Abstract
Purpose
Expression of CXCR4 in cancers has been correlated with poor prognosis and increased metastasis. Quantifying CXCR4 expression non-invasively might aid in prognostication and monitoring therapy. We evaluated a radiolabeled antagonist of CXCR4, 64Cu-AMD3100, as a positron-emitting imaging agent.
Procedures
CXCR4-transfected or non-transfected cell lines were injected into mice to form xenografts. Accumulation of 64Cu-AMD3100 in tumors was analyzed by small-animal PET and biodistribution assays.
Results
64Cu-AMD3100 accumulated in CXCR4-expressing, but not CXCR4-negative, tumors. For CXCR4-expressing tumors, tumor-to-blood and tumor-to-muscle ratios were 23–41 and 50–59, respectively, depending on tumor type. Excess of unlabeled Cu-AMD3100 or AMD3100 significantly reduced 64Cu-AMD3100 accumulation in CXCR4-expressing tumors. Human-absorbed dose calculations predicted a dose limit of 444 MBq.
Conclusions
CXCR4 can be imaged in tumors using 64Cu-AMD3100. Dosimetry studies suggest that imaging in humans is feasible. We conclude that 64Cu-AMD3100 should be investigated as a potential agent for imaging and quantifying CXCR4 in tumors.
Keywords: CXCR4, AMD3100, PET, Tumor, Imaging
Introduction
CXCR4 is a member of the chemokine receptor subfamily of seven transmembrane domain, G-protein coupled receptors, whose sole known natural ligand is CXCL12/SDF-1. CXCR4 is an unusual chemokine receptor in having roles beyond leukocyte recruitment, which include fundamental processes such as the development of the hematopoietic, cardiovascular, and nervous systems during embryogenesis [1]. In the hematopoietic system, CXCR4 serves, among other functions, to control stem cell retention and homing to the bone marrow [2]. More than 23 types of human cancers, including cancers of the breast, prostate, lung, and colon, as well as lymphoma, multiple myeloma, and melanoma, express CXCR4 [3–11]. CXCR4 can be induced by a number of transcription factors important in cancer such as hypoxia-inducible factor 1 (HIF-1) [12], NF-κB [13], and oncoproteins PAX3-FKHR [14] and RET/PTC [15]. A number of recent studies have correlated high levels of CXCR4 expression in cancers with poor prognosis [16,17] and with resistance to chemotherapy, in part by enhancing interactions between cancers and bone marrow stroma [10, 11, 18]. Collectively, the data on CXCR4 in cancer suggest that this receptor increases tumor cell survival and/or growth and/or metastasis, making it a potentially attractive therapeutic target [19–21].
AMD3100/plerixafor has been identified as a specific inhibitor of CXCR4 [22, 23] and is currently approved and marketed as Mozobil™, a stem cell mobilizing agent used in combination with G-CSF [24]. AMD3100/plerixafor and other CXCR4 antagonists might also be used for anti-tumor therapy, as has been proposed for multiple myeloma, mantle cell lymphoma, and breast and prostate cancers [10, 11, 25, 26]. We have recently described the synthesis of 64Cu-AMD3100 (Fig. 1, 64Cu-AMD3100), a tracer based on AMD3100/plerixafor labeled with positron-emitting radionuclide copper-64 (t1/2=12.7 h, electron capture 45%, β− 37.1%, β+ 17.9%) with a specific activity of 11.28 Ci/μmol (417 GBq/μmol), and we have shown that 64Cu-AMD3100 can be used as an imaging agent for positron emission tomography (PET) [27]. Beyond detecting CXCR4-expressing cancers, PET imaging of CXCR4 on tumors would enable non-invasive and repeated quantification of CXCR4 expression on malignant lesions throughout the body, overcoming the sampling bias inherent in performing biopsies on metastases that may be highly heterogeneous.
Fig. 1.
Structures of AMD3100 and 64Cu-AMD3100.
In the experiments described below, we used 64Cu-AMD3100 to evaluate CXCR4 expression by xenografts in mice. We show both by small-animal PET and biodistribution experiments that 64Cu-AMD3100 specifically accumulates in CXCR4-positive but not CXCR4-negative tumors within 1 h after injection of the probe, with tumor-to-blood and tumor-to-muscle ratios of 23–41 and 50–59 respectively, depending on tumor types. From biodistribution studies in normal mice, we calculated the human dosimetry and found that the liver is the limiting organ. These dosimetry data predicted a maximum allowable dose of 444 MBq (12 mCi) for clinical use. Our results suggest that additional work should be done to determine if PET/computed tomography (CT) scanning with 64Cu-AMD3100 could be of value for imaging cancers in humans and for predicting tumor behavior and responses to current and experimental therapies, including therapies targeting CXCR4.
Materials and Methods
Animals
C57BL/6 and athymic (nude) mice were purchased from Taconic and housed under specific pathogen-free conditions. All animal studies were conducted in accordance with the principles and procedures outlined in the National Institutes of Health Guide for the Care and Use of Animals using protocols approved by the NIH Institutional Animal Care and Use Committee.
Cells
3LL Lewis lung carcinoma cells were purchased from the ATCC. 3LL cells were grown in RPMI 1640 (GIBCO); Chinese hamster ovary (CHO) and CHO cells stably transfected to express CXCR4 (CHO-XR4) were a kind gift from Dr. David McDermott (NIAID, NIH, Bethesda, MD). CHO cells were grown in F-12K medium (ATCC). CXCR4 levels on CHO-XR4 and 3LL-XR4 cell lines were evaluated using the FlowCellect kit (Millipore) following the manufacturer’s instructions. All cell culture media were supplemented with 10% fetal bovine serum, 1 mM sodium pyruvate, 2 mM L-glutamine, and non-essential amino acids (GIBCO).
Transfections
3LL cells were transfected with pCDNA3.1 containing DNA encoding human CXCR4 (a kind gift from Dr. David McDermott, NIAID, NIH, Bethesda, MD) and grown in medium supplemented with 1 mg/mL G418 (GIBCO). 3LL cells were transfected using Xtreme GENE transfection reagent (Roche) as directed by the manufacturer, and the resulting CXCR4-expressing cells are referred to as 3LL-XR4. Although these cells retained the resistance to the selecting agent, the expression of CXCR4 diminished in vitro over time. Hence, for in vivo studies, CXCR4-expressing cells were purified using anti-human CXCR4-PE (clone 12G5, R&D Systems) and anti-PE magnetic beads (BD Bioscience) and injected into mice within 5 days. Expression of CXCR4 was evaluated by flow cytometry using anti-human CXCR4-PE.
Flow Cytometry
Cells were blocked in HBSS buffer supplemented with 2% rat serum and 2% mouse serum (Jackson ImmunoResearch labs) on ice for 10 min, and thereafter cells were stained with PE-conjugated anti-human CXCR4 (R&D). Cells from tumors were also stained using fluorescein isothiocyanate (FITC)-labeled anti-mouse MHC-I H-2Dk (BD Bioscience). Flow data were acquired using an LSR-II cytometer (BD Bioscience) and analyzed using FlowJo (TreeStar).
Binding Assay
CHO and CHO-XR4 cells were treated with trypsin and resuspended in phosphate buffered saline containing 50 mM HEPES, 1 mM CaCl2, 5 mM MgCl2, 0.5% (w/v) BSA, and 0.3 mM NaN3. Incubation was done in 200 μL containing 105 cells, 0.065 nM 125I-CXCL12 (Perkin-Elmer Life Sciences), and 0–10,000 nM of unlabeled Cu-AMD3100 for 45 min on a shaker at room temperature. After incubation, cells were spun through 10% sucrose. Cell-bound radioactivity was measured using a gamma counter (1480 Wizard 3, Perkin-Elmer). Binding results were calculated using Prism (GraphPad).
Tumor Models
For producing solid tumors, mice were injected subcutaneously in two sites in the axillae, one site with non-transfected cells and the other with CXCR4-transfected cells. 3LL and 3LL-XR4 cells were injected at 106 cells per site; CHO and CHO-XR4 cells were injected at 107 cells per site. For producing metastatic tumors, mice were injected in the tail vein with 106 CHO-XR4 cells or 106 3LL-XR4 cells.
Analysis of human CXCR4 expression by tumors was done by incubation of the harvested tumors in Liberase-3 (Roche) at 37°C followed by mincing the tumor on a 70-μm strainer (BD Falcon) to obtain single-cell suspensions. Thereafter, cells were washed and stained with anti-human CXCR4-PE and FITC-labeled anti-mouse MHC-I H-2Dk before analysis by flow cytometry. Recipient MHC-I H-2Dk-staining cells were removed from the analysis electronically in order to evaluate expression of CXCR4 by tumor cells.
Chemistry and Radiochemistry
64Cu-AMD3100 and Cu-AMD3100 were synthesized as described [27].
Biodistribution
Twenty-five microcuries (0.925 MBq, total mass of 2 ng or less per mouse) of 64Cu-AMD3100 in a volume of 100 μL was injected via the tail vein. At 1, 2, or 6 h post-injection, blood was withdrawn from the heart under anesthesia, and the mice were killed. Tumors, blood, liver, muscle, and kidneys were removed. All organs were assayed for radioactivity using a gamma counter (1480 Wizard 3, Perkin-Elmer). In blocking experiments, the tracer was injected together with 50 μg of either Cu-AMD3100 or AMD3100/ plerixafor (Sigma).
Small-Animal PET (Micro-PET)
Tumor-bearing mice were anesthetized using isoflurane/O2 (1.5–5% v/v) and injected with 64Cu-AMD3100 or 64Cu-AMD3100 plus Cu-AMD3100 or AMD3100/plerixafor as described above. Micro-PET scans were performed 1, 2, and 6 h after tracer injection using the Advanced Technology Laboratory Animal Scanner (ATLAS) or Focus 120 micro-PET scanner (Siemens Medical Solutions). When using the ATLAS, whole-body scans were done at five bed positions, 8 min per position. Scans with the Focus 120 were done in one bed position for 15 min. All scans were started 1 h after radiotracer injection and recorded with an energy window of 100–700 keV. The images were reconstructed by a two-dimensional ordered subset expectation maximum algorithm, and no correction was applied for attenuation or scatter. Image analysis and movie were done using ImageJ version 1.4 g (NIH) and OsiriX version 2.7.5 software.
Dosimetry
Estimated human-absorbed doses of 64Cu-AMD3100 were calculated by using biodistribution data in non-tumor-bearing C57BL/6 male mice with an average weight of 22.4 g. The mice were injected with 0.925 MBq (25 μCi) of 64Cu-AMD3100 i.v. Five animals were killed at each of the seven time points—1, 2, 6, 12, 19, 28, and 48 h post-injection. Samples of blood, muscle, and bone marrow were taken. The thymus, mesenteric lymph nodes, spleen, liver, kidneys, and the combined small and large intestines were also removed for analysis. Tissue samples and whole organs were weighed, and radioactivity was measured in a gamma counter. Determination of organ doses and the effective dose for a reference human male were made using the OLINDA/EXM program (Vanderbilt University, Nashville, TN). The effective dose is the sum of the doses to individual organs times a tissue-weighting factor for that organ.
Statistical Analysis
Statistical analysis of 64Cu-AMD3100 uptake in tumors was done using Student’s t-test.
Results
Specific Accumulation of 64Cu-AMD3100 in CXCR4-Positive Tumors
CHO cells, a hamster ovarian cancer cell line, and CHO-XR4 cells were evaluated for CXCR4 expression by flow cytometry (Fig. 2a). CHO-XR4 cells had 6.8×105 molecules of CXCR4 expressed on the cell surface (data not shown, see “Materials and Methods”), which is slightly higher than levels reported for transformed cell lines such as Jurkat cells [28]. Competition assays of unlabeled Cu-AMD3100 against radiolabeled CXCL12 revealed an IC50 of 84 nM (Fig. 2b), which is similar to the functional EC50 required for Cu-AMD3100 to inhibit Jurkat cell migration to CXCL12 [27]. Non-transfected CHO cells did not bind radiolabeled CXCL12 (Fig. 2b).
Fig. 2.
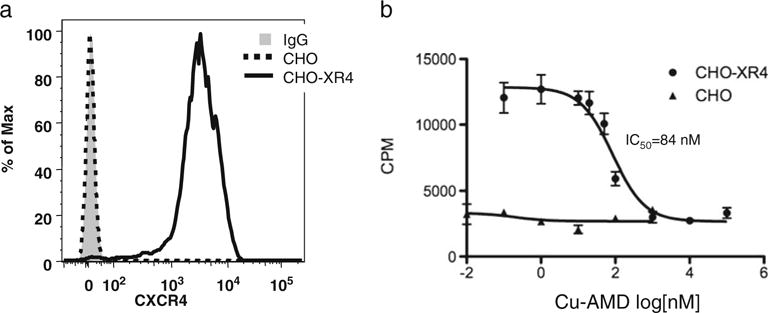
CXCR4 expression and Cu-AMD3100 binding by CHO cells. a Histograms of non-transfected (dashed line) and transfected (solid line) cells stained with anti-CXCR4-PE and compared with transfected cells stained with an isotype control antibody (solid gray fill) by using flow cytometry. b Competition binding of 125I-CXCL12 with unlabeled Cu-AMD3100. Data points are from triplicate samples from two experiments, and error is standard deviation.
The transfected and non-transfected CHO cells were injected subcutaneously in mice, and accumulation of 64Cu-AMD3100 was evaluated at 2 weeks in both CXCR4-positive and CXCR4-negative CHO tumors using small-animal PET (Fig. 3a, Supp. Movie 1). Standardized uptake value (SUV) calculations over time showed accumulation of the tracer in the CXCR4-positive tumors and almost no accumulation in the CXCR4-negative tumors (Fig. 3b). Blocking with excess unlabeled Cu-AMD3100 (50 μg/mouse) diminished the accumulation of the radiolabeled tracer in the CXCR4-positive tumors to the levels seen in the CXCR4-negative tumors.
Fig. 3.
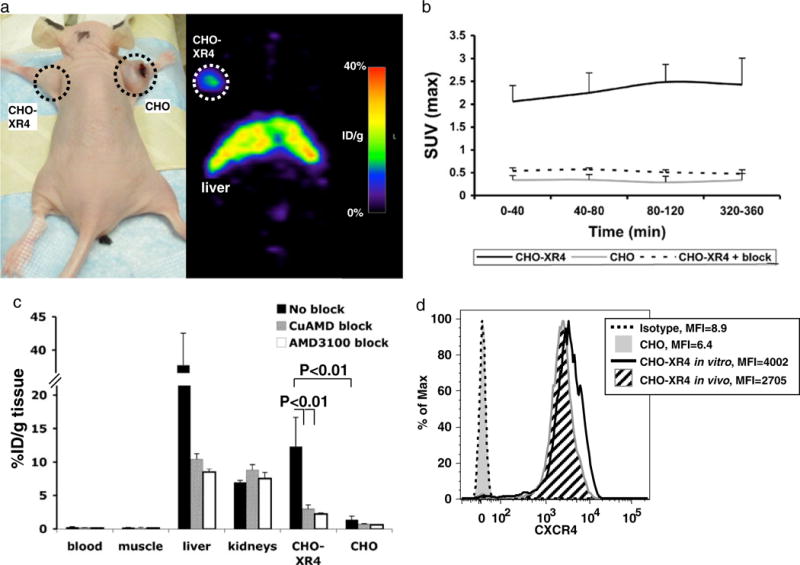
Micro-PET and biodistribution of 64Cu-AMD3100 in CHO-tumor-bearing mice. a Photograph and micro-PET scan of a nude mouse with CXCR4-positive (left) and CXCR4-negative (right) CHO xenografts, injected with 0.925 MBq (25 μCi) of 64Cu-AMD3100 2 weeks after injecting cells and scanned 6 h after injecting the tracer. These results are representative of six mice from two experiments. b Standardized uptake values (SUVmax) were calculated from scans of mice as shown in a, for CXCR4-positive tumors (CHO-XR4, solid line), CXCR4-negative tumors (CHO, gray line), and CXCR4-positive tumors in mice injected with excess unlabeled Cu-AMD3100 (CHO-XR4+ block, dashed line). Each time point is of at least three mice from two experiments. Error bars show positive standard deviations. c Biodistribution of 64Cu-AMD3100 in mice bearing CXCR4-positive (CHO-XR4) and CXCR4-negative (CHO) CHO tumors 2 weeks after injecting the cells and scanned 6 h after injecting the tracer. Data from mice injected without blocking (No block) are in black, with blocking with unlabeled Cu-AMD3100 (CuAMD block) in gray, and with blocking with AMD3100/plerixafor (AMD3100 block) in white. Each group consisted of at least five mice from two experiments. Bracketed horizontal lines indicate significant differences between CHO-XR4 without blockade and the CuAMD3100- and AMD3100-blocked CHO-XR4 and between CHO-XR4 without blockade and CHO without blockade. d Expression of human CXCR4 by cells from CHO-XR4 (angled black lines fill) and CHO (solid gray fill) tumors was evaluated ex vivo by flow cytometry after removing MHC-I H-2Dk-expressing recipient cells from the analysis electronically (see “Materials and Methods”) and compared with the cells grown in vitro (black line) and isotype control staining (dashed line). Mean fluorescent intensities (MFI) are shown. Data are from one CHO-XR4 and one CHO tumor and are representative of results from at least three tumors of each type.
Biodistribution experiments showed high accumulation of 64Cu-AMD3100 in the liver, as we previously reported [27], almost no accumulation in the blood or muscle, and some accumulation in the kidneys (Fig. 3c). Comparison of CXCR4-positive and CXCR4-negative tumors revealed that 64Cu-AMD3100 accumulated only in the CXCR4-positive tumors, reaching 12.3±4%ID/g in the CHO-XR4 tumor after 6 h, and this accumulation could be blocked by an excess of unlabeled Cu-AMD3100. Unlabeled Cu-AMD3100 and the parent molecule AMD3100/plerixafor were equally effective in blocking the accumulation of 64Cu-AMD3100, which rules out a significant contribution of free copper-64 to the micro-PET images. In addition to blocking the accumulation in the CHO-XR4 tumor, accumulation of the tracer was also diminished by both unlabeled reagents in the liver, but not in the kidneys. Isotope levels in the blood and muscle were very low, with a CHO-XR4 tumor/blood ratio of 41.94±14 and tumor/muscle ratio of 59.48±20 (Table 1). CHO-XR4/CHO tumors ratio was 8.82±3.1. Expression of human CXCR4 in the tumors was evaluated by flow cytometry, which revealed that >98% of the CHO-XR4 tumor cells expressed the receptor in vivo. Levels of expression of CXCR4 on CHO-XR4 tumor cells were similar to the levels on CHO-XR4 cells at the time of injection (Fig. 3d).
Table 1.
CXCR4-positive tumor ratio±SD to blood, muscle, and CXCR4-negative tumor (n>5)
| Tumor type | CXCR4+/blood ratio | CXCR4+/muscle ratio | CXCR4+/CXCR4− ratio |
|---|---|---|---|
| CHO | 41.94±14 | 59.48±20 | 8.82±3.1 |
| 3LL | 23.43±6.4 | 50.29±13.8 | 6.68±1.8 |
In order to establish that accumulation of the tracer was dependent only on CXCR4 expression and not related to the species or tissue origin of the CXCR4-expressing cells, we studied 3LL cells, a mouse Lewis lung carcinoma cell line. These cells were transfected with DNA encoding human CXCR4 (3LL-XR4) and had approximately 4×105 molecules of CXCR4 expressed on the cell surface but lost expression of the receptor over time, while retaining the resistance to the selection agent (data not shown). When injected subcutaneously to mice, by 2 weeks after injection, the accumulation of 64Cu-AMD3100 in CXCR4-positive 3LL tumors reached approximately 10%ID/g (Fig. 4a) and was visualized by micro-PET (Fig. 4b), with almost no accumulation in CXCR4-negative tumors, which could not be seen using micro-PET (Fig. 4a). 3LL-XR4 tumor/blood and tumor/muscle ratios were 23.43±6.4 and 50.29±13.8, respectively (Table 1). 64Cu-AMD3100 accumulation in the CXCR4-positive tumors was successfully blocked with excess unlabeled Cu-AMD3100 (Fig. 4b). Of particular interest and despite successful imaging by micro-PET, analysis of cells harvested from the 3LL-XR4 tumors showed that only 60% of the tumor cells had detectable CXCR4, with mean fluorescence intensities (MFIs) that were lower than on the cells at the time of injection, corresponding to approximately 8×104 receptors per cell (Fig. 4c and data not shown).
Fig. 4.
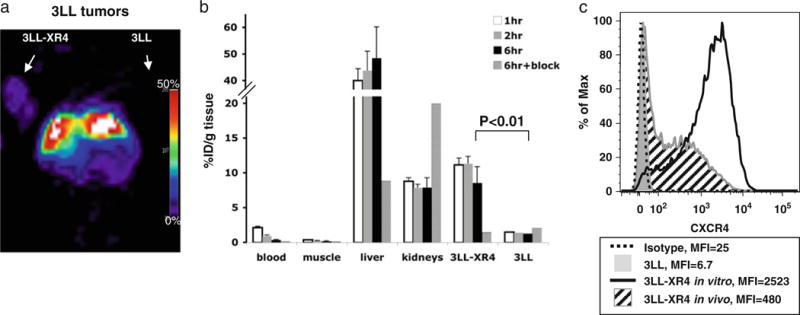
Micro-PET and biodistribution of 64Cu-AMD3100 in lewis lung carcinoma-tumor-bearing mice. Mice with CXCR4-positive and CXCR4-negative 3LL tumors were injected with 0.925 MBq (25 μCi) of 64Cu-AMD3100 2 weeks after injecting cells. a A representative micro-PET image of a mouse bearing 3LL and 3LL-XR4 tumors. Arrows indicate position of the tumors. The 3LL tumor was not seen. b Biodistribution in blood, muscle, liver, kidneys, and tumors was measured after 1, 2, and 6 h. Specificity of tracer accumulation was evaluated by blocking with excess unlabeled Cu-AMD3100 (6 h+block). Each time point contains data from at least five mice from two experiments. Bars show means±SD. Bracketed horizontal line indicates a significant difference between the 6-h samples of 3LL-XR4 without blockade versus 3LL without blockade (black bars). c Expression of human CXCR4 by cells from 3LL-XR4 (angled black lines fill) and 3LL (solid gray fill) tumors was evaluated ex vivo by flow cytometry after removing MHC-I H-2Dk-expressing recipient cells from the analysis electronically (see “Materials and Methods”) and compared with the cells injected initially (black line) and isotype control staining (dashed line). MFIs are shown. Data are from single tumors and are representative of results from at least three tumors of each type.
64Cu-AMD3100 Accumulates in Lung and Liver Tumors
PET technology enables imaging of tumors without limitations as to tissue depth, and PET tracers could work in both subcutaneous as well as internal tumors. In order to evaluate whether 64Cu-AMD3100 is suitable to image tumors at internal sites, mice were injected intravenously with CHO-XR4 cells, which when injected by this route induce lung tumors and in some cases also liver tumors, or with 3LL-XR4 cells, which when injected by this route induce lung tumors. Photographs of representative lung and liver tumors are shown in Fig. 5a. Micro-PET scanning detected CHO-XR4 tumors and 3LL-XR4 tumors in the lungs (Fig. 5b). 3LL-XR4 and CHO-XR4 tumors were undetectable in mice co-injected with 64Cu-AMD3100 and unlabeled AMD3100/plerixafor. In most lung tumor-bearing mice, accumulation of 64Cu-AMD3100 was approximately 5%ID/g tissue (Fig. 5b–d), while in one mouse injected with CHO-XR4 cells, the accumulation reached 20%ID/g (Fig. 5b, right panel). CHO-XR4 tumors dissected from the liver had high accumulation of 64Cu-AMD3100 of 18± 5.3%ID/g. These CHO-XR4 tumors could not be visualized by micro-PET given the high uptake in the surrounding liver, which typically reached 40%ID/g (see Figs. 3 and 4). Biodistribution data showed that accumulation of the 64Cu-AMD3100 in both CHO-XR4 and 3LL-XR4 lung tumors was blocked by co-injecting unlabeled AMD3100/plerixafor (Fig. 5c, d).
Fig. 5.
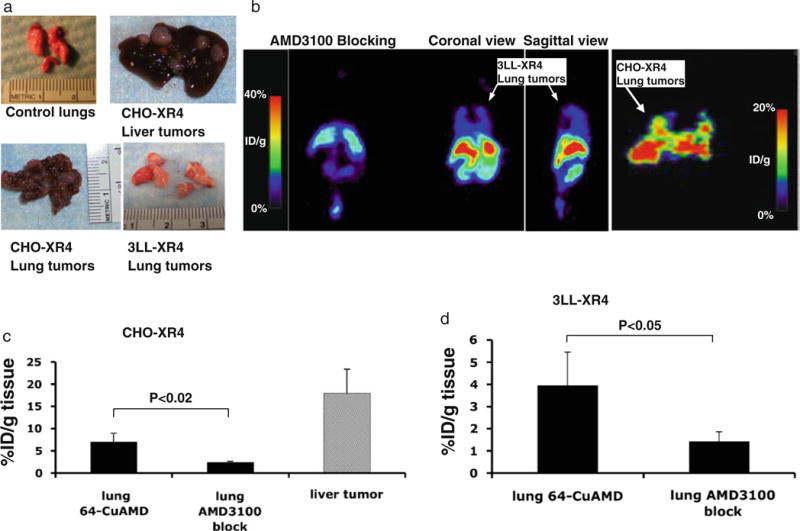
Micro-PET and biodistribution of 64Cu-AMD3100 in lung tumors. a Representative photographs of liver and lung tumors from mice at 3 weeks after intravenous injections with either CHO-XR4 or 3LL-XR4 cells. b Micro-PET images of mice bearing lung tumors after injection of 64Cu-AMD3100, from right to left: CHO-XR4 lung tumors, coronal view; 3LL-XR4 lung tumors, sagittal view; 3LL-XR4 lung tumors, coronal view; and coronal view of mouse with 3LL-XR4 lung tumors also injected with excess unlabeled AMD3100/plerixafor. Data in a and b are from individual CHO-XR4 and 3LL-XR4 mice and are representative of three mice. c Biodistribution of 64Cu-AMD3100 in CHO-XR4 lung and liver tumors or (d) 3LL-XR4 lung tumors, and blocking with excess unlabeled AMD3100/plerixafor. Bars show means±SD. Bracketed horizontal lines indicate significantly lower percent of injected dose per gram in mice also injected with excess unlabeled AMD3100/plerixafor. Data are from two to three mice.
Dosimetry
The human-absorbed dose estimates for normal organs for 64Cu-AMD3100 were calculated from biodistribution in normal C57BL/6 mice. Biodistribution was determined at 1, 2, 6, 19, 28, and 48 h post-injection, and a summary of the human-absorbed dose estimates is shown in Table 2. The dose-critical organ is the liver, with absorbed dose of 1.15 (±0.241)mGy/MBq and an effective dose of 0.057 (±0.012) mGy/MBq. Total body estimated absorbed dose is 0.061± 0.013 mGy/MBq, with an effective dose of 0.108±0.023 mGy/MBq (0.399 rem/mCi).
Table 2.
Human-absorbed radiation doses resulting from administration of 64Cu-AMD3100 calculated from mouse biodistribution (n=5)
| Target organ | Absorbed dose | Effective dose |
|---|---|---|
| Intestine | 2.24E–01±5.05 E–02 | 9.63E–03±2.19E–03 |
| Kidneys | 2.82E–01±5.95E–02 | 1.41E–03±2.98E–04 |
| Liver | 1.15E–00±2.41E–01 | 5.76E–02±1.21E–02 |
| Lungs | 3.75E–02±8.32E–03 | 4.49E–03±9.98E–04 |
| Muscle | 2.14E–02±4.64E–03 | 1.07E–04±2.32E–05 |
| Red marrow | 1.49E–01±3.40E–02 | 1.78E–02±4.08E–03 |
| Spleen | 2.59E–01±5.42E–02 | 1.30E–03±2.71E–04 |
| Thymus | 1.04E–01±2.25E–02 | 5.20E–04±1.13E–04 |
| Total body | 6.11E–02±1.33E–02 | 1.08E–01±2.34E–02 |
Doses are expressed as milligray per megabecquerel, ±SEM
Discussion
There has been significant interest in the expression of CXCR4 by a variety of tumors and blood cancers and in the roles for this chemokine receptor in tumor biology. CXCR4, which was initially identified as a co-receptor for HIV-1 entry, has now been implicated in tumor development, progression, aggressiveness, and metastasis [19]. Negative correlations between CXCR4 expression by tumors and patient survival have suggested that CXCR4 might be used as a prognostic marker and as a target for therapy. As a result, small molecules, peptides, and antibodies have been developed to block this receptor as a therapeutic strategy [29]. Decisions regarding the use of such agents and other therapies, as well as understanding broader aspects of tumor biology, might benefit from information on CXCR4 expression by cancers in various sites and at various stages of disease and treatment.
We have developed a novel CXCR4-specific PET tracer, 64Cu-AMD3100, and reported its radiosynthesis and biodistribution in mice [27]. Here, we show that this tracer is suitable for PET imaging of CXCR4-expressing tumors. Tumors can be visualized within 1 h after 64Cu-AMD3100 administration, with best results achieved at 6 h post-injection. We showed that imaging of CXCR4-positive tumors is independent of the species or tissue origin of the tumor cells. The specificity of the tracer for CXCR4 was established by blocking experiments with unlabeled Cu-AMD3100 and/or AMD3100/plerixafor.
Accumulation of 64Cu-AMD3100 in subcutaneous tumors correlated with levels of CXCR4 expression measured on tumor cells at the time of harvesting for measuring biodistribution. CHO-XR4 tumors, in which all the cells expressed human CXCR4, had the highest accumulation of 64Cu-AMD3100 with 12%ID/g; 3LL-XR4 tumors, in which 60% of the cells in the tumor expressed only moderate levels of human CXCR4, had lower accumulation of the radiotracer but, importantly, could still be visualized by micro-PET.
Comparison of CHO-XR4 tumors in the lung, liver, or subcutaneous locations revealed significant differences in 64Cu-AMD3100 accumulation. CHO-XR4 tumors removed from the liver showed greater accumulation of 64Cu-AMD3100 than subcutaneous tumors, and lung tumors had the lowest accumulation. Whether tissue-specific difference in uptake of 64Cu-AMD3100 resulted from changes in CXCR4 expression or was mediated by other factors, such as efficiency of tracer delivery to tumors at different sites, or other effects of the tumor microenvironments, remains to be determined.
Previous literature on cyclams labeled with copper-64 suggested that some complexes might be less stable in vivo than others, resulting in transchelation of the radioactive copper [30]. To address the possible transchelation of copper-64 in our model, we performed blocking experiments with AMD3100. The unlabeled cyclam blocked most of 64Cu-AMD3100 accumulation in both subcutaneous tumors as well as lung tumors, demonstrating that the 64Cu taken up by tumors was, in fact, bound to AMD3100. Moreover, if the accumulation of 64Cu in the tumor were the result of transchelation, it is likely that 64Cu would have accumulated in the CXCR4-negative tumors. We also addressed the possibility that transchelation contributed to the high accumulation of 64Cu-AMD3100 in the liver by co-injecting mice with large amounts of free, unlabeled copper (50 μg), but we saw no effect on 64Cu-AMD3100 accumulation (data not shown). Although we cannot entirely exclude transchelation of 64Cu in vivo, we conclude that the accumulation of 64Cu in the tumors and liver is AMD3100-dependent and that in the tumors accumulation of 64Cu is also CXCR4-dependent.
There have been four previous studies of labeling CXCR4 ligands in order to detect CXCR4 expression in vivo. One report showed low accumulation of 99mTc-labeled CXCL12/SDF-1 in the heart after myocardial infarction in rats [31]. The other two studies aimed to visualize CXCR4-positive tumors. Hanaoka et al. [32] labeled the small peptide TZ14011 with 111In, which had limited accumulation in tumors. An 125I-labeled derivative of the CXCR4-specific antibody, 12G5, had higher accumulation in CXCR4-positive tumors as compared with 111In- TZ14011, although still only modestly higher than an isotype-identical control antibody [33]. Scanning using the labeled antibody also required a long delay after injection, and detection was limited to large tumors of >200 mm3. Recently, Nimmagadda et al. also showed that 64Cu-labeled AMD3100 could be applied to imaging tumors in mice [34], further validating the potential usefulness of the agent. Our work differs from theirs in two important aspects. Firstly, our 64Cu-AMD3100 was of higher specific activity and synthesized from the AMD3100 free base, which is identical to FDA-approved drug Mozobil™, rather than from the commercially available AMD3100 salt [27, 34]. Secondly, we performed extensive dosimetry in mice in order to establish feasibility in humans (see below).
One potential downside of imaging CXCR4-positive tumors with 64Cu-AMD3100 was the high accumulation that we found in the liver. Although there are some reports of CXCR4 expression in the liver by sinusoidal endothelial cells [35], oval cells [36, 37], and immune cells [38], the expression of the receptor in the liver is low and does not likely fully account for the tracer uptake and retention in this organ. The accumulation in the liver was significantly reduced by excess unlabeled Cu-AMD3100. Therefore, most of the binding in the liver seemed specific, suggesting that 64Cu-AMD3100 may bind to defined sites in the liver other than CXCR4. Nevertheless, accumulation of 64Cu-AMD3100 was also detected in CHO-XR4 tumors mechanically separated from the liver, raising the possibility that CXCR4-positive tumors in the liver might be visualized if PET is done in combination with CT scanning.
One of the implications of the high uptake of 64Cu-AMD3100 in the liver is that this was the dose-critical organ when human-absorbed dose estimates were calculated, based on biodistribution experiments in mice. Our calculations show a predicted total effective dose in humans of 0.108 mGy/MBq (0.399 rem/mCi), which would allow for injecting as much as 12 mCi in order to stay within a 5-rem exposure limit. It is possible that the biodistribution of 64CuAMD3100 will be different in humans, and caution should be exercised in relying on the mouse data for predicting results in patients. Preliminary clinical studies would need to be performed to determine biodistribution and dosimetry in humans. Nonetheless, our data suggest that imaging humans with 64Cu-AMD3100 is feasible.
Conclusions
64Cu-AMD3100, a labeled form of AMD3100/plerixafor, can be used as a PET tracer to image CXCR4 expression by tumors in mice. Dosimetry studies suggest that it may be feasible to administer a dose of 64Cu-AMD3100 that is adequate for visualizing tumors in patients. These results suggest that 64Cu-AMD3100 should be investigated as an agent for imaging human cancers known to express CXCR4. Such imaging may reveal novel information correlating CXCR4 expression with how cancers behave and respond to therapy, including therapy targeting CXCR4, allowing treatment to be customized to the individual patient.
Supplementary Material
Acknowledgments
This work was supported by the intramural research programs of the National Institute of Allergy and Infectious Diseases and the National Institute of Biomedical Imaging and Bioengineering. We wish to thank Dr. David McDermott from NIAID, NIH, for providing reagents and advice.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s11307-010-0466-y) contains supplementary material, which is available to authorized users.
Conflict of Interest Statement
The authors declare that they have no conflict of interest.
References
- 1.Klein RS, Rubin JB. Immune and nervous system CXCL12 and CXCR4: parallel roles in patterning and plasticity. Trends Immunol. 2004;25:306–314. doi: 10.1016/j.it.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 2.Lapidot T, Dar A, Kollet O. How do stem cells find their way home? Blood. 2005;106:1901–1910. doi: 10.1182/blood-2005-04-1417. [DOI] [PubMed] [Google Scholar]
- 3.Muller A, Homey B, Soto H, Ge N, Catron D, Buchanan ME, McClanahan T, Murphy E, Yuan W, Wagner SN, et al. Involvement of chemokine receptors in breast cancer metastasis. Nature. 2001;410:50–56. doi: 10.1038/35065016. [DOI] [PubMed] [Google Scholar]
- 4.Redjal N, Chan JA, Segal RA, Kung AL. CXCR4 inhibition synergizes with cytotoxic chemotherapy in gliomas. Clin Cancer Res. 2006;12:6765–6771. doi: 10.1158/1078-0432.CCR-06-1372. [DOI] [PubMed] [Google Scholar]
- 5.Balkwill F. The significance of cancer cell expression of the chemokine receptor CXCR4. Semin Cancer Biol. 2004;14:171–179. doi: 10.1016/j.semcancer.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 6.Tanaka T, Bai Z, Srinoulprasert Y, Yang BG, Hayasaka H, Miyasaka M. Chemokines in tumor progression and metastasis. Cancer Sci. 2005;96:317–322. doi: 10.1111/j.1349-7006.2005.00059.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taichman RS, Cooper C, Keller ET, Pienta KJ, Taichman NS, McCauley LK. Use of the stromal cell-derived factor-1/CXCR4 pathway in prostate cancer metastasis to bone. Cancer Res. 2002;62:1832–1837. [PubMed] [Google Scholar]
- 8.Vicari AP, Caux C. Chemokines in cancer. Cytokine Growth Factor Rev. 2002;13:143–154. doi: 10.1016/s1359-6101(01)00033-8. [DOI] [PubMed] [Google Scholar]
- 9.Rubin JB, Kung AL, Klein RS, Chan JA, Sun Y, Schmidt K, Kieran MW, Luster AD, Segal RA. A small-molecule antagonist of CXCR4 inhibits intracranial growth of primary brain tumors. Proc Natl Acad Sci USA. 2003;100:13513–13518. doi: 10.1073/pnas.2235846100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Azab AK, Runnels JM, Pitsillides C, Moreau AS, Azab F, Leleu X, Jia X, Wright R, Ospina B, Carlson AL, et al. CXCR4 inhibitor AMD3100 disrupts the interaction of multiple myeloma cells with the bone marrow microenvironment and enhances their sensitivity to therapy. Blood. 2009;113:4341–4351. doi: 10.1182/blood-2008-10-186668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kurtova AV, Tamayo AT, Ford RJ, Burger JA. Mantle cell lymphoma cells express high levels of CXCR4, CXCR5, and VLA-4 (CD49d): importance for interactions with the stromal microenvironment and specific targeting. Blood. 2009;113:4604–4613. doi: 10.1182/blood-2008-10-185827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Staller P, Sulitkova J, Lisztwan J, Moch H, Oakeley EJ, Krek W. Chemokine receptor CXCR4 downregulated by von Hippel–Lindau tumour suppressor pVHL. Nature. 2003;425:307–311. doi: 10.1038/nature01874. [DOI] [PubMed] [Google Scholar]
- 13.Helbig G, Christopherson KW, 2nd, Bhat-Nakshatri P, Kumar S, Kishimoto H, Miller KD, Broxmeyer HE, Nakshatri H. NF-kappa B promotes breast cancer cell migration and metastasis by inducing the expression of the chemokine receptor CXCR4. J Biol Chem. 2003;278:21631–21638. doi: 10.1074/jbc.M300609200. [DOI] [PubMed] [Google Scholar]
- 14.Tomescu O, Xia SJ, Strezlecki D, Bennicelli JL, Ginsberg J, Pawel B, Barr FG. Inducible short-term and stable long-term cell culture systems reveal that the PAX3-FKHR fusion oncoprotein regulates CXCR4, PAX3, and PAX7 expression. Lab Invest. 2004;84:1060–1070. doi: 10.1038/labinvest.3700125. [DOI] [PubMed] [Google Scholar]
- 15.Castellone MD, Guarino V, De Falco V, Carlomagno F, Basolo F, Faviana P, Kruhoffer M, Orntoft T, Russell JP, Rothstein JL, et al. Functional expression of the CXCR4 chemokine receptor is induced by RET/PTC oncogenes and is a common event in human papillary thyroid carcinomas. Oncogene. 2004;23:5958–5967. doi: 10.1038/sj.onc.1207790. [DOI] [PubMed] [Google Scholar]
- 16.Lee HJ, Kim SW, Kim HY, Li S, Yun HJ, Song KS, Kim S, Jo DY. Chemokine receptor CXCR4 expression, function, and clinical implications in gastric cancer. Int J Oncol. 2009;34:473–480. [PubMed] [Google Scholar]
- 17.Oda Y, Tateishi N, Matono H, Matsuura S, Yamamaoto H, Tamiya S, Yokoyama R, Matsuda S, Iwamoto Y, Tsuneyoshi M. Chemokine receptor CXCR4 expression is correlated with VEGF expression and poor survival in soft-tissue sarcoma. Int J Cancer. 2009;124:1852–1859. doi: 10.1002/ijc.24128. [DOI] [PubMed] [Google Scholar]
- 18.Zeng Z, Samudio IJ, Munsell M, An J, Huang Z, Estey E, Andreeff M, Konopleva M. Inhibition of CXCR4 with the novel RCP168 peptide overcomes stroma-mediated chemoresistance in chronic and acute leukemias. Mol Cancer Ther. 2006;5:3113–3121. doi: 10.1158/1535-7163.MCT-06-0228. [DOI] [PubMed] [Google Scholar]
- 19.Gelmini S, Mangoni M, Serio M, Romagnani P, Lazzeri E. The critical role of SDF-1/CXCR4 axis in cancer and cancer stem cells metastasis. J Endocrinol Investig. 2008;31:809–819. doi: 10.1007/BF03349262. [DOI] [PubMed] [Google Scholar]
- 20.Wu XS, Lonsdorf AS, Hwang ST. Cutaneous T-cell lymphoma: roles for chemokines and chemokine receptors. J Investig Dermatol. 2009;129:1115–1119. doi: 10.1038/jid.2009.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Furusato B, Mohamed A, Uhlen M, Rhim JS. CXCR4 and cancer. Pathol Int. 2010;60:497–505. doi: 10.1111/j.1440-1827.2010.02548.x. [DOI] [PubMed] [Google Scholar]
- 22.Bridger GJ, Skerlj RT, Thornton D, Padmanabhan S, Martellucci SA, Henson GW, Abrams MJ, Yamamoto N, De Vreese K, Pauwels R, et al. Synthesis and structure–activity relationships of phenylenebis (methylene)-linked bis-tetraazamacrocycles that inhibit HIV replication. Effects of macrocyclic ring size and substituents on the aromatic linker. J Med Chem. 1995;38:366–378. doi: 10.1021/jm00002a019. [DOI] [PubMed] [Google Scholar]
- 23.Bridger GJ, Skerlj RT, Padmanabhan S, Martellucci SA, Henson GW, Struyf S, Witvrouw M, Schols D, De Clercq E. Synthesis and structure–activity relationships of phenylenebis(methylene)-linked bis-azamacrocycles that inhibit HIV-1 and HIV-2 replication by antagonism of the chemokine receptor CXCR4. J Med Chem. 1999;42:3971–3981. doi: 10.1021/jm990211i. [DOI] [PubMed] [Google Scholar]
- 24.DiPersio JF, Uy GL, Yasothan U, Kirkpatrick P. Plerixafor. Nat Rev Drug Discov. 2009;8:105–106. doi: 10.1038/nrd2819. [DOI] [PubMed] [Google Scholar]
- 25.Porvasnik S, Sakamoto N, Kusmartsev S, Eruslanov E, Kim WJ, Cao W, Urbanek C, Wong D, Goodison S, Rosser CJ. Effects of CXCR4 antagonist CTCE-9908 on prostate tumor growth. Prostate. 2009;69:1460–1469. doi: 10.1002/pros.21008. [DOI] [PubMed] [Google Scholar]
- 26.Singh B, Cook KR, Martin C, Huang EH, Mosalpuria K, Krishnamurthy S, Cristofanilli M, Lucci A. Evaluation of a CXCR4 antagonist in a xenograft mouse model of inflammatory breast cancer. Clin Exp Metastasis. 2010;27:233–240. doi: 10.1007/s10585-010-9321-4. [DOI] [PubMed] [Google Scholar]
- 27.Jacobson O, Weiss ID, Szajek L, Farber JM, Kiesewetter DO. 64Cu-AMD3100—a novel imaging agent for targeting chemokine receptor CXCR4. Bioorg Med Chem. 2009;17:1486–1493. doi: 10.1016/j.bmc.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee B, Sharron M, Montaner LJ, Weissman D, Doms RW. Quantification of CD4, CCR5, and CXCR4 levels on lymphocyte subsets, dendritic cells, and differentially conditioned monocyte-derived macrophages. Proc Natl Acad Sci USA. 1999;96:5215–5220. doi: 10.1073/pnas.96.9.5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burger JA, Peled A. CXCR4 antagonists: targeting the micro-environment in leukemia and other cancers. Leukemia. 2009;23:43–52. doi: 10.1038/leu.2008.299. [DOI] [PubMed] [Google Scholar]
- 30.Boswell CA, Sun X, Niu W, Weisman GR, Wong EH, Rheingold AL, Anderson CJ. Comparative in vivo stability of copper-64-labeled cross-bridged and conventional tetraazamacrocyclic complexes. J Med Chem. 2004;47:1465–1474. doi: 10.1021/jm030383m. [DOI] [PubMed] [Google Scholar]
- 31.Misra P, Lebeche D, Ly H, Schwarzkopf M, Diaz G, Hajjar RJ, Schecter AD, Frangioni JV. Quantitation of CXCR4 expression in myocardial infarction using 99mTc-labeled SDF-1alpha. J Nucl Med. 2008;49:963–969. doi: 10.2967/jnumed.107.050054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hanaoka H, Mukai T, Tamamura H, Mori T, Ishino S, Ogawa K, Iida Y, Doi R, Fujii N, Saji H. Development of a 111In-labeled peptide derivative targeting a chemokine receptor, CXCR4, for imaging tumors. Nucl Med Biol. 2006;33:489–494. doi: 10.1016/j.nucmedbio.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 33.Nimmagadda S, Pullambhatla M, Pomper MG. Immunoimaging of CXCR4 expression in brain tumor xenografts using SPECT/CT. J Nucl Med. 2009;50:1124–1130. doi: 10.2967/jnumed.108.061325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nimmagadda S, Pullambhatla M, Stone K, Green G, Bhujwalla ZM, Pomper MG. Molecular imaging of CXCR4 receptor expression in human cancer xenografts with [64Cu]AMD3100 positron emission tomography. Cancer Res. 2010;70:3935–3944. doi: 10.1158/0008-5472.CAN-09-4396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li W, Gomez E, Zhang Z. Immunohistochemical expression of stromal cell-derived factor-1 (SDF-1) and CXCR4 ligand receptor system in hepatocellular carcinoma. J Exp Clin Cancer Res. 2007;26:527–533. [PubMed] [Google Scholar]
- 36.Mavier P, Martin N, Couchie D, Preaux AM, Laperche Y, Zafrani ES. Expression of stromal cell-derived factor-1 and of its receptor CXCR4 in liver regeneration from oval cells in rat. Am J Pathol. 2004;165:1969–1977. doi: 10.1016/S0002-9440(10)63248-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hatch HM, Zheng D, Jorgensen ML, Petersen BE. SDF-1alpha/CXCR4: a mechanism for hepatic oval cell activation and bone marrow stem cell recruitment to the injured liver of rats. Cloning Stem Cells. 2002;4:339–351. doi: 10.1089/153623002321025014. [DOI] [PubMed] [Google Scholar]
- 38.Wald O, Weiss ID, Galun E, Peled A. Chemokines in hepatitis C virus infection: pathogenesis, prognosis and therapeutics. Cytokine. 2007;39:50–62. doi: 10.1016/j.cyto.2007.05.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


