Abstract
We studied the effects of specific retroviral elements in a first-generation serotype 5 adenoviral (Ad5) vector, AdLTR2EF1α-hEPO. This vector contains 858 base pair (bp) [251-bp envelope sequence plus 607-bp long-terminal repeat (LTR)] from Moloney murine leukemia virus (MoMLV), upstream of the human elongation factor-1α (EF1α) promoter and human erythropoietin (hEPO) cDNA, with the LTR sequence downstream of the polyadenylation signal. We compared expression of AdLTR2EF1α-hEPO with corresponding expressions of two conventional Ad5 vectors, AdEF1α-hEPO and AdCMV-hEPO, in vivo in submandibular glands in rats. Both the conventional vectors yielded low serum hEPO levels by day 7, and little change in hematocrits. In contrast, after receiving AdLTR2EF1α-hEPO, the rats showed elevated hEPO levels and hematocrits for 1–3 months. In vitro studies showed that the integration efficiencies of all the vectors were similar (~10−3). Approximately 0.1% of the vector genomes were present 1 year after delivery in the case of each of the three vectors, primarily as intact linear double-strand DNA. The unique results seen with AdLTR2EF1α-hEPO are partly because of LTR enhancer activity. However, other cis-acting activity, which is not immunomodulatory but nevertheless influences promoter methylation, appears to be involved. A vector such as AdLTR2EF1α-hEPO may prove useful in clinical applications in which extended, but not “permanent,” transgene expression is desirable.
INTRODUCTION
Successful gene therapy requires efficient, nontoxic vectors to provide adequate transgene expression for an appropriate duration of time. Currently, there is no such ideal vector, and considerable effort is being directed at improving existing gene transfer systems. Two commonly used viral vectors, based on adenovirus serotype 5 (Ad5) and Moloney murine leukemia virus (MoMLV), have both advantages and drawbacks. First-generation Ad5 vectors offer ease of production, high titers, efficient transduction of many cell types, and infrequent genomic integration;1–4 on the other hand, they provoke vigorous immune responses resulting in short-term transgene expression (≤14 days).5–7 MoMLV vectors provide long-term transgene expression, but titers are low, transduction requires dividing cells, and genomic integration is uncontrolled, thereby introducing the risk of insertional mutagenesis.8–12 There are no vectors that are generally useful for intermediate-length applications (e.g., 1–3 months).
Earlier, we reported the construction of a hybrid adenoretroviral vector, AdLTR-luc, that is capable of transducing a wide variety of cells and mediating integration of the transgene cassette into genomic DNA with considerable efficiency.13–15 We hypothesized that MoMLV DNA interacts with cellular factors to mediate the integration of the transgene cassette and prolong transgene expression. During the course of studies to test that hypothesis, we discovered that the inclusion of certain retroviral elements in an Ad5 vector (AdLTR2EF1α-hEPO; Figure 1) leads to intermediate-length expression at low doses of the episomally localized vector.
Figure 1. Vector design.
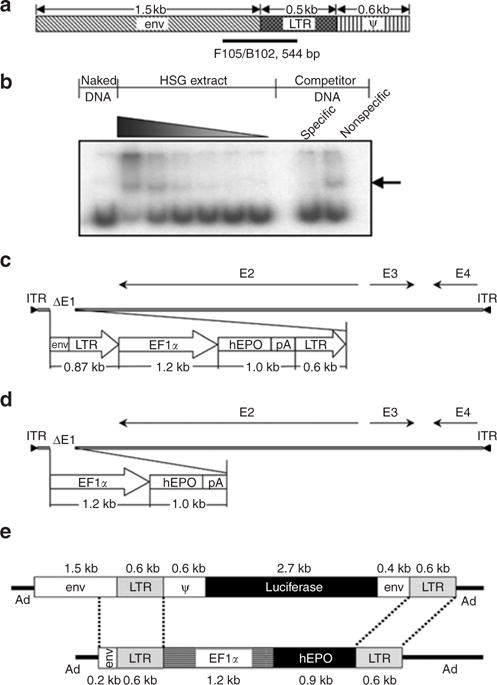
(a) Schematic representation of the 2.7 kilobase (kb) Moloney murine leukemia virus element used in AdLTR-luc,13 and used as a template to amplify 10 different DNA fragments for protein– DNA binding studies with HSG cell nuclear proteins. F105/B102 indicates the position of a 544-base pair (bp) fragment exhibiting the greatest interaction. (b) Autoradiograph of a representative gel shift assay. The interacting DNA–protein complex is denoted by the arrow, which shows that the F105/B102 544-bp fragment has strong and specific binding with increasing concentrations of the HSG cell nuclear protein extract. (c) Schematic depiction of AdLTR2EF1α-hEPO. (d) Schematic depiction of AdEF1α-hEPO. (e) Schematic comparison of the transgene cassettes in AdLTR-luc13 and AdLTR2EF1α-hEPO (reported herein). See text for additional details. Ad, adenovirus; EF1α, elongation factor-1α; env, envelope; hEPO, human erythropoietin; ITR, inverted terminal repeat; LTR, long-terminal repeat; pA, polyadenylation.
RESULTS
Interaction between MoMLV elements and nuclear proteins
Because of the organization of the MoMLV elements used in AdLTR-luc13 (Figure 1), we studied interactions between 10 MoMLV DNA fragments spanning the envelope and long-terminal repeat (LTR) junction with nuclear proteins using DNA gel retardation assays. We observed strong and specific nuclear protein binding with a 544-base pair (bp) fragment from this region (Figure 1a), lesser binding with three immediately adjacent MoMLV sequences (not shown), and the absence of binding to six other DNA fragments tested (not shown).
hEPO expression, in vitro and in vivo
On the basis of these results, we selected an 858-bp DNA fragment from MoMLV for further study (Figure 1; Supplementary Table S1; see also Materials and Methods). The resulting Ad vector was termed AdLTR2EF1α-hEPO. We compared AdLTR2EF1α-hEPO-mediated human erythropoietin (hEPO) expression in A5 cells (10 particles/cell, 24 hours) with that induced by two conventional Ad5 vectors without retroviral elements, namely, AdEF1α-hEPO (Figure 1d) and AdCMV-hEPO.16 The rank order for hEPO production was AdLTR2EF1α-hEPO ≈ AdCMV-hEPO >> AdEF1α-hEPO, with a highly significant approximately sevenfold difference between AdLTR2EF1α-hEPO and AdEF1α-hEPO (data not shown). We extensively tested AdLTR2EF1α-hEPO in vivo following delivery to submandibular glands in rats in six separate experiments, using a total of 148 rats. AdLTR2EF1α-hEPO-mediated hEPO expression was vector dose-dependent (108, 109, 1010 particles/gland; delivery to one gland; n = 5/group, day 7 hEPO expression 16, 91.2, and 93.4 mU/ml serum, respectively). We selected the 109 particles/gland dose for other experiments. Next, we conducted time-course studies with different animal cohorts comparing functional hEPO expression with the three vectors (Figure 2, Table 1). The two conventional Ad5 vectors resulted in very low hEPO levels and little change in hematocrits by day 7. In contrast, rats transduced with AdLTR2EF1α-hEPO had much higher hEPO levels, which generally remained well above baseline for 1–3 months (Figure 2a), as did the hematocrit levels (Figure 2b). Similar results were observed with female rats (not shown). In one rat cohort that was followed for 6 months, AdLTR2EF1α-hEPO-mediated hEPO and hematocrit levels were still significantly elevated in 12 of 20 rats, while only 1 of 18 rats treated with AdEF1α-hEPO showed such a result (Figure 2c and d). Across multiple in vivo experiments, rats treated with AdLTR2EF1α-hEPO had dramatically higher frequencies of elevated serum hEPO levels and hematocrits than rats administered the conventional AdEF1α-hEPO vector (Figure 2e; e.g., elevation in hEPO after 8 weeks was 86% in AdLTR2EF1α-hEPO-treated rats as compared to 3% in those treated with the conventional vector). Interestingly, we found similar copy numbers for all vectors in the targeted glands at 6 months (Table 1). However, only the rats that had been transduced with AdLTR2EF1α-hEPO exhibited any measurable hEPO levels in their sera (Table 1). We conducted a separate experiment to re-examine the early time-course results after administration of AdLTR2EF1α-hEPO and AdEF1α-hEPO. As shown in Figure 3a, transduction with either vector (109 particles/gland) also resulted in comparable vector copies detectable in the targeted glands from days 1–60. After 24 hours, ~10% of the delivered dose was found in the targeted gland, and by day 30, ~1% of the delivered vector dose was present, a level that remained stable (Figure 3a; no significant differences; analysis of variance, followed by Tukey’s test). Serum hEPO levels on day 1 (Figure 3b) from both vectors were not different (P > 0.5). However, by day 7, a dramatic difference was seen between the two vector groups; serum hEPO in AdEF1α-hEPO-treated rats was 0.6 mU/ml as compared to ~20–30 mU/ml in the AdLTR2EF1α-hEPO-treated rats. The latter remained relatively stable until the experiment was stopped. The hematocrit levels reflected the serum hEPO differences (Figure 3c). We next compared these vectors after delivery through the femoral vein (109 particles). As shown in Figure 3d, serum hEPO levels from both vectors were markedly higher (approximately tenfold) than was seen after salivary gland delivery. As with the results after delivery into the salivary gland, intravenous delivery also resulted in much higher AdLTR2EF1α-hEPO-mediated hEPO expression than that from the conventional vector; however, the hematocrit levels after intravenous transduction were similar (Figure 3e).
Figure 2. Effects of vector administration on human erythropoietin (hEPO), hematocrit, and antibody levels.
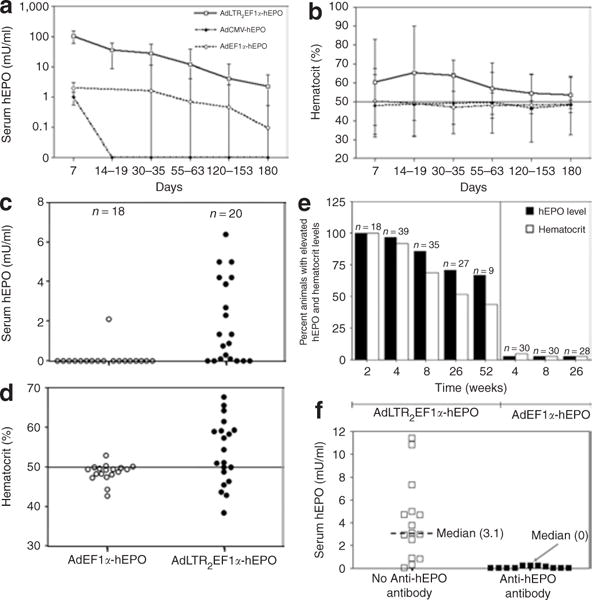
(a) Time-course of hEPO expression after vector (109 particles) delivery to one submandibular gland in each rat. The data shown are the mean values ± SD for results from a total of 82 male rats. The animals received AdLTR2EF1α-hEPO (n = 37), AdEF1α-hEPO (n = 25), or AdCMV-hEPO (n = 5). None of the 15 untreated rats followed over this time-course exhibited any measurable hEPO in their sera (not shown). (b) Time-course of hematocrit levels (mean value ±SD) measured for the same rats as are shown in panel (a). The control rats exhibited hematocrits between 47 and 50 over this duration of time (not shown). (c) Serum hEPO levels measured within a single experiment 6 months after delivery of either AdEF1α-hEPO or AdLTR2EF1α-hEPO. (d) Hematocrit levels of rats in experiment shown in panel (c). (e) Summary of all in vivo experiments shown in this figure after delivery of 109 particles of either AdEF1α-hEPO or AdLTR2EF1α-hEPO to rats, at various time-points. Any measurable hEPO value was considered to be elevated, as were all hematocrit values >50. (f) Differences in serum hEPO levels 6 months after AdLTR2EF1α-hEPO administration, measured in rats with and without serum antibodies to hEPO. Ad, adenovirus; CMV, cytomegalovirus; EF1α, elongation factor-1α; LTR, long-terminal repeat.
Table 1.
Long-term effects of transducing rat submandibular glands with different viral vectors encoding human erythropoietin
| Rat no. | Hematocrit (%) | Serum hEPO (mU/ml) | Anti-hEPO antibody titer | hEPO (×103) | AdE3 (×103) | Ratio (hEPO/AdE3) |
|---|---|---|---|---|---|---|
| AdLTR2EF1α-hEPO | ||||||
| 1 | 61.4 | 3.8 | — | 4.9 | 4.4 | 1.1 |
| 2 | 48.4 | 0 | 1:1,280 | 2.3 | 1.9 | 1.2 |
| 3 | 64.7 | 10.9 | — | 3.7 | 3.0 | 1.2 |
| 5 | 47.8 | 0 | 1:640 | 3.7 | 3.0 | 1.2 |
| Average (±SD) | 3.7 ± 1.2 | 3.0 ± 1.1 | 1.2 ± 0.12 | |||
| AdEF1α-hEPO | ||||||
| 1 | 46.5 | 0 | — | 3.9 | 2.4 | 1.6 |
| 2 | 48.9 | 0 | — | 1.1 | 0.8 | 1.4 |
| 3 | 47.8 | 0 | — | 1.3 | 1.0 | 1.3 |
| 4 | 45.7 | 0 | — | 2.8 | 1.6 | 1.8 |
| 5 | 49.5 | 0 | — | 1.5 | 1.4 | 1.1 |
| Average (±SD) | 2.1 ± 1.2 | 1.4 ± 0.6 | 1.6 ± 0.21 | |||
| AdCMV-hEPO | ||||||
| 1 | 47.1 | 0 | — | 1.1 | 2.7 | 0.4 |
| 2 | 50 | 0 | — | 3.0 | 4.8 | 0.6 |
| 3 | 50 | 0 | — | 3.5 | 4.1 | 0.9 |
| 5 | 45.2 | 0 | — | 1.1 | 1.9 | 0.6 |
| Average (±SD) | 2.1 ± 1.1 | 3.4 ± 1.2 | 0.6 ± 0.12 | |||
Abbreviations: Ad, adenovirus; CMV, cytomegalovirus; EF1α, elongation factor-1α; hEPO, human erythropoietin; LTR, long-terminal repeat.
These results are from gland samples obtained 6 months after transduction with AdLTR2EF1α-hEPO, AdEF1α-hEPO, or AdCMV-hEPO at 109 particles/gland, originally with five rats per group. Missing numbers are of rats that died. “—” means no antibody detected against hEPO in the serum. Vector copies were measured as hEPO and AdE3 sequences. See Materials and Methods for additional details. Paired t-tests were used to compare copy numbers for each vector determined by either hEPO or AdE3 sequences. No significant differences were found for all comparisons (AdLTR2EF1α-hEPO, P = 0.5; AdEF1α-hEPO, P = 0.2; AdCMV-hEPO, P = 0.5).
Figure 3. Early time-course of glandular vector copy number, serum human erythropoietin (hEPO) expression, and hematocrit levels after vector delivery to submandibular glands and femoral veins in rats.
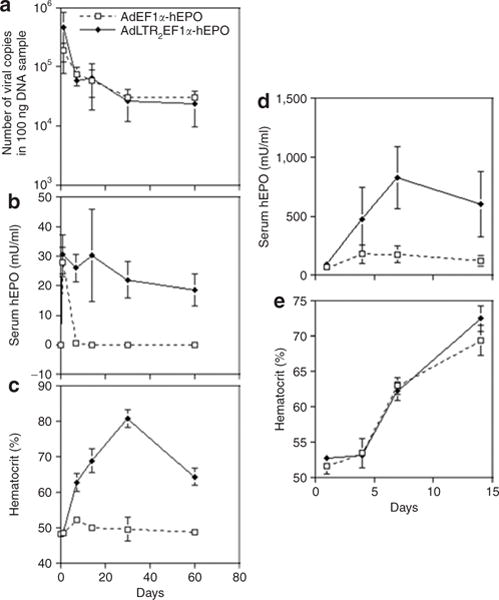
(a) Time-course of vector copy numbers after vector (109 particles) delivery in rats to one submandibular gland in each animal. The animals received either AdLTR2EF1α-hEPO or AdEF1α-hEPO. The data shown are mean values ± SD for glands from three rats. The vector copy number was determined using quantitative PCR and primers for the hEPO cDNA (see Materials and Methods). The data were analyzed using a one-way analysis of variance, and no significant differences over this time-course were found between the two vectors. (b) Time-course of hEPO levels measured for the same rats as are shown in a. The data shown are mean values ± SD. (c) Time-course of hematocrit levels measured for the same rats as are shown in a. The data shown are mean values ± SD. (d) Time-course of hEPO levels after systemic vector (109 particles) delivery to 6 rats through the femoral vein. The animals (n = 3/group) received either AdLTR2EF1α-hEPO or AdEF1α-hEPO. The data shown are mean values ± SD. Note the difference in the y-axis scale from b. (e) Time-course of hematocrit levels measured for the same rats as are shown in d. The data shown are mean values ± SD. Ad, adenovirus; EF1α, elongation factor-1α; LTR, long-terminal repeat.
Assessment of immune responses following vector delivery
We hypothesized that the absence of hEPO in the sera from some of the rats (Table 1, Figure 2) following transduction of glands with AdLTR2EF1α-hEPO was because of antibodies to the transgenic protein. Over multiple experiments, 5 or 6 months after vector delivery, most AdLTR2EF1α-hEPO-treated rats that showed little to no hEPO in their sera (<1 mU/ml; 11/15 rats) had measurable anti-hEPO antibodies in their sera (1:20 to 1:1,280 titers; median = 1:80; Figure 2f and Table 1). However, no serum anti-hEPO antibodies were found in any of the rats transduced with the conventional vector. In order to examine whether the hEPO expression results also could be caused by differences in cellular immune responses, splenocytes were obtained and studied (Figure 4b–e). Splenocytes from rats treated with either of the vectors respond similarly to both concanavalin A and Ad5, but when challenged with recombinant hEPO, splenocytes from AdEF1α-hEPO-treated rats were indistinguishable from those of naive rats. In contrast, splenocytes from rats whose glands were transduced by AdLTR2EF1α-hEPO exhibited increased proliferation, as well as significant production of all three of the cytokines measured (Figure 4b–e). This shows that hEPO activates a cellular immune response in rats.
Figure 4. Immune reactivity following vector administration to salivary glands in rats.
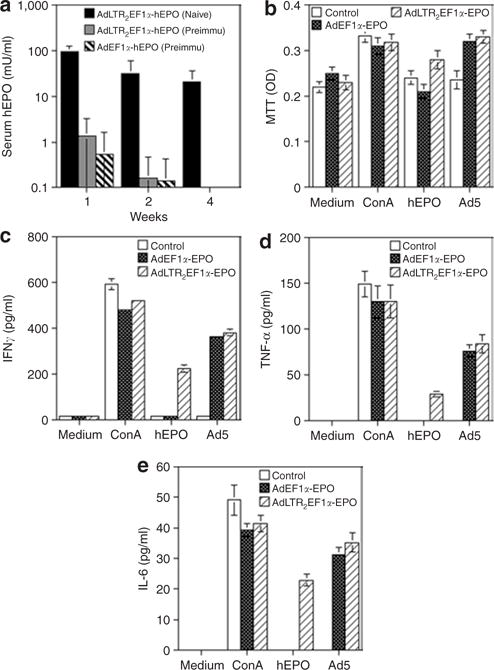
(a) The rats were either pretreated with a conventional Ad5 vector (AdCMV-luc13) by intramuscular injection or not (naive) on day –21. On day 0 the rats were administered either AdLTR2EF1α-hEPO or AdEF1α-hEPO through their submandibular glands, as indicated. The data shown are the mean values ± SD (n = 5/group). (b–e) Spleens were removed 2 months after vector administration. Splenocytes were prepared and used for performing cellular immune response assays. (b) Cellular proliferation was measured using the MTT assay. (c,d) IFN-γ and TNF-α production, i.e., TH1 responses. (e) IL-6 production, i.e., a TH2 response. The data shown in b–e are the mean values ± SD of results from 2 rats/group, each assayed in triplicate. Medium: the negative control, cells incubated in culture medium only. Con A: the positive control, cells incubated in medium supplemented with concanavalin A (10 μg/ml). hEPO: results from cells incubated with 500 mU/ml recombinant hEPO protein. Ad5: results from cells incubated with AdCMV-luc at a multiplicity of infection (MOI) = 10 particles/cell. See text for additional details. Ad, adenovirus; CMV, cytomegalovirus; EF1α, elongation factor-1α; hEPO, human erythropoietin; IFN-γ, interferon-γ; IL-6, interleukin-6; LTR, long-terminal repeat; MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide, TNF-α, tumor necrosis factor-α.
AdLTR2EF1α-hEPO behaves like a conventional Ad5 vector
Our data also suggest that AdLTR2EF1α-hEPO generally behaves like a conventional Ad5 vector. First, when rats had been pre-immunized (–21 days) with an irrelevant Ad5 vector intramuscularly (Figure 4a), it was found that, after subsequent salivary gland transduction with either AdLTR2EF1α-hEPO or AdEF1α-hEPO, none of the rats had any detectable serum hEPO (when compared with naive rats, Figure 4a). Second, all the rats that had been administered 109 particles of AdLTR2EF1α-hEPO produced neutralizing antibodies against Ad5 in their sera, blocking the transduction of 293 cells by AdCMV-luc (data not shown; n = 27; at 5–6 months median titer 1:320).17 Third, when we extracted DNA from the glands of rats previously treated (6 months) with AdLTR2EF1α-hEPO, AdEF1α-hEPO, or AdCMV-hEPO, and then used quantitative PCR (QPCR) to determine vector copies present (Table 1), we found a similar number of vector copies for each treatment group (also in animals followed for 1 year, not shown). Finally, genomic DNA from glands transduced by any one of the three hEPO-encoding vectors demonstrated that each appears to be present in an intact form (Figure 5; below).
Figure 5. Assessment of vector DNA in situ.
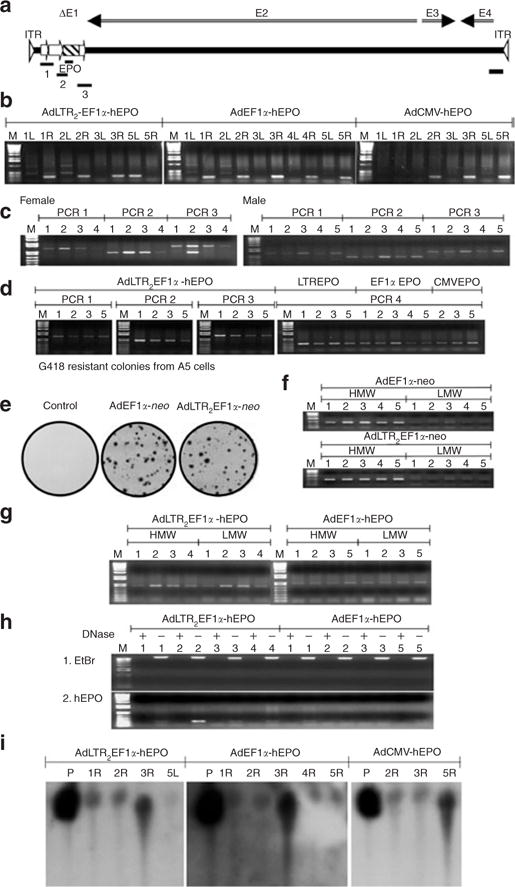
(a) Diagram of PCR products amplified with AdLTR2EF1α-hEPO. The PCR1 amplicon goes from the beginning of the upstream Moloney murine leukemia virus element to the middle of the EF1α promoter. The PCR2 amplicon goes from the EF1α promoter to the middle of the hEPO cDNA. The EPO amplicon is from the middle of the hEPO cDNA. The PCR3 amplicon goes from the hEPO cDNA into the Ad5 E2 region. The PCR4 amplicon is at the 3′-end of the vector in the Ad5 E4 region. (b) Detection of the EPO amplicon in targeted glands 6 months after vector delivery. Note that vectors were delivered to the right (R) submandibular glands of all animals except rat #5 in the AdLTR2EF1α-hEPO group, which received the vector in the left (L) gland. (c) Detection of the amplicons PCR 1, 2, and 3 in targeted glands 1 year after AdLTR2EF1α-hEPO delivery to female and male rats. (d) Detection of the amplicons PCR 1, 2, and 3 in targeted glands 6 months after AdLTR2EF1α-hEPO delivery, and PCR 4 after delivery of AdLTR2EF1α-hEPO (LTREPO), AdEF1α-hEPO (EF1αEPO), and AdCMV-hEPO (CMVEPO). (e) G418-resistant colonies obtained from A5 cells transduced with either AdEF1α-neo or AdLTR2EF1α-neo (neo, neomycin resistance gene). (f) PCR assays (using EF1αF4 and neoB3 primers) with high-molecular-weight (HMW) and low-molecular-weight (LMW) DNA extracts from G418-resistant colonies. (g) Assay for the PCR2 amplicon using HMW and LMW DNA extracts from female rats 1 year after administration of either AdEF1α-hEPO or AdLTR2EF1α-hEPO. (h) Plasmid-safe DNase assays to determine the general vector DNA structure (linear double strand or circular) in rats whose submandibular glands had been transduced with either AdLTR2EF1α-hEPO or AdEF1α-hEPO 1year earlier. Plus means presence of plasmid-safe DNase in the incubation mixture, Minus means without plasmid-safe DNase present. (i) Southern hybridization of undigested DNA samples from targeted glands 6 months after delivery of AdLTR2EF1α-hEPO, AdEF1α-hEPO, or AdCMV-hEPO. P refers to positive control samples from intact vectors spiked into glands from naive rats during genomic DNA extraction. Ad, adenovirus; CMV, cytomegalovirus; hEF1α, human elongation factor-1α; EtBr, ethidium bromide; hEPO, human erythropoietin; LTR, long-terminal repeat.
Assessment of vector DNA in situ
As noted earlier, 6 months after delivery of any of the three vectors significant and similar levels of vector DNA were found in male and female rats (Figure 5). For all three vectors, QPCR analysis showed that there were ~103 particles/100 ng DNA (Table 1), corresponding to ~106 particles/gland, i.e., ~0.1% of the delivered dose. In the AdLTR2EF1α-hEPO-treated rats, it appeared that the entire transgene cassette (Figure 5; PCR products 1, 2, and 3), and much of the vector (PCR product 4), were intact. In order to evaluate the possibility of integration, we tested two vectors encoding the neomycin resistance gene (neo) instead of hEPO (AdEF1α-neo; AdLTR2EF1α-neo) in A5 cells (Figure 5e). We found no differences in the frequency of detection of G418-resistant colonies between these two vectors (~1/103 cells; similar results in HSG cells, not shown). This integration efficiency is comparable to that reported earlier for Ad5 vectors,4 and much less than that seen with AdLTR-luc (i.e., ~5–10%).15 This conclusion was also supported by Southern hybridization analyses of gland samples from all three vector groups (Figure 5i), thereby indicating that intact vector genomes were found in the glands. Subsequently, we also extracted high-molecular-weight (HMW) and low-molecular-weight (LMW) DNA from G418-resistant A5 cell colonies (Figure 5e) and performed PCR and QPCR analyses (Figure 5f, Table 2). Most of the targeted PCR products were in the HMW fraction (HMW:LMW ratio; for AdEF1α-neo, 74.5; for AdLTR2EF1α-neo, 62.3; Table 2), thereby suggesting that in G418-selected colonies both vectors were integrated into the host cell genome. Next, we compared the vector genomes present in HMW and LMW DNA fractions from rat glands 1 year after the administration of either AdEF1α-hEPO or AdLTR2EF1α-hEPO. There appeared to be no difference in the frequency with which these vectors were detected in the HMW and LMW fractions (Figure 5g; HMW:LMW ratio; for AdEF1α-hEPO, 1.1; for AdLTR2EF1α-hEPO, 1.7; Table 2; similar results at 6 months, not shown). Taken together, these results suggest that most of the AdLTR2EF1α-hEPO genomic DNA is found in, and hEPO expression is derived from, a nonintegrated, extrachromosomal locale after the vector was administered in vivo. We also examined, using plasmid-safe adenosine triphosphate–dependent DNase, whether AdEF1α-hEPO and AdLTR2EF1α-hEPO genomes were circular or linear double-stranded. The results indicated that vector DNAs from the glands of female rats at 1year after vector delivery were linear (Figure 5h). Similar results were found in samples from male rats treated with each of the vectors at other time-points (not shown).
Table 2.
Number of vector copies, measured as transgene sequences present (neo or hEPO), in high and low-molecular-weight genomic DNA from G418-resistant A5 cells and female rat submandibular glands after vector administration
| Vector | HMW (×103 ± SD) | LMW (×103 ± SD) | HMW/LMW (±SD) |
|---|---|---|---|
| A5 cells | |||
| AdLTR2EF1α-neo | 143.4 ± 36.9 | 2.3 ± 0.6 | 62.3 ± 19.3 |
| AdEF1α-neo | 149 ± 95.6 | 2.0 ± 0.7 | 74.5 ± 35.8 |
| Submandibular glands | |||
| AdLTR2EF1α-hEPO | 8.7 ± 6.7 | 5.1 ± 4.3 | 1.7 ± 0.18 |
| AdEF1α-hEPO | 8.6 ± 7.1 | 7.8 ± 11.2 | 1.1 ± 0.11 |
Abbreviations: HMW, high-molecular-weight; hEPO, human erythropoietin; LMW, low-molecular-weight; neo, neomycin resistance gene.
HMW and LMW genomic DNA samples were obtained from five colonies each of AdLTR2EF1α-neo and AdEF1α-neo-transduced A5 cells, and from submandibular glands of four female rats transduced with AdLTR2EF1α-hEPO or AdEF1α-hEPO at 109 particles/gland 1 year after administration. The number of vector copies in the HMW and LMW DNA fractions were not different when comparing AdLTR2EF1α-neo and AdEF1α-neo in cloned cells or AdLTR2EF1α-hEPO and AdEF1α-hEPO in submandibular glands (t-test; P ≥ 0.6 for all).
Assessment of possible mechanisms leading to differences in hEPO expression
All data from the AdLTR2EF1α-hEPO-treated rats showed dramatically higher and extended transgene expression as compared to those from rats treated with conventional Ad5 vectors. In order to assess possible mechanisms of action, we followed enhanced green fluorescent protein (EGFP) expression in A5 cells transduced with AdLTR2EF1α-EGFP, AdEF1α-EGFP, or AdCMV-EGFP (Figure 6). The results (upper panels, Figure 6, at day 4) clearly show that AdLTR2EF1α-EGFP-transduced A5 cells exhibit the strongest EGFP expression; rank order: AdLTR2EF1α-EGFP > AdCMV-hEPO > AdEF1α-hEPO. This is consistent with the notion that part of the increased transgene expression was likely caused by retroviral elements playing an enhancing role in this construct and, by implication, with AdLTR2EF1α-hEPO. However, independent of any enhancer effects on initial EGFP expression (i.e., when expression on day 4 was normalized to 100%), expression from the AdLTR2EF1α-EGFP vector displayed very different kinetic behavior. Elevated expression levels in AdLTR2EF1α-EGFP-transduced cells were maintained from ~day 51 through 180 days (Figure 6b; longest duration studied). This represents >170 population doublings of the A5 cells. Conversely, EGFP expression in cells transduced by either of the conventional vectors continued to decline over this period, i.e., by day 180 EGFP expression was <<1% of that of AdLTR2EF1α-EGFP-transduced cells. A possible mechanistic role for the MoMLV elements in the hybrid vectors used (which could account for the difference seen in the kinetic pattern of transgene expression) is to function as insulators, preventing transgene silencing.18 We examined genomic DNA obtained (i) from A5 cells 6 months after transduction with each EGFP-encoding vector shown in Figure 6 and (ii) from rat glands after transduction with the three hEPO-encoding vectors. The results are consistent with our hypothesis, i.e., the MoMLV elements apparently function as insulators, leading to a decrease in the frequency of cytosine methylation in the elongation factor-1α (EF1α) promoter (Supplementary Table S3). When DNA from the AdEF1α-hEPO- and AdEF1α-EGFP-treated samples were examined, 13 methylated cytosines were found in the 125 sequences analyzed from a 295-bp PCR fragment of the 1,272-bp human EF1α promoter. In contrast, when we used DNA obtained from either the AdLTR2EF1α-hEPO- or the AdLTR2EF1α-EGFP-treated samples, no methylated cytosines were found in this same PCR fragment in the 88 sequences that were analyzed. The difference in frequency of detection of methylated cytosines in this fragment of the EF1α promoter was highly significant statistically (Fisher’s exact test; P = 0.000745). Not surprisingly,19 when we analyzed 73 sequences of a 398-bp fragment from the 780-bp cytomegalovirus (CMV) promoter, we found that at least 48 cytosines present in the amplified PCR fragments were methylated.
Figure 6. Time-course of vector transduction of A5 cells in vitro.
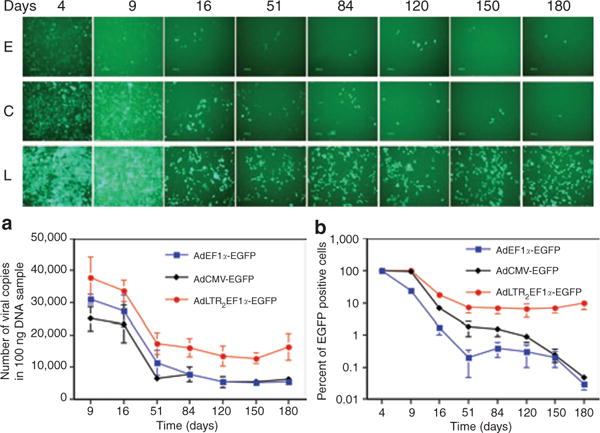
The Ad5 vectors used were AdLTR2EF1α-EGFP, AdEF1α-EGFP, and AdCMV-EGFP. The top panel shows one of five representative fields at each time-point. E: AdEF1α-EGFP-transduced A5 cells. C: AdCMV-EGFP-transduced A5 cells. L: AdLTR2EF1α-EGFP-transduced A5 cells. (a) Time-course of viral vector copy numbers present. (b) Time-course of EGFP positive A5 cells. The data in b were normalized to the maximum EGFP expression seen, i.e., at day 4 (100%). The data shown in panels a and b are the mean values ± SD of results from all five fields. See text for additional details. Ad, adenovirus; CMV, cytomegalovirus; EF1α, elongation factor-1α; EGFP, enhanced-green fluorescent protein; LTR, long-terminal repeat.
DISCUSSION
This study demonstrates that the inclusion of the specific retroviral elements described herein in a first-generation Ad5 vector leads to a vector with novel characteristics that may be potentially useful for gene transfer applications calling for intermediate-length (1–3 months) expression of a transgene. In multiple, separate studies, the prototype vector used, AdLTR2EF1α-hEPO, while generally performing indistinguishably from conventional first-generation Ad5 vectors (e.g., antibody responses and low frequency of genomic integration), led to robust transgene expression both in vitro and in vivo. These findings were consistent during in vivo studies conducted over >3 years, in six separate experiments, in a large number of animals. After delivery to submandibular glands in rats, AdLTR2EF1α-hEPO resulted in significant hEPO expression for at least 2 months in most of the animals. For example, for 35 rats that had been treated 2 months earlier with AdLTR2EF1α-hEPO, the average serum hEPO level was 11.6 ± 4.6 (SEM) mU/ml, while for 30 rats treated with AdEF1α-hEPO this value was 0.6 ± 0.6 mU/ml. Further, the hEPO protein produced was functional. The hematocrits at this time-point were 58.3 ± 2.1 and 47.4 ± 0.9, respectively. That is, the use of the same EF1α promoter in an identical vector, but without the indicated upstream and downstream retroviral sequences, led to much lower hEPO expression, which was short-lived, as is typical for conventional Ad5 vectors.5,6
Additionally, we observed that most of the rats that had been treated with AdLTR2EF1α-hEPO, and showed little or no measurable serum hEPO after 5–6 months, had serum antibodies against hEPO. Furthermore, the hEPO protein was able to stimulate a cellular immune response (increased splenocyte proliferation plus secretion of the cytokines interferon-γ, tumor necrosis factor-α, and interleukin-6) in rats that were administered 109 particles of AdLTR2EF1α-hEPO into the submandibular glands. No such cellular immune responses were observed with splenocytes from AdEF1α-hEPO-treated rats. These immune responses likely were due to the higher levels of transgenic hEPO produced with AdLTR2EF1α-hEPO.20 It is unlikely that such immune responses would occur in humans if they were treated with a similar vector, because the encoded protein would not be foreign. These findings suggest that the prototype AdLTR2EF1α-hEPO and related vectors may be capable of enhanced transgene expression for >1–3 months in humans.
The specific mechanism by which these retroviral elements permit enhanced and extended transgene expression is not yet entirely understood. As is clearly seen from the kinetic results shown in Figure 3a, the PCR and Southern blot results shown in Figure 5, and the QPCR results shown in Table 2, there were no marked differences in the copy numbers, distribution in HMW and LMW DNA fractions, and general intracellular structure of the conventional and modified vectors present in the glands in vivo at early and extended time-points, as well as in nonselected A5 cells in vitro. In the absence of integration, our data indicate that the retroviral elements employed acted partly as enhancers to increase initial expression from the EF1α promoter directly and, in addition, stabilized the expression for a considerable length of time both in vitro (Figure 6b) and in vivo (Figures 2a and 3b). We hypothesized that this stabilization could occur through an insulator function, and we accordingly examined the methylation status of the promoter. We were clearly able to show that the CMV promoter used in one conventional Ad5 construct was methylated in rat submandibular glands and in A5 cells, which, in the light of findings in earlier reports,19 likely led to the transient hEPO expression seen from the use of this vector. Importantly, on the basis of similar analyses of DNA from AdEF1α-hEPO- and AdEF1α-EGFP-treated samples in comparison with samples treated with AdLTR2EF1α-hEPO and AdLTR2EF1α-EGFP, it appears likely that methylation of the EF1α promoter plays a role in the decreased expression seen from this promoter when it is present without the adjacent retroviral elements. Although additional studies are needed to fully elucidate the mechanism responsible for the extended expression seen with the use of AdLTR2EF1α-hEPO [e.g., further examination of the recruitment and/or binding of critical nuclear proteins to transcriptionally-active domains (see Figure 1b)], our results show that the MoMLV elements used in this vector have unique characteristics in addition to and distinct from their well-known enhancer functions.
Interestingly, we found that the adenoviral vector genome was not only retained in slowly dividing submandibular gland cells of rats in vivo, but also in vitro in the rapidly dividing A5 cell line, for 6 months. Indeed, kinetic evaluation showed that vector copy numbers resulting from transduction with the AdEF1α-hEPO or the AdLTR2EF1α-hEPO vectors were quite similar in vivo and in vitro (Figures 3a and 6a), thereby suggesting that vector retention is not related to the presence of MoMLV elements in AdLTR2EF1α-hEPO. While the in vivo result seems not surprising, given the extremely slow rate of cell proliferation in adult rat salivary glands,21 the results in A5 cells were unexpected and difficult to explain on a mechanistic basis.22,23
A vector such as is described herein may prove useful clinically for applications that require extended, but not “permanent” (i.e., resulting from integrated vector) transgene expression. For example, patients undergoing radiation treatment (e.g., 2 Gy/day for 4–6 weeks) for head and neck cancers could benefit from the continuous expression of a gene encoding a free radical metabolizing enzyme such as Mn-superoxide dismutase24 during radiotherapy to prevent radiation-induced mucositis. Similarly, patients undergoing cytotoxic chemotherapy for a variety of cancers could benefit during their treatment phase from the expression of granulocyte colony stimulating factor to prevent neutropenia25 or of keratinocyte growth factor to prevent oral or intestinal mucositis.26 Our data strongly suggest that AdLTR2EF1α-based vectors may allow these possibilities.
MATERIALS AND METHODS
Gel shift assay
Ten DNA fragments (228–740 bp in size) were amplified from a 2.7-kb MoMLV fragment [Figure 1a; containing part of the envelope gene (~1.5 kb)], the 5′ LTR(~0.6 kb), and the packaging sequence (~0.6 kb)]13 using PCR with TurboPfu polymerase (Stratagene, La Jolla, CA). Fragments were 5′-radiolabeled using γ-32P-labeled adenosine triphosphate with T4 polynucleotide kinase. Nuclear protein was extracted from HSG cells and serial dilutions were incubated in binding buffer with 20 fmol of 5′-end-labeled DNA fragments for 20 minutes at room temperature. As a specific competitor for each reaction, we used a molar excess of the unlabeled fragment. The nonspecific competitor used was a DNA fragment that did not bind with the nuclear protein extract. Samples were loaded onto 6% acrylamide DNA retardation gels (Invitrogen, Carlsbad, CA), electrophoresed at 4 °C for 60 minutes and autoradiographed.
Construction of recombinant vectors
The vectors used here were based on the Ad5 genome. E1 deletion was achieved by recombination of the pACCMV-pLpA shuttle plasmid (a gift from C. Newgard) with pJM17 (Microbix Biosystems, Toronto, Canada). pACCMV-pLpA was digested with NotI to delete the CMV promoter and SV40 poly A. An 858-bp DNA fragment (Supplementary Table S1) from MoMLV, containing 167 bp from the 3′-end of the envelope p15E protein coding sequence, plus 84 bp downstream of that sequence, and 607-bp LTR sequence, was added to create a plasmid termed “pACLTR1.” Thereafter, the 607-bp LTR sequence was ligated into pACLTR1 to create the plasmid pACLTR2. The EF1α promoter (1,272 bp) used was excised from pEF-BOS (a gift from S. Gutkind, National Institute of Dental and Craniofacial Research, National Institutes of Health) and ligated into the pACLTR2 to create a plasmid, pACLTR2EF1α. The hEPO cDNA plus SV40 poly A sequence (1,018 bp) was excised from pEAK8-hEPO (a gift from Dr. Y. Terada, Tokyo Medical and Dental University, Japan) and ligated into pACLTR2EF1α to create the plasmid pACLTR2EF1α-hEPO. The recombinant adenoviral vector, AdLTR2EF1α-hEPO (Figure 1c), was generated by homologous recombination of pACLTR2EF1α-hEPO with pJM17 in C7 cells. AdLTR2EF1α-EGFP was constructed similarly, but using an 1,009-bp fragment including the EGFP cDNA and a SV40 poly A sequence excised from pEGFP-N1 (Clontech, Mountain View, CA). AdLTR2EF1α-neo was constructed using a 940-bp fragment including the neomycin resistance cDNA and SV40 poly A sequence excised from pCI-neo (Promega, Madison, WI) instead of the hEPO cDNA and the existing SV40 poly A sequence. AdEF1α-hEPO (Figure 1d), AdCMV-hEPO,16 AdCMV-EGFP,27 AdEF1α-EGFP,27 and AdEF1α-neo were constructed as conventional Ad5 control vectors.28 The titers (particles/ml) of purified viruses were determined using QPCR with primers from the E2 region.27
Cell culture
The A5 cell line was derived from rat submandibular gland29 and grown in McCoy’s 5A medium (Invitrogen). The HSG cell line was obtained from an irradiated human submandibular gland30 and grown in Dulbecco’s modified Eagle’s medium/F12 medium (Invitrogen). The 293 cell line (Microbix Biosystems) is from human embryonic kidney31 and was grown in Eagle’s minimum essential medium (Invitrogen). C7 cells are derived from 293 cells, and stably express the Ad5 preterminal protein and DNA polymerase.32 They were grown in high-glucose Dulbecco’s modified Eagle’s medium (Invitrogen). In order to kinetically compare AdLTR2EF1α-EGFP with two conventional vectors, AdEF1α-EGFP and AdCMV-EGFP, in vitro, A5 cells were transduced with these vectors at a multiplicity of infection of 400 particles/cell in order to achieve 100% transduction. The transduced A5 cells were passaged at a 1:12 split ratio every week for 6 months. At the time-points indicated in Figure 6, the transduced A5 cells were photographed, or counted by hand, or genomic DNA was extracted from them.
In vivo animal experiments
Animal experiments were approved by the National Institute of Dental and Craniofacial Research Animal Care and Use Committee, and the National Institutes of Health Biosafety Committee. Male or female Wistar rats (250–350 g, 3 months old) were anesthetized with ketamine (60 mg/kg) and xylazine (8 mg/kg) intramuscularly. The vectors were administered to the right submandibular gland by retrograde ductal instillation.33 In addition, vectors (109 particles) were also administered into the femoral veins of 6 rats (AdLTR2EF1α-hEPO, n = 3; AdEF1α-hEPO, n = 3). Blood samples were collected from the retro-orbital sinuses at different time-points. Hematocrits were measured using microhematocrit capillary tubes (Fisher Scientific, Pittsburgh, PA).
Measurement of serum hEPO levels
hEPO levels in rat sera were determined with enzyme-linked immunosorbent assay (ELISA) using human erythropoietin ELISA kits from either StemCell Technologies (Vancouver, Canada) or R&D Systems (Minneapolis, MN).
Measurement of serum neutralizing antibodies
In order to determine whether rats that had been administered Ad5 vectors developed anti-Ad5 neutralizing antibodies, their sera were assayed essentially as described.34 In order to determine whether rats developed anti-hEPO antibodies, 25 μl of standard hEPO protein (6.25 mU/ml) from the StemCell ELISA kit was incubated with rat serum (or dilutions) in a total volume of 50 μl at 37 °C. These samples were then assayed using the hEPO ELISA. Titers are reported as the dilution of serum that resulted in 50% inhibition in measuring the hEPO protein level.
Cellular immune response assays
AdLTR2EF1α-hEPO and AdEF1α-hEPO were administered (109 particles) to the right submandibular glands of male Wistar rats by retrograde ductal instillation, and the spleens were removed 2 months later. Splenocytes were freshly isolated as described35 and were plated in either a 24-well plate (4 × 106 cells/well) or a 96-well plate (105 cells/well) using Rosewell Park Memorial Institute 1640 medium for the culture. The cells were incubated at 37 °C in a humidified 5% CO2 atmosphere. After 1 hour, AdCMV-luc13 (multiplicity of infection = 10 particles/cell), concanavalin A (10 μg/ml; Sigma, St Louis, MO), or recombinant hEPO (500 mU/ml; Calbiochem, San Diego, CA) was added to the appropriate wells. After 48 hours, MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] assays were performed in 96-well plates to assess proliferative responses (MTT colorimetric kit; Chemicon International, Temecula, CA). The supernatants from the 24-well plates were collected and analyzed using ELISAs specific for interleukin-6 (Endogen Rat IL-6 Elisa kit; Pierce Biotechnology, Rockford, IL), tumor necrosis factor-α (Rat TNF-α EIA; Alpco, Salem, NH), or interferon-γ (Rat interferon-γ EIA; Alpco).
PCR and QPCR assays
The genomic DNA for most PCRs was extracted using the Wizard Genomic DNA Purification kit (Promega, Madison, WI). HMW genomic DNA was extracted using a Non-Organic DNA Extraction kit (Chemicon, Temecula, CA). LMW genomic DNA was extracted using the Hirt method.36 A total of 400 ng of DNA were used for PCRs and 100 ng for QPCRs. Primers (see Supplementary Table S2 for all primer sequences), LTRF109 and EF1αB1, were used for amplifying PCR1 (Figure 5a). PCR2 was amplified using EF1αF4 and hEPOB6. PCR3 was amplified using hEPOF2 and E2B22. PCR4 was amplified from the 3′-end of E4 region by Ad35up and Ad35down. The hEPO PCR product (Figure 5a) was amplified using hEPOF1 and hEPOB6. The PCR product including the neomycin resistance gene was amplified using EF1αF4 and neoB3. Primers and probes used for QPCR were hEPOTaq1, hEPO-Taq2, and hEPOTaqprobe for the hEPO cDNA; LTRTaq1, LTRTaq2, and LTRTaqprobe for the LTR element; and E3Taq1, E3Taq2, and E3Taqprobe for Ad5 E3. The AdLTR2EF1α-hEPO was treated with EasyXpress Viral Nucleic Acid Release reagent (XpressBio, Thurmont, MD) and used for generating standard curves.
Plasmid-safe DNase assays
Genomic DNA (800 ng) extracted using the Wizard Genomic DNA Purification kit (Promega) was digested with 10 μl of Plasmid-Safe DNase (Epicentre, Madison, WI) plus 20 μl of 25 mmol/l adenosine triphosphate and 10 μl of buffer for 16 hours. Thereafter, 200 ng of DNA sample was subjected to electrophoresis in 1% agarose and PCRs.
G418 resistance assays
Suspensions of A5 or HSG cells were transduced with AdLTR2EF1α-neo or AdEF1α-neo at 1 particle/cell at 37 °C for 1 hour. The cells were next placed in 100 mm dishes at 106 cells/dish with growth medium plus 250 μg/ml of G418 for 24 hours. After 24 hours, the concentration of G418 was increased to 500 μg/ml, and after 48 hours to 1,000 μg/ml, and colonies were counted at 14–16 days.
Southern hybridization
The genomic DNA used in the Southern hybridization analyses was extracted using Wizard Genomic DNA Purification Kits (Promega). Thirty microliters of undigested genomic DNA, obtained from rat submandibular glands 6 months after vector administration, was electrophoresed in a 1% agarose gel. The positive controls were non-transduced rat submandibular gland samples spiked with each Ad5 vector (109 particles) during the DNA extraction procedure. Nucleic acids were then transferred to a nylon membrane and the blots were hybridized with an [α(-32P]dCTP-radiolabeled 580-bp human EPO probe.
DNA methylation assay
The genomic DNA used in the DNA methylation assays was obtained as described earlier. Each genomic DNA sample (300 ng; from A5 cells, or rat submandibular glands, 6 months after vector transduction) was processed using the MethylSEQr Bisulfite Conversion Kit (Applied Biosystems), which converts nonmethylated cytosines to uracils. Methyl Primer Express Software (Applied Biosystems, Foster City, CA) was used for designing primers (Supplementary Table S2) for PCR and sequencing. The primers, EF1BSPF1 and EF1BSPR1, were used for amplifying a 295-bp PCR amplicon from the 1,272-bp hEF1α promoter for sequencing. The primers CMVBSPF1C, CMVBSPF1T, and CMVBSPR1 were used for amplifying a 398-bp PCR amplicon from the 780-bp CMV promoter for sequencing.
Supplementary Material
Table S1. Sequence of Moloney murine leukemia virus elements utilized.
Table S2. Primer sequences used in this paper.
Table S3. Summary of promoter methylation analyses in transduced glands and cells.
Acknowledgments
The Division of Intramural Research, National Institute of Dental and Craniofacial Research, National Institutes of Health, provided all support for this research. We thank Gabor Illei for his help with the statistical analysis used for the methylation results.
References
- 1.Greber UF, Suomalainen M, Stidwill RP, Boucke K, Ebersold MW, Helenius A. The role of the nuclear pore complex in adenovirus DNA entry. EMBO J. 1997;16:5998–6007. doi: 10.1093/emboj/16.19.5998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Greber UF, Webster P, Weber J, Helenius A. The role of the adenovirus protease on virus entry into cells. EMBO J. 1996;15:1766–1777. [PMC free article] [PubMed] [Google Scholar]
- 3.Schaack J, Ho WY, Freimuth P, Shenk T. Adenovirus terminal protein mediates both nuclear matrix association and efficient transcription of adenovirus DNA. Genes Dev. 1990;4:1197–1208. doi: 10.1101/gad.4.7.1197. [DOI] [PubMed] [Google Scholar]
- 4.Harui A, Suzuki S, Kochanek S, Mitani K. Frequency and stability of chromosomal integration of adenovirus vectors. J Virol. 1999;73:6141–6146. doi: 10.1128/jvi.73.7.6141-6146.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang Y, Li Q, Ertl HC, Wilson JM. Cellular and humoral immune responses to viral antigens create barriers to lung-directed gene therapy with recombinant adenoviruses. J Virol. 1995;69:2004–2015. doi: 10.1128/jvi.69.4.2004-2015.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Adesanya M, Redman RS, Baum BJ, O’Connell BC. Immediate inflammatory responses to adenovirus-mediated gene transfer in rat salivary glands. Hum Gene Ther. 1998;7:1085–1093. doi: 10.1089/hum.1996.7.9-1085. [DOI] [PubMed] [Google Scholar]
- 7.Jooss K, Chirmule N. Immunity to adenovirus and adeno-associated viral vectors: implications for gene therapy. Gene Ther. 2003;10:955–963. doi: 10.1038/sj.gt.3302037. [DOI] [PubMed] [Google Scholar]
- 8.Springett GM, Moen RC, Anderson S, Blaese RM, Anderson WF. Infection efficiency of T lymphocytes with amphotropic retroviral vectors is cell cycle dependent. J Virol. 1989;63:3865–3869. doi: 10.1128/jvi.63.9.3865-3869.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miller DG, Adam MA, Miller AD. Gene transfer by retrovirus vectors occurs only in cells that are actively replicating at the time of infection. Mol Cell Biol. 1990;10:4239–4242. doi: 10.1128/mcb.10.8.4239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harel J, Rassart E, Jolicoeur P. Cell cycle dependence of synthesis of unintegrated viral DNA in mouse cells newly infected with murine leukemia virus. Virology. 1981;110:202–207. doi: 10.1016/0042-6822(81)90022-2. [DOI] [PubMed] [Google Scholar]
- 11.Hacein-Bey-Abina S, Von Kalle C, Schmidt M, McCormack M, Wulffraat N, Leboulch P, et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- 12.Baum C, Kustikova O, Modlich U, Li Z, Fehse B. Mutagenesis and oncogenesis by chromosomal insertion of gene transfer vectors. Hum Gene Ther. 2006;17:253–263. doi: 10.1089/hum.2006.17.253. [DOI] [PubMed] [Google Scholar]
- 13.Zheng C, Baum BJ, Iadarola MJ, O’Connell BC. Genomic integration and gene expression by a modified adenoviral vector. Nat Biotechnol. 2000;18:176–180. doi: 10.1038/72628. [DOI] [PubMed] [Google Scholar]
- 14.Zheng C, Baum BJ. Long-term expression after infection by the hybrid vector AdLTR-luc is from integrated transgene. Biochem Biophys Res Commun. 2002;291:34–40. doi: 10.1006/bbrc.2002.6401. [DOI] [PubMed] [Google Scholar]
- 15.Zheng C, Wang J, Baum BJ. Integration efficiency of a hybrid adenoretroviral vector. Biochem Biophys Res Commun. 2003;300:115–120. doi: 10.1016/s0006-291x(02)02801-2. [DOI] [PubMed] [Google Scholar]
- 16.Voutetakis A, Bossis I, Kok MR, Zhang W, Wang J, Cotrim AP, et al. Salivary glands as a potential gene transfer target for gene therapeutics of some monogenetic endocrine disorders. J Endocrinol. 2005;185:363–372. doi: 10.1677/joe.1.06171. [DOI] [PubMed] [Google Scholar]
- 17.Kagami H, Atkinson JC, Michalek SH, Handelman B, Yu S, Baum BJ, et al. Repetitive adenovirus administration to the parotid gland: role of immunological barriers and induction of oral tolerance. Hum Gene Ther. 1998;9:305–313. doi: 10.1089/hum.1998.9.3-305. [DOI] [PubMed] [Google Scholar]
- 18.Fu H, Wang L, Lin CM, Singhanial S, Bouhassira EE, Aladjem MI. Preventing gene silencing with human replicators. Nat Biotechnol. 2006;24:572–576. doi: 10.1038/nbt1202. [DOI] [PubMed] [Google Scholar]
- 19.Brooks AR, Harkins RN, Wang P, Qian HS, Liu P, Rubanyi GM. Transcriptional silencing is associated with extensive methylation of the CMV promoter following adenoviral gene delivery to muscle. J Gene Med. 2004;6:395–404. doi: 10.1002/jgm.516. [DOI] [PubMed] [Google Scholar]
- 20.Zirger JM, Liu C, Barcia C, Castro MG, Lowenstein PR. Immune regulation of transgene expression in the brain: B cells regulate an early phase of elimination of transgene expression from adenoviral vectors. Viral Immunol. 2006;19:508–517. doi: 10.1089/vim.2006.19.508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Redman RS. Proliferative activity by cell type in developing rat parotid gland. Anat Rec. 1995;241:529–540. doi: 10.1002/ar.1092410411. [DOI] [PubMed] [Google Scholar]
- 22.Marienfeld U, Haack A, Thalheimer P, Schneider-Rasp S, Brackmann H-H, Poller H. “Autoreplication” of the vector genome in recombinant adenoviral vectors with different E1 region deletions and transgenes. Gene Ther. 1999;6:1101–1113. doi: 10.1038/sj.gt.3300928. [DOI] [PubMed] [Google Scholar]
- 23.Ehrhardt A, Xu H, Kay MA. Episomal persistence of recombinant adenoviral vector genomes during the cell cycle in vivo. J Virol. 2003;77:7689–7695. doi: 10.1128/JVI.77.13.7689-7695.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo H, Seixas-Silva JA, Jr, Epperly MW, Gretton JE, Shin DM, Bar-Sagi D, et al. Prevention of radiation-induced oral cavity mucositis by plasmid/liposome delivery of the human manganese superoxide dismutase (SOD2) transgene. Radiat Res. 2003;159:361–370. doi: 10.1667/0033-7587(2003)159[0361:porioc]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 25.Aapro MS, Cameron DA, Pettengell R, Bohlius J, Crawford J, Ellis M, et al. EORTC guidelines for the use of granulocyte-colony stimulating factor to reduce the incidence of chemotherapy-induced febrile neutropenia in adult patients with lymphomas and solid tumors. Eur J Cancer. 2006;42:2433–2453. doi: 10.1016/j.ejca.2006.05.002. [DOI] [PubMed] [Google Scholar]
- 26.Borges L, Rex KL, Chen JN, Wei P, Kaufman S, Scully S, et al. A protective role for keratinocyte growth factor in a murine model of chemotherapy and radiotherapy-induced mucositis. Int J Radiat Oncol Biol Phys. 2006;66:254–262. doi: 10.1016/j.ijrobp.2006.05.025. [DOI] [PubMed] [Google Scholar]
- 27.Zheng C, Baum BJ. Evaluation of viral and mammalian promoters for use in gene delivery to salivary glands. Mol Ther. 2005;12:528–536. doi: 10.1016/j.ymthe.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 28.Becker TC, Noel RJ, Coats WS, Gomez-Foix AM, Alam T, Gerard RD, et al. Use of recombinant adenovirus for metabolic engineering of mammalian cells. Methods Cell Biol. 1994;43:161–189. doi: 10.1016/s0091-679x(08)60603-2. [DOI] [PubMed] [Google Scholar]
- 29.Brown AM, Rusnock EJ, Sciubba JJ, Baum BJ. Establishment and characterization of an epithelial cell line from the rat submandibular gland. J Oral Pathol Med. 1989;18:206–213. doi: 10.1111/j.1600-0714.1989.tb00764.x. [DOI] [PubMed] [Google Scholar]
- 30.Shirasuna K, Sato M, Miyasaki T. A neoplastic epithelial duct cell line established from an irradiated human salivary gland. Cancer. 1981;48:745–752. doi: 10.1002/1097-0142(19810801)48:3<745::aid-cncr2820480314>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 31.Graham FL, Smiley J, Russel WC, Nairn R. Characteristics of a human cell lines transformed by DNA from human adenovirus type 5. J Gen Virol. 1977;36:59–74. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- 32.Amalfitano A, Chamberlain JS. Isolation and characterization of packaging cell lines that coexpress the adenovirus E1, DNA polymerase, and preterminal proteins: implications for gene therapy. Gene Ther. 1997;4:258–263. doi: 10.1038/sj.gt.3300378. [DOI] [PubMed] [Google Scholar]
- 33.Baum BJ, Wellner RB, Zheng C. Gene transfer to salivary glands. Int Rev Cytol. 2002;213:93–146. doi: 10.1016/s0074-7696(02)13013-0. [DOI] [PubMed] [Google Scholar]
- 34.Sprangers MC, Lakhai W, Koudstaal W, Verhoeven M, Koel BF, Vogels R, et al. Quantifying adenovirus-neutralizing antibodies by luciferase transgene detection: addressing preexisting immunity to vaccine and gene therapy vectors. J Clin Microbiol. 2003;41:5046–5052. doi: 10.1128/JCM.41.11.5046-5052.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kruisbeek AM. In vitro assays for mouse lymphocytic function. In: Coligan JE, Bierer BE, Margulies DH, Shevach EM, Strober W, editors. Current Protocols in Immunology. John Wiley & Sons, Inc; 2004. pp. 3.1.3.1–3.1.3.2. [Google Scholar]
- 36.Hirt B. Mouse cellular DNA enclosed in polyoma viral capsids (pseudovirions) Proc Natl Acad Sci USA. 1967;58:1381–1388. doi: 10.1073/pnas.58.4.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Sequence of Moloney murine leukemia virus elements utilized.
Table S2. Primer sequences used in this paper.
Table S3. Summary of promoter methylation analyses in transduced glands and cells.


