Highlights
-
•
High H19 expression in primary tumors is an independent predictor of short overall survival in CRC patients.
-
•
RB1-E2F and CDK8-β-catenin signaling are essential in mediating the oncogenic activity of H19 in CRC.
-
•
Combined analysis of H19 and its targets further improved the prediction power on overall survival of CRC patients.
Long noncoding RNAs (lncRNAs) are transcripts at least 200 nucleotides long that do not code for proteins. The clinical relevance of lncRNAs in colorectal cancer (CRC) is largely unknown. Here we identified that H19 expression in primary tumors is an independent prognostic predictor of poor prognosis of CRC patients and further proved its oncogenic role. To characterize the mechanisms, we profiled gene expression changes following H19 modulation in CRC cell lines and analyzed gene expression association in clinical datasets. Our data revealed important cancer-signaling pathways, including the RB1-E2F and the CDK8-β-catenin signaling, underlying H19 function.
Keywords: H19, RB1, E2F, CDK8, β-Catenin, Colorectal cancer
Abstract
The clinical significance of long noncoding RNAs (lncRNAs) in colorectal cancer (CRC) remains largely unexplored. Here, we analyzed a large panel of lncRNA candidates with The Cancer Genome Atlas (TCGA) CRC dataset, and identified H19 as the most significant lncRNA associated with CRC patient survival. We further validated such association in two independent CRC cohorts. H19 silencing blocked G1-S transition, reduced cell proliferation, and inhibited cell migration. We profiled gene expression changes to gain mechanism insight of H19 function. Transcriptome data analysis revealed not only previously identified mechanisms such as Let-7 regulation by H19, but also RB1-E2F1 function and β-catenin activity as essential upstream regulators mediating H19 function. Our experimental data showed that H19 affects phosphorylation of RB1 protein by regulating gene expression of CDK4 and CCND1. We further demonstrated that reduced CDK8 expression underlies changes of β-catenin activity, and identified that H19 interacts with macroH2A, an essential regulator of CDK8 gene transcription. However, the relevance of H19-macroH2A interaction in CDK8 regulation remains to be experimentally determined. We further explored the clinical relevance of above mechanisms in clinical samples, and showed that combined analysis of H19 with its targets improved prognostic value of H19 in CRC.
1. Introduction
Colorectal cancer (CRC) remains the most frequently diagnosed cancer, and a leading cause of cancer-related mortality worldwide (Torre et al., 2015). In the United States, it is estimated that 132,700 new cases and 49,700 deaths occur in 2015 (Siegel et al., 2015). Discovering novel biomarkers and therapeutic targets is of outmost importance to improve CRC outcome (Toiyama et al., 2016, Abbasi et al., 2015). The ENCODE project revealed widespread transcription of long noncoding RNAs (lncRNAs) that possess at least 200 nucleotides in length but do not code for proteins (Morris and Mattick, 2014, Derrien et al., 2012). Accumulating evidence suggests that lncRNAs have essential roles in cancer initiation and progression, and deregulated lncRNA expression is found in a variety of cancer types (Ling et al., 2015, Spizzo et al., 2012, Tsai et al., 2011). However, the clinical relevance of lncRNAs in CRC remains largely unexplored.
In this study, we analyzed The Cancer Genome Atlas (TCGA) CRC dataset (Cancer Genome Atlas Network, 2012) for clinical relevance of lncRNA expression, and identified H19 as the lncRNA most significantly associated with survival. As one of the first imprinted genes to be discovered, H19 is exclusively transcribed from the maternally inherited allele, and plays key role in embryonic development (Monnier et al., 2013). Deregulated expression of H19 has been reported in diverse human malignancies (Raveh et al., 2015). Studies on H19 function in cancer show both oncogenic and tumor suppressive effects (Medrzycki et al., 2014, Barsyte-Lovejoy et al., 2006, Hao et al., 1993, Juan et al., 2000). The reported H19 activities are diverse, including interaction with epigenetic regulators (Monnier et al., 2013, Giovarelli et al., 2014, Zhou et al., 2015, Luo et al., 2013), regulation of microRNAs as competing endogenous RNA (ceRNA) (Yan et al., 2015, Imig et al., 2015, Kallen et al., 2013, Liang et al., 2015), or production of miR-675 as a primary transcript (Keniry et al., 2012, Venkatraman et al., 2013, Zhu et al., 2014, Shi et al., 2014, Tsang et al., 2010).
We performed functional studies examining H19 activity and mechanism in CRC. With an unbiased approach to analyze essential networks mediating H19 function, we found that H19 binds to the tumor suppressor retinoblastoma 1 (RB1), and affects the RB-E2F signaling that controls G1-S transition (Polager and Ginsberg, 2009). H19 also regulates transcription of cyclin-dependent kinase 8 (CDK8), an oncogenic driver in CRC, and consequently affects β-catenin activity (Firestein et al., 2008). We finally validated the clinical relevance and explored the prognostic value of these findings.
2. Materials & Methods
2.1. Patient Samples
For the TCGA dataset, we obtained and analyzed gene expression and clinical data from the Cancer Genome Atlas Project (TCGA; http://tcga-data.nci.nih.gov/) for CRC patients containing a total of 534 CRC tumor samples. We used two additional CRC cohorts including 295 cases for validation. Cohort 2 comprises 178 CRC samples obtained from the Mie University, Mie, Japan. Cohort 3 contains 117 colon cancer samples obtained from the Masaryk Memorial Cancer Institute, Czech Republic. The clinicopathological features of CRC cases are detailed in Table S1. Tissue samples were obtained from fresh surgical specimens frozen in liquid nitrogen and stored at − 80 °C. All the samples were obtained with the patients' informed consent and were histologically confirmed. The Institutional research and ethics committee approved this study.
2.2. RNA Immunoprecipitation (RIP)
We used the Magna RIP Kit (Millipore) according to the manufacturer's protocol. Cells were prepared in RIP lysis buffer, and RNA-protein complexes were immunoprecipitated using corresponding antibodies. RNA was purified using phenol:chloroform:isoamyl alcohol and treated with Dnase to remove genomic DNA, followed by reverse transcription-PCR or real-time PCR analysis. Control amplification was carried out on input RNA before immunoprecipitation.
2.3. Chromatin Immunoprecipitation (ChIP)
We used the EZ ChIP Kit (Millipore) according to the manufacturer's protocol. Chromatin was prepared following crosslinking with formaldehyde solution. DNA was sheared to size of 200–700 bp by sonication. The samples were pre-cleared with agarose beads and subsequently immunoprecipitated using 5 μg of antibodies.
2.4. Affymetrix HTA 2.0 Array
We used Affymetrix Human Transcriptome Array 2.0 for genetic changes after knocking down H19, CTNNB1 and CDK8. Five hundred nanograms total RNA of each cell line (in duplicate) was provided to the Sequencing & Non-coding RNA Program at M.D. Anderson Cancer Center for performing the microarray. Raw data was extracted with console Affymetrix® Transcriptome Analysis Console (TAC) Software. Comprehensive function and pathway, including upstream regulator prediction, were performed by Ingenuity Pathway Analysis (IPA) (Qiagen). The microarray data was deposited in Gene Expression Omnibus (GEO) database (accession numbers: GSE87431).
2.5. Statistical Analysis
We analyzed the association of gene expression or other clinical parameters (sex, age, tumor location, tumor stage) with survival using a Cox proportional hazard model. In the multivariable regression model, only the factors that were statistically significant in univariable analyses were included. After determining the cut-off value that allows most significant split with log-rank test in the TCGA dataset, we used the same cut-off value to generate Kaplan-Meier plots for all cohorts. The Spearman's rank correlation was applied for the strength of association between tested genes. The differences between groups were analyzed using 2-way ANOVA analysis, t-test, or nonparametric test, with the GraphPad software. Graphics represent the mean ± standard deviation from at least two independent experiments repeated in triplicate, unless otherwise stated. Statistical significance was considered if P < 0.05.
Additional methods, including Cell Culture, RNA interference experiments, Proliferation Assay, Cell Scratch Assay, Cell-Cycle Assay, Click-iT EdU Assay, Reverse Transcription Quantitative RT-PCR Analysis, Western Blot, Plasmid and Virus Generation, and Northern Blot are available in Supplemental Experimental Procedures. Reagent, primer, and antibody information is available in Table S3–5.
3. Results
3.1. H19 Is an Independent Predictor of CRC Survival
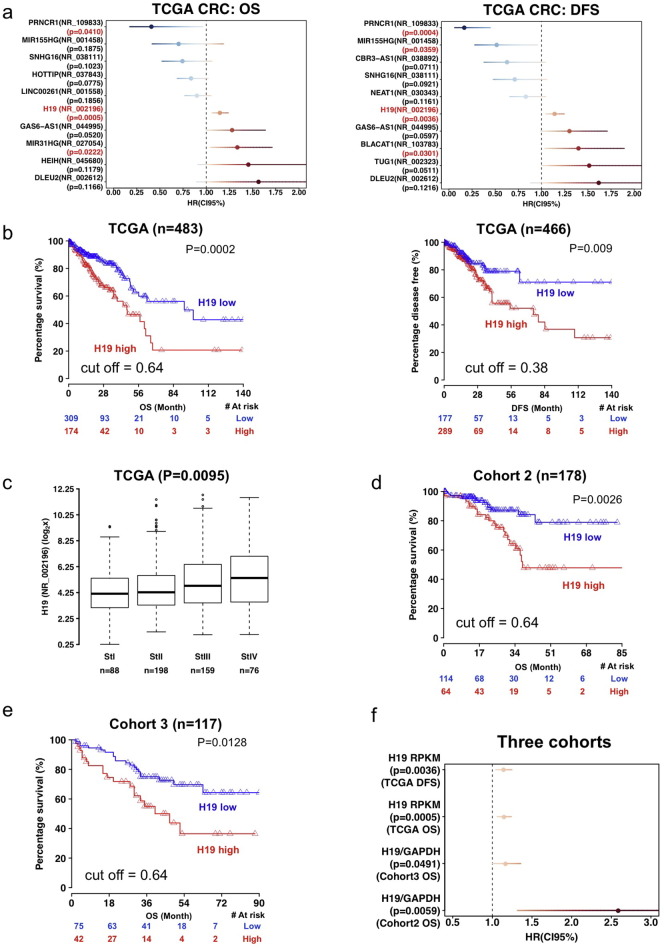
We analyzed 534 patients from the TCGA CRC dataset for a panel of 84 cancer-related lncRNAs (listed on the QIAGEN Human Cancer PathwayFinder RT2 lncRNA PCR Array). We excluded lncRNAs that were detectable in less than half of the samples, and obtained expression data for 37 lncRNAs. Detailed clinical parameters were available for 483 of these CRC cases (Table S1). The TCGA analysis revealed that high H19 levels, high MIR31HG levels, and low PRNCR1 levels were significantly associated with shorter overall survival (OS), and High H19, high BLACAT1, low MIR155HG, and low PRNCR1 correlated with shorter disease free survival (DFS), respectively (Fig. 1a and b). As the most significant lncRNA associated with OS (P = 0.0005), H19 does not correlate with the status of microsatellite stability (Fig. S1a), but shows steady increase (P = 0.0095) as tumor stage advances (Fig. 1c). Remarkably, the association of H19 with survival was independent of tumor stages [Hazard ratio (HR) 1.11, 95% confidence interval (CI95%) 1.03–1.21, P = 0.011 for OS; and HR 1.11, CI95% 1.01–1.21, P = 0.027 for DFS]. H19-derived microRNA (Tsang et al., 2010) miR-675-5p was undetectable in TCGA samples, while miR-675-3p expression correlated with H19 levels (r = 0.65, P < 0.0001) (Fig. S1b), but was not a predictor for either OS (0.0856) or DFS (P = 0.1189). In two independent cohorts of 178 CRC cases (cohort 2) and 117 colon cancer cases (cohort 3), respectively, patients with high H19 expression in primary tumors, measured by quantitative real time polymerase chain reaction (qRT-PCR), displayed significantly shorter OS (P = 0.0026 and P = 0.0128 for these two cohorts, respectively) (Fig. 1d, e and f). We could not perform multivariate analysis for the second cohort because all cases reported as ‘dead’ were from the stage III–IV group. Multivariate analysis of the third cohort revealed that H19 is an independent prognostic marker (HR 1.28, CI95% 1.08–1.50, P = 0.0034).
Fig. 1.
H19is an independent prognostic marker for CRC. (a) TCGA analysis of lncRNA and overall survival (OS) or disease free survival (DFS). Top 10 lncRNAs ranked by P value are listed. In red: P < 0.05. (b)H19 association with CRC OS and DFS in the TCGA cohort. (c)H19 and CRC stage. Data were presented as box-and-whisker plot (10th, 25th, median, 75th and 90th percentile) and compared with Kruskal-Wallis test. (d, e)H19 and OS in other cohorts. (f)H19 association with OS and DFS in three cohorts. Log-rank test was used for analysis in a and f. Hazard ratio (HR), Median, 95% confidence interval (CI95%) were indicated. See also Fig. S1.
3.2. H19 Loss of Function Dramatically Reduces CRC Malignant Phenotypes
We detected a wide range of H19 expression in CRC cell lines: H19 was abundantly expressed in DLD1 and HCT116, but barely detected in HT29 and SW480 cell lines (Fig. S2a and b). With northern blotting, we detected a band of ~ 2.5 kb (size of the reported H19 transcript according to NR_002196.2), with stronger signal in tumors than non-cancerous mucosae, and high and modest signal in DLD1 and HCT116, respectively (Fig. S2c). In contrast, miR-675 expression is very low with Ct value larger than 30 in DLD1 and HCT116 cell lines (Fig. S2b). Therefore, we selected these two models for the H19 knock-down experiments, where the influence of miR-675 would be minimal.
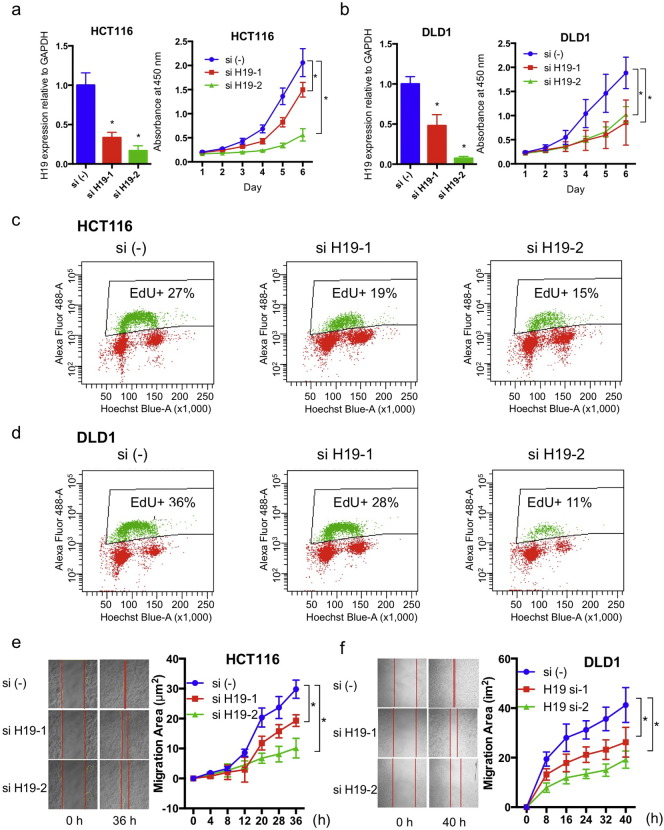
Silencing H19 expression (Fig. S2d) significantly reduced cell proliferation, as determined by CCK-8 assay, in both HCT116 (Fig. 2a) and DLD1 cells (Fig. 2b). Propidium iodide (PI) staining showed a clear reduction of cells in the S phase, and accumulation of cells in the G1 phase in both cell lines (Fig. S2e). Concordantly, with Click-iT EdU assay we identified a sharp reduction of DNA synthesis in H19 knockdown cells, with nearly one third and one half drop of EdU-positive cells with si-H19-1 and si-H19-2, respectively (Fig. 2c and d). Because of the association of H19 with tumor stage (Fig. 1c), we tested the effect of H19 on cell motility using the scratch assay. To avoid the interference of cell proliferation, we treated cells with mitomycin C prior to this assay. H19 knockdown caused a noticeable reduction of cancer cell migration, in a manner dependent on silencing effect, in both HCT116 (Fig. 2e) and DLD1 (Fig. 2f) cells. Together, these cellular functional studies clearly showed that H19 plays essential oncogenic role in CRC cells.
Fig. 2.
H19loss of function reduces CRC aggressiveness. (a, b)H19 effect on proliferation, determined with cell counting kit 8 assay. (c, d)H19 effect on DNA synthesis in CRC cells, determined with Click-iT EdU assay. (e, f)H19 effect on cell migration, determined with cell scratch assay. Typical images on left, and quantifications on right. Difference was determined using 2-way ANOVA with GraphPad software. * P < 0.05. See also Fig. S2.
3.3. H19 Regulates RB1-E2F1 Activity
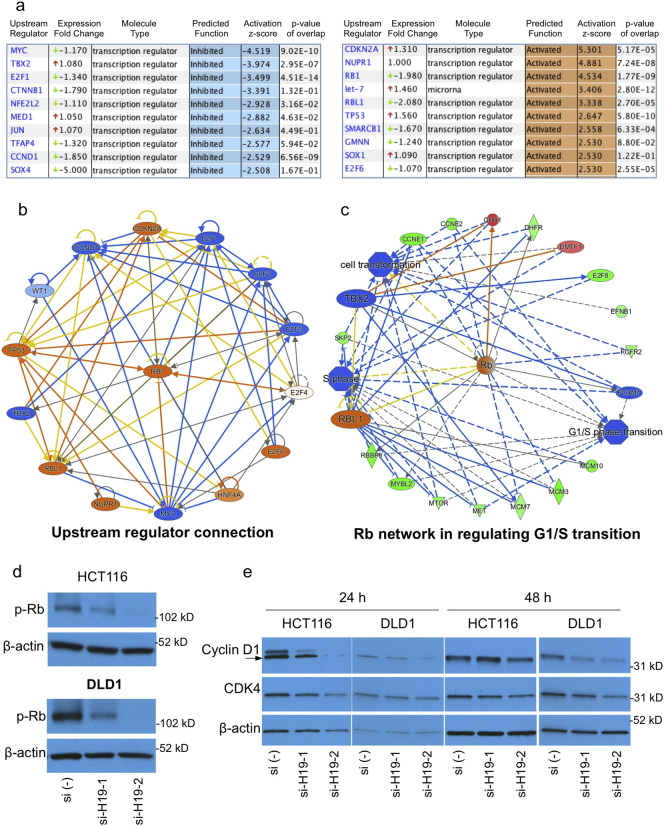
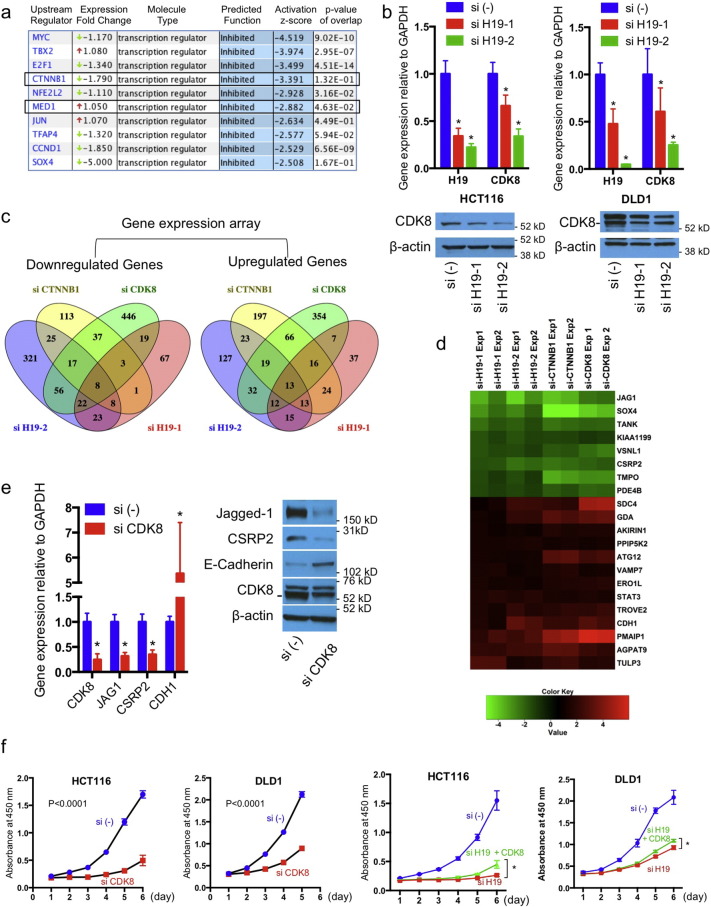
We profiled global gene expression changes following H19 knockdown by using gene expression microarray (GEM). Analyzing with Ingenuity Pathway Analysis (IPA, Qiagen), we found that cell cycle control of chromosomal replication ranks as the top canonical pathway significantly affected, consistent with the observed phenotype (Fig. S3a). IPA predicted MYC, TBX2, E2F1, CTNNB1, NFE2L2, MED1, JUN, TFAP4, CCND1, and SOX4 as top transcription regulators, ranked by activation z-score, to be functionally inhibited, and CDKN2A, NUPR1, RB1, let-7, RBL1, TP53, SMARCB1, GMNN, SOX1, E2F6 as regulators to be functionally activated (Fig. 3a). The prediction of MYC and let-7 is in consensus with reports that H19 antagonizes let-7 to regulate MYC (Yan et al., 2015, Kallen et al., 2013). Among all these, E2F1 is the most significant regulator predicted to control 87 of the 734 mRNAs changed after H19 knockdown (Fig. 3a). The E2F family includes multiple transcription factors, where E2F1-3 are cell cycle activators, and E2F4-8 are cell cycle repressors (Chen et al., 2009). Interestingly, the negative regulator of E2F signaling, RB1 is predicted to regulate 90 mRNAs and be functionally activated (Fig. 3a). RB1 is functionally connected with most of the predicted regulators, including CDKN2A, TBX2, E2F1, E2F2, E2F3, E2F6, CCND1, and TP53 (Fig. 3b), and is essential in regulating G1-S transition (Polager and Ginsberg, 2009), by affecting transcription of genes including CCNE1, CCNE2 and SKP2 (Fig. 3c). Consistently, IPA analysis of HCT116 si-H19-1, DLD1 si-H19-1, and DLD1 si-H19-2 samples all predicted that E2F transcriptional activity was inhibited (Fig. S3b, c and d). However, the RNA (Fig. 3a) and protein expression levels (Fig. S3e and f for protein) of RB1 and E2F1 were not affected by si-H19 treatment, suggesting a mechanism independent of transcription and translation. Phosphorylation of RB1 protein is a critical step in dissociation of RB1-E2F1 complex, which leads to E2F1 activation (Dick and Rubin, 2013). Consistently with above findings of H19 on E2F1 function, H19 silencing dramatically reduced RB1 phosphorylation at ser608 (Fig. 3d). Concordantly, H19 silencing reduced both RNA and protein expression of CDK4 and cyclin D1, two essential upstream factors controlling RB1 phosphorylation (Dick and Rubin, 2013) (Fig. 3e). Interestingly, we detected an interaction of H19 with RB1 protein using RNA immunoprecipitation (RIP) experiments (Fig. S3g): the biological relevance of such interaction is not clear. Together, these data suggest that H19 regulates RB1-E2F signaling to control cell proliferation.
Fig. 3.
H19regulates RB1-E2F signaling. (a) Predicted transcriptional regulators and microRNAs (z-score > 2.5 or <− 2.5, respectively), determined with Ingenuity Pathway Analysis of microarray data (si-H19-2 versus scrambled control). (b) Predicted connection of most significant upstream regulators. (c) Predicted RB1 network in regulation of G1-S transition. (d)H19 silencing reduces RB1 phosphorylation without affecting the level of unphosphorylated form. (e)H19 silencing reduces the expression levels of CDK4 and cyclin D1. See also Fig. S3.
3.4. H19 Regulates β-Catenin Activity via Modulating CDK8 Expression
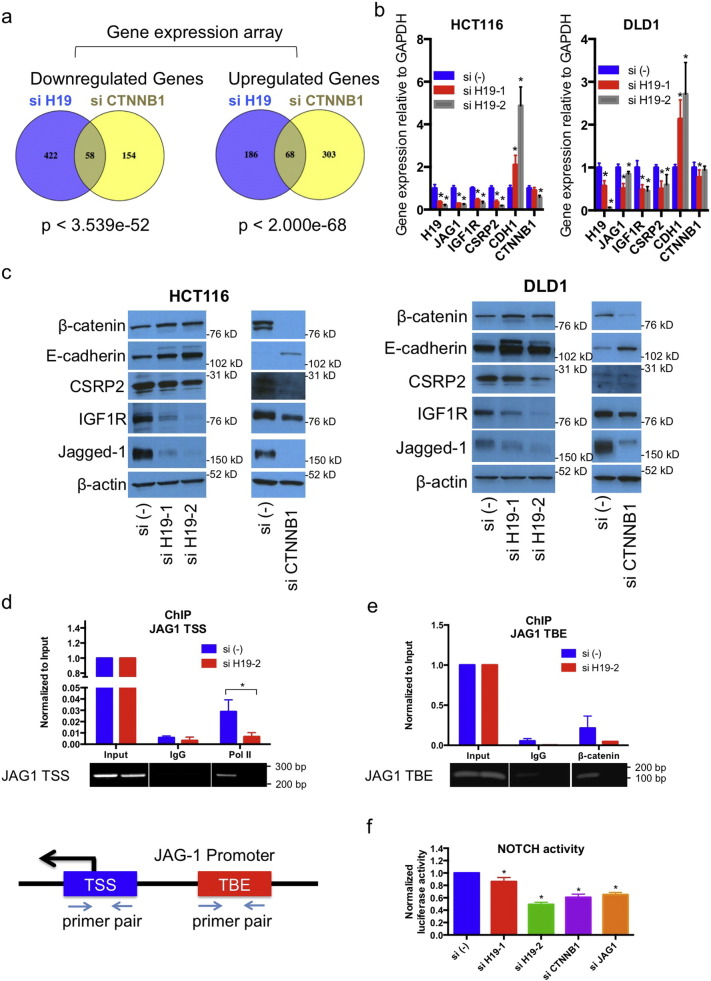
Aberrant activation of Wnt signaling is an essential mechanism of CRC (Anastas and Moon, 2013). The prediction of CTNNB1 (β-catenin) as upstream regulator (Fig. 3a) prompted us to examine H19 involvement in β-catenin activity. GEM analysis of mRNAs with > 2 fold changes (P < 0.05) revealed that almost one fourth of genes regulated by siCTNNB1 are modulated by si-H19 in the same direction (Fig. 4a and Fig. S4). We validated the GEM data by qRT-PCR and western blot with a high concordance: JAG1, IGF1R, and CSRP2 were noticeably reduced, and CDH1 expression was increased by both H19 siRNAs (Fig. 4b and c). We selected NOTCH ligand JAG1, the most downregulated gene in HCT116 cells, to test whether the changes are mediated through β-catenin. Chromatin immunoprecipitation (ChIP) revealed a significant drop in polymerase II binding to the transcription start site (TSS) of JAG1 gene (Fig. 4d), and a reduction of β-catenin binding to TCF-binding element (TBE) (Rodilla et al., 2009) after H19 silencing (Fig. 4e). NOTCH activity measured with Cignal RBP-Jk reporter assay was significantly reduced in cells treated with H19, CTNNB1 or JAG1 siRNAs (Fig. 4f). Taken together, these results suggest an intrinsic connection of H19 with β-catenin activity.
Fig. 4.
H19regulates β-catenin activity. (a) Genes regulated by H19 and β-catenin in HCT116 cells defined as “overlapping” genes (for significance: http://nemates.org/MA/progs/overlap_stats.html). (b, c)H19 effect on overlapping genes and CTNNB1. (d) Polymerase II binding to transcription start site (TSS) of JAG1 gene. (e) β-Catenin binding to TCF-binding site (TBE) upstream of JAG1 gene. Upper: quantification by qRT-PCR; lower: PCR gel image. (f) NOTCH luciferase assay. * P < 0.05. See also Fig. S4.
We did not find significant reduction on total or nuclear β-catenin protein levels by H19 knockdown (Figs. 4b, and c, and S5a). On the GEM, CDK8, a CRC oncogenic driver that regulates β-catenin activity (Firestein et al., 2008), ranks as the most regulated gene by si-H19 (reduced 2 fold and 4 fold, in si-H19-1 and si-H19-2 samples, respectively), but not by si-CTNNB1. CDK8 controls mediator-polymerase II interaction (Allen and Taatjes, 2015, Tsai et al., 2013), and thus CDK8 changes could impair functions of β-catenin and Med1 as predicted (Fig. 5a). Further supporting this hypothesis, IPA analysis of HCT116 si-H19–1, DLD1 si-H19-1, and DLD1 si-H19-2 samples consistently suggested that functions of β-catenin and Med1 are inhibited (Fig. S5b, c and d). We validated the effect of H19 on CDK8 expression with qRT-PCR and western blot (Fig. 5b). Additionally, we profiled genes changed > 2 fold (P < 0.05) by si-H19-1, si-H19-2, si-CTNNB1, and si-CDK8, and identified 8 overlapping genes that were downregulated, and 13 genes that were upregulated (Fig. 5c and d). We validated the microarray finding by showing a clear reduction of JAG1, CSRP2, and KIAA1199, and a marked increase of CDH1 and PPIP5K2 (Figs. 4c, 5e, and S5e). Similar to H19 knockdown, CDK8 siRNA dramatically reduced CRC cell proliferation (Fig. 5f). CDK8 overexpression partially rescued the phenotype triggered by H19 knockdown (Fig. 5f). These data together suggest that CDK8 is a molecular target mediating the oncogenic function of H19.
Fig. 5.
H19 regulates CDK8 transcription levels. (a) Predicted functional change of CTNNB1 and MED1. (b)H19 on CDK8 levels. (c) Venn diagram of overlapping genes regulated > 2 fold in HCT116 cells by siRNAs against each of CTNNB1, H19, and CDK8.(d) Heat map representation of Log2 transformed fold changes of overlapping genes. (e) CDK8 effect on overlapping genes in HCT116 cells. (f) CDK8 effect on cell proliferation. The statistical significance between groups was determined using 2-way ANOVA analysis with GraphPad software. * P < 0.05. See also Fig. S5.
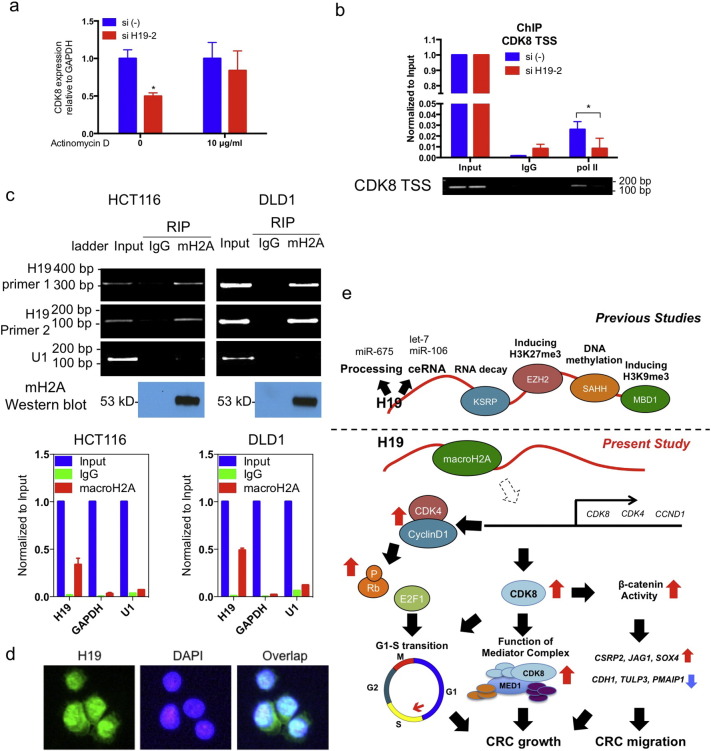
3.5. A Mechanistic Link between H19 and CDK8 Expression
Pretreatment of cells with actinomycin D, an inhibitor of DNA transcription (Siboni et al., 2015), attenuated the H19 effect on CDK8, suggesting a transcriptional regulation (Fig. 6a). Concordantly, H19 silencing significantly reduced polymerase II binding to the CDK8 TSS (Fig. 6b). The mechanisms underlying CDK8 transcription are not well documented. We exclude the possibility that CDK8 is regulated by promoter methylation since CDK8 RNA expression was not induced by treatment of CRC cells with the de-methylation agent 5-Aza-2′-deoxycytidine (data not shown). Recent studies revealed that the histone variant macroH2A is a strong repressive regulator of CDK8 expression (Kapoor et al., 2010, Xu et al., 2015). H19 silencing does not change the protein expression levels of macroH2A (Fig. S6). However, with RIP experiment we detected a strong physical association of H19 RNA with macroH2A protein (Fig. 6c), indicating a potential macroH2A involvement in the regulation of CDK8 transcription by H19. Together with the fact that H19 is expressed in both cytosol and nucleus (Fig. 6e), our data provide additional information on H19 regulation network that controls CRC malignancies (Fig. 6e).
Fig. 6.
Mechanistic link between H19 and CDK8 expression. (a)CDK8 expression after exposure to siH19 for 12 h as determined by qRT-PCR in presence or absence of actinomycin D. (b)H19 silencing reduces polymerase II binding of transcription start site of CDK8 by chromatin immunoprecipitation (ChIP) experiments. (c)H19 interacts with macroH2A protein, by RNA immunoprecipitation (RIP). (d) In situ hybridization shows H19 expression in both nucleus and cytosol of HCT116 cells. (e) A scheme of H19 action: we provided additional insights of H19 on Rb-E2F1 signaling and CDK-β-catenin activity. We propose that H19 interacts with macroH2A, and this may consequently lead to de-repression of genes including CDK8, CDK4, and CCND1. Increased CDK4-cyclin D1 complex phosphorylates Rb to disrupt Rb-E2F1 interaction, leading to E2F1 activation. The increase of CDK8 expression enhances the function of mediator complex including MED1, and facilitates the gene regulation by β-catenin. These downstream targets could work in a synergistic way of promoting cell proliferation and increasing cell motility in CRC. See also Fig. S6.
3.6. Clinical Significance of H19 and its Molecular Targets in CRC
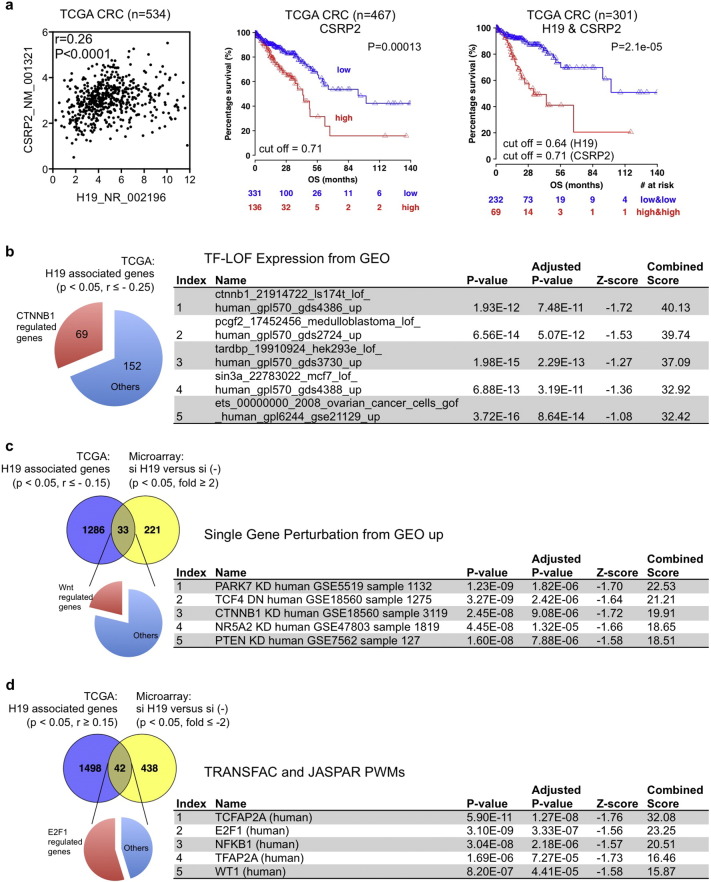
We support the CDK8-β-catenin mechanism identified from cell line models with the TCGA CRC data. CSRP2, one of the 8 overlapping genes downregulated by si-H19–1, si-H19–2, si-CTNNB1, and si-CDK8 (Fig. 5d), shows positive association with H19 expression in TCGA CRC samples (n = 534, r = 0.26, P < 0.0001) (Fig. 7a). Among the 13 upregulated genes (Fig. 5d), GDA (r = − 0.18, P < 0.0001), PPIP5K2 (r = − 0.15, P = 0.0004), AGPAT9 (r = − 0.15, P = 0.0004), and CDH1 (r = − 0.15, P = 0.0006) show inverse correlation with H19 expression (Fig. S7a). CSRP2 (HR = 1.51, CI95% 1.17–1.96, P = 0.001762) is associated with shorter OS (Fig. 7a), while CDH1 level is associated with longer OS (HR = 0.69, CI95% 0.51–0.93, P = 0.0145) (Fig. S7a). Combined analysis of H19 with CSRP2 or CDH1 are powerful independent prognostic factors (HR = 2.53, CI95% 1.39–4.61, P = 0.00242 for H19-high/CSRP2-high versus H19-low/CSRP2-low; HR = 3.96, CI95% 1.85–8.45, P = 0.00039 for H19-high/CDH1-low versus H19-low/CDH1-high) for OS, comparable to the prognostic power of stage (HR = 2.66, CI95% 1.45–4.9, P = 0.00169; and HR = 2.56, CI95% 1.43–4.59, P = 0.0015, respectively) (Figs. 7a and S7a, and Table S2). We found a positive expression association between H19 and CDK8 in cohort 2 comprising 178 CRC cases (r = 0.25, P = 0.0008), but not in the TCGA CRC dataset (P = 0.1529) (Fig. S7b and c). Interestingly, in the 138 rectal cancer cases of the TCGA set, H19 and CDK8 expression levels are also positively correlated (r = 0.17, P = 0.0308) (Fig. S7d), and the same was confirmed in an independent analysis of 71 TCGA rectal cancer cases by TANRIC open-access resource (r = 0.49, P = 0.000017) (Li et al., 2015) (http://ibl.mdanderson.org/tanric/_design/basic/query.html) (Fig. S7d). Analysis of the 138 TCGA rectal cancer cases showed that combined analysis of H19 and CDK8 expression stratified patients into different risk groups (P = 0.0185), with the subgroup expressing both high H19 and CDK8 displaying shortest DFS (Fig. S7e).
Fig. 7.
Clinical significance of H19 and its molecular targets in CRC. (a) Expression correlation of H19 and CSRP2, and their association with overall survival (OS) in TCGA CRC dataset. (b) Enrichment analysis of genes showing significantly inverse expression correlation with H19 in TCGA CRC dataset. Genes with spearman's rank correlation coefficient less or equal to − 0.25 were analyzed using Enrichr at http://amp.pharm.mssm.edu/Enrichr/. (c) Genes showing inverse correlation with H19 in both TCGA CRC data and microarray data were subject to enrichment analysis using Enrichr. To maximize the overlapping genes, we used genes with spearman's rank correlation coefficient ≤− 0.15 in TCGA dataset, and used the microarray data of HCT116 si-H19-2. (d) Genes showing positive correlation with H19 in both TCGA data and microarray data were subject to enrichment analysis using Enrichr. To maximize the overlapping genes, we used genes with spearman's rank correlation coefficient ≥ 0.15 in TCGA dataset. See also Fig. S7 and Table S2.
To gain further mechanistic insight, we analyzed expression correlation of all protein coding genes with H19 in the TCGA CRC data set and performed enrichment analysis using Enrichr (http://amp.pharm.mssm.edu/Enrichr/) (Chen et al., 2013, Kuleshov et al., 2016). Interestingly, CTNNB1 function ranked at top according to the score combining both p value and z score: 69 out of the 221 genes that show significant inverse association with H19 (r ≤− 0.25) in TCGA dataset are inversely correlated with CTNNB1 function in the previously published Gene Expression Omnibus (GEO) data (Mokry et al., 2012) (Fig. 7b). We compared the TCGA association data with the si-H19 microarray data, and identified 33 genes (r ≤− 0.15 in TCGA data, fold ≥ 2 in array data) (Fig. 7c) and 42 genes (r ≥ 0.15 in TCGA data, fold ≤− 2 in array data) (Fig. 7d) in common. The overlapping genes showing inverse correlation with H19 are enriched for Wnt target genes (either by TCF7L2 dominant negative or CTNNB1 knockdown) in the single gene perturbation database (Mokry et al., 2012) (Fig. 7c). With the transcription factor binding profile database TRANScription FACtor (TRANSFAC) and JASPAR, E2F1 is predicted as the top transcription factor in regulation of the overlapping genes showing positive association with H19 (Fig. 7d). Together, these data concordantly support the clinical relevance of our mechanism findings with cell models.
4. Discussion
Despite the fact that many lncRNAs are expressed at low levels, H19 expression is highly abundant and readily detected. Echoing a very recent study reporting H19 association with CRC survival in 83 CRC cases (Han et al., 2016), our findings with 778 CRC cases with multiple cohorts strongly suggest that H19 could be a useful biomarker for CRC survival. It would be valuable to test further whether H19 levels associate with therapeutic response, and this may help identify patients that could benefit from adjuvant chemotherapy (Dalerba et al., 2016).
One advantage of our study is that we used an unbiased approach for mechanism elucidation. With such approach, we discovered not only the known H19 targets including let-7 and MYC (Kallen et al., 2013), but also several additional mechanisms underlying the oncogenic activity of H19 in CRC. First, H19 regulates RB1/E2F signaling by modulating RB1 function. This provides a detailed explanation for the cell cycle regulation by H19. Second, CDK8, an oncogenic driver in CRC, is transcriptionally regulated by H19. Third, we detected significant overlap between genes regulated by H19 and β-catenin. Although H19 effect on Wnt signaling has been reported (Luo et al., 2013, Wang et al., 2016, Liang et al., 2016), we showed a different mechanism, by which H19 regulates CDK8 expression, and consequently affects β-catenin activity. Accordingly, we speculate that H19 may be specifically involved in the canonical consensus molecular subtypes 2 (CMS2) of CRC that shows high Wnt and MYC activity (Guinney et al., 2015). Importantly, the essential findings on E2F1 signaling and β-catenin activity are supported by the clinical data analysis.
The regulatory effect of H19 on RB1-E2F1 can be traced to its transcriptional regulation of CDK4 and cyclin D1, two immediate upstream regulator of RB1 phosphorylation (Dick and Rubin, 2013). This matches with the array prediction of CDKN2A (p16), an inhibitor of CDK4-cyclin D1 function, to be functionally upregulated (Dick and Rubin, 2013). Interestingly, we also found interaction of H19 with RB1. The activity of RB1 is tightly controlled by its interaction partners: for instance, binding of RB1 by human papillomavirus E7 oncoprotein inactivates the suppressive effect of RB1 on E2F transcription activators by interrupting RB1-E2F binding in human cervical cancer (Dyson et al., 1989). However, the relevance of RB1 interaction with H19 needs to be further investigated.
CDK8 oncogene is an essential driver of CRC malignancies with multiple functions (Guinney et al., 2015, Roychowdhury et al., 2011). Together with its partner cyclin C, MED12, and MED13, CDK8 form a subcomplex that controls mediator-polymerase II interaction to initiate gene transcription (Allen and Taatjes, 2015, Tsai et al., 2013). Whether CDK8 regulates Wnt target genes through this mediator complex is not clearly addressed. However, studies suggest that CDK8 possibly stabilizes β-catenin interaction with the promoter of Wnt targets to achieve regulatory control (Firestein et al., 2008). The reported association of CDK8 expression with β-catenin activation in 470 CRC samples reinforced the intrinsic functional connection between CDK8 and β-catenin (Firestein et al., 2010). In our study, CDK8 reduction offered a sound explanation for the predicted decrease in β-catenin and MED1 function following H19 knockdown.
As part of the essential transcription machinery, CDK8 is important in regulating expression of other genes (Allen and Taatjes, 2015, Tsai et al., 2013). However, regulators of CDK8 transcription are not well defined, except the findings that the histone variant macroH2A represses CDK8 expression (Kapoor et al., 2010, Xu et al., 2015). MacroH2A has emerged as a critical epigenetic regulator of cancer, and aberrant expression of macroH2A has been observed in many cancer types including CRC (Cantarino et al., 2013). Recently, macroH2A was also shown to repress transcription of CDK4 and CCND1, as well as CDK8 (Lei et al., 2014, Yang et al., 2015). We speculate that the interaction of H19 with macroH2A protein could sequester the suppressive effect of macroH2A on transcription of above genes, and might represent the initial mechanism of H19 function. Further studies are warranted to address this critical gene regulation mechanism. We also acknowledge the limitation of our study with actinomycin D treatment, where we evaluated CDK8 expression at an early time point of 12 h in respect with the quick drop of CDK8 level in such experiments. Therefore, although we believe that transcription regulation should at least partially contribute to the observed CDK8 changes, we cannot rule out the possibility that H19 regulates degradation of CDK8 RNA, for instance, by microRNAs.
Our data revealed that CSRP2 and CDH1 are H19 targets with clinical significance in CRC survival. CDH1 is a well-known tumor suppressor, involved in cell migration and metastasis (Luo et al., 2013). Although H19 was reported to repress CDH1 expression via interaction with EZH2 (Luo et al., 2013), our findings of CDH1 regulation by H19, and by both of H19 targets CDK8 and β-catenin, suggest a different mechanism on CDH1 regulation. CSRP2 is a rarely studied gene (Hoffmann et al., 2016), and in this study we identified its clear association with CRC survival. Interestingly, our unpublished data showed that E2F1 knockdown also dramatically reduces and increases CSRP2 and CDH1 mRNA expression, respectively. Regulation of these two genes by both E2F1 and β-catenin may explain why they are robust H19 targets in both experimental conditions and clinical samples. Addition of CSRP2 or CDH1 or CDK8 with H19 further improves the prognostic value of H19, indicating that they are downstream targets mediating H19 function.
In conclusion, we demonstrate the clinical and biological significance of H19 in CRC, and discovered the H19 action on RB1-E2F pathway and CDK8-β-catenin signaling. We conjecture that H19, abundantly expressed in primary CRC, could be detected in body fluids and developed into a biomarker for CRC diagnosis or prognosis. The strategies employed in this study might be useful in identifying other lncRNA candidates with translational potential.
Author Contributions
G.A.C obtained funding and supervised the study. H.L. designed this study. H.L., M.O., C.I., and G.A.C. discussed and prepared the data. H.L. drafted the manuscript, and G.A.C. provided critical comments on the manuscript. M.O., J.A.A., S.K., J.R.G., C.I., and M.R. further commented on the manuscript. H.L. revised and finalized the manuscript. M.O., H.L., D.M., M.P., M.G., V.S., K.S, G.A.B, F.V., and O.S. performed the wet-lab experiments, and X.Z. performed the ISH experiments. M.R. provided and C.I. analyzed TCGA data. C.I., H.L., M.O. and D.M. participated in the patient data analysis. C.I. and H.L. performed statistical analysis. A.G., Y.T., M.K., and O.S. provided the patient specimens. All authors read and approved the manuscript's content.
Acknowledgements
Dr. Calin is The Alan M. Gewirtz Leukemia & Lymphoma Society Scholar. Work in Dr. Calin's laboratory is supported in part by the NIH/NCI grants 1UH2TR00943-01 and 1 R01 CA182905-01, the UT MD Anderson Cancer Center SPORE in Melanoma grant from NCI (P50 CA093459), AIM at Melanoma Foundation and the Miriam and Jim Mulva research funds, the UT MD Anderson Cancer Center Brain SPORE (2P50CA127001), a Developmental Research award from Leukemia SPORE, a CLL Moonshot Flagship project, a 2015 Knowledge GAP MDACC grant, and the Estate of C. G. Johnson, Jr., Dr. Slaby is supported by the Ministry of Education, Youth and Sports of the Czech Republic under the project CEITEC 2020 (LQ1601). Dr. Pichler's research is supported by an Erwin-Schroedinger Scholarship of the Austrian Science Funds (project no. J3389-B23) and in part by funds of the Oesterreichische Nationalbank (Anniversary Fund, project number: 14869). These study sponsors did not play role in the collection, analysis, and interpretation of data.
Footnotes
Supplementary data to this article can be found online at doi:10.1016/j.ebiom.2016.10.026.
Contributor Information
Hui Ling, Email: hling@mdanderson.org.
George A. Calin, Email: gcalin@mdanderson.org.
Appendix A. Supplementary data
Supplementary material
References
- Abbasi O., Mashayekhi F., Mirzajani E., Fakhriyeh Asl S., Mahmoudi T., Saeedi Saedi H. Soluble VEGFR1 concentration in the serum of patients with colorectal cancer. Surg. Today. 2015;45:215–220. doi: 10.1007/s00595-014-0886-4. [DOI] [PubMed] [Google Scholar]
- Allen B.L., Taatjes D.J. The Mediator complex: a central integrator of transcription. Nat. Rev. Mol. Cell Biol. 2015;16:155–166. doi: 10.1038/nrm3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anastas J.N., Moon R.T. WNT signalling pathways as therapeutic targets in cancer. Nat. Rev. Cancer. 2013;13:11–26. doi: 10.1038/nrc3419. [DOI] [PubMed] [Google Scholar]
- Barsyte-Lovejoy D., Lau S.K., Boutros P.C. The c-Myc oncogene directly induces the H19 noncoding RNA by allele-specific binding to potentiate tumorigenesis. Cancer Res. 2006;66:5330–5337. doi: 10.1158/0008-5472.CAN-06-0037. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Network Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantarino N., Douet J., Buschbeck M. MacroH2A—an epigenetic regulator of cancer. Cancer Lett. 2013;336:247–252. doi: 10.1016/j.canlet.2013.03.022. [DOI] [PubMed] [Google Scholar]
- Chen H.Z., Tsai S.Y., Leone G. Emerging roles of E2Fs in cancer: an exit from cell cycle control. Nat. Rev. Cancer. 2009;9:785–797. doi: 10.1038/nrc2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E.Y., Tan C.M., Kou Y. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinf. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalerba P., Sahoo D., Paik S. CDX2 as a prognostic biomarker in stage II and stage III colon cancer. N. Engl. J. Med. 2016;374:211–222. doi: 10.1056/NEJMoa1506597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien T., Johnson R., Bussotti G. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick F.A., Rubin S.M. Molecular mechanisms underlying RB protein function. Nat. Rev. Mol. Cell Biol. 2013;14:297–306. doi: 10.1038/nrm3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson N., Howley P.M., Munger K., Harlow E. The human papilloma virus-16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science. 1989;243:934–937. doi: 10.1126/science.2537532. [DOI] [PubMed] [Google Scholar]
- Firestein R., Bass A.J., Kim S.Y. CDK8 is a colorectal cancer oncogene that regulates beta-catenin activity. Nature. 2008;455:547–551. doi: 10.1038/nature07179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firestein R., Shima K., Nosho K. CDK8 expression in 470 colorectal cancers in relation to beta-catenin activation, other molecular alterations and patient survival. Int. J. Cancer. 2010;126:2863–2873. doi: 10.1002/ijc.24908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovarelli M., Bucci G., Ramos A. H19 long noncoding RNA controls the mRNA decay promoting function of KSRP. Proc. Natl. Acad. Sci. U. S. A. 2014;111:E5023–E5028. doi: 10.1073/pnas.1415098111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guinney J., Dienstmann R., Wang X. The consensus molecular subtypes of colorectal cancer. Nat. Med. 2015;21:1350–1356. doi: 10.1038/nm.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han D., Gao X., Wang M. Long noncoding RNA H19 indicates a poor prognosis of colorectal cancer and promotes tumor growth by recruiting and binding to eIF4A3. Oncotarget. 2016 doi: 10.18632/oncotarget.8063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Y., Crenshaw T., Moulton T., Newcomb E., Tycko B. Tumour-suppressor activity of H19 RNA. Nature. 1993;365:764–767. doi: 10.1038/365764a0. [DOI] [PubMed] [Google Scholar]
- Hoffmann C., Mao X., Dieterle M. CRP2, a new invadopodia actin bundling factor critically promotes breast cancer cell invasion and metastasis. Oncotarget. 2016 doi: 10.18632/oncotarget.7327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imig J., Brunschweiger A., Brummer A. miR-CLIP capture of a miRNA targetome uncovers a lincRNA H19-miR-106a interaction. Nat. Chem. Biol. 2015;11:107–114. doi: 10.1038/nchembio.1713. [DOI] [PubMed] [Google Scholar]
- Juan V., Crain C., Wilson C. Evidence for evolutionarily conserved secondary structure in the H19 tumor suppressor RNA. Nucleic Acids Res. 2000;28:1221–1227. doi: 10.1093/nar/28.5.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallen A.N., Zhou X.B., Xu J. The imprinted H19 lncRNA antagonizes let-7 microRNAs. Mol. Cell. 2013;52:101–112. doi: 10.1016/j.molcel.2013.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor A., Goldberg M.S., Cumberland L.K. The histone variant macroH2A suppresses melanoma progression through regulation of CDK8. Nature. 2010;468:1105–1109. doi: 10.1038/nature09590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keniry A., Oxley D., Monnier P. The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r. Nat. Cell Biol. 2012;14:659–665. doi: 10.1038/ncb2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuleshov M.V., Jones M.R., Rouillard A.D. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei S., Long J., Li J. MacroH2A suppresses the proliferation of the B16 melanoma cell line. Mol. Med. Rep. 2014;10:1845–1850. doi: 10.3892/mmr.2014.2482. [DOI] [PubMed] [Google Scholar]
- Li J., Han L., Roebuck P. TANRIC: an interactive open platform to explore the function of lncRNAs in cancer. Cancer Res. 2015;75:3728–3737. doi: 10.1158/0008-5472.CAN-15-0273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang W.C., Fu W.M., Wong C.W. The lncRNA H19 promotes epithelial to mesenchymal transition by functioning as miRNA sponges in colorectal cancer. Oncotarget. 2015;6:22513–22525. doi: 10.18632/oncotarget.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang W.C., Fu W.M., Wang Y.B. H19 activates Wnt signaling and promotes osteoblast differentiation by functioning as a competing endogenous RNA. Sci. Rep. 2016;6:20121. doi: 10.1038/srep20121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling H., Vincent K., Pichler M. Junk DNA and the long non-coding RNA twist in cancer genetics. Oncogene. 2015;34:5003–5011. doi: 10.1038/onc.2014.456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M., Li Z., Wang W., Zeng Y., Liu Z., Qiu J. Long non-coding RNA H19 increases bladder cancer metastasis by associating with EZH2 and inhibiting E-cadherin expression. Cancer Lett. 2013;333:213–221. doi: 10.1016/j.canlet.2013.01.033. [DOI] [PubMed] [Google Scholar]
- Medrzycki M., Zhang Y., Zhang W. Histone h1.3 suppresses h19 noncoding RNA expression and cell growth of ovarian cancer cells. Cancer Res. 2014;74:6463–6473. doi: 10.1158/0008-5472.CAN-13-2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokry M., Hatzis P., Schuijers J. Integrated genome-wide analysis of transcription factor occupancy, RNA polymerase II binding and steady-state RNA levels identify differentially regulated functional gene classes. Nucleic Acids Res. 2012;40:148–158. doi: 10.1093/nar/gkr720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monnier P., Martinet C., Pontis J., Stancheva I., Ait-Si-Ali S., Dandolo L. H19 lncRNA controls gene expression of the Imprinted Gene Network by recruiting MBD1. Proc. Natl. Acad. Sci. U. S. A. 2013;110:20693–20698. doi: 10.1073/pnas.1310201110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris K.V., Mattick J.S. The rise of regulatory RNA. Nat. Rev. Genet. 2014;15:423–437. doi: 10.1038/nrg3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polager S., Ginsberg D. p53 and E2f: partners in life and death. Nat. Rev. Cancer. 2009;9:738–748. doi: 10.1038/nrc2718. [DOI] [PubMed] [Google Scholar]
- Raveh E., Matouk I.J., Gilon M., Hochberg A. The H19 Long non-coding RNA in cancer initiation, progression and metastasis — a proposed unifying theory. Mol. Cancer. 2015;14:184. doi: 10.1186/s12943-015-0458-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodilla V., Villanueva A., Obrador-Hevia A. Jagged1 is the pathological link between Wnt and Notch pathways in colorectal cancer. Proc. Natl. Acad. Sci. U. S. A. 2009;106:6315–6320. doi: 10.1073/pnas.0813221106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roychowdhury S., Iyer M.K., Robinson D.R. Personalized oncology through integrative high-throughput sequencing: a pilot study. Sci. Transl. Med. 2011;3:111ra121. doi: 10.1126/scitranslmed.3003161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Wang Y., Luan W. Long non-coding RNA H19 promotes glioma cell invasion by deriving miR-675. PLoS One. 2014;9 doi: 10.1371/journal.pone.0086295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siboni R.B., Nakamori M., Wagner S.D. Actinomycin D specifically reduces expanded CUG repeat RNA in myotonic dystrophy models. Cell Rep. 2015;13:2386–2394. doi: 10.1016/j.celrep.2015.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2015. CA Cancer J. Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- Spizzo R., Almeida M.I., Colombatti A., Calin G.A. Long non-coding RNAs and cancer: a new frontier of translational research? Oncogene. 2012;31:4577–4587. doi: 10.1038/onc.2011.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toiyama Y., Tanaka K., Inoue Y., Mohri Y., Kusunoki M. Circulating cell-free microRNAs as biomarkers for colorectal cancer. Surg. Today. 2016;46:13–24. doi: 10.1007/s00595-015-1138-y. [DOI] [PubMed] [Google Scholar]
- Torre L.A., Bray F., Siegel R.L., Ferlay J., Lortet-Tieulent J., Jemal A. Global cancer statistics, 2012. CA Cancer J. Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- Tsai M.C., Spitale R.C., Chang H.Y. Long intergenic noncoding RNAs: new links in cancer progression. Cancer Res. 2011;71:3–7. doi: 10.1158/0008-5472.CAN-10-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai K.L., Sato S., Tomomori-Sato C., Conaway R.C., Conaway J.W., Asturias F.J. A conserved Mediator-CDK8 kinase module association regulates Mediator-RNA polymerase II interaction. Nat. Struct. Mol. Biol. 2013;20:611–619. doi: 10.1038/nsmb.2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang W.P., Ng E.K., Ng S.S. Oncofetal H19-derived miR-675 regulates tumor suppressor RB in human colorectal cancer. Carcinogenesis. 2010;31:350–358. doi: 10.1093/carcin/bgp181. [DOI] [PubMed] [Google Scholar]
- Venkatraman A., He X.C., Thorvaldsen J.L. Maternal imprinting at the H19-Igf2 locus maintains adult haematopoietic stem cell quiescence. Nature. 2013;500:345–349. doi: 10.1038/nature12303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Wu X., Liu Y. Long noncoding RNA H19 inhibits the proliferation of fetal liver cells and the Wnt signaling pathway. FEBS Lett. 2016;590:559–570. doi: 10.1002/1873-3468.12078. [DOI] [PubMed] [Google Scholar]
- Xu D., Li C.F., Zhang X. Skp2-macroH2A1-CDK8 axis orchestrates G2/M transition and tumorigenesis. Nat. Commun. 2015;6:6641. doi: 10.1038/ncomms7641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L., Zhou J., Gao Y. Regulation of tumor cell migration and invasion by the H19/let-7 axis is antagonized by metformin-induced DNA methylation. Oncogene. 2015;34:3076–3084. doi: 10.1038/onc.2014.236. [DOI] [PubMed] [Google Scholar]
- Yang P., Yin K., Zhong D., Liao Q., Li K. Inhibition of osteosarcoma cell progression by MacroH2A via the downregulation of cyclin D and cyclindependent kinase genes. Mol. Med. Rep. 2015;11:1905–1910. doi: 10.3892/mmr.2014.2903. [DOI] [PubMed] [Google Scholar]
- Zhou J., Yang L., Zhong T. H19 lncRNA alters DNA methylation genome wide by regulating S-adenosylhomocysteine hydrolase. Nat. Commun. 2015;6:10221. doi: 10.1038/ncomms10221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu M., Chen Q., Liu X. lncRNA H19/miR-675 axis represses prostate cancer metastasis by targeting TGFBI. FEBS J. 2014;281:3766–3775. doi: 10.1111/febs.12902. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material