Abstract
Endogenous DNA damage arises frequently, particularly apurinic (AP) sites. These must be dealt with by cells in order to avoid genotoxic effects. DNA polymerase θ is a newly identified enzyme encoded by the human POLQ gene. We find that POLQ has an exceptional ability to bypass an AP site, inserting A with 22% of the efficiency of a normal template, and continuing extension as avidly as with a normally paired base. POLQ preferentially incorporates A opposite an AP site and strongly disfavors C. On nondamaged templates, POLQ makes frequent errors, incorporating G or T opposite T about 1% of the time. This very low fidelity distinguishes POLQ from other A-family polymerases. POLQ has three sequence insertions between conserved motifs in its catalytic site. One insert of ∼22 residues into the tip of the polymerase thumb subdomain is predicted to confer considerable flexibility and additional DNA contacts to affect enzyme fidelity. POLQ is the only known enzyme that efficiently carries out both the insertion and extension steps for bypass of AP sites, commonly formed as endogenous genomic lesions.
Keywords: AP site, DNA damage, DNA polymerase, POLQ, thymine glycol
Introduction
Mammalian cells have about 15 distinct DNA polymerases, but the functions of only a few of them are well defined (Goodman, 2002; Hübscher et al, 2002). Some of these DNA polymerases help cells tolerate endogenous DNA damage. Such enzymes are of particular importance, because DNA damage can block progression of replication forks. About 10 000 apurinic (AP) sites arise per cell per day through endogenous hydrolytic loss of purine bases (Lindahl, 1993; Nakamura et al, 1998). Many further AP sites arise as intermediates during removal by base excision repair of dUTP incorporated into DNA (Guillet and Boiteux, 2003). Enzymes that could efficiently bypass AP sites encountered during DNA replication could be valuable in promoting cellular survival and minimizing chromosomal breakage.
DNA polymerases are divided into families denoted A, B, X, and Y based on amino-acid sequence relationships. Members of the A-family have a wide distribution in nature. The most extensively studied A-family polymerase, Escherichia coli pol I, is a high-fidelity enzyme that assists in maturation of Okazaki fragments during DNA replication and also participates in gap-filling during base excision repair, nucleotide excision repair, and repair of DNA interstrand crosslinks. We recently identified the DNA polymerase activity of an A-family enzyme found in mammalian cells, POLQ (Seki et al, 2003). There are no yeast homologs, but a POLQ homolog in Drosophila nuclei called Mus308 apparently functions in a pathway of DNA crosslink repair or tolerance. This is suggested because fly mus308 mutants are sensitive to DNA crosslinking agents (Harris et al, 1996). POLQ and Mus308 have a characteristic three-domain organization with a helicase-like domain in the N-terminal portion, a less conserved central domain, which is largely derived from one large exon, and a C-terminal A-family polymerase domain (Seki et al, 2003). Mouse Polq was recently proposed as a candidate gene defective in the chaos1 mouse mutant, which displays chromosome segregation abnormalities and radiation sensitivity in reticulocytes (Shima et al, 2003). The identity of Polq and chaos1 has been confirmed by direct disruption of Polq in the mouse and by correction of the phenotype with the Polq gene (Shima et al, 2004).
To understand the functions of human POLQ, we have explored its fidelity in DNA synthesis and ability to perform translesion synthesis when encountering DNA lesions. Although residues in the polymerase catalytic site are highly homologous to high-fidelity A-family DNA polymerases, we report here that POLQ has extremely low fidelity, comparable to some Y-family polymerases. Moreover, POLQ can perform translesion DNA synthesis at an AP site and a thymine glycol (Tg). Unusually, POLQ both inserts a base opposite an AP site and efficiently extends the misincorporated nucleotide, making it the most efficient known polymerase for AP-site bypass.
Results
Low fidelity of POLQ
These experiments utilized full-length recombinant POLQ, a 2590-amino-acid protein including both the DNA polymerase and helicase-like domains (Figure 1A). As POLQ is an A-family polymerase, it was anticipated that it would perform relatively accurate DNA synthesis. Fidelity was analyzed on a 30-mer template with a 16-mer DNA primer (Figure 1B). The results suggested an unexpected tendency of POLQ to misincorporate bases and extend from mispaired termini. There was frequent misincorporation when the first template base was a T (Figure 1C, lanes 1–4). With dATP as the only deoxynucleotide present, there was not only efficient incorporation of A opposite the first two T bases in the template, but further extension by incorporation opposite noncomplementary bases (Figure 1C, lane 1). Incorporation of G and T across from template T and further extension past noncomplementary bases was also observed (Figure 1C, lanes 3 and 4), but incorporation of C was not apparent (Figure 1C, lane 2). When the first template base was changed to A, G, or C, other misincorporation events took place (Figure 1C, lanes 5–16). With template C, only a low level of incorporation was catalyzed by POLQ (Figure 1C, lanes 13–16), with the correct nucleotide dGTP the best utilized and extended (Figure 1C, lane 15).
Figure 1.
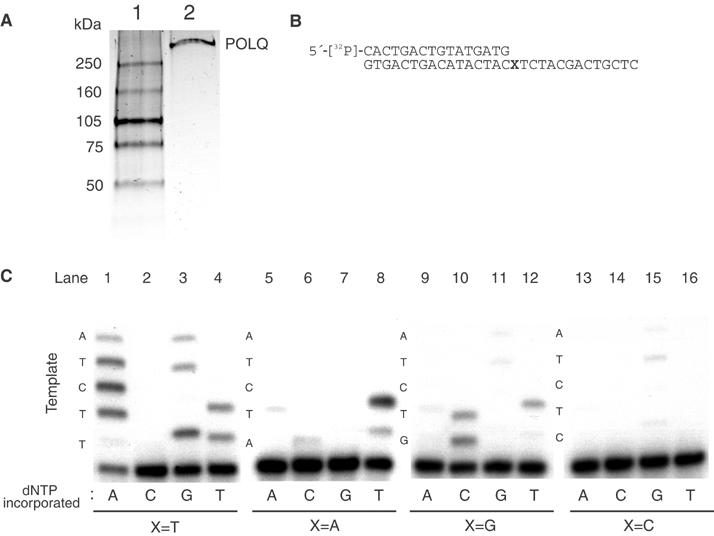
POLQ is an error-prone enzyme. (A) Full-length recombinant human POLQ was purified as described and 75 ng was electrophoresed on an SDS–polyacrylamide gel and silver-stained (lane 2). Molecular weight markers are shown in lane 1. (B) Template used for the primer extension assay. 5′-32P-labeled 16-mer was annealed to 30-mer DNA. The first template base is denoted by X. (C) POLQ frequently misincorporates residues. The first template base (X) was changed from T (lanes 1–4) to A (lanes 5–8), G (lanes 9–12), and C (lanes 13–16). Template bases are indicated on the left.
These experiments indicated that POLQ catalyzes considerable misincorporation, dependent on sequence context. This was surprising because most DNA polymerases with very low fidelity are classified in the Y-family and include human polymerases η, ι, κ, and Rev1 (Goodman, 2002). Consequently, we determined the fidelity of POLQ by making kinetic measurements (Table I). As a control, the values for human pol η and exonuclease-deficient E. coli pol I Klenow fragment (exo-free pol I Kf) were measured. For pol η and pol I, the Km and Vmax values for incorporation of dATP opposite template dT were similar to values previously reported (Polesky et al, 1990; Kusumoto et al, 2002) and were similar, within a few fold, to the corresponding values for human POLQ (Table I). Measurements were obtained with other incoming nucleotides, and from the ratio Vmax/Km (Creighton et al, 1995), a misincorporation frequency (finc) was calculated for POLQ. Compared to a value of 1.0 for correct utilization of dATP opposite template T, these were 1.7 × 10−3 for dCTP, 2.0 × 10−2 for dGTP, and 7.5 × 10−3 for dTTP (Table I). These frequencies are remarkably high for an A-family polymerase, and about 10–100 times higher than the misincorporation frequency of ∼10−5–10−4 found for exonuclease-defective E. coli pol I or Thermus aquaticus (Taq) polymerase (Minnick et al, 1999). The misincorporation frequencies are similar to those found for Y-family DNA polymerases (Goodman, 2002).
Table 1.
Fidelity of POLQ on normal DNA and at an AP site
| DNA substrate | dNTP | Km (μM) | Vmax (nM/min) | Vmax/Km | finc |
|---|---|---|---|---|---|
| Insertion opposite T | dATP | 7.0±1.4 | 7.5±0.3 | 1.1 | 1 |
| 5′-ATG | dCTP | 220±110 | 0.42±0.13 | 1.9 × 10−3 | 1.7 × 10−3 |
| -TACTTC | dGTP | 110±22 | 2.4±0.1 | 2.2 × 10−2 | 2.0 × 10−2 |
| dTTP | 660±150 | 5.5±1.7 | 8.3 × 10−3 | 7.5 × 10−3 | |
| Pol η (insertion opposite T) | dATP | 4.4±0.2 | 3.4±0.7 | 0.77 | |
| Exo-free pol I Kf (insertion opposite T) | dATP | 3.1±0.9 | 1.7±0.2 | 0.55 | |
| Insertion opposite AP site (X) | dATP | 9.5±1.8 | 2.3±0.6 | 0.24 | 0.22 |
| 5′-ATG | dCTP | UDa | |||
| -TACXTC | dGTP | 52±3.0 | 1.4±0.2 | 2.7 × 10−2 | 2.5 × 10−2 |
| |
dTTP |
510±22 |
1.3±0.5 |
2.5 × 10−3 |
2.3 × 10−3 |
| Extension with dATP from N opposite AP site (X) |
Base (N) opposite AP site: |
Km (μM) |
Vmax (nM/min) |
Vmax/Km |
fext |
| 5′-ATGN | A | 2.8±0.6 | 3.2±0.4 | 1.1 | 1.0 |
| -TACXTC | C | 41±0.5 | 1.5±0.1 | 3.7 × 10−2 | 3.4 × 10−2 |
| G | 350±140 | 0.4±0.1 | 1.1 × 10−3 | 1.0 × 10−3 | |
| T | 8.2±1.6 | 2.9±0.8 | 0.35 | 0.32 | |
| UD: undetectable. | |||||
One possible mechanism of misincorporation is primer–template slippage, as has been observed for the Y-family enzyme DNA pol κ (Ohashi et al, 2000a). In this mechanism, a template base loops out and allows a second, complementary template base to align with incoming nucleotide (see footnote to Table II). To examine this possibility, the first template base was fixed as T and the second template base was varied systematically. Each of the four incoming nucleotides was then tested separately. If a primer–template slippage mechanism is in operation, an incoming nucleotide would be best ‘misincorporated' if the second template base were complementary. For example, with incoming dGTP, apparent incorporation across from the first template T would arise from slippage and pairing when the second template base was C, and would be much more efficient than if the second template base were A, G, or T. We found, however, that misincorporation was not influenced by complementarity with the second template base (Table II). Total extension with dGTP was ∼12% when the second template base was C, but was similar or higher (11–21%) when the second base was A, G, or T. An analogous result was obtained with the other incoming dNTPs (Table II). It therefore appeared that frequent misincorporation was due to low insertion fidelity of the enzyme, not mediated by a slippage mechanism.
Table 2.
Primer/template slippage does not determine misincorporation by POLQ
| Incoming dNTP | Second template base | % of primer extended |
|---|---|---|
| dGTP | A | 11–16 |
| C | 9.2–14 | |
| G | 11–15 | |
| T | 15–21 | |
| dTTP | A | 8.5–14 |
| C | 4.6–10 | |
| G | 8.3–12 | |
| T | 11–17 | |
| dCTP | A | 1.4–2.1 |
| C | 1.4–1.5 | |
| G | 2.1–2.5 | |
| T | 1.0–1.4 | |
| dATP | A | 26–31 |
| C | 24–25 | |
| G | 27–29 | |
| T | 36–38 | |
| To test whether primer/template slippage was involved in misincorporation, the second template base, denoted X below, was varied. The fraction (%) of primer extended in two separate experiments is shown in the table. The second template base complementary to incoming deoxynucleotide is highlighted in bold type. | ||
| As an example for incoming dGTP, where X=C, only part of the substrate is shown: | ||
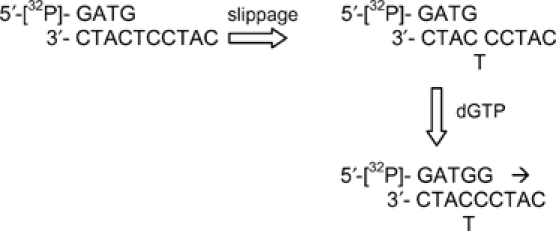 | ||
POLQ efficiently bypasses an AP site and Tg in DNA
The error-prone DNA synthesis and the lack of 3′ to 5′ exonuclease proofreading activity (Seki et al, 2003) prompted us to test the ability of POLQ to replicate past DNA adducts. The bypass activity of POLQ was assessed using a cyclobutane pyrimidine dimer (CPD), a 6-4 photoproduct (6-4 PP), an abasic (AP) site, 5R- and 5S-diastereoisomers of Tg, and the major adduct produced by cis-diaminedichloroplatinum (II) (cisplatin) (Figures 2 and 3). A primer with a 3′ terminus located just before the lesion was used. As controls, the translesion activities of exo-free pol I Kf and pol η were also tested.
Figure 2.
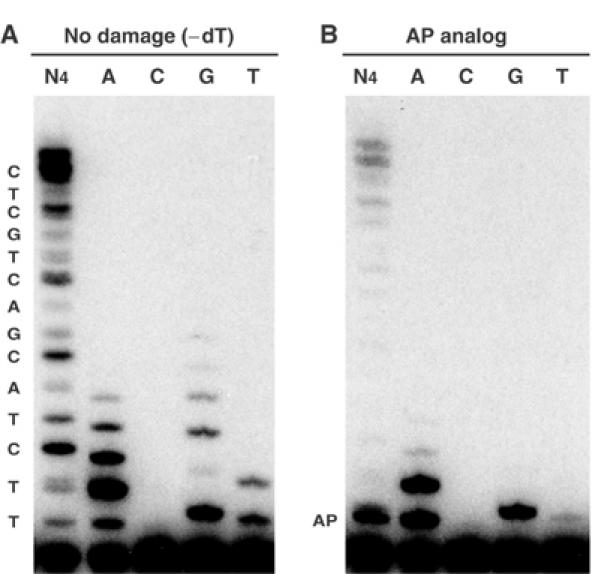
Nucleotide preference for bypass of an AP site. A 5′-32P-labeled 16-mer primer was annealed to (A) 30-mer template DNA or (B) a template containing an AP site. Either all four dNTPs (N4) or each deoxynucleotide separately at 100 μM was utilized in reactions as indicated.
Figure 3.
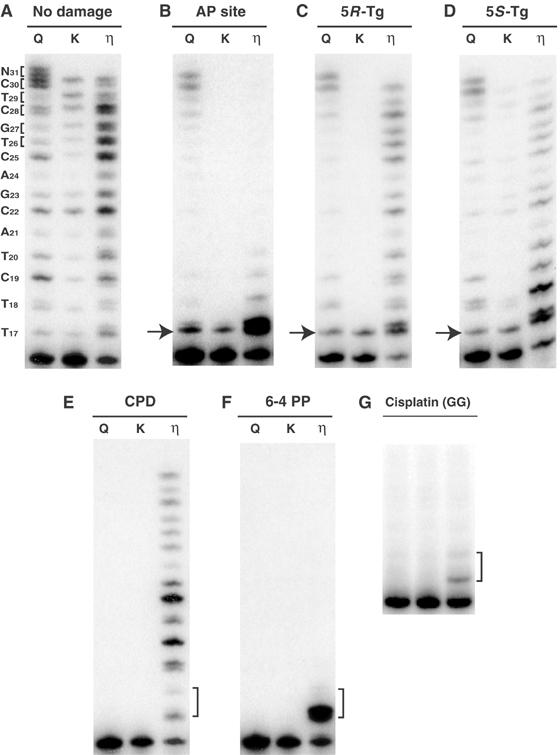
POLQ bypasses AP sites and Tg. (A–G) A 5′-32P-labeled 16-mer was annealed with 30-mer template DNA containing lesions and incubated with either human POLQ (Q), exonuclease-defective pol I Klenow fragment (K), or pol η (η). (A) Undamaged control, (B) AP analog, (C) 5R-Tg, (D) 5S-Tg, (E) T–T CPD, (F) T–T 6-4 PP, and (G) 1,2-d(GpG) cisplatin adduct. The position of the adduct is indicated by an arrow (B–D) or bracket (E–G).
All three enzymes could extend the primer to the end of a nondamaged 30-mer template (Figures 2A and 3A). There were several notable features of POLQ extension on nondamaged DNA. POLQ extended 1 nt further than did pol η or exo-free pol I Kf, indicating that the enzyme can perform efficient nontemplated addition at a blunt end in a way similar to the A-family enzyme Taq polymerase (Clark et al, 1987; Clark, 1988). Second, many of the bands representing extension with POLQ were doublets, particularly apparent after extension past C25 (Figure 3A). Some bands resulting from extension with pol η were also doublets. This reflects frequent misincorporation, giving rise to a mixture of DNA fragments with slightly different mobilities. Third, in the extension reaction with POLQ, several bands were more intense than the rest (Figure 3A), and these occurred each time the polymerase incorporated a base opposite template C (positions 19, 22, 25, 28). This suggests that POLQ tends to stall after incorporation at template C.
POLQ was able to bypass an AP site, by both inserting a base opposite the site and extending from the inserted base to the end of the template (Figures 2B and 3B). The efficiency of AP site bypass was much higher than that of pol η, which can incorporate a deoxynucleotide opposite an AP site but stalls and has only a weak ability to extend after incorporation (Figure 3B) (Masutani et al, 2000; Haracska et al, 2001). POLQ also bypassed an AP site when a shorter 14-mer primer was used that allowed incorporation opposite two normal template bases before the enzyme encountered the AP site (not shown).
POLQ could also bypass Tg lesions in DNA, common adducts produced by reactive oxygen species (Figure 3C and D). As reported before, pol η extended efficiently from the 5R-Tg diastereoisomer (Figure 3C), and somewhat less well from the 5S-Tg diastereoisomer (Figure 3D) (Kusumoto et al, 2002). POLQ bypassed both types of Tg lesions with similar efficiency. Exo-free pol I Kf could bypass 5S-Tg with low efficiency but not 5R-Tg (Figure 3C and D). POLQ was unable to insert deoxynucleotides opposite a UV-induced CPD (Figure 3E). Human pol η, on the other hand, is defined by its ability to bypass this adduct. POLQ also did not insert a deoxynucleotide opposite a 6-4 PP, although pol η was able to insert one dNTP opposite this adduct as reported previously (Figure 3F) (Masutani et al, 2000). Neither a 1,2-d(GpG) adduct (Figure 3G) nor a 1,3-d(GpTpG) adduct (not shown) was bypassed by POLQ.
As efficient AP-site bypass activity is a unique feature of POLQ, we investigated the incorporation of deoxynucleotides at an AP site and found that A was the most efficiently inserted and extended base (Figure 2B). The Km for incorporation of A opposite the AP site was 9.5 μM, close to the Km for incorporation of A opposite template T (Table I). The relative efficiency of incorporation (finc) of A against an AP site was 22% of that of a nondamaged template. G and T were also inserted, with 10- and 100-fold lower efficiency, respectively. Incorporation of C was too rare to be detected, even with high concentrations of deoxynucleotide. POLQ was also very efficient in extending a primer with a base opposite an AP site (Table I). Remarkably, the Km and Vmax values for POLQ extension from an A residue opposite an AP site were within a few fold of the values for insertion of A opposite T with normal base pairing (Table I). When the primer-terminal base opposite the AP site was T, POLQ also worked well in primer extension (fext=0.32). A primer-terminus with C opposite the AP site was poorly extended on this template (fext=0.034).
The Y-family enzyme DNA polymerase pol κ can carry out some bypass of an AP site via a misalignment mechanism, resulting in a −1 deletion during extension and a shorter DNA product than with nondamaged DNA (Ohashi et al, 2000b). In contrast, the bypass by POLQ took place directly and not by misalignment, as shown by several observations. First, the extension products formed by POLQ on a template containing an AP site (or Tg) were of the same length as those on a nondamaged template, extending to 31 nt (Figures 2 and 3). Second, POLQ could efficiently extend a T residue opposite an AP site (fext=0.32) even when the next template base was a T (Table I). This could not occur if extension utilized misaligned pairing to the next template base as occurs with pol κ (Ohashi et al, 2000b). Finally, experiments were performed on templates where the base following the AP site was changed from T to G, A, or C (Figure 4). Regardless of the sequence context, an A was also the best-incorporated residue opposite an AP site, a G the next best incorporated, and a T or C incorporated very poorly or not at all, consistent with the kinetic measurements in Table I. The absence of a marked effect of sequence context shows that misincorporation opposite the AP site takes place directly, rather than by a misalignment mechanism involving pairing with the template base immediately following the AP site.
Figure 4.
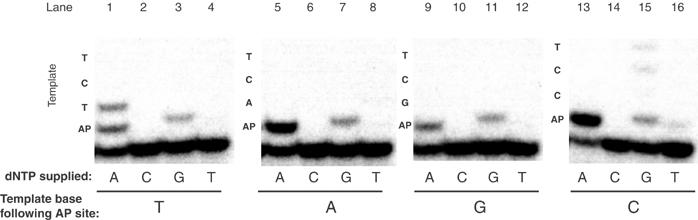
Incorporation opposite an AP site takes place directly rather than by template slippage. A 5′-[32P]-labeled 16-mer was annealed to 30-mer templates containing an AP site at the next template position as in Figures 2A and 3A. Four templates were used, having either T, A, G, or C as the next template base following the AP site. Each of the four possible reaction mixtures containing only a single dNTP was tested. After a 10 min incubation at 37°C, an A was preferentially incorporated opposite the AP site, regardless of the following sequence context.
Sequence inserts in the POLQ catalytic domain
Like other DNA polymerases, enzymes of the A-family have a structure that can be envisioned as a clasping right hand with palm, thumb, and finger domains. In the DNA polymerase domain of POLQ, all six conserved motifs of A-family polymerases are present in the primary protein sequence, and are well conserved with the same motifs in other A-family members (Seki et al, 2003). Yet the other known A-family polymerases are high-fidelity enzymes without significant ability to bypass AP sites. How can the properties of POLQ be rationalized with the presence of an A-family polymerase catalytic site? A sequence alignment of POLQ/Mus308 family polymerases with other A-family member polymerases reveals that POLQ from human and mouse cells has three extra spans of sequence in the polymerase catalytic domain (Figures 5A and 6B). One sequence of ∼22 amino acids (designated Insert 1 in Figures 4 and 5) lies between Motifs 1 and 2. A longer sequence, designated Insert 2, is located between Motifs 2 and 3. Insert 3, close to the C-terminus, is located between Motifs 5 and 6.
Figure 5.
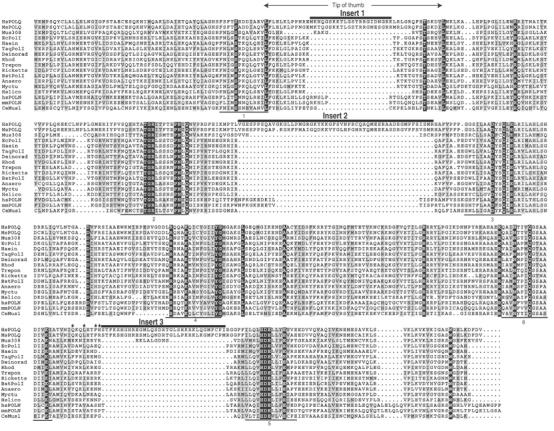
Sequence insertions in the catalytic site of POLQ. Sequence alignment of the polymerase catalytic region of POLQ/Mus308 family members and other prokaryotic A-family polymerases is shown. Identical amino acids are indicated with dark shading and similar amino acids with light shading. The alignment was carried out using the Clustal X program. The locations of the inserts and the conserved DNA polymerase motifs 1–6 are indicated. A putative PCNA binding motif is denoted by asterisks. EcPolI: E. coli pol I; TaqPolI: Thermus aquaticus pol I; BstPolI: Bacillus stearothermophilus pol I; Anaero: Anaerocellum thermophilum pol I; Ricketts: Rickettsia prowazekii pol I; Haein: Haemophilus influenzae pol I; Trepon: Treponema pallidum pol I; Deinorad: Deinococcus radiodurans pol I; Rhod: Rhodothermus marinus pol I; Myctu: Mycobacterium tuberculosis pol I; Helico: Helicobacter pylori pol I; HsPOLQ: human POLQ; MsPOLQ: mouse POLQ; hsPOLN: human POLN; mmPOLN: mouse POLN; Mus308: Drosophila melanogaster Mus308; CeMus1: Caenorhabditis elegans Mus1.
Figure 6.
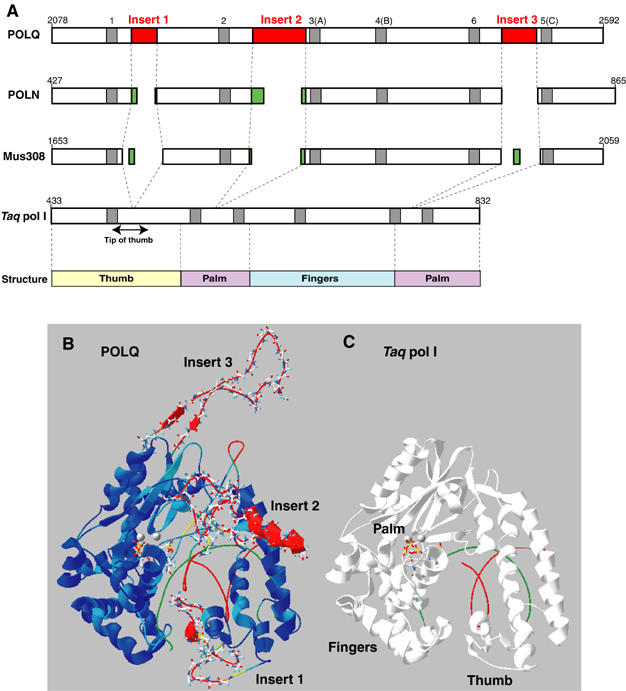
Sequence insertions in the catalytic site of POLQ. (A) The DNA polymerase catalytic region of POLQ is shown in relation to the other A-family polymerases human POLN, Drosophila Mus308, and Taq pol I. The locations of the inserts are shown in red and the conserved DNA polymerase motifs 1–6 are indicated as gray regions. Nonconserved differences in the insert regions are indicated in green. The bottom line shows coding regions for the main finger, palm, and thumb structural domains. (B, C) A computationally derived model for the polymerase domain of POLQ–DNA (B) obtained with the Amber94 potential, as compared to the known structure of the closed form of Taq pol I (C). The polymerase domain of Taq pol I (1QTM) is shown in white, while the modeled closed structure of human POLQ is colored blue to red, in the increasing order of r.m.s. deviation from Taq pol I. In both diagrams, the DNA template strand is green, and the primer strand is red. Gray spheres show the location of two Mg2+ cations, and an incoming ddTTP is positioned nearby in the structure. Inserts 1–3 of POLQ are shown in a ball-and-stick mode. The model is not meant to propose a detailed structure for the inserts, but instead to provide insight into the potential of Insert 1 to make contact with template DNA.
To gain some idea of the location of these inserts, an energy-minimized structural model for the POLQ–DNA complex was obtained, using as modeling template the closed form of Taq pol I bound to a 12-mer replicating DNA (Figure 6B and C). To model inserts, the Molecular Operating Environment package utilizes a library of pentapeptide rotamers and sequence-specific structural motifs extracted from the Protein Data Bank (PDB). The underlying method is a combination of the segment-matching procedure of Levitt (1992) and modeling of insertions and deletions according to Fechteler et al (1995). The approach is to combine available structural motifs for generating the possible conformations of groups of residues in insert regions and to select the conformers that minimize the intra- and intermolecular energy in the context of other segments of the protein and substrate (double-stranded DNA and ddTTP in the case of the Taq structure). The resulting model is not meant to be interpreted as a detailed atomic structure, but it does provide an indication of the location of the inserts in the protein and their potential to interact with DNA.
Insert 1 is located at the tip of the thumb subdomain. This area of other A-family polymerases is highly flexible and known to interact with the duplex template–primer (Beese et al, 1993). The thumb appears to influence DNA binding, processivity, and frameshift fidelity by tracking the minor groove of the duplex (Minnick et al, 1996; Doublie et al, 1998; Cannistraro and Taylor, 2004). Deletion of this region in E. coli pol I causes significant reduction of DNA binding, reduction of processivity, and enhanced usage of misaligned primer/template (Minnick et al, 1996). The extended Insert 1 in POLQ is predicted to make close contacts with the major groove of double-stranded DNA, adjacent to the nucleotide incorporation site. One factor contributing to this prediction is that within the 36 amino acids in the loop at the tip of the thumb domain including Insert 1, there are six Arg and five Lys residues, which favor interaction with the negatively charged DNA backbone. The remaining 25 amino acids contain 14 nonpolar residues, which may favor docking into the major groove of DNA. Several residues in this model of Insert 1 are close enough to form hydrogen bonds with the sugar–phosphate backbone. All three inserts in the model consist largely of coil, with a short helical region likely in Insert 2. Regardless of their detailed conformation, the residues of Inserts 2 and 3 seem likely to be located beyond the range of probable interaction with DNA (Figure 6B and C). They may serve, for example, as parts of interaction domains with other proteins.
Discussion
POLQ has unusual properties of fidelity and ability to bypass DNA damage
Although several human DNA polymerases in the Y-family (pol η, pol ι, and REV1) have some capacity to incorporate a base opposite an AP site, they cannot efficiently extend the primer-terminus after doing so (Johnson et al, 2000; Masutani et al, 2000; Zhang et al, 2001; Lawrence, 2002; Vaisman et al, 2002). This has prompted the prevailing model that a multistep reaction is necessary for efficient bypass, where a switch to a second DNA polymerase, such as pol ζ, is required to extend the primer after insertion (Johnson et al, 2000). Yeast pol ζ indeed extends primer ends opposite AP sites, but with very low efficiency (Johnson et al, 2000). However, POLQ is unusual among all known DNA polymerases in its ability to efficiently carry out both the insertion and extension reactions of AP site bypass, so that a two-step mechanism is unnecessary. The biochemical features of POLQ result in preferential incorporation of A during AP-site bypass (Table III). A is the most efficiently incorporated base opposite an AP site and a primer ending with an A residue opposite an AP site is the best primer extended by POLQ. It is interesting that POLQ is extremely poor at incorporating dCTP opposite an AP site, completely opposite to the preferred reaction of REV1 at an AP site (Lawrence, 2002). Moreover, POLQ poorly inserts C opposite an AP site and extends a primer end having C opposite such a site. This might be termed an ‘Anti-C rule' (Table III) and could ensure that POLQ works for bypass of AP sites in situations quite separate from those where REV1 may be employed.
Table 3.
POLQ AP-site bypass and extension efficiency
| dNTP | Insertion | Extension | Relative finc |
|---|---|---|---|
| dATP | 0.22 | 1.0 | 0.22 |
| dCTP | UD | 3.4 × 10−2 | — |
| dGTP | 2.5 × 10−2 | 3.7 × 10−4 | 9.3 × 10−6 |
| dTTP | 2.3 × 10−3 | 0.32 | 7.4 × 10−4 |
| The relative finc value was calculated as the product of the insertion and extension values from Table 2. For POLQ, this calculation is meaningful because the efficiency of extension from an incorporated A opposite an AP site is the same as extension of A opposite an undamaged template. UD: undetectable. | |||
Maga et al (2002) partially purified a DNA polymerase activity from HeLa cell nuclear extracts, provisionally identifying it as pol θ. This was based on reactivity of one of several bands of <100 kDa in the preparation with an antibody raised against a fragment of pol θ. The fidelity of DNA synthesis performed by this enzyme preparation was high and a 3′–5′ exonuclease activity was present (Maga et al, 2002). The features of this activity are quite different from human POLQ, which is an ∼250 kDa protein (Seki et al, 2003; Shima et al, 2003) with low fidelity (this study) and no exonuclease activity (Seki et al, 2003) or conserved exonuclease motifs in the sequence.
Role of POLQ-specific insertions in the catalytic fold
POLQ is distinguished from other A-family DNA polymerases by its ability to efficiently bypass AP sites and Tg, and by the presence of insertions in the DNA polymerase catalytic domain. Although more detailed biochemical analyses and structural studies are needed, it is possible that these two features are connected. Insert 1 is somewhat analogous to the loop on the tip of the thumb of T7 DNA polymerase that binds thioredoxin and acts as a processivity element (Bedford et al, 1997). It is possible that Insert 1 is important for the ability of POLQ to bind and extend the primer end opposite an AP site.
POLQ has relatively low fidelity, in contrast to related enzymes such as pol I and Taq. Insert 1 may potentially interfere with the nucleotide recognition process by directly participating in DNA–protein interactions or indirectly altering the structure of the subdomain that accommodates incoming nucleotides. A Gaussian network analysis (Bahar et al, 1997, 1999) of the collective dynamics of POLQ polymerase domain reveals that in a low-frequency mode, Insert 1 is the most mobile region of the entire polymerase domain (not shown). While high mobility is usually a property that facilitates substrate recognition, it may also give rise to a loose association at the binding site, thereby decreasing the cooperativity and efficiency of functional changes in conformation. One possibility is that the low fidelity of POLQ is not due to a decrease in its DNA binding affinity, but to a lack of cooperativity in transmitting signals between different parts of the protein. It may be important to remember, however, that full-length POLQ also contains a helicase-like protein domain. The role of this domain has not yet been determined. It may participate, for example, in binding to specific DNA templates (Seki et al, 2003).
It has been assumed that vertebrate POLQ may be functionally similar to the Mus308 protein of Drosophila (Sharief et al, 1999; Seki et al, 2003). However, the fly Mus308 sequence does not have the area identified as Insert 1 (Figures 5 and 6), and Inserts 2 and 3 in POLQ are also distinct from Mus308. This raises the possibility that Mus308 and POLQ are not functional orthologs. Mus308 has been genetically implicated in DNA crosslink repair, while this study shows that the POLQ enzyme efficiently bypasses AP sites. It is possible that the recently recognized POLN (Marini et al, 2003) is instead the vertebrate ortholog of the DNA polymerase portion of Mus308, as the sequence of POLN also lacks Inserts 1–3.
Implications of the unusual properties of POLQ for function in vivo
The preference of some DNA polymerases to incorporate A opposite noncoding or miscoding lesions has been referred to as the ‘A-rule' (Strauss, 2002), and a recent study indicates that human cells normally follow an A-rule when encountering an AP site (Avkin et al, 2002). POLQ may be a major enzyme for such bypass in mammalian cells as it both inserts a base opposite an AP site and efficiently extends the misincorporated nucleotide, making it the most proficient known polymerase for AP-site bypass. Although POLQ is an error-prone DNA polymerase, the incorporation of A opposite an AP site is 10- to 100-fold more efficient than misincorporation of a base opposite T. Insertion of A opposite an AP site would be the correct choice if such a site arose as an intermediate in base excision repair of incorporated dUTP, as appears to occur frequently (Guillet and Boiteux, 2003). Insertion of A opposite an AP site arising from spontaneous hydrolytic depurination would be mutagenic, and is probably not the most likely in vivo function for POLQ. The ability of POLQ to bypass Tg adducts may also be of physiological importance, as these major DNA adducts can block replicative DNA polymerases (Kusumoto et al, 2002).
POLQ is expressed in many human tissues, cell lines, and cancer cells (Seki et al, 2003; Kawamura et al, 2004), and is represented by ESTs from various tissue types. Thus, it may have a function in maintaining normal genomic integrity by allowing bypass of lesions. A need for such bypass may arise in emergency situations, such as when a DNA replication fork encounters an unrepaired lesion. This is underlined by the identity of murine POLQ with the mouse chaos1 mutant, noted in Introduction. These mice have elevated levels of spontaneous and mutagen-induced micronuclei in red blood cells, indicative of increased chromosome breakage or errors in chromosome segregation (Shima et al, 2003). As we have shown here, POLQ can efficiently bypass AP sites and Tg lesions, and it is possible that lack of POLQ bypass activity could cause replication fork collapse or breakage, leading to the instability seen in the chaos1 model. As there are also some indications for preferred expression of POLQ in lymphoid tissues (Kawamura et al, 2004), another potential function is during the somatic hypermutation of antibody genes (Neuberger et al, 2003), which generates AP sites as a mutagenic intermediate.
Materials and methods
Purification of POLQ
Recombinant POLQ was purified as reported (Seki et al, 2003), with minor changes. Instead of Buffer A, cells were lysed in 40 ml of 100 mM Tris–HCl (pH 8.0), 0.6 M (NH4)2SO4, 10% glycerol, 0.5% Nonidet P-40, 10 mM EDTA, 5 mM DTT, 1 mM PMSF, 50 mM N-acetyl-leucyl-leucyl-norleucinal (ALLN), and EDTA-free protease inhibitor mix (Roche Diagnostics). After centrifugation, POLQ was purified from the supernatant by FLAG and Ni2+-agarose affinity chromatography (Seki et al, 2003).
DNA polymerase assays
Reaction mixtures for dNTP misincorporation experiments were incubated at 37°C for 10 min and contained 300 fmol of 5′-32P-labeled 16-mer primer annealed to 30-mer template DNA, 113 fmol of POLQ, and 100 μM of each dNTP (unless otherwise noted) in 20 mM Tris–HCl (pH 8.75), 4% glycerol, 80 μg/ml BSA, 8 mM MgCl2, and 0.1 mM EDTA. Assay conditions for recombinant POLQ were optimized for pH and temperature before undertaking these experiments. At pH 8.75 and 37°C, the DNA polymerase activity of POLQ was four times higher than the previously used conditions (Seki et al, 2003) of pH 7.5 and 30°C. The overall properties of POLQ were not changed, as for example avid bypass activity for an AP site was also observed at pH 7.5. For translesion synthesis, the same amounts of templates containing specific lesions were used. The amounts of pol η (0.125 ng) and exo-free pol I Kf (2.5 × 10−5 U, with pol I reaction buffer, New England Biolabs) were chosen to yield activity similar to POLQ. Purification and assay of pol η were as described (Kusumoto et al, 2002). For steady-state kinetics (Table I), 1 pmol of primer/template was used and the procedure was as detailed elsewhere (Creighton et al, 1995; Kusumoto et al, 2002). This procedure utilizes extensive titration of nucleotide concentration and time, and produces results valid for a considerable range of enzyme concentrations. For the primer–template slippage assay (Table II), the amount of substrate was reduced to 150 fmol and the dNTP concentrations increased to 1 mM in order to increase the possibility of slippage.
Oligonucleotide substrates
Primer oligonucleotides were purchased from Bio-Synthesis or Sigma GenoSys, purified, and 5′-labeled using polynucleotide kinase and [γ-32P]dATP. Oligonucleotides containing a single thymine–thymine CPD (T–T CPD), T–T 6-4 PP, AP site, or Tg were synthesized as described (Kusumoto et al, 2002). The oligonucleotide containing a 1,2-d(GpG) cisplatin intrastrand adduct was as described by Szymkowski et al (1992). Primers were annealed to these oligonucleotides to create substrates for bypass assays (lesions highlighted by bold letters) as follows:
CPD and (6-4)PP substrate:
 |
Selection of a template from the PDB for 3D homology modeling
The protein sequences of 27 pol I family members were aligned, all containing the DNA polymerase domain with the previously defined six conserved motifs (Marini et al, 2003). Among these 27 sequences, those whose crystal structures were available in the PDB and whose sequence identity with respect to human POLQ was higher than 35% within the polymerase domain were selected as template candidates. E. coli pol I, Taq pol I, and Bacillus stearothermophilus pol I were found to match these two criteria. Taq pol I has 36% sequence identity with POLQ within the DNA polymerase domain, and both open (native protein alone) and closed (complexed with DNA double strand) structures available in the PDB. Consequently, the ddTTP-trapped closed form of Taq pol I (PDB code: 1QTM) (Li et al, 1999) with 12-mer double-stranded DNA, and the open form of Taq pol I (PDB code: 1TAQ) (Kim et al, 1995) were chosen as templates for structural homology modeling of the open and closed structures of POLQ. The 5′ nuclease domains of both chains were removed in silico. The polymerase domain of human POLQ has 515 residues, while that of the template Taq pol I has 400. The length difference is mainly contributed by the three inserts connecting conserved motifs of HsPOLQ: Insert 1 between Motifs 1 and 2 (22 residues), Insert 2 between Motifs 2 and 3 (52 residues), and Insert 3 between Motifs 6 and 5 (33 residues).
The MOE package (Molecular Operating Environment, Chem Comp Group Inc.) (http://www.chemcomp.com/Corporate_Information/MOE_Bioinformatics.html) was used for modeling of the inserts. For this purpose, MOE uses a library of pentapeptide rotamers, or sequence-specific structural motifs, extracted from the PDB. The underlying methodology is a combination of a segment-matching procedure (Levitt, 1992) and modeling of insertions/deletions (Fechteler et al, 1995). The approach is to combine the structural motifs generating possible conformations of groups of residues in the loop regions and to select the conformers that minimize the intra- and intermolecular energy in the context of other protein segments and substrate (here the double-stranded DNA 12-mer and ddTTP). The method of Levitt has been applied to a series of proteins ranging in size from 46 to 323 residues to obtain an all-atom-root-mean-square deviation of 0.93–1.73 Å. Fechteler et al (1995) have shown that the search of representative protein fragments using geometric scoring criteria can assist in predicting three-dimensional structures in insertion and deletion regions of proteins. Among the three insertions of POLQ, Insert 1 is the shortest (22 residues), and it is conceivable that its preferred conformation(s) can be estimated to a reasonable approximation by this segment-matching methodology, as used in MOE. The model for Insert 1 shown in Figure 6B is one of 10 predicted by MOE, which vary in detailed atomic structure with a root mean square (r.m.s.) deviation (for Insert 1 only) of 5.77 Å. Notably, all of the models show insertion into the DNA major groove, and this tendency is consistent with the abundance of positively charged residues in Insert 1. Interpretation of secondary structure used the algorithm of SWISS-Model (Swiss-PDB Viewer) (Guex et al, 1999).
Refinement by energy minimization
The MOE package was then used to build the models for the human POLQ open and closed structures. Calculations were performed using two different force fields, AMBER94 (Weiner et al, 1984, 1986) and MMFF94s (Halgren, 1996), to assess the sensitivity and robustness of the results with respect to the choice of intramolecular potentials. For each target structure (open or closed form of POLQ), 10 intermediate structures were generated by MOE. The Cartesian average model was created by averaging the coordinates of the 10 intermediates in each case, which led to two open and two closed form structures using the two different force fields. The structures found with the two sets of potentials were in close agreement, except for the relative orientation of Insert 2 in the two DNA-bound forms: the r.m.s. deviation in the backbone coordinates of the two open forms was 2.3 Å, and that of the two closed forms without Insert 2 was 1.7 Å. While the conformation of Insert 2 differed in the two models, none of its atoms was closer to double-stranded DNA and incoming nucleotides than 7.5 Å for both models.
The resulting four structures were subjected to structural refinement to correct any bond lengths and angles that violated the conventional ranges of accessible values (e.g. if the peptide bond significantly departed from the trans state). A constrained energy minimization was performed for each structure for this procedure, during which Inserts 1–3 and the corresponding regions of the original templates were allowed to undergo conformational changes while the remaining structural elements (essentially the core and surrounding conserved secondary structural elements) were held fixed at their original state. A total of 500 steps of steepest descent and 100 steps of the conjugated gradient method were performed for each energy minimization run. The procedure was repeated with decreasing strengths of bond stretch, angle bend, and torsion potentials that penalize the unrealistic bond lengths and angles, until no further change in conformation or decrease in energy was observed.
The computations performed for obtaining the closed structure considered the complex formed with the bound DNA. The DNA segment was incorporated into the models after structural alignment with the cognate template, 1QTM. The same energy minimization procedure as the one described above for the open structure was then applied to the DNA polymerase–DNA complex to generate the most stable conformations. The r.m.s. deviation in the backbone coordinates for the POLQ–DNA complex between the initial homology model and its energy-minimized form was only 0.55 Å, indicating that the model was stable and closely retained its conformation during energy minimization.
Acknowledgments
We thank Dr Birgitte Wittschieben for discussion and advice on protein purification, Dr Dror Tobi for advice on protein modeling, and Dr Tom Ellenberger, Dr Luis Breiba and Dr Patrick Moore for comments on the manuscript. This work was supported by grants NIH CA101980 (to RDW), NIH P20 GM065805 (to IB), and by a grant to CM from the US–Japan Cooperative Cancer Research Program of the Japan Society for the Promotion of Science (JSPS).
References
- Avkin S, Adar S, Blander G, Livneh Z (2002) Quantitative measurement of translesion replication in human cells: evidence for bypass of abasic sites by a replicative DNA polymerase. Proc Natl Acad Sci USA 99: 3764–3769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahar I, Atilgan AR, Erman B (1997) Direct evaluation of thermal fluctuations in proteins using a single-parameter harmonic potential. Fold Des 2: 173–181 [DOI] [PubMed] [Google Scholar]
- Bahar I, Erman B, Jernigan RL, Atilgan AR, Covell DG (1999) Collective motions in HIV-1 reverse transcriptase: examination of flexibility and enzyme function. J Mol Biol 285: 1023–1037 [DOI] [PubMed] [Google Scholar]
- Bedford E, Tabor S, Richardson CC (1997) The thioredoxin binding domain of bacteriophage T7 DNA polymerase confers processivity on Escherichia coli DNA polymerase I. Proc Natl Acad Sci USA 94: 479–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beese LS, Derbyshire V, Steitz TA (1993) Structure of DNA polymerase I Klenow fragment bound to duplex DNA. Science 260: 352–355 [DOI] [PubMed] [Google Scholar]
- Cannistraro VJ, Taylor JS (2004) DNA–thumb interactions and processivity of T7 DNA polymerase in comparison to yeast polymerase eta. J Biol Chem 279: 18288–18295 [DOI] [PubMed] [Google Scholar]
- Clark JM (1988) Novel non-templated nucleotide addition reactions catalyzed by procaryotic and eucaryotic DNA polymerases. Nucleic Acids Res 16: 9677–9686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark JM, Joyce CM, Beardsley GP (1987) Novel blunt-end addition reactions catalyzed by DNA polymerase I of Escherichia coli. J Mol Biol 198: 123–127 [DOI] [PubMed] [Google Scholar]
- Creighton S, Bloom LB, Goodman MF (1995) Gel fidelity assay measuring nucleotide misinsertion, exonucleolytic proofreading, and lesion bypass efficiencies. Methods Enzymol 262: 232–256 [DOI] [PubMed] [Google Scholar]
- Doublie S, Tabor S, Long AM, Richardson CC, Ellenberger T (1998) Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 Å resolution. Nature 391: 251–258 [DOI] [PubMed] [Google Scholar]
- Fechteler T, Dengler U, Schomburg D (1995) Prediction of protein three-dimensional structures in insertion and deletion regions: a procedure for searching data bases of representative protein fragments using geometric scoring criteria. J Mol Biol 253: 114–131 [DOI] [PubMed] [Google Scholar]
- Goodman MF (2002) Error-prone repair DNA polymerases in prokaryotes and eukaryotes. Annu Rev Biochem 71: 17–50 [DOI] [PubMed] [Google Scholar]
- Guex N, Diemand A, Peitsch MC (1999) Protein modelling for all. Trends Biochem Sci 24: 364–367 [DOI] [PubMed] [Google Scholar]
- Guillet M, Boiteux S (2003) Origin of endogenous DNA abasic sites in Saccharomyces cerevisiae. Mol Cell Biol 23: 8386–8394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halgren T (1996) The Merck force field. J Comp Chem 17: 490 [Google Scholar]
- Haracska L, Washington MT, Prakash S, Prakash L (2001) Inefficient bypass of an abasic site by DNA polymerase eta. J Biol Chem 276: 6861–6866 [DOI] [PubMed] [Google Scholar]
- Harris PV, Mazina OM, Leonhardt EA, Case RB, Boyd JB, Burtis KC (1996) Molecular cloning of Drosophila mus308, a gene involved in DNA cross-link repair with homology to prokaryotic DNA polymerase I genes. Mol Cell Biol 16: 5764–5771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hübscher U, Maga G, Spadari S (2002) Eukaryotic DNA polymerases. Annu Rev Biochem 71: 133–163 [DOI] [PubMed] [Google Scholar]
- Johnson RE, Washington MT, Haracska L, Prakash S, Prakash L (2000) Eukaryotic polymerases iota and zeta act sequentially to bypass DNA lesions. Nature 406: 1015–1019 [DOI] [PubMed] [Google Scholar]
- Kawamura K, Ahar RB, Eimiya MS, Hiyo MC, Ada AW, Kada SO, Atano MH, Tokuhisa T, Imura HK, Atanabe SW, Onda IH, Akiyama SS, Agawa MT, O-Wang J (2004) DNA polymerase theta is preferentially expressed in lymphoid tissues and upregulated in human cancers. Int J Cancer 109: 9–16 [DOI] [PubMed] [Google Scholar]
- Kim Y, Eom SH, Wang J, Lee DS, Suh SW, Steitz TA (1995) Crystal structure of Thermus aquaticus DNA polymerase. Nature 376: 612–616 [DOI] [PubMed] [Google Scholar]
- Kusumoto R, Masutani C, Iwai S, Hanaoka F (2002) Translesion synthesis by human DNA polymerase eta across thymine glycol lesions. Biochemistry 41: 6090–6099 [DOI] [PubMed] [Google Scholar]
- Lawrence CW (2002) Cellular roles of DNA polymerase zeta and Rev1 protein. DNA Repair (Amst) 1: 425–435 [DOI] [PubMed] [Google Scholar]
- Levitt M (1992) Accurate modeling of protein conformation by automatic segment matching. J Mol Biol 226: 507–533 [DOI] [PubMed] [Google Scholar]
- Li Y, Mitaxov V, Waksman G (1999) Structure-based design of Taq DNA polymerases with improved properties of dideoxynucleotide incorporation. Proc Natl Acad Sci USA 96: 9491–9496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl T (1993) Instability and decay of the primary structure of DNA. Nature 362: 709–715 [DOI] [PubMed] [Google Scholar]
- Maga G, Shevelev I, Ramadan K, Spadari S, Hubscher U (2002) DNA polymerase theta purified from human cells is a high-fidelity enzyme. J Mol Biol 319: 359–369 [DOI] [PubMed] [Google Scholar]
- Marini F, Kim N, Schuffert A, Wood RD (2003) POLN, a nuclear PolA family DNA polymerase homologous to the DNA cross-link sensitivity protein Mus308. J Biol Chem 278: 32014–32019 [DOI] [PubMed] [Google Scholar]
- Masutani C, Kusumoto R, Iwai S, Hanaoka F (2000) Mechanisms of accurate translesion synthesis by human DNA polymerase eta. EMBO J 19: 3100–3109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minnick DT, Astatke M, Joyce CM, Kunkel TA (1996) A thumb subdomain mutant of the large fragment of Escherichia coli DNA polymerase I with reduced DNA binding affinity, processivity, and frameshift fidelity. J Biol Chem 271: 24954–24961 [DOI] [PubMed] [Google Scholar]
- Minnick DT, Bebenek K, Osheroff WP, Turner RM Jr, Astatke M, Liu L, Kunkel TA, Joyce CM (1999) Side chains that influence fidelity at the polymerase active site of Escherichia coli DNA polymerase I (Klenow fragment). J Biol Chem 274: 3067–3075 [DOI] [PubMed] [Google Scholar]
- Nakamura J, Walker VE, Upton PB, Chiang SY, Kow YW, Swenberg JA (1998) Highly sensitive apurinic/apyrimidinic site assay can detect spontaneous and chemically induced depurination under physiological conditions. Cancer Res 58: 222–225 [PubMed] [Google Scholar]
- Neuberger MS, Harris RS, Di Noia J, Petersen-Mahrt SK (2003) Immunity through DNA deamination. Trends Biochem Sci 28: 305–312 [DOI] [PubMed] [Google Scholar]
- Ohashi E, Bebenek K, Matsuda T, Feaver WJ, Gerlach VL, Friedberg EC, Ohmori H, Kunkel TA (2000a) Fidelity and processivity of DNA synthesis by DNA polymerase kappa, the product of the human DINB1 gene. J Biol Chem 275: 39678–39684 [DOI] [PubMed] [Google Scholar]
- Ohashi E, Ogi T, Kusumoto R, Iwai S, Masutani C, Hanaoka F, Ohmori H (2000b) Error-prone bypass of certain DNA lesions by the human DNA polymerase kappa. Genes Dev 14: 1589–1594 [PMC free article] [PubMed] [Google Scholar]
- Polesky AH, Steitz TA, Grindley ND, Joyce CM (1990) Identification of residues critical for the polymerase activity of the Klenow fragment of DNA polymerase I from Escherichia coli. J Biol Chem 265: 14579–14591 [PubMed] [Google Scholar]
- Seki M, Marini F, Wood RD (2003) POLQ (Pol theta), a DNA polymerase and DNA-dependent ATPase in human cells. Nucleic Acids Res 31: 6117–6126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharief FS, Vojta PJ, Ropp PA, Copeland WC (1999) Cloning and chromosomal mapping of the human DNA polymerase theta (POLQ), the eighth human DNA polymerase. Genomics 59: 90–96 [DOI] [PubMed] [Google Scholar]
- Shima N, Hartford SA, Duffy T, Wilson LA, Schimenti KJ, Schimenti JC (2003) Phenotype-based identification of mouse chromosome instability mutants. Genetics 163: 1031–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima N, Munroe RJ, Schimenti JC (2004) The mouse genomic instability mutation chaos1 is an allele of Polq that exhibits genetic interaction with Atm. Mol Cell Biol (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss BS (2002) The ‘A' rule revisited: polymerases as determinants of mutational specificity. DNA Repair (Amst) 1: 125–135 [DOI] [PubMed] [Google Scholar]
- Szymkowski DE, Yarema KJ, Essigmann JE, Lippard SJ, Wood RD (1992) An intrastrand d(GpG) platinum crosslink in duplex M13 DNA is refractory to repair by human cell extracts. Proc Natl Acad Sci USA 89: 10772–10776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaisman A, Frank EG, McDonald JP, Tissier A, Woodgate R (2002) pol iota-dependent lesion bypass in vitro. Mutat Res 510: 9–22 [DOI] [PubMed] [Google Scholar]
- Weiner SJ, Kollman PA, Case D, Singh U, Ghio C, Alagona G, Profeta S, Weiner P (1984) A new force field for molecular mechanical simulation of nucleic acids and proteins. J Am Chem Soc 106: 765 [Google Scholar]
- Weiner SJ, Kollman PA, Nguyen D, Case D (1986) An all atom force field for simulations of proteins and nucleic acids. J Comp Chem 7: 230. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Yuan F, Wu X, Taylor JS, Wang Z (2001) Response of human DNA polymerase iota to DNA lesions. Nucleic Acids Res 29: 928–935 [DOI] [PMC free article] [PubMed] [Google Scholar]


