Abstract
As an approach to the study of rRNA synthesis in Gram-positive bacteria, we characterized the regulation of the Bacillus subtilis rrnB and rrnO rRNA promoters. We conclude that B. subtilis and Escherichia coli use different strategies to control rRNA synthesis. In contrast to E. coli, it appears that the initiating NTP for transcription from B. subtilis rRNA promoters is GTP, promoter strength is determined primarily by the core promoter (−10/−35 region), and changes in promoter activity always correlate with changes in the intracellular GTP concentration. rRNA promoters in B. subtilis appear to be regulated by changes in the initiating NTP pools, but in some growth transitions, changes in rRNA promoter activity are also dependent on relA, which codes for ppGpp synthetase. In contrast to the situation for E. coli where ppGpp decreases rRNA promoter activity by directly inhibiting RNA polymerase, it appears that ppGpp may not inhibit B. subtilis RNA polymerase directly. Rather, increases in the ppGpp concentration might reduce the available GTP pools, thereby modulating rRNA promoter activity indirectly.
Keywords: B. subtilis, GTP concentrations, ppGpp, promoters, rRNA transcription
Introduction
Ribosomal RNA synthesis is the rate-limiting step in ribosome synthesis in both Escherichia coli and Bacillus subtilis (Henkin, 2002; Paul et al, 2004b). rRNA promoters are tightly regulated with nutritional conditions to accommodate the cell's changing translational requirements while preventing overinvestment of biosynthetic resources in energetically costly ribosome synthesis.
Each of the seven rRNA operons in E. coli contains two promoters, rrn P1 and rrn P2. The core (−10/−35) region in rrn P1 promoters is preceded by upstream (UP) elements that increase promoter activity ∼20- to 50-fold by binding the C-terminal domains of the two α subunits of RNA polymerase (RNAP) (Ross et al, 1993; Hirvonen et al, 2001). This region is preceded by binding sites for the transcription factor Fis that increase promoter activity an additional three- to eight-fold, depending on the operon (Hirvonen et al, 2001). Fis does not activate rrn P2 promoters, and UP elements play a much smaller role in P2 than in P1 activity (Murray et al, 2003a).
Most regulation of E. coli rrn P1 and P2 promoter activity is attributable to the effects of small molecule effectors, ppGpp and NTPs, whose concentrations change at specific times in growth and alter rRNA transcription (Murray et al, 2003b; Murray and Gourse, 2004). (ppGpp refers collectively here to both ppGpp and its precursor pppGpp.)
Transcription initiation begins with the interaction of RNAP and the promoter to form an initial closed complex. This step is followed by the formation of several kinetic intermediates, culminating in open complex(es) in which the transcription initiation site is exposed. RNA synthesis starts with the incorporation of incoming NTPs and the transition to an elongation complex capable of processive transcription.
E. coli rRNA promoters form open complexes with extraordinarily short half-lives compared to most other promoters (Gourse, 1988; Murray and Gourse, 2004). We have proposed that this is the kinetic property that makes rRNA promoters sensitive to changing concentrations of their initiating NTP (iNTP); increasing NTP concentration could increase transcription simply by mass action (Gaal et al, 1997; Barker and Gourse, 2001). Alternatively, in theory the iNTP(s) could induce a conformational change in RNAP to facilitate transcription (Lew and Gralla, 2004). In any case, increasing concentrations of the iNTP directly stimulate E. coli rrn P1 and rrn P2 promoter activity in vitro and in vivo (Murray et al, 2003b; Murray and Gourse, 2004).
ppGpp is made in response to amino-acid starvation and some other nutritional stresses (Cashel et al, 1996). ppGpp binds directly to RNAP and increases the rate of open complex collapse at all promoters (Barker et al, 2001). We have proposed that rRNA transcription is specifically inhibited by ppGpp, at least in part, because this step is rate determining for rRNA promoters (Barker et al, 2001). In support of this model, studies on a large number of mutant rrnB P1 promoters indicate that there is a strict correlation between formation of a short-lived open complex and effects of ppGpp and the iNTP on transcription in vitro and in vivo (Barker, 2001; Barker et al, 2001; Barker and Gourse, 2001).
It has also been proposed that ppGpp competes directly with iNTP binding (Jores and Wagner, 2003). The recent X-ray structure of ppGpp in complex with RNAP suggests that ppGpp binds close to, but not overlapping, the catalytic center (Artsimovitch et al, 2004), but direct effects of ppGpp on NTP binding have not been ruled out. There may also be cases where ppGpp inhibits transcription by a mechanism different from effects on open complex collapse or competition for NTP addition (Potrykus et al, 2002).
Here we address whether bacteria evolutionarily distant from E. coli, such as the spore-forming Gram-positive bacterium B. subtilis, use strategies similar to E. coli to regulate rRNA synthesis. B. subtilis rRNA synthesis has been studied in the past (e.g. Testa and Rudner, 1975; Deneer and Spiegelman, 1987; Wellington and Spiegelman, 1993; Henkin, 2002), but the detailed molecular mechanisms responsible for regulation remain unclear. B. subtilis contains 10 rrn operons (Henkin, 2002). Six operons appear to contain tandem P1–P2 promoters, and four only a single promoter. When the B. subtilis rrnB P1 and P2 promoters were measured in E. coli, rrn P2 activity changed with growth rate more than rrn P1 activity (Deneer and Spiegelman, 1987).
We report here the properties and regulation of the B. subtilis rrnB and rrnO promoters in their natural host. We demonstrate that the rrn P1 promoters display more pronounced changes with growth rate and phase than the rrn P2 promoters, and that DNA sequences upstream of the core promoters contribute much less to promoter activity than in E. coli rRNA promoters. Transcription from these B. subtilis promoters initiates with GTP and is regulated by changes in GTP and ppGpp concentrations in vivo. However, in contrast to the situation in E. coli, ppGpp appears to regulate rRNA promoter activity indirectly by affecting GTP pools.
Results
B. subtilis rRNA promoters initiate with GTP
As a first step in studying B. subtilis rRNA transcription, we constructed lacZ fusions to the P1 and P2 promoters from two rRNA operons, rrnB and rrnO, and from a control promoter, Pveg, that is expressed constitutively during vegetative growth (Fukushima et al, 2003). rrnB was chosen because it had been investigated extensively previously (Deneer and Spiegelman, 1987; Wellington and Spiegelman, 1993). rrnO was chosen as representative of the rRNA promoter class differing in sequence from rrnB. The fusions were integrated in single copy at the B. subtilis amyE locus (see Supplementary material).
The DNA sequences of rrnB and rrnO P1 and P2 and Pveg are shown in Figure 1A. Primer extension from RNAs synthesized in vivo identified start sites 8 bp downstream of the presumptive rrn P1 –10 hexamers and 7 bp downstream of the presumptive rrn P2 –10 hexamers, either in constructs in which the two promoters were in their natural tandem configuration or in which they were separated from each other (Figure 1B–D and data not shown). All four rrn promoters initiated with GTP. The veg transcript initiated with ATP (data not shown), as reported previously (e.g. Fukushima et al, 2003). For experiments described below, rrn P1 and Pveg promoters were created with A and G at position +1 (nontemplate strand), respectively, and their transcription start sites were verified by primer extension (data not shown). When appropriate, the identity of the +1 position is indicated in the promoter name (e.g. rrnB P1+1A).
Figure 1.
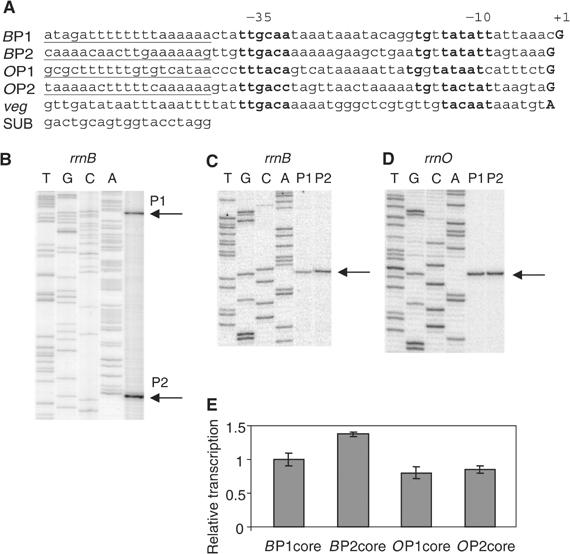
(A) Sequences of B. subtilis promoter constructs used in this study. Putative −10 and −35 hexamers and the +1 positions are in bold. Putative UP elements are underlined. Core promoter constructs contain native sequence from 3 bp upstream of the −35 element (−39 in rrnB P1) to +1. The arbitrary triplet TCT was inserted adjacent to the +1 position, followed by the HindIII site, to avoid positioning an A next to +1. In indicated core promoter constructs, the SUB sequence (Rao et al, 1994) was substituted for the same length of native sequence upstream of the −35 element. The veg promoter is described in the text. (B) Primer extension mapping of start sites from cells grown in rich medium containing a B. subtilis rrnB P1–P2 tandem promoter construct (RLG6930; contains rrnB sequence from −248 upstream of BP1 to +8 of BP2). (C, D) Primer extension mapping of start sites from isolated B. subtilis rrnB and rrnO P1 and P2 promoter constructs (−39 to +1 for BP1, −38 to +1 for BP2, OP1, and OP2 (RLG7554, RLG7553, RLG7369, and RLG7370, respectively). Since the isolated P1 and P2 promoters make the same RNA in these fusions, the primer extension products migrate to the same position in the gel. Arrows indicate start sites. Sequencing ladders are shown for P1 promoters only. (E) Relative activities of rrnB and rrnO core promoters. Cells containing the core promoter constructs used in (C, D) were grown concurrently in LB to OD600 ∼0.3, and promoter activities were measured by primer extension, normalized to cell density and to the same RNA recovery marker (see Materials and methods). Activities are in arbitrary units, relative to the rrnB P1 core promoter.
Since the rrnB and rrnO P1 and P2 core promoter constructs made the exact same RNA, their activities could be compared directly by quantitative primer extension (Materials and methods). The four core promoters had similar activities in LB medium (Figure 1E). These results, in conjunction with the small effects of their upstream regions (see below), indicate that rrnB and rrnO promoter activities are similar in rich medium. This result does not support the conclusion of an earlier study employing lacZ fusions that the rrnB promoters are much weaker than the rrnO promoters (Okamoto and Vold, 1992; see below and Materials and methods).
Sequences upstream of the B. subtilis rrn P1 and P2 core promoters increase transcription much less than in E. coli
The RNAP αCTD(s) interact with sequences upstream of the core promoter in many B. subtilis operons, increasing transcription (e.g. Fredrick et al, 1995; Helmann, 1995; Meijer and Salas, 2004). We initially used promoter-lacZ fusions with different upstream end points, measuring β-galactosidase activities to estimate the effects of upstream sequences in rRNA operons. However, we found that β-galactosidase activities consistently underestimated rRNA promoter activities at the highest growth rates (data not shown; see Materials and methods). Therefore, we used quantitative primer extension to measure promoter activities directly from the same fusions (Figure 2A). In contrast to the situation with E. coli rrn P1 promoters, the native sequence upstream of the −35/−10 region of rrnB P1 increased expression only two- to three-fold (Figures 1A, 2A, and B) relative to constructs containing plasmid-derived sequences or the G+C-rich ‘SUB' sequence (which does not bind the E. coli αCTD; Rao et al, 1994) fused to the core promoter. Only small increases were observed when the native rrnB P1 sequence was extended as far upstream as −352 (Figure 2B). Even smaller effects of upstream sequences (<2-fold) were observed for rrnB P2, rrnO P1, and rrnO P2 (Figure 2A–C). Consistent with the ∼2-fold effect of the B. subtilis rrnB P1 upstream sequence on promoter activity in vivo, we detected only slight stimulation (<2-fold) of this promoter by its upstream sequence with B. subtilis RNAP in vitro (Figure 2D).
Figure 2.

Contribution of sequences upstream of the −35 element to B. subtilis rRNA promoter activity. (A) Representative primer extension bands from RNAs synthesized in cells grown in LB medium from B. subtilis rrnB P1 and rrnB P2 (promoter end points: BP1 core: −39 to +1, RLG7554; BP1 SUB: −39 to +1, RLG7372; BP1 UP: −58 to +1, RLG7373; BP1 long: −352 to +1, RLG7584; BP2 SUB: −38 to +1, RLG7374; BP2 UP: −57 to +1, RLG7375). The test (T) and recovery marker (RM) reverse transcripts are indicated (see also Materials and methods). (B) Effects of upstream sequences on transcription from BP1 and BP2 (constructs described in panel A, normalized to the activity of BP1 SUB). (C) Effects of upstream sequences on transcription from rrnO P1 and rrnO P2 (OP1 core: −38 to +10, RLG7027; OP1 UP: −77 to +10, RLG7028; OP1 long: −227 to +10, RLG7030; OP2 core: −38 to +10, RLG6937; OP2 UP: −77 to +10, RLG7029). Promoter activities are normalized to OP1 core. (D, E) Effect of upstream sequences on in vitro transcription from (D) B. subtilis rrnB P1 using B. subtilis (B.s.) RNAP and supercoiled templates containing B.s. BP1 SUB (−39 to +1, pRLG7599) or B.s. BP1 UP (−58 to +1, pRLG7598) or (E) E. coli rrnB P1 using E. coli (E.c.) RNAP and supercoiled templates containing E.c. BP1 SUB (−41 to +50, pRLG2230) or E.c. BP1 UP (−66 to +50, pRLG6214). Promoter activities are from quantitation (phosphorimager units) of the in vitro transcripts shown at the top of the panels. The fold effect of the B. subtilis rrn P1 upstream sequence varied from 1.3- to 1.6-fold, while the fold effect of the E. coli rrn P1 upstream sequence varied from 15- to 30-fold. (F) Effect of E. coli rrnB P1 UP element on transcription in vivo in E. coli. Transcripts were measured by primer extension from RNAs transcribed from E. coli rrnB P1 SUB (−39 to +50, RLG3097) and E. coli rrnB P1 UP (−66 to +50, RLG3074) constructs in single copy in the E. coli chromosome. Activities are normalized to the core promoter construct. (G) Effect of a B. subtilis hag promoter UP element on transcription in B. subtilis using the same methods as in (A–C). B. subtilis promoters (hag core, −43 to +4, RLG7391; hag promoter with upstream sequences=hag UP, −96 to +4, RLG7392) were integrated in the B. subtilis chromosome. The activity of hag UP is normalized to hag core. For the experiments in panels A–C and F, total RNA was extracted in early exponential phase (OD600 ∼0.3) from cells grown in LB for at least four doublings. In (G), RNA was extracted from cells in late exponential phase (OD600∼2.0) when the hag promoter is most active.
As controls to verify that these methods would likely have detected stimulation by rrn upstream sequences, we measured the effects of previously characterized UP elements using purified RNAP in vitro and by primer extension in vivo. The E. coli rrnB P1 UP element stimulated its core promoter ∼30-fold using E. coli RNAP in vitro (Figure 2E) and ∼45-fold by primer extension with RNA extracted from E. coli cells (Figure 2F), consistent with previous observations (Rao et al, 1994). Furthermore, our methods detected a 14-fold stimulatory effect of the B. subtilis hag UP element in vivo (Figure 2G), in good agreement with a previous report (Fredrick et al, 1995).
Taken together, these results suggest that classical transcription activators and UP elements do not make large contributions to the activities of B. subtilis rRNA promoters. At least under the conditions tested, B. subtilis rrnB and rrnO promoter strength derives primarily from the core promoter.
rrn P1 promoters display more pronounced changes in activity with changing nutritional conditions than rrn P2 promoters
E. coli rRNA promoter activity increases proportionally with growth rate (Paul et al, 2004b). Regulation of B. subtilis rRNA promoter activity was estimated by growing cells at different steady-state growth rates (determined by the nutritional composition of the medium) and measuring transcription from rrnB P1 and P2 promoter constructs by primer extension. Since the constructs made the exact same RNA transcript, differences in transcription reflected only differences in promoter activity and not some potential difference in RNA half-life (see also Supplementary material). Each promoter activity is plotted as a function of growth rate, normalized at the lowest growth rate, to facilitate visualization of differences in regulation (Figure 3). Contrary to the conclusion reached previously (from measurements of B. subtilis rrnB promoters in E. coli; Deneer and Spiegelman, 1987), rrnB P1 increased much more with growth rate than rrnB P2. Similar results were obtained for the rrnO P1 versus rrnO P2 promoters, and for the rrnB P1 versus rrnB P2 promoters in their natural tandem configuration (data not shown). Thus, regulation of rrn promoter activities in a heterologous host may not mimic the native situation.
Figure 3.
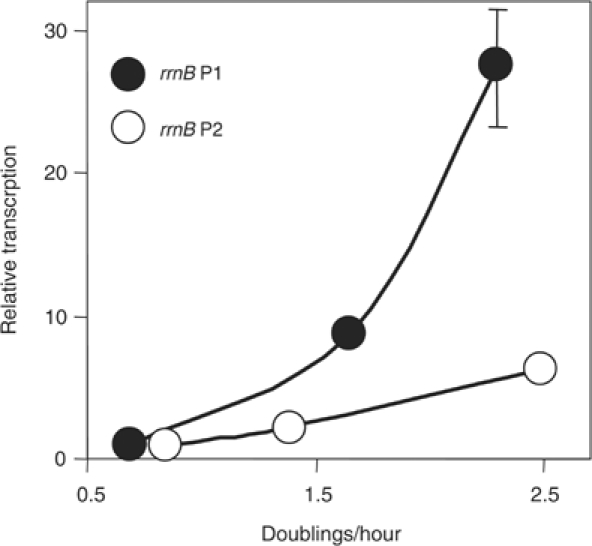
Growth rate-dependent control of B. subtilis rRNA promoters (rrnB P1: −39 to +1, RLG7554; rrnB P2: −38 to +1, RLG7553). Promoter activity (arbitrary units) was measured by primer extension from RNA extracted from cells grown in different media. Slowest to fastest growth rate: (i) MOPS, 1% glucose, phenylalanine and tryptophan; (ii) MOPS, 1% glucose, 20 amino acids; (iii) LB. To facilitate comparison of slopes, promoter activities were normalized to the activity of its own promoter at the lowest growth rate.
Although the full-length P1 promoters and full-length P2 promoters account for similar amounts of rRNA transcription in rich medium (Figure 2B), at slow growth rates the P2 promoters are less inhibited than the P1 promoters and thus account for the majority of rRNA synthesis (as in E. coli; Murray et al, 2003a; Murray and Gourse, 2004). The growth rate-dependence curves derived from measuring total RNA:protein ratios (data not shown) were very similar to those derived from the analysis of the individual promoters, suggesting that regulation of the rrnB and rrnO promoters is typical for B. subtilis rRNA operons. The rrnB P2 and rrnO P2 promoters also exhibited much less pronounced changes in activity than their respective P1 promoters during outgrowth from, and entry into, stationary phase (data not shown), further suggesting that B. subtilis rrn P1 promoters are regulated more than their respective P2 promoters.
B. subtilis rrn P1 promoters require high concentrations of their iNTP but appear insensitive to ppGpp in vitro
We next investigated potential mechanisms for regulation of B. subtilis rRNA promoter activity. Changing concentrations of the iNTP and ppGpp account for much of the regulation of rRNA transcription in E. coli (Murray et al, 2003b; Murray and Gourse, 2004). In support of this conclusion, E. coli rRNA promoters require higher concentrations of the iNTP for transcription than other promoters in vitro, and ppGpp moderately but specifically inhibits transcription from rRNA promoters in vitro (Barker et al, 2001). Therefore, we measured the effects of iNTP and ppGpp concentrations in vitro on the more regulated of the two B. subtilis rrn promoters, rrn P1, using purified B. subtilis RNAP (Figures 4 and 5).
Figure 4.
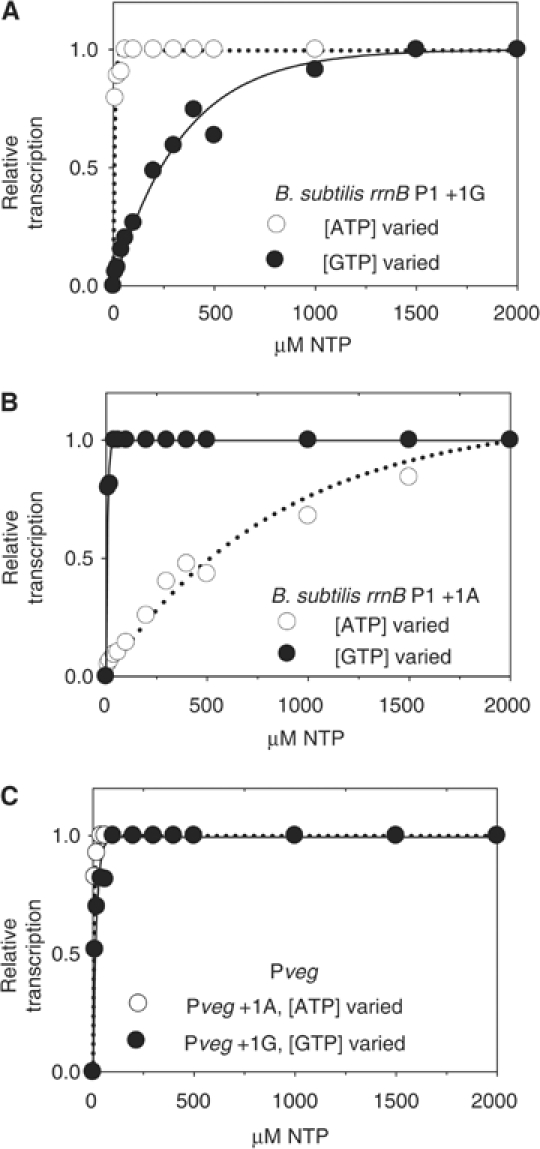
Effects of changing NTP concentration on rrnB P1 promoter activity in vitro. B. subtilis promoters on supercoiled templates were transcribed with B. subtilis RNAP and normalized to transcription at the highest NTP concentration (2000 μM). Transcription (arbitrary units) from (A) rrnB P1+1G (pRLG7596), (B) rrnB P1+1A (pRLG7597), and (C) Pveg+1A (pRLG7595) or +1G (pRLG7558) at varying ATP or GTP concentration.
Figure 5.
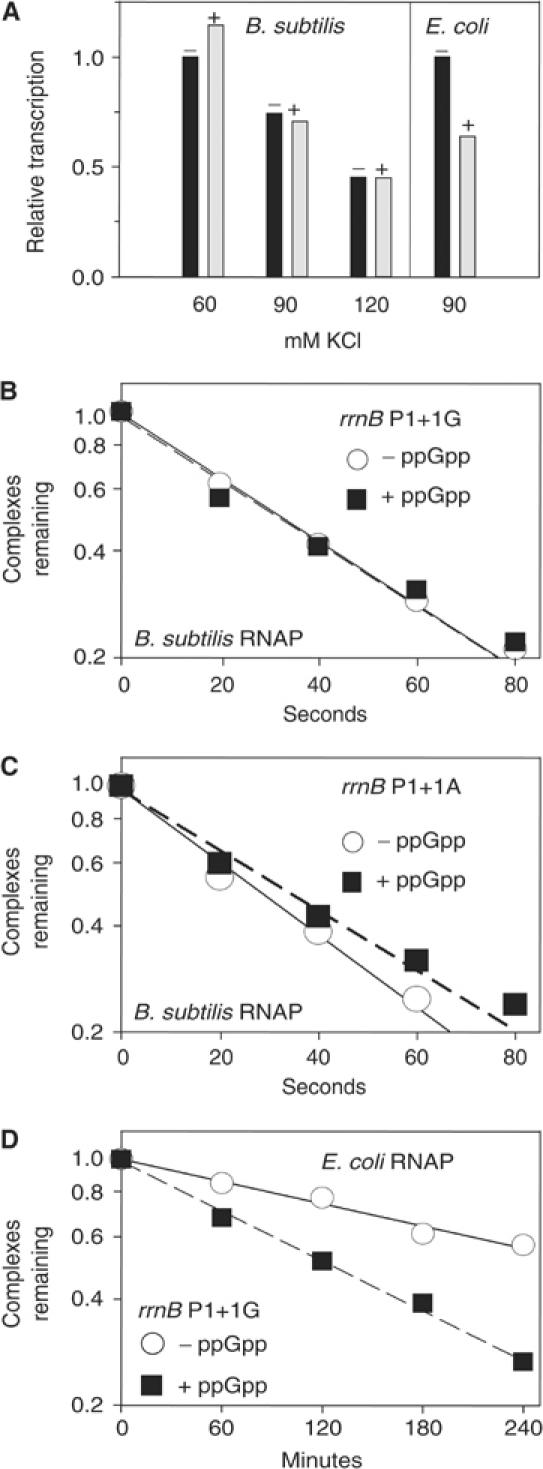
Effect of ppGpp on transcription by B. subtilis and E. coli RNAP in vitro. (A) Single round transcription from B. subtilis rrnB P1+1G (pRLG7596) with B. subtilis RNAP and from E. coli rrnB P1 (pRLG6555) with E. coli RNAP. (+), ppGpp added at 0.5 mM. (B–D) Effect of ppGpp on open complex lifetime. Y-axis, fraction of competitor-resistant complexes. Representative experiments are shown; absolute values of the half-lives varied by only ∼5% in different experiments. (B) B. subtilis rrnB P1+1G (pRLG7596) with B. subtilis RNAP. (C) B. subtilis rrnB P1+1A (pRLG7597) with B. subtilis RNAP. (D) B. subtilis rrnB P1+1G (pRLG7596) with E. coli RNAP.
Relative to the control, Pveg+1G (Figure 4C), rrnB P1+1G required high levels of GTP, but not ATP, for maximal transcription (Figure 4A), and relative to Pveg+1A (Figure 4C), rrnB P1+1A required high levels of ATP and not GTP (Figure 4B). rrnO P1+1G and +1A displayed NTP dependences similar to those of rrnB P1 (data not shown). Thus, like E. coli rRNA promoters, B. subtilis rRNA promoters displayed iNTP dependences in vitro characteristic of regulation of promoter activity by changes in NTP concentrations in vivo. The identity of the +1 position in B. subtilis rrn P1 promoters was important for specifying the NTP to which the promoter responded, but it was not responsible for the high iNTP concentration requirement (see Discussion).
Complexes containing B. subtilis rRNA promoters and E. coli RNAP are inhibited by ppGpp in vitro (Wellington and Spiegelman, 1993). To our surprise, rrnB P1+1G (or rrnB P1+1A, data not shown) with B. subtilis RNAP was not inhibited by ppGpp in vitro using single round or multiple round transcription assays at any of several salt concentrations (Figure 5A and data not shown). ppGpp also failed to inhibit B. subtilis RNAP transcribing a tandem B. subtilis rrnB P1 and P2 construct (data not shown). The same ppGpp preparation inhibited E. coli rrnB P1 ∼2-fold using E. coli RNAP (Figure 5A and data not shown).
Inhibition of E. coli rRNA promoter activity by ppGpp in vitro requires using solution conditions in which the half-life of the promoter complex is rate determining. However, ppGpp binds to E. coli RNAP and decreases the half-lives of all open complexes, even at promoters where this lifetime is not rate determining for transcription (Barker et al, 2001). Hence, the ability of ppGpp to interact with RNAP can be detected from the effects on this lifetime, even under solution conditions where inhibition of transcription is not observed.
ppGpp did not decrease the half-lives of open complexes formed on either B. subtilis rrnB P1+1G or rrnB P1+1A (Figure 5B and C), consistent with its lack of an effect on transcription from these promoters. However, ppGpp did reduce the lifetimes of complexes containing E. coli RNAP, either using E. coli rRNA promoters (Barker et al, 2001) or using B. subtilis rrnB P1+1G and the same solution conditions (Figure 5D; see also Wellington and Spiegelman, 1993). We tentatively conclude that purified B. subtilis RNAP is either insensitive, or at least not as sensitive, to ppGpp as E. coli RNAP. Interestingly, the open complexes formed using B. subtilis rrnB P1+1G (or rrnB P1+1A, data not shown) with E. coli RNAP were longer-lived than those formed with B. subtilis RNAP.
rrnB P1 promoter can ‘sense' changes in NTP concentrations in vivo
We next examined rRNA promoter responses to three different kinds of downshifts (Figure 6) and an upshift (Figure 7). Similar approaches were instrumental in elucidating the differential roles of changing concentrations of NTPs and ppGpp in regulation of rRNA transcription in E. coli (Murray et al, 2003b; Murray and Gourse, 2004).
Figure 6.
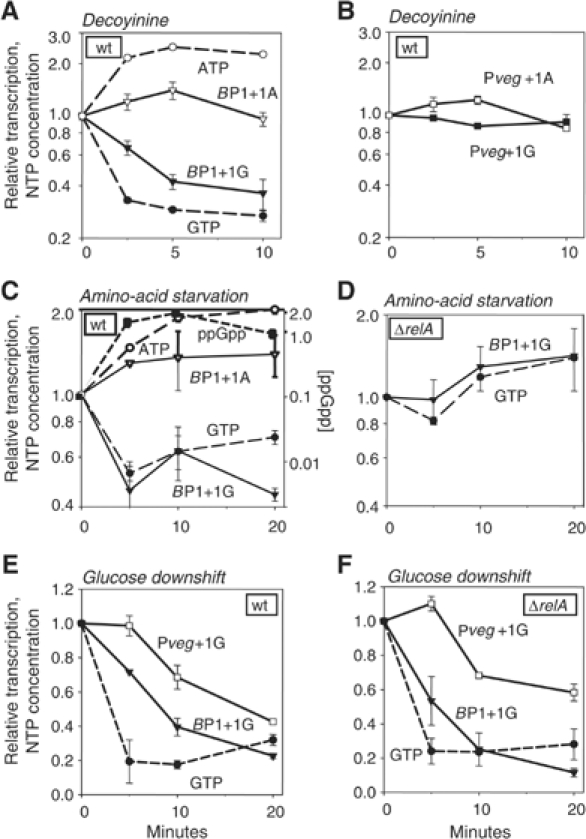
Correlation between GTP concentration and B. subtilis rrnB P1 promoter activity following three kinds of downshifts. NTP concentrations (dashed lines) and promoter activities (solid lines) are normalized to 1 at time 0. The GTP concentration was 45, 18, and 46% of the ATP concentration at time 0 in panels A, C, and E, respectively. Promoter activities were measured by primer extension from a wild-type strain: rrnB P1+1G (RLG7554), rrnB P1+1A (RLG7585), Pveg+1G (RLG7555), Pveg+1A (RLG7376), or from a ΔrelA strain: rrnB P1+1G (RLG7580), Pveg+1G (RLG7581). (A, B) Changes in promoter activity and NTP concentration after decoyinine addition. Cells were grown in a medium containing MOPS, 1% glucose, and 20 amino acids (50 μg/ml each). Decoyinine (final concentration 0.5 mg/ml) was added to exponentially growing cells at time 0 (OD600∼0.3). (C) Effect of amino-acid starvation on B. subtilis rrnB P1 promoter activity. Cells were grown in a medium containing MOPS, 0.4% glucose, and six amino acids (FILMVW). Serine hydroxamate (1.5 mg/ml final concentration) was added to exponentially growing cells at time 0 (OD600∼0.25). The ppGpp concentration is presented relative to the GTP concentration. Note the different scale for ppGpp. (D) Effect of amino-acid starvation on B. subtilis rrnB P1 promoter activity in a ΔrelA strain. Conditions are as in (C). (E) Effect of glucose deprivation on B. subtilis rrnB P1 promoter activity. Cells were grown in a medium containing MOPS, 0.2% glucose, and 20 amino acids. α-Methyl glucoside (final concentration 2%) was added to exponentially growing cells at time 0 (OD600∼0.3). The decrease in ATP concentration was ∼2-fold by 5 min (data not shown). (F) Effect of glucose deprivation on B. subtilis rrnB P1 promoter activity in a ΔrelA strain. Conditions are as in (E).
Figure 7.
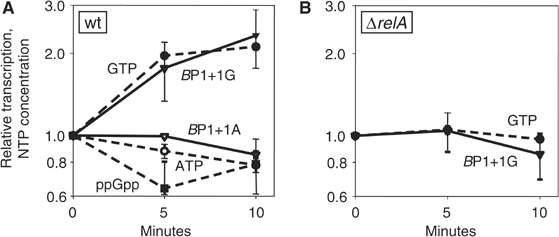
Effect of amino-acid upshift on B. subtilis rrnB P1 promoter activity. Cells were grown in MOPS, 0.2% glucose, and six amino acids (FILMVW, 50 μg/ml each). The remaining 14 amino acids were added to exponentially growing cells at time 0 (OD600∼0.25) to a final concentration of 50 μg/ml each. Promoter activity was measured by primer extension from rrnB P1+1G (RLG7554) and rrnB P1+1A (RLG7585) in (A) and from rrnB P1+1G (RLG7580) in (B) (ΔrelA). NTP concentrations are normalized to 1.0 at time zero. The GTP concentration was ∼16% of the ATP concentration and ppGpp was ∼10% of the GTP concentration at time 0.
In order to determine whether regulation by the concentration of the iNTP in vivo is direct, we used the drug decoyinine to uncouple cellular GTP and ATP concentrations. Decoyinine inhibits GMP synthetase, thereby decreasing GTP but not ATP (Mitani et al, 1977). After decoyinine treatment, the activity of rrnB P1+1G (Figure 6A), but not Pveg+1G (Figure 6B), decreased ∼3-fold within the first 5 min, correlating with the decrease in GTP concentration (Figure 6A). The observed rate of decrease in rrnB P1+1G activity was close to the maximal possible rate, since the half-life of the reporter RNA was ∼4 min when measured by primer extension following rifampicin treatment (data not shown; see Materials and methods).
The ATP concentration did not decrease following decoyinine treatment (in fact, it increased slightly; see also Lopez et al, 1979). rrnB P1+1A (and Pveg+1A) were not inhibited by decoyinine, a qualitatively different response from that of rrnB P1+1G (Figure 6A and B). Thus, a G at the +1 position, as well as some property specific to rRNA promoters, is required for the response of rrn P1 promoters to the decrease in GTP concentration. This result is consistent with the model that B. subtilis rRNA promoters can be controlled directly by the cellular concentration of their iNTP, GTP.
ppGpp is required for stringent control of B. subtilis rrn P1 promoter activity, but only when the promoter initiates with GTP
The in vitro experiments described above suggested that the mechanism of inhibition of transcription by ppGpp might be different in B. subtilis than in E. coli. However, necessary components might have been missing from the in vitro reactions, and/or the solution conditions might have been inappropriate for detecting effects of ppGpp on B. subtilis RNAP. Therefore, we investigated inhibition of rRNA promoter activity following amino-acid starvation in vivo (stringent control), when the regulator responsible for controlling rRNA promoter activity in E. coli is ppGpp and not the iNTP (Cashel et al, 1996; Murray et al, 2003b; Paul et al, 2004a). Serine hydroxamate (SHX), a competitive inhibitor of aminoacylation of serine tRNA, was used to induce a stringent response. SHX addition resulted in a ⩾10-fold increase in ppGpp, an ∼2-fold decrease in GTP, and a corresponding decrease in rrnB P1+1G activity (Figure 6C) (for technical reasons, the magnitude of this decrease in rRNA promoter activity following SHX treatment was somewhat smaller than reported for E. coli; Paul et al, 2004a). RelA is the sole ppGpp synthetase in B. subtilis (Wendrich and Marahiel, 1997). There was no decrease in GTP levels or rrnB P1+1G activity in a ΔrelA mutant (Figure 6D). Thus, ppGpp was required for the observed inhibition of rrnB P1+G.
Although ppGpp was responsible for the decrease in rrnB P1+1G activity after amino-acid starvation, rrnB P1+1A activity was not inhibited and even increased slightly, correlating with a slight increase in ATP concentration. Thus, unlike the situation in E. coli, where rRNA promoters are inhibited by ppGpp irrespective of the identity of the iNTP (Murray et al, 2003b), B. subtilis rRNA promoters appear to require +1G to respond to ppGpp, suggesting that ppGpp's effect might be indirect.
The concentration of the iNTP and not ppGpp controls rrn P1 promoter activity during a carbon source downshift in B. subtilis
B. subtilis RNAP was not affected by ppGpp in vitro, and B. subtilis rrnB P1+1A activity was not inhibited by ppGpp following amino-acid starvation in vivo. Furthermore, GTP is consumed in the biosynthesis of ppGpp (Cashel et al, 1996), and it was reported previously that ppGpp decreases GTP levels in B. subtilis by inhibiting IMP dehydrogenase (Lopez et al, 1981). Together, these results suggested that ppGpp might elicit its effect on B. subtilis rRNA transcription indirectly by reducing GTP concentrations. To investigate this model further, we studied the effect of glucose deprivation in B. subtilis, another condition where increases in ppGpp concentrations rather than decreases in NTP concentrations directly regulated rRNA promoter activity in E. coli (Murray et al, 2003b).
Addition of α-methyl glucoside, a competitive inhibitor of glucose uptake, induces synthesis of ppGpp in E. coli, while NTPs remain relatively constant (Murray et al, 2003b). In B. subtilis, however, we did not detect an increase in ppGpp concentrations under the conditions tested (minimal medium supplemented with glucose and 20 amino acids); ppGpp concentrations remained below detection throughout the time course of the experiment (data not shown). In contrast to the situation in E. coli, GTP concentrations dropped ∼5-fold (Figure 6E). At the same time, rrnB P1+1G activity decreased by about the same magnitude, whereas the activity of the control promoter, Pveg+1G, decreased less, and this decrease was delayed (Figure 6E). rrnB P1+1A and Pveg+1A decreased with kinetics similar to their respective +1G versions, correlating with a two-fold decrease in ATP concentration (data not shown).
Since we were not able to detect ppGpp under these conditions, we repeated the experiment in a ΔrelA strain to confirm that a potential increase in ppGpp levels was not responsible for the observed decrease in rRNA promoter activity. The absence of relA did not affect rrnB P1+1G promoter behavior following α-methyl glucoside treatment (Figure 6F). We conclude that B. subtilis rRNA promoter activity correlates with changes in GTP concentration during mild carbon deprivation, and that under these conditions ppGpp has no direct or indirect effect on rrnB P1+1G promoter activity.
Decreasing the ppGpp concentration during an upshift appears to increase rRNA promoter activity indirectly by increasing the GTP concentration
We also addressed whether ppGpp's effects on B. subtilis rRNA promoters might be indirect during a nutritional upshift. In E. coli, decreases in ppGpp concentration account for increases in rRNA promoter activity under these conditions, independent of the identity of the +1 position (Murray et al, 2003b; data not shown). In B. subtilis, ppGpp concentration decreased following amino-acid upshift, and GTP, but not ATP, increased two-fold (Figure 7A; see also Lopez et al, 1981). Corresponding with the concentrations of GTP and ATP, rrnB P1+1G activity increased two-fold, but rrnB P1+1A did not (Figure 7A). The increase in GTP concentration and the corresponding increase in rrnB P1+1G promoter activity were dependent on the decrease in ppGpp concentration, since the effects were abolished in a ΔrelA strain (Figure 7B). These results further support a model in which increasing GTP concentrations directly stimulate rrnB P1+1G promoter activity, and the effect of ppGpp on rRNA promoter activity is indirect.
Discussion
B. subtilis and E. coli appear to use different strategies to control rRNA synthesis
We systematically examined the properties and regulation of promoters from two of the 10 B. subtilis rRNA operons. Our primary conclusions are as follows: (i) B. subtilis rrn core promoters are intrinsically strong; that is, upstream sequences contribute much less to rRNA promoter activity than in E. coli rRNA promoters. (ii) Regulation of B. subtilis rRNA transcription can occur from direct effects of changes in the concentration of GTP, the initiating nucleotide, on rRNA promoter activity. (iii) When ppGpp inhibits B. subtilis rRNA promoter activity, it may do so indirectly by reducing GTP pools. Thus, although B. subtilis and E. coli rRNA promoters employ the same small regulatory molecules, they appear to use different strategies to control rRNA synthesis. A schematic diagram (undoubtedly oversimplified) emphasizing these differences is shown in Figure 8 and discussed further below. Characterization of the promoters from the other eight operons will be required to confirm that these characteristics are typical of B. subtilis rRNA promoters. In this context, we note that additional rRNA operons have been studied by R Rudner (City University of NY, personal communication).
Figure 8.
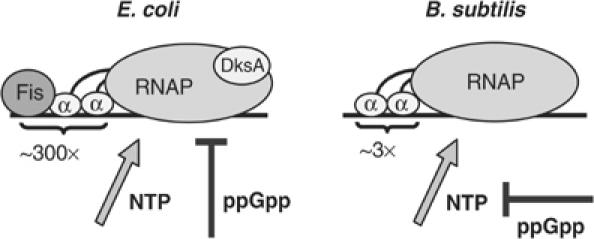
Schematic diagram illustrating mechanisms contributing to rrn P1 promoter activity in E. coli versus B. subtilis. The transcription factor Fis and RNAP αCTD binding to UP element DNA account for the unusually high activity of rrn P1 promoters from E. coli, but not B. subtilis. Changing NTP and ppGpp concentrations regulate rRNA promoter activities in both bacteria, but in B. subtilis ppGpp may inhibit rRNA transcription indirectly by reducing GTP levels. For the sake of simplicity (and since its effect on B. subtilis rRNA promoters was not examined), H-NS is not pictured.
Role of upstream sequences in B. subtilis rRNA promoters
Stimulation of rrn core promoter activity by upstream sequences in B. subtilis is moderate compared to that in E. coli. No fis gene was apparent from examination of the B. subtilis genome sequence, consistent with the lack of evidence for an activator (Figure 2). Most or all of the modest effect of the rrn upstream sequences could derive from αCTD–UP element interactions. UP elements are common in B. subtilis promoters (Fredrick et al, 1995; Helmann, 1995; Meijer and Salas, 2004).
Binding of B. subtilis RNAP to rrn core promoters apparently is efficient enough that αCTD binding has relatively small additional impact on recruitment of RNAP to the promoter. However, rrn UP elements might stimulate weaker core promoters if fused as chimeric constructs. B. subtilis rRNA core promoters contain −10 and −35 hexamers with excellent matches to the proposed consensus sequences for recognition by regions 4.2 and 2.3–2.4 of the σ subunit of RNAP, and in contrast to E. coli rRNA core promoters, they also contain sequences characteristic of extended −10 elements, have A+T-rich sequences between the −10 hexamer and the transcription start site, and some have consensus −10/−35 spacer lengths (17 bp). These properties likely account for the intrinsic strength of B. subtilis rRNA promoters.
GTP sensing by rRNA promoters
Our data suggest that B. subtilis rRNA promoters are regulated directly by the concentration of the iNTP, namely GTP (Figure 8). Changes in GTP concentrations have also been reported to regulate gene expression in several other systems in B. subtilis, but by completely different mechanisms than that described here (Ochi et al, 1982; Ratnayake-Lecamwasam et al, 2001; Inaoka and Ochi, 2002; Inaoka et al, 2003).
We found that GTP concentrations changed with growth phase and nutritional conditions in B. subtilis. rRNA promoter activities correlated with these changes in vivo and were dependent on the concentration of GTP in vitro. rRNA promoter activity also correlated with a decrease in GTP concentration during entry into stationary phase (data not shown). Furthermore, a B. subtilis rrnB P1 promoter mutant requiring lower GTP levels for maximal transcription in vitro than wild-type rrnB P1 displayed much less pronounced changes in activity with growth rate and phase than the wild-type promoter (data not shown).
The detailed mechanism underlying regulation of B. subtilis rRNA promoters by GTP concentration remains to be determined. Future studies will define the B. subtilis rRNA promoter sequences required for regulation and whether open complex half-life is a crucial determinant of sensitivity to the iNTP concentration. The G+C-rich sequence element between the −10 element and the transcription start site, referred to as the discriminator region (Travers, 1984), is essential for regulation of E. coli rRNA promoters by the iNTP and by ppGpp (Barker and Gourse, 2001; Murray et al, 2003b; Murray and Gourse, 2004). However, this element is A+T rich in B. subtilis rRNA promoters. Future studies will address how the promoter remains sensitive to the iNTP concentration despite the A+T-rich character of its discriminator sequence. Interestingly, the B. subtilis rRNA promoters formed longer-lived open complexes with E. coli RNAP than with B. subtilis RNAP when challenged with heparin (Figure 5D and data not shown). This may simply reflect a greater sensitivity of B. subtilis RNAP to heparin (Whipple and Sonenshein, 1992).
Role of ppGpp in control of rRNA transcription
The ubiquity of relA in bacteria and the recent discovery of ppGpp in chloroplasts (Givens et al, 2004; Takahashi et al, 2004) attest to ppGpp's importance as a regulator of gene expression. It is possible that ppGpp affects B. subtilis RNAP directly under conditions not examined here, but it is also conceivable that effects of ppGpp on B. subtilis rRNA promoters are always mediated by changes in GTP concentration. Further studies will be required to address this issue. The reduction in GTP concentration from increases in ppGpp might be ascribable both to consumption of GTP during ppGpp biosynthesis and to direct inhibition of IMP dehydrogenase, the first enzyme in GTP biosynthesis (Lopez et al, 1981). Considering the evolutionary conservation of at least some of the amino-acid residues in RNAP that contact ppGpp (Artsimovitch et al, 2004), it is surprising that B. subtilis RNAP appears insensitive to ppGpp. However, we note that B. subtilis and E. coli RNAP differ in other important aspects of gene regulation as well (Mencia et al, 1998; Artsimovitch et al, 2000).
Selection for G at the +1 position in B. subtilis rRNA promoters
The mechanism of regulation of rRNA transcription may have driven the evolution of B. subtilis rRNA promoter sequence. Comparisons of the sequences downstream from the likely −10 elements from both promoters in all 10 B. subtilis rRNA operons (data not shown), and extrapolation from the primer extension results with rrnB and rrnO (Figure 1), suggest that all B. subtilis rRNA promoters initiate with GTP. Since ATP concentration increases slightly in at least some conditions where an increase in rRNA transcription would be disadvantageous (e.g. amino-acid starvation), and ATP concentration decreases slightly in at least some conditions where a decrease in rRNA transcription would be disadvantageous (e.g. upshift), the apparent choice of G residues at the +1 position in rRNA promoters is likely not a chance event. The fact that the activity of the rrnB P1+1A variant increased slightly under conditions where ATP concentration increased slightly (Figure 6A and C) suggests that there might be promoters with kinetic characteristics in common with rRNA promoters that have evolved with A residues at the +1 position in order to respond positively to changes in ATP concentration in these conditions.
DksA homologs in B. subtilis
DksA was recently identified as a transcription factor crucial for regulation of rRNA promoters in E. coli (Figure 8; Paul et al, 2004a). DksA binds in the secondary channel of E. coli RNAP (Perederina et al, 2004), stabilizes interactions of RNAP with ppGpp, and decreases open complex lifetime, putting rRNA promoters into a kinetic range in vivo where they are susceptible to changes in ppGpp and NTP concentrations (Paul et al, 2004a). Although there are no B. subtilis ORFs with strong similarity to dksA, we singly deleted the three most homologous ORFs, yteA, yocK, and ylyA. We detected no differences in rrnB P1 promoter activity under a variety of growth conditions in these mutant strains (data not shown). Either there is no B. subtilis DksA homolog, or these gene products have redundant functions (masking effects of single disruptions), or a factor analogous to DksA is not recognizable from sequence analysis. We cannot exclude the possibility that a factor is needed to mediate the interaction of RNAP with ppGpp, but the failure of ppGpp to exert an effect on transcription in vivo from rrnB P1 promoters lacking a G at the +1 position, even under conditions where the relA gene is required for regulation, suggests that effects of ppGpp may be indirect.
Concluding remarks
In both E. coli and B. subtilis, the sequences of rRNA promoters, the properties of RNAP, the responses of NTPs and ppGpp concentrations to changes in growth conditions, and the responses of rRNA promoters to changes in these small molecules have all coevolved to achieve similar responses in rRNA expression. However, each organism has solved the requirements for proper rRNA promoter strength and regulation in its own unique manner. As has become apparent from studies on the mechanisms of regulation of other genes in B. subtilis and E. coli, there is more than one strategy for solving a similar regulatory problem (e.g. Henkin and Yanofsky, 2002; Yanofsky, 2003).
Materials and methods
Strains, strain construction, media and growth conditions, primer extension, protein purification, and methods for determination of mRNA half-life, NTP concentration, and ppGpp concentration are provided in Supplementary material.
Reporters of promoter activity
Promoter constructs were fused to lacZ, but activities were assayed by primer extension, rather than by β-galactosidase activity. When measured by primer extension, rrn P1 promoter activities increased proportionally to the growth rate, as expected (Figure 3), correlating with the increase in RNA:protein ratios observed with increasing growth rate (data not shown). At the highest growth rates (i.e. in cells grown in rich medium), β-galactosidase activities consistently underestimated rRNA promoter activities. Apparently, transcription initiation is not reported accurately by β-galactosidase activity in B. subtilis for promoters that initiate transcription at such high frequency. Previous conclusions about the relative activities of different promoters using lacZ fusions with different RNA leaders (e.g. Okamoto and Vold, 1992) might be compromised further by differences in mRNA half-life or translation efficiency.
RNA extraction
Preparation of RNA did not include centrifugation steps prior to cell lysis and extraction with phenol, limiting the potential for decay of the short-lived reporter mRNA. A recovery marker RNA (RM) was added at the time of extraction, controlling for differences in degradation during extraction and for variation between samples at later steps. The RM RNA was made from B. subtilis strain RLG6943 or E. coli strain RLG1100 from a template containing a binding site for the same primer used for extension of the test promoter transcripts, but differed from the test transcript in length at the 5′ end, allowing extension products to be distinguishable on gels (Josaitis et al, 1995). Typically, 1 ml of cells was pipetted directly into 2 ml phenol/chloroform (1:1) and 0.25 ml lysis buffer (50 mM Tris–HCl pH 8.0, 500 mM LiCl, 50 mM EDTA pH 8.0, 5% SDS). After brief vortexing, the RM (∼20 μl) was added, followed by immediate sonication. Water was added to increase the aqueous volume to 6 ml to prevent precipitation of salts, followed by two extractions with phenol/chloroform, two precipitations with ethanol, and suspension of the pellet in 20–50 μl 10 mM Tris–HCl, pH 8.0.
In vitro transcription
Plasmid templates (EcoRI–HindIII promoter fragments in pRLG770) are listed in Supplementary Table S1. For Figure 4, multiple round transcription was performed (10 μl reactions, 15 min, 30°C, 10 nM RNAP, 1 nM supercoiled plasmid template, 40 mM Tris–HCl pH 8.0, 10 mM MgCl2, 1 mM DTT, 0.1 μg/ml BSA, and 150 mM KCl). CTP, GTP, and ATP were 100 μM (except when ATP or GTP was varied from 10 to 2000 μM), and UTP was 10 μM plus 2 μM [α-32P]UTP. For Figure 2, ATP, CTP, and GTP were 200 μM, and a range of RNAP concentrations was tested to determine a subsaturating concentration at which potential effects of the UP element would not be obscured. Reactions were initiated with RNAP, allowed to proceed for 15 min at 30°C, terminated by addition of an equal volume of formamide loading buffer (95% formamide, 20 mM EDTA pH 8.0), electrophoresed on 7 M urea–5.5% polyacrylamide gels, and quantified by phosphorimaging.
For the single round assays testing effects of ppGpp (0.5 mM; TriLink Inc.) in Figure 5, RNAP was incubated with DNA for 10 min at 30°C at the indicated KCl concentrations. In Figure 5A, transcription was initiated with 200 μM each ATP, CTP, GTP, 10 μM UTP, 2 μM [α-32P]UTP, and 600 nM dsDNA competitor containing a consensus promoter, as described (Paul et al, 2004a). In Figure 5B–D, the fraction of RNAP–promoter complexes remaining at various times after addition of 3 μg/ml heparin was determined by in vitro transcription as described (Barker et al, 2001), except that the KCl concentration was 30 mM and the NTP concentrations were 400 μM ATP and GTP, 200 μM CTP, 10 μM UTP, and 2 μM [γ-32P]UTP. No transcript was detected when plasmid and heparin were preincubated before RNAP addition.
Supplementary Material
Supplementary material
Acknowledgments
We thank T Gaal, W Ross, M Barker, H Murray, T Henkin, G Spiegelman, and R Rudner for helpful discussions and comments on the manuscript. This work was supported by RO1 GM37048 from the National Institutes of Health and by a Hatch grant from the United States Department of Agriculture.
References
- Artsimovitch I, Patlan V, Sekine S, Vassylyeva MN, Hosaka T, Ochi K, Yokoyama S, Vassylyev DG (2004) Structural basis for transcription regulation by alarmone ppGpp. Cell 117: 299–310 [DOI] [PubMed] [Google Scholar]
- Artsimovitch I, Svetlov V, Anthony L, Burgess RR, Landick R (2000) RNA polymerases from Bacillus subtilis and Escherichia coli differ in recognition of regulatory signals in vitro. J Bacteriol 182: 6027–6035, Erratum (2001) J Bacteriol 183: 1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker MM (2001) PhD Thesis, University of Wisconsin-Madison
- Barker MM, Gaal T, Josaitis CA, Gourse RL (2001) Mechanism of regulation of transcription initiation by ppGpp. I. Effects of ppGpp on transcription initiation in vivo and in vitro. J Mol Biol 305: 673–688 [DOI] [PubMed] [Google Scholar]
- Barker MM, Gourse RL (2001) Regulation of rRNA transcription correlates with nucleoside triphosphate sensing. J Bacteriol 183: 6315–6323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cashel M, Gentry DR, Hernandez VH, Vinella D (1996) The stringent response. In Escherichia Coli and Salmonella Typhimurium, Neidhardt FC (ed) pp 1458–1496. Washington, DC: ASM Press [Google Scholar]
- Deneer HG, Spiegelman GB (1987) Bacillus subtilis rRNA promoters are growth rate regulated in Escherichia coli. J Bacteriol 169: 995–1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredrick K, Caramori T, Chen YF, Galizzi A, Helmann JD (1995) Promoter architecture in the flagellar regulon of Bacillus subtilis: high-level expression of flagellin by the sigma D RNA polymerase requires an upstream promoter element. Proc Natl Acad Sci USA 92: 2582–2586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushima T, Ishikawa S, Yamamoto H, Ogasawara N, Sekiguchi J (2003) Transcriptional, functional and cytochemical analyses of the veg gene in Bacillus subtilis. J Biochem (Tokyo) 133: 475–483 [DOI] [PubMed] [Google Scholar]
- Gaal T, Bartlett MS, Ross W, Turnbough CL Jr, Gourse RL (1997) Transcription regulation by initiating NTP concentration: rRNA synthesis in bacteria. Science 278: 2092–2097 [DOI] [PubMed] [Google Scholar]
- Givens RM, Lin MH, Taylor DJ, Mechold U, Berry JO, Hernandez VJ (2004) Inducible expression, enzymatic activity, and origin of higher plant homologues of bacterial RelA/SpoT stress proteins in Nicotiana tabacum. J Biol Chem 279: 7495–7504 [DOI] [PubMed] [Google Scholar]
- Gourse RL (1988) Visualization and quantitative analysis of complex formation between E. coli RNA polymerase and an rRNA promoter in vitro. Nucleic Acids Res 16: 9789–9809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmann JD (1995) Compilation and analysis of Bacillus subtilis sigma A-dependent promoter sequences: evidence for extended contact between RNA polymerase and upstream promoter DNA. Nucleic Acids Res 23: 2351–2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henkin TM (2002) Ribosomes, protein synthesis factors, and tRNA synthetases. In Bacillus Subtilis and its Closest Relatives: from Genes to Cells, Sonenshein AL (ed) pp 313–322. Washington, DC: ASM Press [Google Scholar]
- Henkin TM, Yanofsky C (2002) Regulation by transcription attenuation in bacteria: how RNA provides instructions for transcription termination/antitermination decisions. BioEssays 24: 700–707 [DOI] [PubMed] [Google Scholar]
- Hirvonen CA, Ross W, Wozniak CE, Marasco E, Anthony JR, Aiyar SE, Newburn VH, Gourse RL (2001) Contributions of UP elements and the transcription factor FIS to expression from the seven rrn P1 promoters in Escherichia coli. J Bacteriol 183: 6305–6314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaoka T, Ochi K (2002) RelA protein is involved in induction of genetic competence in certain Bacillus subtilis strains by moderating the level of intracellular GTP. J Bacteriol 184: 3923–3930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaoka T, Takahashi K, Ohnishi-Kameyama M, Yoshida M, Ochi K (2003) Guanine nucleotides guanosine 5′-diphosphate 3′-diphosphate and GTP co-operatively regulate the production of an antibiotic bacilysin in Bacillus subtilis. J Biol Chem 278: 2169–2176 [DOI] [PubMed] [Google Scholar]
- Jores L, Wagner R (2003) Essential steps in the ppGpp-dependent regulation of bacterial ribosomal RNA promoters can be explained by substrate competition. J Biol Chem 278: 16834–16843 [DOI] [PubMed] [Google Scholar]
- Josaitis CA, Gaal T, Gourse RL (1995) Stringent control and growth-rate-dependent control have nonidentical promoter sequence requirements. Proc Natl Acad Sci USA 92: 1117–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lew SJ, Gralla JD (2004) Mechanism of stimulation of ribosomal promoters by binding of the +1 and +2 nucleotides. J Biol Chem 279: 19481–19485 [DOI] [PubMed] [Google Scholar]
- Lopez JM, Dromerick A, Freese E (1981) Response of guanosine 5′-triphosphate concentration to nutritional changes and its significance for Bacillus subtilis sporulation. J Bacteriol 146: 605–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez JM, Marks CL, Freese E (1979) The decrease of guanine nucleotides initiates sporulation of Bacillus subtilis. Biochim Biophys Acta 587: 238–252 [DOI] [PubMed] [Google Scholar]
- Meijer WJ, Salas M (2004) Relevance of UP elements for three strong Bacillus subtilis phage F29 promoters. Nucleic Acids Res 32: 1166–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mencia M, Monsalve M, Rojo F, Salas M (1998) Substitution of the C-terminal domain of the Escherichia coli RNA polymerase alpha subunit by that from Bacillus subtilis makes the enzyme responsive to a Bacillus subtilis transcriptional activator. J Mol Biol 275: 177–185 [DOI] [PubMed] [Google Scholar]
- Mitani T, Heinze JE, Freese E (1977) Induction of sporulation in Bacillus subtilis by decoyinine or hadacidin. Biochem Biophys Res Commun 77: 1118–1125 [DOI] [PubMed] [Google Scholar]
- Murray HD, Appleman JA, Gourse RL (2003a) Regulation of the Escherichia coli rrnB P2 promoter. J Bacteriol 185: 28–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray HD, Gourse RL (2004) Unique roles of the rrn P2 rRNA promoters in Escherichia coli. Mol Microbiol 52: 1375–1387 [DOI] [PubMed] [Google Scholar]
- Murray HD, Schneider DA, Gourse RL (2003b) Control of rRNA expression by small molecules is dynamic and nonredundant. Mol Cell 12: 125–134 [DOI] [PubMed] [Google Scholar]
- Ochi K, Kandala J, Freese E (1982) Evidence that Bacillus subtilis sporulation induced by the stringent response is caused by the decrease in GTP or GDP. J Bacteriol 151: 1062–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto K, Vold BS (1992) Activity of ribosomal and tRNA promoters of Bacillus subtilis during sporulation. Biochimie 74: 613–618 [DOI] [PubMed] [Google Scholar]
- Paul BJ, Barker MM, Ross W, Schneider DA, Webb C, Foster JW, Gourse RL (2004a) DksA: a critical component of the transcription initiation machinery that potentiates the regulation of rRNA promoters by ppGpp and the initiating NTP. Cell 118: 311–322 [DOI] [PubMed] [Google Scholar]
- Paul BJ, Ross W, Gaal T, Gourse RL (2004b) rRNA transcription in Escherichia coli. Annu Rev Genet 38: 749–770 [DOI] [PubMed] [Google Scholar]
- Perederina A, Svetlov V, Vassylyeva MN, Artsimovitch I, Yokoyama S, Vassylyev D (2004) Regulation through the secondary channel—structural framework for ppGpp-DksA synergism during transcription. Cell 118: 297–309 [DOI] [PubMed] [Google Scholar]
- Potrykus K, Wegrzyn G, Hernandez VJ (2002) Multiple mechanisms of transcription inhibition by ppGpp at the lambda p(R) promoter. J Biol Chem 277: 43785–43791 [DOI] [PubMed] [Google Scholar]
- Rao L, Ross W, Appleman JA, Gaal T, Leirmo S, Schlax PJ, Record MT Jr, Gourse RL (1994) Factor independent activation of rrnB P1. An ‘extended' promoter with an upstream element that dramatically increases promoter strength. J Mol Biol 235: 1421–1435 [DOI] [PubMed] [Google Scholar]
- Ratnayake-Lecamwasam M, Serror P, Wong KW, Sonenshein AL (2001) Bacillus subtilis CodY represses early-stationary-phase genes by sensing GTP levels. Genes Dev 15: 1093–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross W, Gosink KK, Salomon J, Igarashi K, Zou C, Ishihama A, Severinov K, Gourse RL (1993) A third recognition element in bacterial promoters: DNA binding by the alpha subunit of RNA polymerase. Science 262: 1407–1413 [DOI] [PubMed] [Google Scholar]
- Takahashi K, Kasai K, Ochi K (2004) Identification of the bacterial alarmone guanosine 5′-diphosphate 3′-diphosphate (ppGpp) in plants. Proc Natl Acad Sci USA 101: 4320–4324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Testa D, Rudner R (1975) Synthesis of ribosomal RNA during sporulation in Bacillus subtilis. Nature 254: 630–632 [DOI] [PubMed] [Google Scholar]
- Travers AA (1984) Conserved features of coordinately regulated E. coli promoters. Nucleic Acids Res 12: 2605–2618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellington SR, Spiegelman GB (1993) The kinetics of formation of complexes between Escherichia coli RNA polymerase and the rrnB P1 and P2 promoters of Bacillus subtilis. Effects of guanosine tetraphosphate on select steps of transcription initiation. J Biol Chem 268: 7205–7214 [PubMed] [Google Scholar]
- Wendrich TM, Marahiel MA (1997) Cloning and characterization of a relA/spoT homologue from Bacillus subtilis. Mol Microbiol 26: 65–79 [DOI] [PubMed] [Google Scholar]
- Whipple FW, Sonenshein AL (1992) Mechanism of initiation of transcription by Bacillus subtilis RNA polymerase at several promoters. J Mol Biol 223: 399–414 [DOI] [PubMed] [Google Scholar]
- Yanofsky C (2003) Using studies on tryptophan metabolism to answer basic biological questions. J Biol Chem 278: 10859–10878 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material


