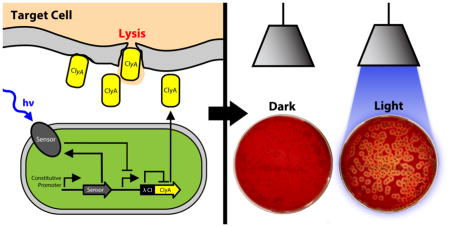
Abstract
By delivering payloads in response to specific exogenous stimuli, smart bacterial therapeutics have the potential to overcome many limitations of conventional therapies, including poor targeting specificity and dosage control in current cancer treatments. Although not yet explored as a trigger for bacterial drug delivery, light is an ideal induction mechanism because it offers fine spatiotemporal control and is easily and safely administered. Using recent advances in optogenetics, we have engineered two strains of Escherichia coli to secrete a potent mammalian cytotoxin in response to blue or red light. The tools in this study demonstrate the initial feasibility of light-activated bacterial therapeutics for applications such as tumor cytolysis, and their modular nature should enable simple substitution of other payloads of interest.
Graphical Abstract
INTRODUCTION
Synthetic biology has the potential to create novel therapeutic strategies to improve the treatment of diseases such as cancer. Conventional cancer treatment regimens involving chemotherapy, radiotherapy, or surgical excision are often limited by low targeting specificity, inadequate dosage control, or damage to healthy tissue, but these obstacles could be overcome by the development of ‘smart’ living therapeutics through synthetic biology approaches1. In particular, bacteria hold promise as potential therapeutic agents for cancer due to the relative ease of engineering genetic circuitry within them, the innate propensity of certain strains to target tumors, and their ability to actively penetrate solid tumors, as opposed to drugs which rely on passive diffusion1,2. Bacteria have been engineered with genetic circuits that allow for tumor cell invasion or therapeutic delivery in response to stimuli such as L-arabinose, hypoxia, cell density, γ-irradiation, and acetylsalicylic acid2.
In contrast to these stimuli, light can enable safe triggering of gene expression with fine spatiotemporal control without reliance on passive diffusion. Fiber optic systems enabling targeted and accurate light delivery into tumors are already used for photodynamic therapy3. Optogenetic control of drug delivery has been demonstrated in mammalian cells with blue light4, but this approach has not yet been demonstrated in bacteria or extended to light inputs of different wavelengths. In this study we exploited bacterial optogenetic components to enable light-activated delivery of a cytolytic payload using E. coli. We achieved blue light-mediated cytolysis of mammalian cells with a common laboratory strain of E. coli and replicated these findings with a minimally-immunogenic strain that is being explored for clinical applications. Since the longer wavelength of red light makes it more advantageous for therapeutic applications of optogenetics, we also constructed a plasmid (pMars) which allows for red light-controlled expression of a gene of interest, and successfully utilized it to control cytolysis. Our results provide proof of concept that optogenetic control of cytotoxicity can be achieved in bacteria with multiple wavelengths of light.
RESULTS AND DISCUSSION
We selected the pore-forming toxin cytolysin A (ClyA) as the payload for our system because it is readily secreted via outer membrane vesicles5 and it has been shown to reduce tumor growth when expressed by bacteria in vivo6. To characterize this toxin, we first codon optimized and synthesized the clyA gene for expression in E. coli and this gene was cloned into the pET26b(+) expression vector with a C-terminal His tag for protein purification (ClyA: BBa_K811000, ClyA-His: BBa_K811002). To verify that the synthesized product can lyse cancer cells, purified ClyA (0.3 nM) was added to cultured SKBR3 cells and cytotoxicity was determined using the commercial Cytotox-Fluor Cytotoxicity Assay (Promega). ClyA exhibited a significant 6-fold increase in cytotoxicity over the negative control vehicle (Figure 1A, p < 0.001) and a 1.2-fold increase in cytotoxicity over the digitonin positive control (p < 0.001), verifying that ClyA can potently lyse cancer cells. For more readily visualizing mammalian cell lysis by ClyA-producing bacteria, the remaining experiments were performed using classical blood agar assays.
Figure 1.
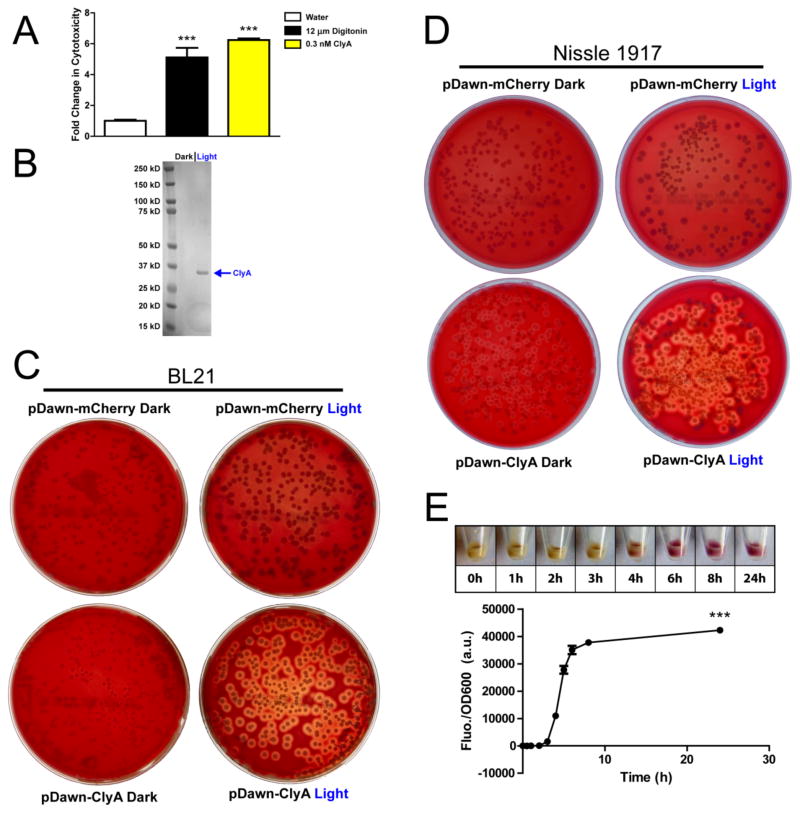
Blue light-activated cytolysis in E. coli BL21 and Nissle 1917 using pDawn. (A) Synthesized and purified ClyA lyses SKBR3 cancer cells in vitro. Purified ClyA (0.3 nM) resulted in a 6-fold increase in cytotoxicity, significant (p < 0.001) over both the negative control vehicle (water) and the positive control (digitonin, 12 μM) (n = 6 samples per group). (B) BL21(pDawn-ClyA) shows specific, blue light-dependent expression of the ~34 kDa ClyA protein. (C) In contrast to BL21(pDawn-mCherry), BL21(pDawn-ClyA) exhibits blue light-dependent cytolysis on blood agar. (D) Blue light-dependent cytolysis on blood agar is seen from Nissle 1917(pDawn-ClyA) but not from Nissle 1917(pDawn-mCherry). (E) Time course for mCherry protein expression from BL21(pDawn-mCherry) using 480 nm light. Light-dependent mCherry expression in BL21 cultures (n = 3 per time point) is significantly higher (p < 0.001) at all illumination times beyond 3 h (asterisks omitted at 4, 5, 6, and 8 hour time points for clarity). All data are presented as mean ± SD.
The pDawn plasmid, a well-characterized optogenetic tool7, was used to achieve blue (480 nm) light-activated gene expression. To obtain light-dependent ClyA expression, the clyA gene was ligated into pDawn while adding an N-terminal His tag (His-ClyA: BBa_K811001), creating the plasmid pDawn-ClyA. E. coli BL21(DE3) cells were transformed with pDawn-ClyA and induced with blue light (or kept in the dark) at 25°C for 24 hours. ClyA was purified and a 34-kDa band was visible only in the blue light condition, verifying light-dependent production of ClyA (Figure 1B). To visualize light-activated cytolysis, we subsequently plated pDawn-ClyA-bearing bacteria onto blood agar plates. To further demonstrate efficacy in a clinically-relevant strain, we also transformed pDawn-ClyA into E. coli Nissle 1917, a nonpathogenic and minimally immunogenic strain which is already used in clinical applications8 and has tumor-targeting capabilities in mice9. By illuminating plates with blue light, we achieved inducible cytotoxicity with both BL21(DE3) and Nissle 1917, as evidenced by the clear zones of lysis surrounding only those colonies which were both transformed with pDawn-ClyA and illuminated with blue-light (Figures 1C,D). When bacteria were transformed instead with the pDawn plasmid containing the non-cytotoxic protein mCherry, no light-dependent cytolysis was observed.
A key advantage of light-activated cytotoxicity is that the amount of protein produced can be finely tuned by controlling the illumination time. In a preliminary demonstration, we switched the output of our pDawn plasmid to mCherry and demonstrated control over total protein production by varying illumination time (Figure 1E). In the future, this tool could be calibrated based on the potency of ClyA to precisely produce the minimum amount of protein needed for cytolysis of a given tissue volume.
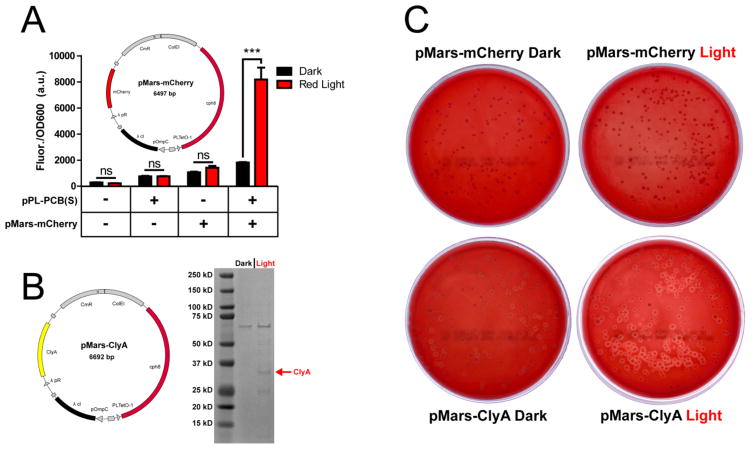
For eventual in vivo applications, the ideal light-activated therapeutic would be triggered by wavelengths longer than 480 nm because blue light is relatively poor at penetrating tissue. We thus sought to construct a new tool for red light-activated payload delivery. We developed a new plasmid backbone, pMars, which uses the well-characterized cph810 red-light sensor to activate gene expression with 650 nm light. A pMars-mCherry plasmid (red-light activated mCherry expression, Figure 2A) or a pMars-ClyA plasmid (red-light activated ClyA expression, Figure 2B) was transformed into E. coli JT2. The pPL-PCB(S) plasmid, which encodes the chromophore synthesis genes necessary to functionalize cph810, was co-transformed with the above plasmids. Red light- and chromophore-dependent mCherry expression was demonstrated with pMars-mCherry (Figure 2A, p < 0.001) and red light-induced expression of ClyA was achieved with pMars-ClyA (Figure 2B). JT2 is a knockout strain that is not optimized for protein expression and purification and we did indeed observe lower purity of His-tagged ClyA recovered from JT2 (Figure 2B) compared to BL21(DE3) (Figure 1B), as evidenced by additional ~25-kDa and ~9-kDa light-dependent bands (hypothesized to be either cleavage-products of the 34-kDa ClyA protein or endogenous JT2 proteins that are co-purified with ClyA) and a ~67-kDa constitutive band (hypothesized to be an endogenous JT2 protein that interacts with the purification column). Nevertheless, the level of ClyA expression achieved with this strain was sufficient for eliciting red light-mediated cytotoxicity, with clear zones of lysis surrounding pMars-ClyA bearing colonies plated onto blood agar (Figure 2C). Given the natively low on:off ratio of the cph8 sensor10, successful light-induced cytolysis is likely due to the strong potency of the ClyA toxin (Figure 1A). In our experiments, the better performance of pDawn relative to pMars is due to the properties of the strains and circuits themselves, because no diffusion barriers were placed between the light sources and the bacterial colonies.
Figure 2.
Red light-activated cytolysis in E. coli JT2 using pMars. (A) The red light-activated system was first tested with pMars-mCherry to ensure functioning of components. Specific, red light-induced expression of mCherry was measured by fluorescence (p < 0.001 between dark and light) and this effect was dependent on the presence of both the chromophore synthesis plasmid pPL-PCB(S) and pMars-mCherry (p > 0.05 between dark and light in first three conditions). (B) Red light-activated system with pMars-ClyA demonstrates weak light-dependent expression of the ClyA protein (~34 kDa). (C) JT2(pMars-ClyA) demonstrate red light-activated cytolysis on blood agar plates, while this effect is not observed for JT2(pMars-mCherry). All data are presented as mean ± SD.
Engineered bacteria hold promise as potential therapeutic agents, and one of the factors that will determine the success of this approach is the choice of induction mechanism for therapeutic delivery. While other triggering methods have been explored previously2, here we have introduced light-activated bacterial drug delivery, which is uniquely suited for achieving precise dosage control not only temporally but also spatially. Optogenetic control of mammalian cell cytotoxicity was demonstrated using two engineered strains of E. coli, including clinically-relevant Nissle 1917. Blue or red light was used to successfully induce the desired cytolytic response, although further optimization of the red-light-activated system will be necessary to improve its dynamic range and reduce its leakiness, especially for in vivo applications. More generally, this approach could be used for light-activated expression of molecular payloads other than ClyA, including cytokines or other cytotoxic agents for specific cancer applications2.
Supplementary Material
Acknowledgments
The authors acknowledge Daphne Ng and Najaf Shah for helpful experimental discussions, Sevile Mannickarottu for providing laboratory space, and the iGEM competition for providing a platform for undergraduate synthetic biology research. The following materials were generously gifted: SKBR3 cells by Matthew Lazzara (University of Pennsylvania); the pDawn plasmid by Andreas Möglich (Humboldt University of Berlin); the pPL-PCB(S) plasmid by J. Clark Lagarias (University of California, Davis); and the strain JT2 by Jeff Tabor (Rice University). This work was supported by the University of Pennsylvania and a National Science Foundation CAREER Award (#CBET-1055231) to CAS. The 2012 Penn iGEM Team wiki can be found at http://2012.igem.org/Team:Penn, and the URL’s for the BioBricks described in this work are as follows: http://parts.igem.org/Part:BBa_K811000, http://parts.igem.org/Part:BBa_K811001, http://parts.igem.org/Part:BBa_K811002.
Footnotes
AUTHOR INFORMATION
MSM and AV conceived and developed the project idea in consultation with JSM and CAS. MSM, AV, PQ, AA, and JYL designed and performed experiments. MSM, AV, JSM, and CAS wrote the manuscript. JSM, MG, and CAS supervised all aspects of the project.
References
- 1.Chen YY, Smolke CD. From DNA to targeted therapeutics: bringing synthetic biology to the clinic. Sci Transl Med. 2011;3:1–5. doi: 10.1126/scitranslmed.3002944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Forbes NS. Engineering the perfect (bacterial) cancer therapy. Nat Rev Cancer. 2010;10:785–94. doi: 10.1038/nrc2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown SB, Brown EA, Walker I. The present and future role of photodynamic therapy in cancer treatment. Lancet Oncology. 2004;4:497–508. doi: 10.1016/S1470-2045(04)01529-3. [DOI] [PubMed] [Google Scholar]
- 4.Ye H, Daoud-El Baba M, Peng RW, Fussenegger M. A synthetic optogenetic transcription device enhances blood-glucose homeostasis in mice. Science. 2011;332:1565–8. doi: 10.1126/science.1203535. [DOI] [PubMed] [Google Scholar]
- 5.Wai SN, Lindmark B, Söderblom T, Takade A, Westermark M, Oscarsson J, Jass J, Richter-Dahlfors A, Mizunoe Y, Uhlin BE. Vesicle-mediated export and assembly of pore-forming oligomers of the enterobacterial ClyA cytotoxin. Cell. 2003;115:25–35. doi: 10.1016/s0092-8674(03)00754-2. [DOI] [PubMed] [Google Scholar]
- 6.Jiang SN, Phan TX, Nam TK, Nguyen VH, Kim HS, Bom HS, Choy HE, Hong Y, Min JJ. Inhibition of tumor growth and metastasis by a combination of Escherichia coli-mediated cytolytic therapy and radiotherapy. Mol Ther. 2010;18:635–42. doi: 10.1038/mt.2009.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ohlendorf R, Vidavski RR, Eldar A, Moffat K, Möglich A. From Dusk till Dawn: One-Plasmid Systems for Light-Regulated Gene Expression. J Mol Biol. 2012;416:534–42. doi: 10.1016/j.jmb.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 8.Schultz M. Clinical use of E. coli Nissle 1917 in inflammatory bowel disease. Inflamm Bowel Dis. 2008;14:1012–8. doi: 10.1002/ibd.20377. [DOI] [PubMed] [Google Scholar]
- 9.Brader P, Stritzker J, Riedl CC, Zanzonico P, Cai S, Burnazi EM, Ghani ER, Hricak H, Szalay AA, Fong Y, Blasberg R. Escherichia coli Nissle 1917 facilitates tumor detection by positron emission tomography and optical imaging. Clin Cancer Res. 2008;14:2295–302. doi: 10.1158/1078-0432.CCR-07-4254. [DOI] [PubMed] [Google Scholar]
- 10.Tabor JJ, Levskaya A, Voigt CA. Multichromatic control of gene expression in Escherichia coli. J Mol Biol. 2011;405:315–24. doi: 10.1016/j.jmb.2010.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.