review
Highlights
-
•
DNMT1, DNMT3A and DNMT3B polymorphisms are associated with gastric cancer (GC) risk, in which rs16999593 and rs1550117 might contribute to GC and rs1569686 could be a protective factor against carcinogenesis.
-
•
Significant DNMTs polymorphisms were particularly found in people from Chinese Jiangsu province.
-
•
PCR-RFLP was a sensitive method to discover significant DNMTs polymorphisms.
Research in context Genetic factors play a crucial role in GC risk. DNMTs genes were associated with occurrence or decrease of various tumors. But the effects of DNMTs on gastric cancer (GC) were not clear. Now our results proved that two and one variations in DNMTs were associated with GC, indicating a range of effects from the increased to the reduced. By using these variations as biomarkers, it is feasible to estimate the risk of acquiring GC and thus formulate timely preventive strategy.
1. Introduction
In 2012, 951,000 new gastric cancer (GC) cases and 723,000 deaths were estimated worldwide, making it the fifth most common tumor (Ferlay et al., 2015, Torre et al., 2015). GC is a complex disease arising from environmental and genetic factors. However in individuals infected with H. pylori, defined as a definite gastric carcinogen (Yang, 2006), only a few eventually develop into GC, which suggested that host genetic factors may play a crucial role in the susceptibility of GC (Saeki et al., 2013).
The epigenetics is believed to be important in the development of cancers, which was defined as a stably heritable changes through modifying gene expression without DNA sequence alterations (Esteller, 2008). The most common epigenetic phenomenon is DNA methylation that refers to a methyl group is conferred to the 5′ carbon of a cytosine in a CpG dinucleotide. It is catalyzed by a family of DNA methyltransferases (DNMTs) mainly consisting of three activated forms: DNMT1, DNMT3A and DNMT3B. DNMT1 is thought to be a maintenance DNA methyltransferase which principally maintains CpG methylation, involving in embryonic development and somatic cells survival (Brown and Robertson, 2007) and it is encoded by DNMT1 gene which locates on chromosome 19p13.2 (Jiang et al., 2012a). DNMT3A and DNMT3B are considered as de novo methyltransferases which are required for the establishment of embryonic methylation patterns, mainly occurring during gametogenesis and early development (Okano et al., 1999) and they are encoded by DNMT3A and DNMT3B genes locating on chromosome 2p23 and 20q11.2 respectively (Yang et al., 2012).
There is considerable evidence that a number of abnormal changes in single nucleotide polymorphisms (SNPs) of DNMTs (DNMT1, DNMT3A and DNMT3B), which could cause DNA hypo-methylation or hyper-methylation (Gao et al., 2011, Fu et al., 2010, Harder et al., 2008, Zhao and Bu, 2012), are correlated to tumor occurrence or decrease (Luo et al., 2015, Chang et al., 2014, Mostowska et al., 2013, Kullmann et al., 2013, Sun et al., 2012, Xiang et al., 2010, Kanai et al., 2003) such as head and neck cancer, and colorectal cancer (Zhu et al., 2015, Duan et al., 2015). However, the associations between DNMTs SNPs and GC risk were still conflicting (Jiang et al., 2012a, Yang et al., 2012). Therefore, for the first time, the effects of DNMTs polymorphisms on the susceptibility to GC were systematically and comprehensively estimated.
2. Materials and Methods
2.1. Search
We did a literature search of PubMed, MEDLINE, Embase, Sinomed, CNKI, and WanFang databases to identify relevant studies up to June 1, 2016, using the search strategy: (stomach OR gastric) AND (neoplasms OR tumors OR cancers OR carcinomas) AND (DNMT1 OR DNMT3A OR DNMT3B OR DNMTs OR DNA methyltransferases). The languages were limited to English and Chinese. The search strategy for PubMed was listed in Appendix A.
2.2. Selection Criteria
All studies included in the meta-analysis were accorded with the following inclusion criteria: (a). study focused on the association of DNMTs polymorphisms and GC risk; (b). case-control or cohort studies. In addition, exclusion criteria were as follows: (a). reviews or meta-analysis; (b). overlapped articles or studies with overlapping data.
2.3. Data Extraction
Two investigators independently extracted the following data: first author, year of publication, province/country of origin, ascertainment of cases, source of controls, genotyping methods, DNMT genes, SNPs, number of cases and controls, and value of HWE. To ensure accuracy of the data, inconsistencies were discussed with another reviewer until reach a consensus.
2.4. Quality Assessment
The quality of each study was assessed according to the quality assessment criteria (Table S1) (Thakkinstian et al., 2011, Xue et al., 2015), in which the overall quality scores ranged from 0 to 15. Studies with scores ≥ 9 were regarded as high quality studies; otherwise, studies were considered to have a low quality.
2.5. Data Analysis
Stata software (version 12.0; Stata Corporation, College Station, TX) was used to perform all analysis. We used four types of genetic models (Lieb et al., 2006): homozygote model (homozygous rare vs. homozygous frequent allele), heterozygote model (heterozygous vs. homozygous frequent allele), dominant model (homozygous rare + heterozygous vs. homozygous frequent allele) and recessive model (homozygous rare vs. heterozygous + homozygous frequent allele). Association between DNMTs polymorphisms and the GC risk was evaluated by pooled odds ratios (OR), 95% confidence interval (95% CI) and P value of Z test (POR). If 95%IC across 1 or POR < 0.05, a significant association existed. Then if OR or 95%IC < 1, the mutant gene was a protective factor; otherwise, it was a risk factor. Heterogeneity was analyzed using the P value of Q test (Phet) and I2. If Phet < 0.1 or I2 > 50%, a significant heterogeneity existed. And then a sensitivity analysis and a subgroup analysis were performed. Sensitivity analysis was conducted through omitting one study by turns (Lu et al., 2016), if the 95%CI markedly deviated from the original interval or the I2 largely decreased, this study was an originator of heterogeneity.
3. Results
3.1. Literature Search and Study Characteristics
A total of 350 records were identified through database searching. After removing duplicates, 274 records were screened on details of the abstracts. In those 249 publications were excluded because 5 were meta-analysis and the other 244 were not related to DNMTs SNPs and GC risk. Then 25 full-text articles were obtained to be assessed, in which 5 articles were excluded because 1 was duplicate publication and 4 did not contain information on DNMTs SNPs and GC risk. Ultimately, 20 eligible studies (Jiang et al., 2012a, Yang et al., 2012, Yan et al., 2015, Khatami et al., 2009, Wu et al., 2014, Cao et al., 2013, Wu et al., 2012, Fan et al., 2010, Liu, 2009, Zhang et al., 2014, Hu et al., 2010, Zhang, 2008, Liu, 2008, Wang et al., 2005, Aung et al., 2005, Wang et al., 2015a, Jiang et al., 2013, Jiang et al., 2012b, Cao et al., 2012, Chang et al., 2010)were included in the qualitative synthesis, and 7 of them could not be quantitatively synthesized (3 studies respectively reported a different SNP (Wu et al., 2014, Wu et al., 2012, Liu, 2008), 4 studies were conference abstracts (Jiang et al., 2013, Jiang et al., 2012b, Cao et al., 2012, Chang et al., 2010)), so 13 studies involving 3959 GC cases and 5992 healthy controls were finally included in the meta-analysis (Fig. 1). Among the 20 studies, 18 studies were for Chinese population (respectively from Jiangsu, Jiangxi, Hebei, Shandong, Jilin and Heilongjiang provinces of China), 1 study was for Iranian population (from Fars and Tork) and another one was for Japanese population (from Hiroshima and Yamaguchi). According to the quality assessment criteria (Table S1), scores of the 13 studies (included in the meta-analysis) were 4–12 and 8 studies were with high quality scores (Xue et al., 2015). The main characteristics of the 13 studies were listed in Table 1.
Fig. 1.
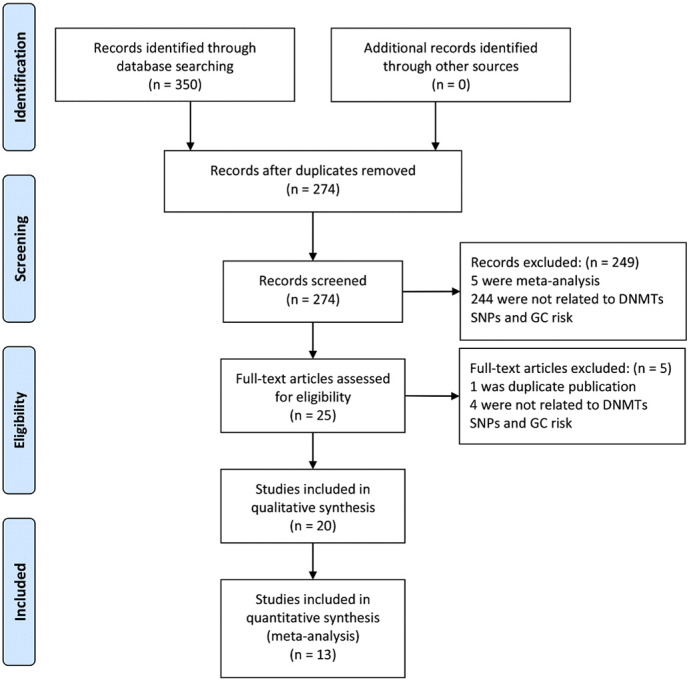
Flow chart of study selection process.
Table 1.
Characteristics of 13 studies included in the meta-analysis.
| Study | Province/Country | Ascertainment of cases | Source of controls | Genotyping methods | Gene | SNPs | Sample size (cases/controls) | HWE (controls) | Score |
|---|---|---|---|---|---|---|---|---|---|
| Yan et al., 2015 | Shandong/China | Histological | HB | Sequencing | DNMT1 | rs16999593 | 310/420 | 0.469 | 9 |
| rs2228611 | 0.423 | ||||||||
| Yang et al., 2012 | Jiangxi/China | Histological | HB | MassArray | DNMT1 | rs16999593 | 242/294 | 0.120 | 9 |
| rs2228611 | 0.068 | ||||||||
| rs8101866 | 0.747 | ||||||||
| DNMT3A | rs1550117 | 0.444 | |||||||
| rs13420827 | |||||||||
| Jiang et al., 2012a, Jiang et al., 2012b | Jilin/China | Histological | HB | TaqMan | DNMT1 | rs16999593 | 447/961 | 0.910 | 9 |
| rs8101866 | |||||||||
| Khatami et al., 2009 | Fars/Iran, Tork/Iran | Histological | HB | PCR-RFLP | DNMT1 | rs2228611 | 200/200 | 0.187 | 9 |
| Cao et al., 2013 | Jilin/China | Histological | HB | TaqMan | DNMT3A | rs1550117 | 447/961 | 0.658 | 9 |
| rs13420827 | 0.833 | ||||||||
| Fan et al., 2010 | Jiangsu/China | Histological | HB/PB | PCR-RFLP | DNMT3A | rs1550117 | 208/346 | 0.205 | 12 |
| Liu, 2009 | Jiangsu/China | NA | NA | PCR-RFLP | DNMT3B | rs2424913 | 308/189 | 0.942 | 6 |
| rs1569686 | 313/350 | > 0.05 | |||||||
| Wang et al., 2015a, Wang et al., 2015b | Jilin/China | Histological | HB | TaqMan | DNMT3B | rs1569686 | 447/961 | 0.001 | 7 |
| Zhang et al., 2014 | Heilongjiang/China | NA | NA | PCR-RFLP | DNMT3B | rs1569686 | 50/60 | 0.389 | 4 |
| Hu et al., 2010 | Jiangsu/China | Histological | HB/PB | PCR-RFLP | DNMT3B | rs2424913 | 259/262 | 0.926 | 12 |
| rs1569686 | 0.901 | ||||||||
| Zhang, 2008 | Jiangsu/China | NA | HB | PCR-RFLP | DNMT3B | rs2424913 | 156/156 | 0.968 | 6 |
| rs1569686 | 0.001 | ||||||||
| Wang et al., 2005 | Hebei/China | Histological | HB/PB | PCR-RFLP | DNMT3B | rs2424913 | 212/294 | 0.654 | 12 |
| Aung et al., 2005 | Hiroshima/Japan, Yamaguchi/Japan | Histological | HB | PCR-RFLP | DNMT3B | rs2424913 | 152/247 | 1.000 | 6 |
NA, not available; HB, hospital based; PB, population based; PCR-RFLP, polymorphism chain reaction-restriction fragment length polymorphism; DNMT genes, deoxyribonucleic acid methyltransferase genes; SNPs, single nucleotide polymorphisms; HWE, Hardy-Weinberg equilibrium.
3.2. Meta-analysis and Systematic Review
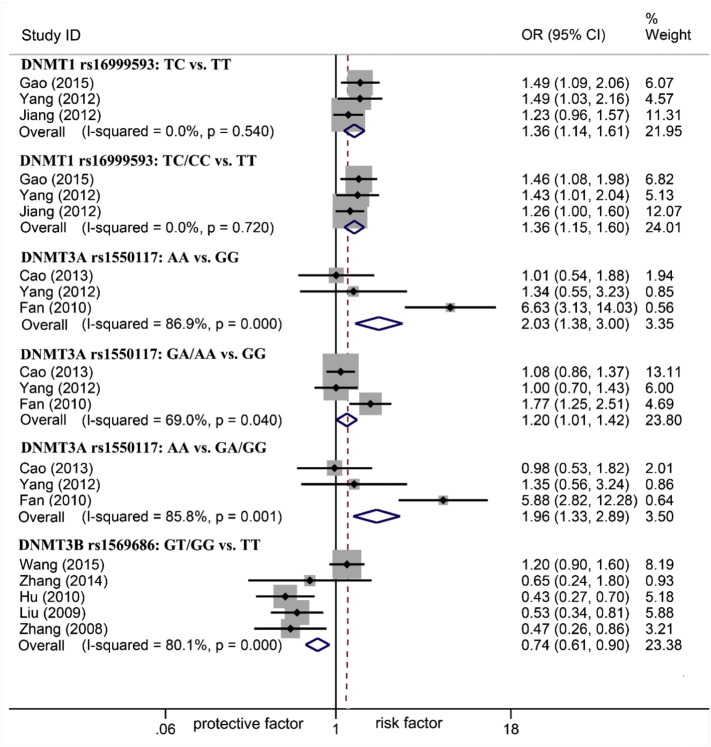
The associations between DNMTs polymorphisms and gastric carcinogenesis were shown in Table 2 and the statistically significant associations (only Chinese population were discovered in significant associations) were represented in Fig. 2. In terms of DNMT1 and DNMT3A, GC risk increased. For rs16999593, there was an association under heterozygote and dominant models (TC vs. TT: OR 1.36, 95%CI 1.14–1.61; TC/CC vs. TT: OR 1.36, 95%CI 1.15–1.60) but not homozygote and recessive models (CC vs. TT: OR 1.36, 95%CI 0.93–1.99; CC vs. TC/TT: OR 1.22, 95%CI 0.84–1.78). For rs1550117, the increased GC risk was discovered under homozygote, dominant and recessive models (AA vs. GG: OR 2.03, 95%CI 1.38–3.00; GA/AA vs. GG: OR 1.20, 95%CI 1.01–1.42; AA vs. GA/GG: OR 1.96, 95%CI 1.33–2.89) but not heterozygote model (GA vs. GG: OR 1.12, 95%CI 0.93–1.33). Conversely, GC risk decreased in DNMT3B. For rs1569686, the association was found under dominant model (GT/GG vs. TT: OR 0.74, 95%CI 0.61–0.90) but not heterozygote, homozygote and recessive models (GT vs. TT: OR 0.88, 95%CI 0.69–1.13; GG vs. TT: OR 0.96, 95%CI 0.46–2.01; GG vs. GT/TT: OR 0.97, 95%CI 0.46–2.02). Except all of the above, for rs2228611, rs8101866, rs13420827 and rs2424913, no significant associations were observed among all of the genetic models. Lastly, for SNPs not able to be quantitatively synthesized, the systematic review presented their associations with GC (Table 3). Three SNPs rs36012910, rs7560488 and rs6087990 (Wu et al., 2014, Wu et al., 2012, Liu, 2008) were reported associated with GC and others not.
Table 2.
Meta-analysis of association between DNMTs SNPs and gastric cancer risk.
| SNPs | N (cases/controls) | OR (95%CI) | PORa | I2 | Phetb |
|---|---|---|---|---|---|
| DNMT1 rs16999593 | |||||
| TC vs. TTc | 949/1609 | 1.36 (1.14,1.61) | 0.001 | 0.0% | 0.540 |
| CC vs. TTd | 654/1202 | 1.36 (0.93,1.99) | 0.117 | 0.0% | 0.743 |
| TC/CC vs. TTe | 999/1675 | 1.36 (1.15,1.60) | 0.000 | 0.0% | 0.720 |
| CC vs. TC/TTf | 999/1675 | 1.22 (0.84,1.78) | 0.303 | 0.0% | 0.635 |
| DNMT1 rs2228611 | |||||
| GA vs. GGc | 656/804 | 1.09 (0.88,1.36) | 0.408 | 0.0% | 0.732 |
| AA vs. GGd | 427/537 | 0.87 (0.60,1.27) | 0.478 | 11.0% | 0.325 |
| GA/AA vs. GGe | 752/912 | 1.05 (0.86,1.29) | 0.622 | 0.0% | 0.987 |
| AA vs. GA/GGf | 752/912 | 0.97 (0.71,1.32) | 0.829 | 56.9% | 0.098 |
| DNMT1 rs8101866 | |||||
| TC vs. TTc | 643/1159 | 0.99 (0.81, 1.21) | 0.926 | 48.2% | 0.165 |
| CC vs. TTd | 411/751 | 0.80 (0.55,1.17) | 0.252 | 0.0% | 0.452 |
| TC/CC vs. TTe | 686/1255 | 0.96 (0.80, 1.16) | 0.662 | 0.0% | 0.324 |
| CC vs. TC/TTf | 686/1255 | 0.80 (0.55,1.17) | 0.252 | 13.1% | 0.283 |
| DNMT3A rs1550117 | |||||
| GA vs. GGc | 839/1548 | 1.12 (0.93,1.33) | 0.229 | 0.0% | 0.436 |
| AA vs. GGd | 605/1102 | 2.03 (1.38,3.00) | 0.000 | 86.9% | 0.000 |
| GA/AA vs. GGe | 1104/1892 | 1.20 (1.01,1.42) | 0.038 | 69.0% | 0.040 |
| AA vs. GA/GGf | 896/1601 | 1.96 (1.33,2.89) | 0.001 | 85.8% | 0.001 |
| DNMT3A rs13420827 | |||||
| CG vs. CCc | 656/1206 | 0.84 (0.68,1.03) | 0.090 | 44.3% | 0.180 |
| GG vs. CCd | 495/851 | 1.16 (0.73,1.85) | 0.523 | 0.0% | 0.423 |
| CG/GG vs. CCe | 689/1255 | 0.87 (0.72,1.06) | 0.171 | 0.0% | 0.336 |
| GG vs. CG/CCf | 689/1255 | 1.23 (0.78,1.95) | 0.371 | 0.0% | 0.320 |
| DNMT3B rs2424913 | |||||
| CT vs. TTc | 1086/1053 | 0.66 (0.32,1.36) | 0.258 | 0.0% | 0.992 |
| CC vs. TTd | 1075/1032 | 3.02 (0.12,74.69) | 0.500 | – | – |
| CT/CC vs. TTe | 1087/1053 | 0.71 (0.35,1.44) | 0.346 | 0.0% | 0.849 |
| CC vs. CT/TTf | 1087/1053 | 3.02 (0.12,74.69) | 0.500 | – | – |
| DNMT3B rs1569686 | |||||
| GT vs. TTc | 745/1262 | 0.88 (0.69,1.13) | 0.320 | 83.7% | 0.002 |
| GG vs. TTd | 644/1072 | 0.96 (0.46,2.01) | 0.923 | 3.1% | 0.310 |
| GT/GG vs. TTe | 1225/1789 | 0.74 (0.61,0.90) | 0.003 | 80.1% | 0.000 |
| GG vs. GT/TTf | 756/1283 | 0.97 (0.46,2.02) | 0.930 | 0.0% | 0.394 |
The bolds pointed to models that had statistically significant associations with gastric cancer.
P value of the Z-test for odds ration test.
P value of the Q-test for heterogeneity test.
Heterozygote model (heterozygous vs. homozygous frequent allele).
Homozygote model (homozygous rare vs. homozygous frequent allele).
Dominant model (homozygous rare + heterozygous vs. homozygous frequent allele).
Recessive model (homozygous rare vs. heterozygous + homozygous frequent allele).
Fig. 2.
Forest plot of DNMT1, DNMT3A and DNMT3B polymorphisms associated with GC risk.
Table 3.
Systematic review of associations between DNMTs SNPs and gastric cancer risk.
| Study | Country | Sample size (cases/controls) | Gene | SNPs | OR (95%CI) |
|
|---|---|---|---|---|---|---|
| Heterozygote model | Homozygote model | |||||
| Yang et al., 2012 | China | 242/294 | DNMT1 | rs2114724 C > T | 1.16 (0.81, 1.68) | 0.62 (0.30, 1.27) |
| Jiang et al., 2012a, Jiang et al., 2012b | China | 447/961 | DNMT1 | rs10420321 A > G | 0.96 (0.66, 1.41) | 1.17 (1.88,1.55) |
| Jiang et al., 2012a, Jiang et al., 2012b | China | 447/961 | DNMT1 | rs8111085 T > C | 1.08 (0.88, 1.43) | 1.18 (0.82, 1.69) |
| Jiang et al., 2012a, Jiang et al., 2012b | China | 447/961 | DNMT1 | rs2288349 G > A | 0.93 (0.71, 1.22) | 0.81 (0.50, 1.33) |
| Khatami et al., 2009 | Iran | 200/200 | DNMT1 | rs721186 G > A | 1.12 (0.06, 16.0) | – |
| Khatami et al., 2009 | Iran | 200/200 | DNMT1 | rs13784 G > A | – | – |
| Khatami et al., 2009 | Iran | 200/200 | DNMT1 | rs11488 A > T | – | – |
| Wu et al., 2012 | China | 340/251 | DNMT3A | rs36012910 A > G | 2.44 (1.37, 4.33) | 1.00 (0.98, 1.01) |
| Yang et al., 2012 | China | 242/294 | DNMT3A | rs13428812 A > G | 0.93 (0.64, 1.35) | 1.11 (0.58, 2.12) |
| Yang et al., 2012 | China | 242/294 | DNMT3A | rs11887120 T > C | 0.96 (0.63, 1.47) | 1.26 (0.76, 2.07) |
| Wu et al., 2014 | China | 405/408 | DNMT3A | rs7560488 T > C | 1.73 (1.24, 2.41) | 2.50 (1.01, 6.23) |
| Wang et al., 2015a, Wang et al., 2015b | China | 447/961 | DNMT3B | rs6119954 G > A | 1.00 (0.76, 1.31) | 1.37 (0.88, 2.13) |
| Wang et al., 2015a, Wang et al., 2015b | China | 447/961 | DNMT3B | rs4911107 A > G | 0.86 (0.26, 2.88) | 0.76 (0.23, 2.46) |
| Wang et al., 2015a, Wang et al., 2015b | China | 447/961 | DNMT3B | rs4911259 G > T | 0.86 (0.26, 2.89) | 0.76 (0.23, 2.45) |
| Wang et al., 2015a, Wang et al., 2015b | China | 447/961 | DNMT3B | rs8118663 A > G | 1.28 (0.95, 1.72) | 1.32 (0.91, 1.91) |
| Yang et al., 2012 | China | 242/294 | DNMT3B | rs2424908 T > C | 0.98 (0.66, 1.45) | 1.05 (0.64, 1.71) |
| Liu, 2008 | China | 313/350 | DNMT3B | rs6087990 C > T | – | 1.46 (1.07, 2.01) |
SNPs, single nucleotide polymorphisms; heterozygote model (heterozygous vs. homozygous frequent allele); homozygote model (homozygous rare vs. homozygous frequent allele).
The bolds pointed to SNPs that had statistically significant associations with gastric cancer.
3.3. Heterogeneity Analysis (Sensitivity and Subgroup Analysis)
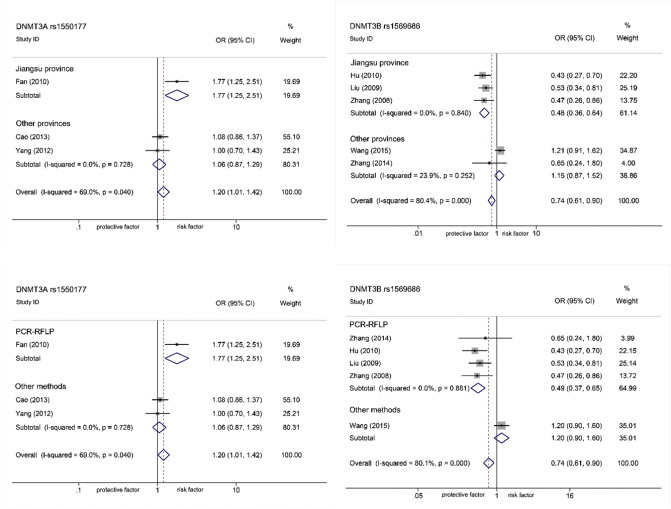
There was obvious heterogeneity in rs1550117 (AA vs. GG I2 86.9%, Phet 0.000; GA/AA vs. GG: I2 69.0%, Phet 0.040; AA vs. GA/GG: I2 85.8%, Phet 0.001) and rs1569686 (GT vs. TT: I2 83.7%, Phet 0.002; GT/GG vs. TT: I2 80.1%, Phet 0.000). A sensitivity analysis was conducted to explore which study primarily influenced the pooled ORs (Table S2, Fig. S1–S2). For rs1550117, the heterogeneity was mostly caused by a study (Fan et al., 2010), since when it was removed, 95%IC changed in direction of association (OR 1.06, 95%CI 0.87–1.29) and heterogeneity went to zero (I2 0%, Phet 0.73). Likewise, for rs1569686, Wang et al. (2015b) was found to be the major originator after excluded (95%IC didn't change in direction but heterogeneity went to zero: OR 0.49, 95%CI 0.37–0.65, I2 0%, Phet 0.88). We compared characteristics of the two studies to the other's. Two factors were screened out to explain the heterogeneity: population areas (Jiangsu province or others) and genotyping methods (PCR-RFLP or others). Then a subgroup analysis was performed (Fig. 3). Population areas: for Jiangsu population, rs1550117 and rs1569686 were associated with GC (OR 1.77, 95%CI 1.25–2.51; OR 0.48, 95%CI 0.36–0.64), but for others (Jiangxi, Jilin and Heilongjiang provinces) no associations were found (OR 1.06, 95%CI 0.87–1.29; OR 1.15, 95%CI 0.87–1.52). Genotyping methods: by PCR-RFLP, rs1550117 and rs1569686 were detected associated with GC (OR 1.77, 95%CI 1.25–2.51; OR 0.49, 95%CI 0.37–0.65) but by others (TaqMan and MassArray) significant associations were not discovered (OR 1.06, 95%CI 0.87–1.29; OR 1.20, 95%CI 0.90–1.60).
Fig. 3.
Forest plot of subgroup analysis on DNMT3A rs1550117 and DNMT3B rs1569686 polymorphisms (dominant model) by population area and genetic methods. Population area (Jiangsu province and other provinces: Jiangxi, Jilin and Heilong Jiang provinces, in China) (A); Genetic methods (PCR-RFLP and other methods: TaqMan and MassArray) (B).
4. Discussion
Of the seven SNPs, two (rs16999593 and rs1550117) and one (rs1569686) were significantly associated with GC risk indicating a range of effects from the increased (DNMT1 and DNMT3A) to the reduced (DNMT3B).
4.1. DNMT1
Our results proved rs16999593 as a potential biomarker for GC susceptibility which was exactly consistent with the results on other types of cancers, such as breast cancer and prostate cancer (Tao et al., 2015, He et al., 2014). In addition, we did not find rs2228611 associated with GC, but it was recently reported that patients carrying the mutant genotypes significantly lived longer than those bearing the wild, indicating that rs2228611 might be a positive prognostic marker for GC survival (Jia et al., 2016).
4.2. DNMT3A and DNMT3B
In terms of rs1550117, our findings opposed a previous meta-analysis and we could attribute this contradiction to differences in using homozygote models (Liu et al., 2015). For rs1569686, we consider it as a protective factor for gastric carcinogenesis and similar results were discovered in head and neck cancer, lung cancer and colorectal cancer (Duan et al., 2015, Zhang et al., 2015, Xia et al., 2015, Zhu et al., 2012). However, another study argued it was associated with poor prognosis in GC cases (Wang et al., 2015a). Maybe it played different roles in pathogenesis and prognosis. Particularly, we found in Jiangsu, a high GC incidence area of China (Liu et al., 2007), mutant rs1550117 doubled the risk and mutant rs1569686 lowered by a half of it. Also, even though some studies discovered TaqMan was more specific and sensitive than PCR-RFLP to detect polymorphisms or virus (Martinez-Trevino et al., 2016, Campsall et al., 2004), we found PCR-RFLP was so far a best method for risk detection in GC. Regarding rs2424913, we didn't find it associated with GC in Chinese. A review reported it could significantly decrease cancers in African but not Asian (Duan et al., 2015). It was speculated whether rs2424913 enabled African to catch GC rather than other populations. Although some meta-analysis studies demonstrated that rs6087990 might confer protection against overall cancers (Duan et al., 2015, Zhang et al., 2015), but it represented an opposite effect on GC as our systematic review showed (Liu, 2008).
4.3. Strengths and Limitations
Previous meta-analysis studies primarily evaluated associations between a few SNPs and cancers without classification, such as GC (Zhu et al., 2015, Duan et al., 2015, Liu et al., 2015, Zhang et al., 2015, Xia et al., 2015). The major strengths of our study was its comprehensive and systematic focus on GC and SNPs from three main types of DNMTs, 17 SNPs in total. Also, some mistakes in previous results were corrected in our study (Liu et al., 2015). At the same time, there were some limitations. Firstly, significant heterogeneities were observed for a few genetic models. Although a sensitivity analysis and a subgroup analysis were performed to clarify sources, we cannot find all potential factors. Second the meta-analysis findings were currently restricted to Chinese population pending results from other populations in future studies.
5. Conclusion
Our meta-analysis suggested that DNMT1 rs16999593 and DNMT3A rs1550117 could contribute to GC and that DNMT3B rs1569686 might function as a protective factor against gastric carcinogenesis. By using these significant SNPs as biomarkers, it is feasible to estimate the risk of catching GC and thus formulate timely preventive strategy.
Author Contributions
F.G.H., Q.S. and H.J.L conceived and designed the study. H.J.L., W.L., S.S.L. and S.Q.Z. took full responsibility for data collecting and accuracy. H.J.L. and W.L. performed the meta-analysis and systematic review, and drafted the manuscript. W.B.W., J.L.R., and Q.L. helped revise the manuscript.
Funding
This work was supported by Natural Science Foundation of China (NSFC, NO: 81473624) and Key Specialty Foundation of The State Administration of Traditional Chinese Medicine (NO:ZJ0901ZL020). The sponsor had no role in study design, data collection, data analysis, data interpretation, or writing of the report.
Conflicts of Interest
The authors have no conflicts of interest.
Footnotes
Supplementary data to this article can be found online at doi:10.1016/j.ebiom.2016.10.028.
Contributor Information
Fenggang Hou, Email: fghou555@126.com.
Qi Shi, Email: stevenshi_qi@hotmail.com.
Appendix A. The Search Strategy for PubMed Comprised the Following
-
1.
Colorectal neoplasms[mesh]
-
2.
DNMT1[tiab]
-
3.
DNMT3A [tiab]
-
4.
DNMT3B[tiab]
-
5.
DNMTs[tiab]
-
6.
DNA methyltransferases[tiab]
-
7.
(2 or 3 or 4 or 5 or 6)
-
8.
1 and 7
Appendix A. Supplementary data
Table of quality assessment
Table of Meta-sensitivity analysis
References
- Aung P.P. No evidence of correlation between the single nucleotide polymorphism of DNMT3B promoter and gastric cancer risk in a Japanese population. Oncol. Rep. 2005;14(5):1151–1154. [PubMed] [Google Scholar]
- Brown K.D., Robertson K.D. DNMT1 knockout delivers a strong blow to genome stability and cell viability. Nat. Genet. 2007;39(3):289–290. doi: 10.1038/ng0307-289. [DOI] [PubMed] [Google Scholar]
- Campsall P.A. Detection and genotyping of Varicella-zoster virus by TaqMan allelic discrimination real-time PCR. J. Clin. Microbiol. 2004;42(4):1409–1413. doi: 10.1128/JCM.42.4.1409-1413.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X. Single nucleotide polymorphisms of DNA methyltransferase 1 gene is associated with the risk of gastric atrophy and Helicobacter pylori infection. J. Gastroenterol. Hepatol. 2012;27:143. [Google Scholar]
- Cao X.Y. DNMT3a rs1550117 polymorphism association with increased risk of Helicobacter pylori infection. Asian Pac. J. Cancer Prev. 2013;14(10):5713–5718. doi: 10.7314/apjcp.2013.14.10.5713. [DOI] [PubMed] [Google Scholar]
- Chang S.C. One-carbon metabolism related gene polymorphisms and the risk of stomach cancer in a Chinese population. Am. J. Epidemiol. 2010;171:S67. [Google Scholar]
- Chang S.C. Single nucleotide polymorphisms of one-carbon metabolism and cancers of the esophagus, stomach, and liver in a Chinese population. PLoS One. 2014:9(10). doi: 10.1371/journal.pone.0109235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan F. Systematic evaluation of cancer risk associated with DNMT3B polymorphisms. J. Cancer Res. Clin. Oncol. 2015;141(7):1205–1220. doi: 10.1007/s00432-014-1894-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M. Epigenetics in cancer. N. Engl. J. Med. 2008;358(11):1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- Fan H. A functional polymorphism in the DNA methyltransferase-3A promoter modifies the susceptibility in gastric cancer but not in esophageal carcinoma. BMC Med. 2010;8:12. doi: 10.1186/1741-7015-8-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferlay J. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer. 2015;136(5):E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- Fu H.Y. Arsenic trioxide inhibits DNA methyltransferase and restores expression of methylation-silenced CDKN2B/CDKN2A genes in human hematologic malignant cells. Oncol. Rep. 2010;24(2):335–343. doi: 10.3892/or_00000864. [DOI] [PubMed] [Google Scholar]
- Gao Q. Deletion of the de novo DNA methyltransferase Dnmt3a promotes lung tumor progression. Proc. Natl. Acad. Sci. U. S. A. 2011;108(44):18061–18066. doi: 10.1073/pnas.1114946108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harder J. Quantitative promoter methylation analysis of hepatocellular carcinoma, cirrhotic and normal liver. Int. J. Cancer. 2008;122(12):2800–2804. doi: 10.1002/ijc.23433. [DOI] [PubMed] [Google Scholar]
- He B.S., Pan Y.Q., Zhu C.B. Polymorphisms of DNA methyltransferases and the risk of prostate cancer. Zhonghua Nan Ke Xue. 2014;20(12):1077–1081. [PubMed] [Google Scholar]
- Hu J. DNMT3B promoter polymorphism and risk of gastric cancer. Dig. Dis. Sci. 2010;55(4):1011–1016. doi: 10.1007/s10620-009-0831-3. [DOI] [PubMed] [Google Scholar]
- Jia Z. Polymorphisms of the DNA methyltransferase 1 gene predict survival of gastric cancer patients receiving tumorectomy. Dis. Markers. 2016;2016:8578064. doi: 10.1155/2016/8578064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J. Polymorphisms of the DNA methyltransferase 1 associated with reduced risks of Helicobacter pylori infection and increased risks of gastric atrophy. PLoS One. 2012;7(9) doi: 10.1371/journal.pone.0046058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J. Role of polymorphisms of DNA methyltransferases in risks of gastric cancer and atrophic gastritis. Eur. J. Cancer. 2012;48:S3. [Google Scholar]
- Jiang J. Polymorphisms of DNA methyltransferase 3a associated with risk of helicobacter pylori infection in gastric cancer. Eur. J. Cancer. 2013;49:S18. [Google Scholar]
- Kanai Y. Mutation of the DNA methyltransferase (DNMT) 1 gene in human colorectal cancers. Cancer Lett. 2003;192(1):75–82. doi: 10.1016/s0304-3835(02)00689-4. [DOI] [PubMed] [Google Scholar]
- Khatami F. Lack of effects of single nucleotide polymorphisms of the DNA methyltransferase 1 gene on gastric cancer in Iranian patients: a case control study. Asian Pac. J. Cancer Prev. 2009;10(6):1177–1182. [PubMed] [Google Scholar]
- Kullmann K. DNMT1 genetic polymorphisms affect breast cancer risk in the central European Caucasian population. Clin. Epigenetics. 2013;5(1) doi: 10.1186/1868-7083-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieb W. Association of angiotensin-converting enzyme 2 (ACE2) gene polymorphisms with parameters of left ventricular hypertrophy in men. Results of the MONICA Augsburg echocardiographic substudy. J. Mol. Med. (Berl.) 2006;84(1):88–96. doi: 10.1007/s00109-005-0718-5. [DOI] [PubMed] [Google Scholar]
- Liu D. Correlation analysis of DNMT3B − 283 SNP and the susceptibility of gastric cancer. Inner Mongolia J. Tradit. Chin. Med. 2008;27(11):187–188. [Google Scholar]
- Liu D. Southeast University; 2009. Associated Study on the Promoter SNPs of DNMT3A/3B With Genetic Susceptibility to Gastric Cancer and Esophageal Cancer. [Google Scholar]
- Liu A. Case-control analysis on stomach cancer in a high cancer incidence area of Jiangsu province. Chin. J. Public Health. 2007;23(5):575–576. [Google Scholar]
- Liu C.H. DNMT3A − 448A > G polymorphism and cancer risk: a meta-analysis. Genet. Mol. Res. 2015;14(2):3640–3649. doi: 10.4238/2015.April.17.14. [DOI] [PubMed] [Google Scholar]
- Lu L. Prognostic and clinicopathological value of Gli-1 expression in gastric cancer: a meta-analysis. Oncotarget. 2016 doi: 10.18632/oncotarget.12011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y. The association of DNA methyltransferase 1 gene polymorphisms with susceptibility to childhood acute lymphoblastic leukemia. Biomed. Pharmacother. 2015;73:35–39. doi: 10.1016/j.biopha.2015.05.001. [DOI] [PubMed] [Google Scholar]
- Martinez-Trevino D.A. Rapid detection of the GSTM3 A/B polymorphism using real-time PCR with TaqMan probes. Arch. Med. Res. 2016 doi: 10.1016/j.arcmed.2016.04.002. [DOI] [PubMed] [Google Scholar]
- Mostowska A. DNMT1, DNMT3A and DNMT3B gene variants in relation to ovarian cancer risk in the Polish population. Mol. Biol. Rep. 2013;40(8):4893–4899. doi: 10.1007/s11033-013-2589-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano M. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99(3):247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- Saeki N. Genetic factors related to gastric cancer susceptibility identified using a genome-wide association study. Cancer Sci. 2013;104(1):1–8. doi: 10.1111/cas.12042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M.Y. Association of DNMT1 and DNMT3B polymorphisms with breast cancer risk in Han Chinese women from South China. Genet. Mol. Res. 2012;11(4):4330–4341. doi: 10.4238/2012.September.26.1. [DOI] [PubMed] [Google Scholar]
- Tao R. The possible role of EZH2 and DNMT1 polymorphisms in sporadic triple-negative breast carcinoma in southern Chinese females. Tumor Biol. 2015;36(12):9849–9855. doi: 10.1007/s13277-015-3754-y. [DOI] [PubMed] [Google Scholar]
- Thakkinstian A. Systematic review and meta-analysis of the association between complement component 3 and age-related macular degeneration: a HuGE review and meta-analysis. Am. J. Epidemiol. 2011;173(12):1365–1379. doi: 10.1093/aje/kwr025. [DOI] [PubMed] [Google Scholar]
- Torre L.A. Global cancer statistics, 2012. CA Cancer J. Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- Wang Y.M. Single nucleotide polymorphism in DNA methyltransferase 3B promoter and its association with gastric cardiac adenocarcinoma in North China. World J. Gastroenterol. 2005;11(23):3623–3627. doi: 10.3748/wjg.v11.i23.3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C. Polymorphism of DNA methyltransferase 3b and association with development and prognosis in gastric cancer. PLoS One. 2015;10(8) doi: 10.1371/journal.pone.0134059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C. Association between polymorphisms of DNMT3b gene and its expression level in gastic cancer tissue and gastric cancer. J. Jilin Univ. (Med. Ed.) 2015;41(2):368–373. [Google Scholar]
- Wu Q. DNMT3A rs36012910 A > G polymorphism and gastric cancer susceptibility in a Chinese population. Mol. Biol. Rep. 2012;39(12):10949–10955. doi: 10.1007/s11033-012-1996-y. [DOI] [PubMed] [Google Scholar]
- Wu H. A novel functional TagSNP Rs7560488 in the DNMT3A1 promoter is associated with susceptibility to gastric cancer by modulating promoter activity. PLoS One. 2014;9(3) doi: 10.1371/journal.pone.0092911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Z. Quantitative assessment of the association between DNMT3B-579G > T polymorphism and cancer risk. Cancer Biomark. 2015;15(5):707–716. doi: 10.3233/CBM-150512. [DOI] [PubMed] [Google Scholar]
- Xiang G. Association of DNMT1 gene polymorphisms in exons with sporadic infiltrating ductal breast carcinoma among Chinese Han women in the Heilongjiang Province. Clin. Breast Cancer. 2010;10(5):373–377. doi: 10.3816/CBC.2010.n.049. [DOI] [PubMed] [Google Scholar]
- Xue W. Association between PLCE1 rs2274223 A > G polymorphism and cancer risk: proof from a meta-analysis. Sci. Rep. 2015;5:7986. doi: 10.1038/srep07986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan G., Yanxin C., Xiangjun J. Association of Dnmt1 single-nucleotide polymorphisms and risk of gastric cancer. Chi. J. Oncol. Prev. Treat. 2015;7(6):394–397. [Google Scholar]
- Yang L. Incidence and mortality of gastric cancer in China. World J. Gastroenterol. 2006;12(1):17–20. doi: 10.3748/wjg.v12.i1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X.X. Risk-association of DNA methyltransferases polymorphisms with gastric cancer in the Southern Chinese population. Int. J. Mol. Sci. 2012;13(7):8364–8378. doi: 10.3390/ijms13078364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S. Jiamusi University; 2008. Correlation Analysis of SNP in DNMT3B Promoter Region and Gastric Cancer Risk. [Google Scholar]
- Zhang S. Association of DNMT3B SNP (− 579G/T) with the risk of gastric cancer in Jiamusi. Heilongjiang Med. Pharm. 2014;37(5):82–83. [Google Scholar]
- Zhang Y. Association of DNMT3B − 283 T > C and − 579 G > T polymorphisms with decreased cancer risk: evidence from a meta-analysis. Int. J. Clin. Exp. Med. 2015;8(8):13028–13038. [PMC free article] [PubMed] [Google Scholar]
- Zhao C., Bu X. Promoter methylation of tumor-related genes in gastric carcinogenesis. Histol. Histopathol. 2012;27(10):1271–1282. doi: 10.14670/HH-27.1271. [DOI] [PubMed] [Google Scholar]
- Zhu S. DNMT3B polymorphisms and cancer risk: a meta analysis of 24 case-control studies. Mol. Biol. Rep. 2012;39(4):4429–4437. doi: 10.1007/s11033-011-1231-2. [DOI] [PubMed] [Google Scholar]
- Zhu J. Polymorphism of DNA methyltransferase 3B − 149C/T and cancer risk: a meta-analysis. Med. Oncol. 2015;32(1):399. doi: 10.1007/s12032-014-0399-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table of quality assessment
Table of Meta-sensitivity analysis