Abstract
Clathrin-mediated endocytosis involves the assembly of a network of proteins that select cargo, modify membrane shape and drive invagination, vesicle scission and uncoating. This network is initially assembled around adaptor protein (AP) appendage domains, which are protein interaction hubs. Using crystallography, we show that FxDxF and WVxF peptide motifs from synaptojanin bind to distinct subdomains on α-appendages, called ‘top' and ‘side' sites. Appendages use both these sites to interact with their binding partners in vitro and in vivo. Occupation of both sites simultaneously results in high-affinity reversible interactions with lone appendages (e.g. eps15 and epsin1). Proteins with multiple copies of only one type of motif bind multiple appendages and so will aid adaptor clustering. These clustered α(appendage)-hubs have altered properties where they can sample many different binding partners, which in turn can interact with each other and indirectly with clathrin. In the final coated vesicle, most appendage binding partners are absent and thus the functional status of the appendage domain as an interaction hub is temporal and transitory giving directionality to vesicle assembly.
Keywords: AP180, clathrin-AP2 adaptors, eps15, epsin1, synaptojanin
Introduction
Clathrin-mediated endocytosis is a major pathway for synaptic vesicle recycling following exocytosis. This pathway must assemble functional synaptic vesicles of the correct size and with the correct complement of fusion molecules, calcium sensors and pumps, and it must do this after each exocytic event. Clathrin, adaptors and accessory proteins are the key components of this process, and their versatility is illustrated by their use in a wide variety of processes (for reviews, see Kirchhausen, 2000; Slepnev and De Camilli, 2000; Brodsky et al, 2001; Takei and Haucke, 2001; Evans and Owen, 2002; Conner and Schmid, 2003).
The term ‘clathrin-mediated endocytosis' emphasises a central organising role for clathrin. Self-assembly of clathrin around the nascent vesicle provides a rigid scaffold and is also believed to recruit many of the cargo adaptors and other components necessary for completion of vesicle invagination, scission and uncoating. In the brain, the major cargo adaptor enriched in nerve terminals and found in purified coated vesicles is the AP2 adaptor complex (see scheme in Figure 1B; Pearse and Robinson, 1984; Maycox et al, 1992). Adaptor protein (AP) complexes all have appendage domains that bind to other endocytic components, grouped together under the name ‘accessory proteins'. This group covers all the proteins used in clathrin-mediated endocytosis apart from clathrin and cargo adaptors.
Figure 1.
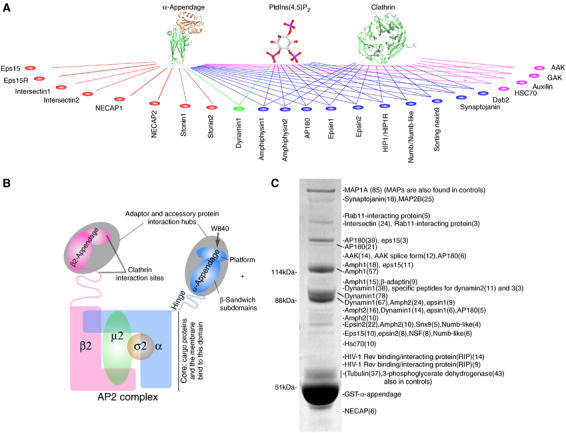
Proteomic analysis of AP2 α-appendage interactions. (A) Scheme showing all α-appendage binding partners, highlighting those that also interact with other interaction hubs in clathrin-mediated endocytosis. Colours separate interaction groups. (B) Scheme of an AP2 adaptor complex with its two appendage domains. The α-appendage is the main accessory protein interaction hub. (C) Protein from brain extracts bound to the α-appendage and analysed by tandem mass spectrometry. Numbers in brackets indicate the number of peptides sequenced to confirm the identity of the band. More details of interacting proteins are given in Supplementary Table 1.
Deep-etch visualisation of AP2 adaptors (Heuser and Keen, 1988) shows a central core flanked by two appendage domains. The appendages are connected to the core by flexible links of up to 6 nm. If fully unfolded, the linker sequences could extend up to 50 nm from the core. However, for entropic and statistical reasons, unfolded domains with no tertiary structure tend to be compact in solution. Thus, while appendages may be able to sample a wide radius around the core adaptor complex for endocytic components, they will always contract back to the core, pulling binding proteins into the ‘assembly-zone'. In this paper, we investigate the mode of binding partner interactions with the α-appendage.
Most adaptor appendages have two subdomains: a β-sandwich and a platform subdomain (see scheme in Figure 1B). When we originally crystallised the α-appendage of the AP2 complex, we showed that peptides containing DPF sequences could bind to the platform subdomain with an affinity of around 100 μM (Owen et al, 1999). A more extensive interaction using a variation of this motif, FxDxF (where x represents any amino acid), was later found and bound to the same W840 site on the α-appendage platform (Brett et al, 2002). The γ-adaptin and GGA appendages do not have the platform subdomain yet they still bind proteins, albeit a more limited set, via their β-sandwich subdomains. (Collins et al, 2003; Lui et al, 2003; Miller et al, 2003; Mills et al, 2003). It would be surprising if this same conserved β-sandwich subdomain in other appendages did not also have ligand interaction sites. In this paper, we show such an interaction site for the recently identified WxxF motifs on the β-sandwich subdomain of the α-appendage (WVxF in intersectin2 and synaptojanin, stonin and NECAP; Ritter et al, 2003; Jha et al, 2004; Walther et al, 2004). Given at least two independent peptide binding sites on the α-appendage that recognise at least four different types of peptide motifs (DPF/W, FxDxF, WVxF and a new FxxFxxL motif), we can now explain why multiple low-affinity peptide interactions give rise to nM protein interactions and how these high-affinity interactions are still readily reversible. Also, with multiple possible modes of interaction with the appendage, we consider the possibility that subsets of proteins can simultaneously bind the same appendage.
There are well over 20 proteins implicated in clathrin-coated vesicle (CCV) assembly. Many interact with multiple partners creating a complicated network of possible interactions. While traditionally these cellular processes have been considered as linear sequences of events, this is now recognised as being over simplistic. In this paper, we consider CCV assembly as the result of networks of protein interactions. The low affinities of many interactions in clathrin-mediated endocytosis give transient complexes a dynamic instability, which will mean that the network can be assembled by many different pathways and that the network is constantly sampling its environment, for example, for the presence of cargo in membranes. Examples of these low-affinity interactions are DPF/W motifs binding to the α-appendage, NPF motifs to EH domains and LLDLD motifs to the clathrin terminal domain (Owen et al, 1999; Kim et al, 2001; Miele et al, 2004). Thus, stability is only achieved by many such weak interactions. The prediction of dynamic instability in this network is evidenced by rapid exchange of components after photobleaching (Wu et al, 2003).
When all possible protein interactions are mapped, we can recognise interaction hubs, which are interaction nodes with a disproportionately large number of protein interactions. Within clathrin-mediated endocytosis, the α-appendage, clathrin terminal domain and PtdIns(4,5)P2 can be considered as hubs (Figure 1A). Many α-appendage partners interact with membranes and thus the whole process is firmly membrane anchored.
Appendage binding proteins often have multiple copies of the same motif concentrated in regions of no predicted secondary structure (Kalthoff et al, 2002; Mills et al, 2003). We call these regions ‘motif-domains' (MDs). For example, there are eight DPW motifs in the epsin1 MD. We have determined the number of appendages that can simultaneously bind an individual MD and thus we can predict whether a protein will prefer to bind to multiple appendages simultaneously, as they are found in forming coated pits. Thus, in assembly-zones, where there are clustered appendages on a two-dimensional surface, multiple low-affinity appendage interactions can work like Velcro™, in search of multiple sticking points. This will mean that the range of accessory proteins bound will depend on the avidity of each component for the network (where avidities are the combined affinities of all the network interactions). Combining the structural information and our affinity measurements with experiments on the dynamics of coat assembly, we now have a basis for predicting which groups of ligands will be recruited by the AP complexes at certain points during the vesicle retrieval cycle. This leads us to a view of AP appendage domains as the coordinators in time and space of accessory protein recruitment during CCV formation.
Results
Competition between binding partners for the α-appendage
In a representation of protein interactions in clathrin-mediated endocytosis, the AP2 α-appendage is a hub. We set out to characterise how the appendage can interact with multiple ligands and how the relative abundance of bound ligands is determined. We therefore characterised protein ligands using liquid chromatography tandem mass spectrometry (LC-MS/MS) and confirmed this using Western blotting to recognise both major and minor bands. A complete analysis of a wild-type appendage pull-down is shown in Figures 1C and 2A. Most of these ligands are already known, but a number of new ligands including the HIV-1 Rev interacting protein (RIP), important for nuclear export of viral RNA and infectivity (Fritz et al, 1995), are discussed in Supplementary Table 1. Most binding partners found by LC-MS/MS are enriched on α-appendage beads over brain extract (Figure 2A), with the exception of Hsc70, dynamin and numb-like. Dynamin's interaction was previously shown to be largely indirect and via amphiphysin (Owen et al, 1999). Some of the more minor interactors are very strongly enriched (AAK, auxilin, Dab2, eps15, epsin1 and synaptojanin170). All these enriched proteins have multiple copies of short α-appendage interaction motifs (Figure 2B), clustered in an MD.
Figure 2.
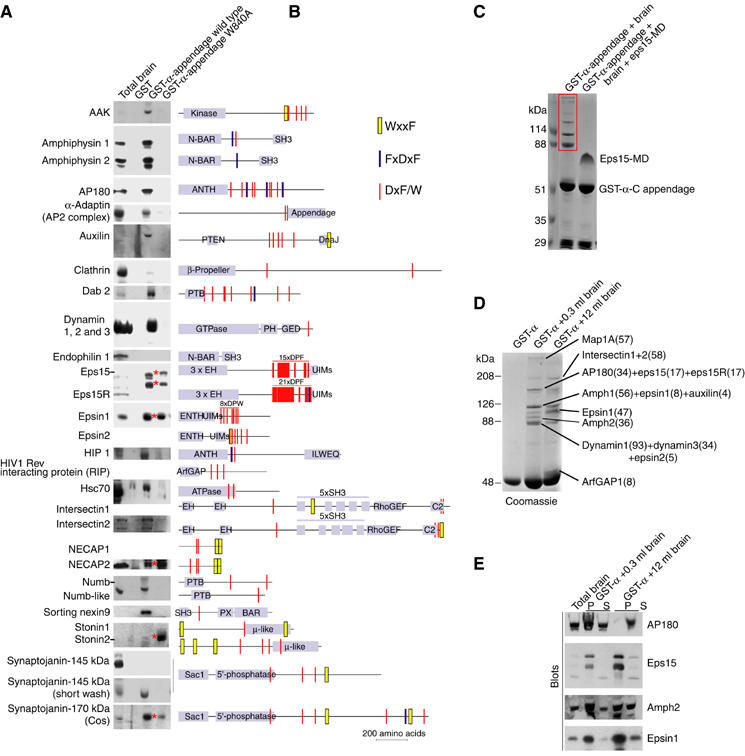
Protein interactions with AP2 α-appendage domain. (A) Protein from brain extracts bound to the α-appendage and the α mutant W840A. The red stars indicate proteins that are not displaced completely by the α-W840A mutation implying an additional interaction mode. 100 μg of GST fusion protein were used in 10 mg of rat brain extract and 6 μg of fusion protein were loaded for blotting. Given the number of interacting partners for the α-appendage, this excess of GST fusion protein allows us to sample all possible interacting partners without saturating the fusion proteins with higher affinity ligands. Bead-bound proteins were washed for 10 min. (B) Domains of α-appendage ligands showing the predicted α-binding motifs clustered in many cases into MDs. These are not all functional motifs but some have been tested in Figures 4 and 5. Note that for eps15 a polyclonal antibody was used and that only the upper two forms detected by the antibody bind to the appendages. As there is very little of the 170 kDa form of synaptojanin in the brain, we probed the extract from a 90 mm dish of COS cells. We have used a pan-dynamin antibody and by mass spectrometry we found dynamins I, II and III bound in the α-appendage complex. Intersectins 1 and 2, epsin2, NECAP and sorting nexin9 were identified as α-appendage ligands in mass spectrometry of these samples (see Figure 1C). (C) A Coomassie-stained gel showing a five-fold molar excess of eps15 MD in brain extract competes off all ligands from the α-appendage. (D) GST-α-appendage interactions in 0.3 and 12 ml of brain extract. The major proteins detected by Coomassie staining that change intensity have been identified by LC-MS/MS and the number of peptides is in parentheses. (E) Blots of samples in (D). AAK: adaptor-associated kinase; Amph: amphiphysin; Dab2: disabled protein2; HIP1: Huntingtin interacting protein1; UIMs: ubiquitin interacting motifs; PTB: phosphotyrosine binding domain; BAR: Bin/amphiphysin/Rvs homology domain; PX: phox homology domain.
The α-appendage platform subdomain is already characterised as containing the binding site for DxF-like motifs, and for the special cases of this motif (DPF, DPW and FxDxF, see arrow in Figure 1B). A mutant of this site (W840A) reduced or abolished almost all ligand interactions (Figure 2A) and thus the appendage cannot act as a protein interaction scaffold able to independently and simultaneously interact with multiple partners with high affinity. This was confirmed by using the MD from eps15 (eps15-MD), which competed off all the visible α-ligands (Figure 2C). This effectively means that the single α-appendage may work as a protein interaction hub by sampling its ligands. The intensity of Coomassie-stained ligands in Figure 1C will be mostly due to the relative abundance in brain extracts and to the affinities. Thus, we should be able to detect the higher affinity α-ligands in brain extract by increasing the extract to appendage domain ratio, whereas with a more limiting extract to appendage ratio, lower affinity ligands will be more readily visible. We see using LC-MS/MS (Figure 2D) and blotting (Figure 2E) that the amounts of intersectin, epsin1 and eps15 bound to the α-appendage increase with more extract, while dynamin and AP180 decrease and thus epsin1, eps15 and intersectin are predicted to have higher affinities. In Figure 2A, we also note that synaptojanin170, eps15, NECAP and epsin1 interactions are not completely abolished by α-W840A and the stonin2 interaction increases (see stars). This implies another mode of interaction alongside the W840A site.
A second binding site on the α-appendage for WVxF motifs centred around F740 and G742
Stonin2, whose interaction increases with α-W840A, has multiple WxxF motifs, which are known to bind to the α-appendage (Walther et al, 2004). WxxF motifs are also found in synaptojanin, NECAPs and other endocytic proteins (see Figure 2B). We solved the 1–9 Å resolution structure of the α-appendage bound to WVxF and FxDxF peptides (Syj-P3 and Syj-P1, respectively) found next to each other in the 170 kDa form of human synaptojanin (Figure 3). The binding sites for these peptides are the only regions on the α-appendage that are conserved in plants and from yeast to man (Supplementary Movie 1 and Supplementary Figure 1). The Syj-P3 peptide (sequence: NPKGWVTFEEEE) binds to an extensive surface on the β-sandwich subdomain, which we call the ‘side site'. There are two shallow pockets for the W and F of the peptide (Figure 3E). The core of the W5 pocket is formed by G742 and F8 is π stacked to F740 with space for F8 created by G725 lying immediately beneath. If these glycines were replaced by larger amino acids, the space in the pocket would be severely restricted. The acidic C-terminus of the peptide contributes to the strength of the interaction by hydrogen bonding. The main specificity determinants on the peptide are W, V and F, where W is hydrogen bonded, F must be large and hydrophobic and the V position would preferably be a small, uncharged residue because of its close proximity to F740 (Figure 3E, and see Supplementary Figure 2 for a more detailed description). Thus, the core motif is WVxF, which is the sequence found in NECAP, stonin, intersectin and synaptojanin. This binding site is on the opposite side of the β-sandwich subdomain to the previously identified γ-appendage binding site for DFxDF motifs (Supplementary Figure 3). A DPW peptide was previously shown to bind at the side site (Brett et al, 2002) around F740, although the orientation is different with a tryptophan in the F8 pocket (Supplementary Figure 4).
Figure 3.
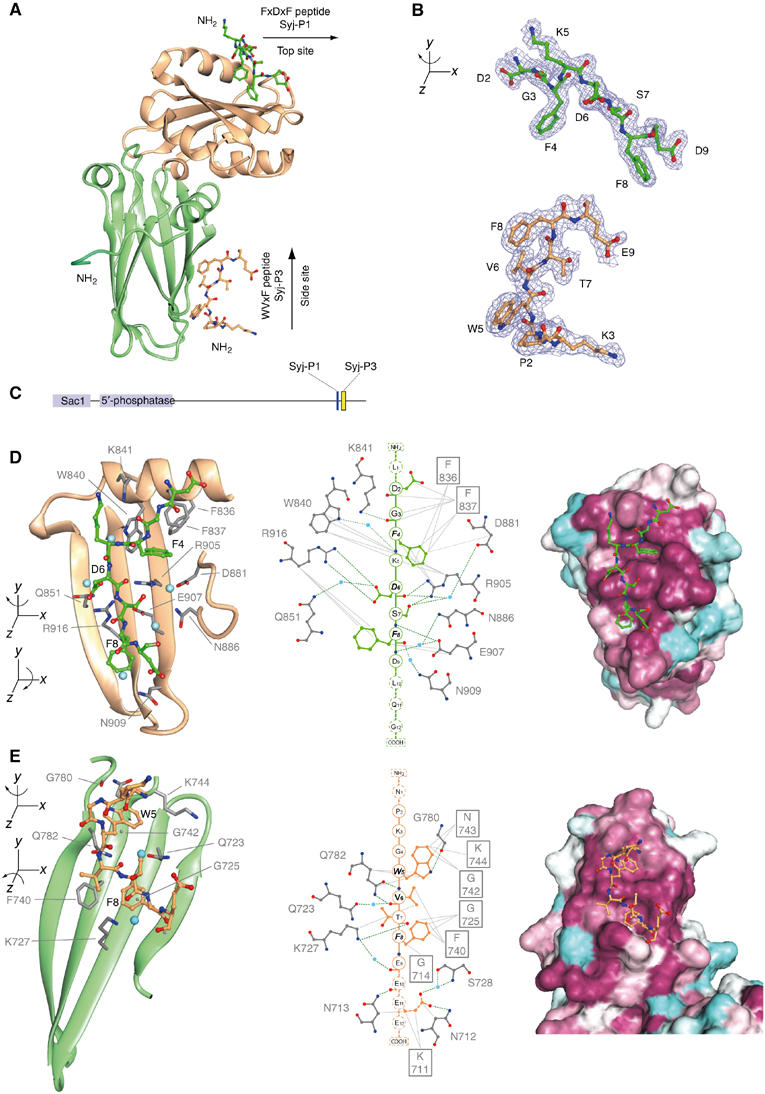
α-Adaptin appendage bound to WVxF and FxDxF motifs from synaptojanin. (A) Ribbon diagram showing bound peptides. The β-sandwich subdomain is coloured green and the platform subdomain is gold. The dark green residues at the N-terminus are from the vector. Syj-P3 and Syj-P1 bound to the α-appendage have been deposited with the Protein Data Bank (PDB ids: 1w80 and r1w80sf). (B) Density profiles for the peptides. (C) Scheme of the position of P1 and P3 peptides in synaptojanin170. (D, E) Details of peptide binding sites showing critical α-appendage residues and coordinated waters (blue) involved in binding. Peptides displayed as a linear chain are in the centre panel showing hydrogen bonding potential (dashed green lines) and hydrophobic (grey lines) interactions. The most crucial peptide residues are in bold italics and the residues for which there is little or no density are dotted. On the right are surface representations around the peptide binding sites, coloured according to sequence conservation (maroon (well conserved) through white to light blue (not conserved)). Binding pockets are more easily visible in this representation.
The WVxF sequence (Syj-P3) in synaptojanin is preceded closely by an FxDxF sequence, which is known to interact with the hydrophobic pocket centred around W840 on the platform subdomain of α-appendage. We made this as a peptide (Syj-P1: LDGFKDSFDLQG) and soaked it into the crystals containing Syj-P3. Syj-P1 occupied the W840 site (top site; see Figure 3A, B and D) in the same orientation as the recently presented structure of the α-appendage bound to the FxDxF motif from amphiphysin (Brett et al, 2002).
From the structure, we make the following observations: synaptojanin170 will not be able to use both WVxF and FxDxF motifs simultaneously to bind to a single α-appendage because the motifs are very close to each other and in reverse orientations. There are 11 amino acids between the visible ends of the peptides in our structure, which is insufficient to stretch the >50 Å distance from the C-terminus of the top peptide to the N-terminus of the side peptide. Instead, the motifs could interact with two different α-appendages if they were extremely close together, as may occur at a late stage of coated pit assembly or even following clathrin uncoating during disassembly. We also learn that there is no structural crosstalk between binding sites; therefore, the cooperativity of interactions seen below cannot be accounted for by changes in the architecture of the appendage.
Structure-based mutagenesis
Based on the structure, we made multiple mutations (Figure 4) and identified interacting proteins from pull-downs by LC-MS/MS. It is striking that mutants of the side site, F740D and G742D, have a weak phenotype compared to α-W840A. We blotted for NECAP, which has a WVxF motif, using a range of mutants around the side site and found that α-F740D and α-G742D completely abolish the interaction while α-W840A does not affect the interaction (Figure 4B). Using shorter wash pulses, we can still see some ligands bound to W840A (Figure 4C), which are completely abolished with α-F740D+W840A. Taken together, these results show that NECAP interacts predominantly with the side site (thus the DxF motifs in the sequence in Figure 2B do not contribute greatly to the interaction) and this site also makes a less dominant contribution to many other protein interactions (Figure 4C). Stonin2 with its three WVxF motifs also interacts predominantly with the side site (Figure 2A and data not shown). The stronger phenotype of the top site mutant in these experiments is likely due to the frequent presence of multiple copies of top site motifs and fewer copies of side site motifs in MDs, and the contributions of the side site to individual appendage affinities may be masked by the high avidity of top site interactions with α-appendages on beads.
Figure 4.
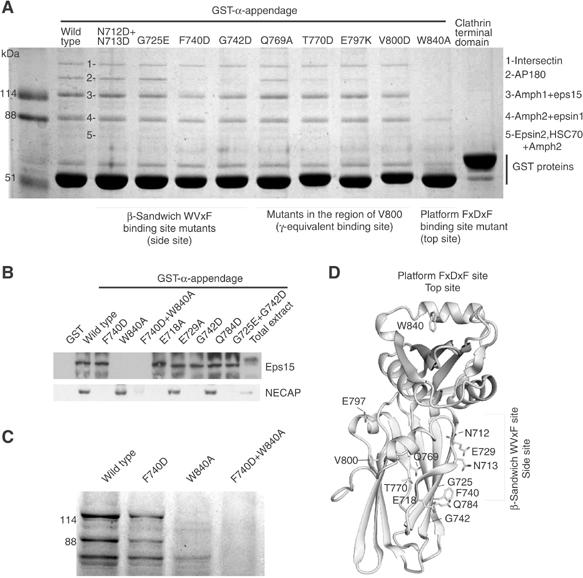
Top and side interaction surfaces on the α-appendage. (A) Coomassie analysis of ligand binding to α-appendage mutants. The results of a mass spectrometry analysis of the major Coomassie bands 1–5 are given on the right. (B) We found that NECAP does not bind to α-F740D and thus we probed some mutants around this site along with the top site mutant W840A. (C) Coomassie analysis of mutants with less stringent washing than in (A) leaving visible Coomassie bands in α-W840A that are absent in the double top and side site mutant. (D) Positions of our α-appendage mutants.
Specificity of α-appendage binding motifs
The Syj-P1 peptide (human) bound to the α-appendage with an affinity of 29 μM (Figure 5). The same peptide from rat, Syj-P2, where FxDNF replaces FxDSF, had a 4.5 μM affinity, which is similar to the 2.5 μM affinity for a 12-mer from amphiphysin (Amph-P2, containing FFxDNF). Deletion of the first phenylalanine from Amph-P2 results in a 10-fold decrease in affinity, consistent with the coordination of this phenylalanine by F837 and our earlier finding that α-F837A reduced the binding of amphiphysin and AP180 to the α-appendage (Owen et al, 1999). As expected from the structure, the D in the FxDxF is crucial, with substitution to R abolishing the α-interaction.
Figure 5.
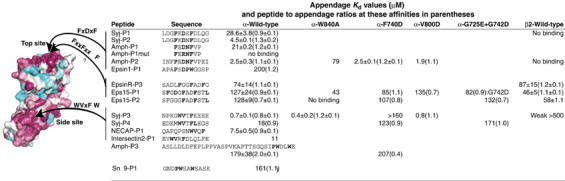
Affinities of peptides measured by ITC. Dissociation constants for the interactions of peptides and α-appendages as determined by ITC at 10°C. Where we have more than one determination, we give the average±range. The stoichiometries are in parentheses. The Amph-P3 peptide is from amphiphysin2 and contains two adaptor binding sequences, one for the platform and the other for the β-sandwich subdomain, neither of which have been previously shown. A stoichiometry of 2:1 means one peptide cannot stretch between the two appendage sites. The surface conservation to the left is colour coded: burgandy for high conservation through white to blue/jade for residues not conserved. Syj: synaptojanin; Amph: amphiphysin; Snx9: sorting nexin9. Further ITC parameters are found in Supplementary Table 3.
Many of the major α-appendage ligands (amphiphysin, eps15, epsins) do not contain WVxF motifs, while other ligands contain several (see Figure 2B). The WVxF peptide (Syj-P3) present only in the 170 kDa form of synaptojanin bound to the α-appendage with an affinity of 0.7 μM (Figure 5). The synaptojanin WVxF motif present in the 145 kDa and the 170 kDa form (Syj-P4) bound with an affinity of 18 μM, and this lower affinity may be why its contribution was previously missed. Comparing these peptides to NECAP-P1 (which is immediately followed by the acidic C-terminus of the protein) suggests that affinities are strengthened by negative charges at the C-terminus of the motif. WVxF motifs are specific for the side site as Syj-P3 bound to α-W840A with a wild-type affinity, but to α-F740D with a substantially weakened affinity. A similar sequence motif (PWxxW) is known to bind to the clathrin terminal domain (Lundmark and Carlsson, 2003; Miele et al, 2004) and thus we tested peptides from sorting nexin9 and from amphiphysin, which bound to the α-appendage with relatively weak affinities (Snx9-P1: 161 μM; Amph-P3: 179 μM).
A number of peptides that contain FxxFxxF/L motifs also bind to the α-appendage (Figure 5), including EpsinR-P3, Eps15-P1 and Eps15-P2 (from human, human and mouse, respectively). The epsinR-P3 peptide was previously shown to bind to the γ-appendage β-sandwich subdomain with an affinity of 110 μM (Mills et al, 2003). This site is not conserved on the α-appendage (Supplementary Figure 3); given that W840A abolished the interaction of Eps15-P2, whereas side site mutant had no effect, we conclude that this motif can bind to the top site.
If ligands are to interact with isolated appendages, then the concentrations must be similar or higher than the Kd values observed for the peptides. Most ligands are present in brain extract at lower concentrations (data not shown), and therefore micromolar affinities are insufficient to achieve significant interactions without additional contributions.
Clustering of α-appendages
Brain extract pull-down experiments using appendage domains on beads are informative but crude, in that ligands with more than one appendage interaction motif should bind with a higher avidity and additional layers of interactions between ligands may lead to interaction networks. We simplified this using isothermal titration calorimetry (ITC) where we measured the affinity of individual protein adaptor binding domains (MDs) for appendages. We define high affinity as 1 μM or tighter, an affinity where one can easily purify complexes by gel filtration. We define medium affinity as up to 20 μM.
Eps15-MD has an affinity of 21 nM for a first α-appendage with 2–3 additional binding sites of 10–20 μM, as determined by ITC (Figure 6A, and see Figure 6B for how the first appendage interaction almost completely saturates before the medium-affinity interactions become visible). The mutation F740D abolished the high-affinity interaction leaving only the medium-affinity interactions. G742D abolished the high-affinity site and only left one medium-affinity site of 17 μM (Supplementary Table 3). α-W840 weakened the high-affinity interaction by 100-fold. Thus, to achieve a high-affinity interaction, eps15 occupies both sites of the α-appendage. These results imply that there should be a motif in eps15 with a 1.2 μM affinity for the side site, but there are no WVxF motifs; so there is an uncharacterised motif that interacts with the side site. The binding of β2-adaptin to eps15 was similar to α-adaptin (Figure 6A), with low affinities and fewer sites.
Figure 6.
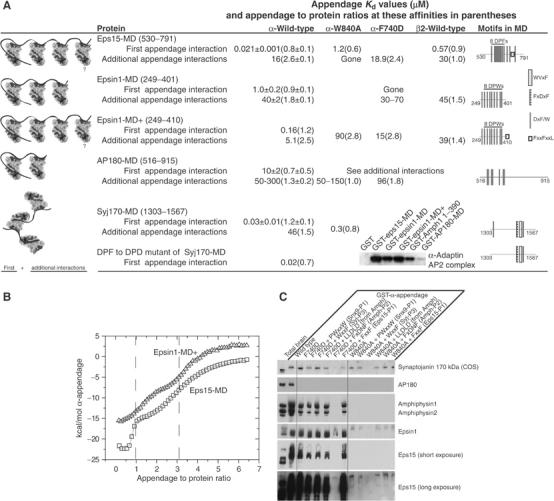
Affinities of proteins measured by ITC. (A) Dissociation constants and other binding parameters for the interactions of MD with α-appendages at 10°C measured by ITC. Schematic diagrams of the appendage MD interactions are on the left and schematics of the MD constructs used are on the right; numbers refer to amino acids. Inset: pull-downs from brain extract with some of the same MDs showing that affinities measured by ITC match the amounts of AP2 bound. Further ITC parameters are found in Supplementary Table 4. (B) Typical ITC data showing separation of high- and medium-affinity binding sites in eps15-MD and epsin1-MD+. (C) Peptide competition for ligand binding from brain extract to GST-α-appendage mutants. All peptides were used at 100 μM. The sequence of LLDLD peptides from amphiphysin1 is KEETLLDLDFDPF. This peptide and Amph-P3 bind to clathrin terminal domains. Other peptide sequences are found in Figure 5. As in Figure 2A, COS cell extract was used to monitor the interaction of synaptojanin170. Beads were washed for at least 10 min.
The MD of epsin1 contains eight DPW motifs and again several α-appendages bind simultaneously. The first α-appendage interacts with an affinity of 1 μM and the affinity of two others is approximately 40 μM (Figure 6A). As with eps15, α-F740D (and α-W840A, see below) abolished the high-affinity interaction. Again the top and side sides are occupied to give the first high-affinity appendage interaction. The additional low-affinity interactions are not affected by α-F740D and are likely due to the use of DPW motifs in the top site.
By increasing the length of the construct to include an additional FxxFxxL (epsin1-MD+), the affinities of both sites increased six- to eight-fold. A peptide covering this motif (EPDEFSDFDRLR) bound to α-appendage with only a millimolar affinity. Thus, a very low-affinity interaction motif can make a very significant contribution to the overall affinity of MD when used in combination with motifs that bind to other sites. It is interesting that the affinities of all appendage interactions are increased by this additional motif, and therefore we wonder if there might be an overall conformational rearrangement of the MD that is lost with α-W840A where the FxxFxxL motif is proposed to bind.
In these experiments, we show that both eps15 and epsin1 bind to multiple α- and β-appendages. The MD of eps15 has 12 DPF motifs and that of epsin1 has eight DPW motifs. It is probable that for steric reasons only some of these motifs can be occupied when saturated with α-appendage (3–4 for eps15-MD and three for epsin1-MD). For eps15, the medium-affinity interactions are abolished by α-W840A and preserved by α-F740D; therefore, these are single-site interactions. We measured the affinities of many DPF peptides from this domain and all are in the region of 100 μM. As one can predict, the presentation of multiple motifs increases the affinity, because when one appendage becomes bound then the probability of rebinding is higher due to a local concentration of binding motifs.
Given that the MDs from eps15 and epsin1 can bind simultaneously to at least three appendages, this will mean that these proteins will have a much higher compound affinity (avidity) for clustered AP2 complexes, which could equally be a concentrating mechanism for AP2s.
AP180-MD contains seven DxF motifs. We observed two appendage interactions with dissociation constants of medium to low affinity. Mutation of either W840 or F740 leaves only low-affinity interactions. A weak interaction with AP2 is also seen in the direct pull-downs from brain extract (see inset in Figure 6A).
The C-terminal domain of synaptojanin170 (Syj170-MD) contains the FxDxF and WVxF motifs whose peptides were bound to the α-appendage in Figure 3 as well as other DxF motifs. This domain has one high-affinity appendage interaction due to use of top and side binding sites and at least one medium-affinity appendage interaction. The high-affinity interaction is not due to the use of the N-terminal DPF motif as a mutant has no effect (Figure 6A) and we propose that it is due to the use of both FxDxF and WVxF motifs on different appendages.
From the peptide affinity measurements, we chose a number of peptides to use in competition studies for ligand binding to the α-appendage (Figure 6C). Given the high avidity of ligands for the α-appendage when using bead-bound protein, we did competition studies with α-W840A and α-F740D. Amphiphysin and eps15 binding to α-F740D were reduced and the interactions were abolished by the addition of an FxDxF peptide. This shows that these proteins use both sites, despite the absence of WVxF sequences as also shown in Figure 6A for eps15. There is no visible reduction of epsin1 binding to α-F740D (probably due to the avidity of the remaining three appendage interactions; see Figure 6A), and the FxDxF peptide reduces but does not abolish this interaction, consistent with the avidity afforded by its multiple DxF/W motifs. Synaptojanin170 can bind to both mutants and is abolished by F740D+Amph-P2 or by W840A+Syj-P3. Thus, this protein uses the top site in α-F740D or the side site in α-W840A, and unlike eps15 and epsin1 the interactions of both motifs are independent of each other, implying that both do not interact with the same appendage (see also the structure prediction). The 170 kDa isoform of synaptojanin has an additional WVxF motif ∼520 residues upstream of the FxDxF motif. Using these two motifs, synaptojanin may be predicted to interact with the top and side sites of a single α-appendage; however, our results do not support this.
Dynamics of coated pit assembly
We next examined accessory protein binding to clustered appendages on beads and to isolated appendages to mimic the competing adaptor interactions in assembly-zones and free AP2 complexes in the cytosol (Figure 7A). In the Coomassie visualisation, there are clear changes in the proteins bound (see arrows), although there was some loss in the recapture of isolated appendages. By blotting, we conclude that amphiphysin, AP180 and eps15 all bind more weakly to isolated appendages compared to clustered appendages, while epsin1 does not change and perhaps binds better. This shows that the dynamics of appendage interactions in solution favours epsin1 binding. This is consistent with in vivo data, where epsin1 seems to have a strong ability to target AP2 adaptors to the plasma membrane and lipid binding mutants deplete adaptors from the plasma membrane (Ford et al, 2002). The β-appendage does not interact strongly with any of the proteins in solution but has a significant interaction with AP180 and epsin1 when bound to the beads. We conclude that there is a change in ligand preference when adaptors move from the cytosol and are clustered on the membrane.
Figure 7.
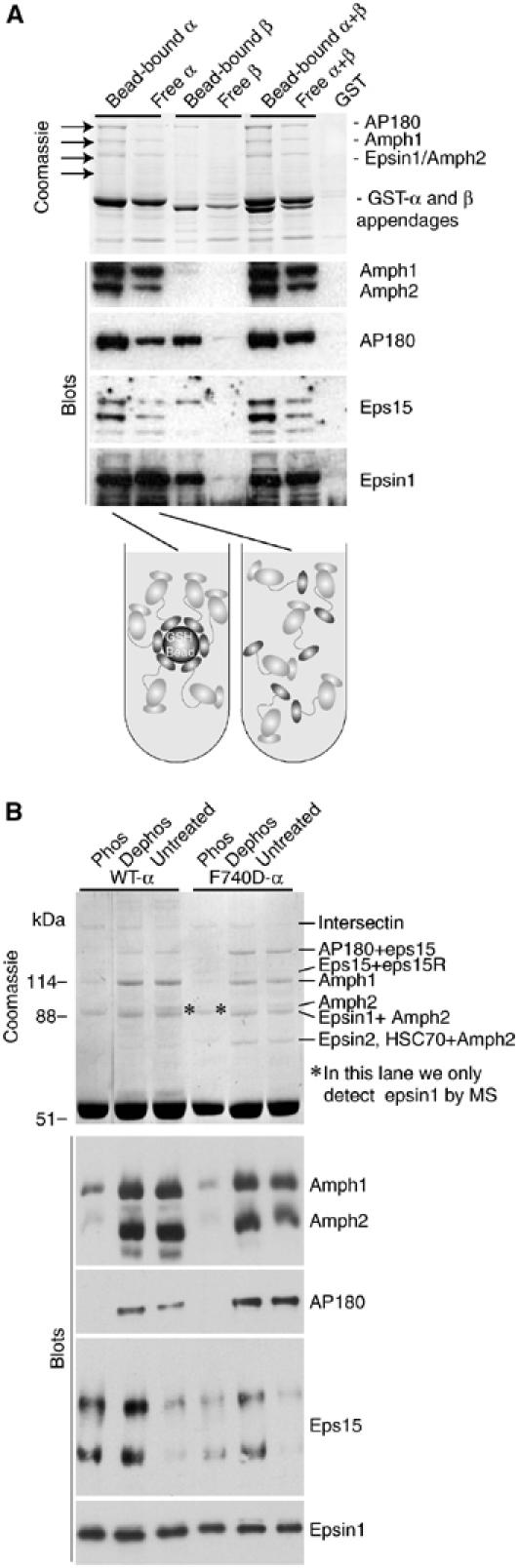
Influence of clustering and phosphorylation on α-appendage interactions. (A) Free appendages versus clustered appendages have different ligand preferences. As an approximation to free appendages, we have incubated GST-appendages in brain extract and subsequently captured these by pouring the extract over a filter with GSH beads on top. This was compared with GST-appendages previously bound to beads and then incubated in brain extract. The Coomassie gel shows the major changes taking place and some proteins have been blotted. The identities of the Coomassie bands have not been determined by mass spectrometry. (B) α-Appendages incubated in phosphorylated, dephosphorylated and untreated brain extracts change their ligand preferences. The identities of dominant proteins by LC-MS/MS are given beside the Coomassie gel.
Phosphorylation has previously been reported to modify AP interactions (Slepnev et al, 1998; Chen et al, 1999) and dephosphorylation of endocytic component in vivo is critical in regulating clathrin-mediated endocytosis at the synapse (Marks and McMahon, 1998). In our mass spectrometry analysis, we found in vivo phosphorylation sites in most of the major accessory proteins, having not treated the brain extracts to induce phosphorylation (data not shown). Thus, we used the α-appendage in samples of brain extract that were either untreated or incubated under conditions to promote phosphorylation (Mg-ATP and phosphatase inhibitors) or dephosphorylation (alkaline phosphatase). Coomassie staining showed that phosphorylation reduced the interactions of most ligands. Blotting confirmed this for amphiphysins, AP180 and eps15 but not for epsin1 (Figure 7B). The effects of dephosphorylation on the interaction of eps15 with the α-appendage imply that basal endogenous phosphorylation of this protein inhibits its interaction, while with amphiphysin and AP180 additional phosphorylation is needed to inhibit the interaction. The effect of basal phosphorylation on α-interactions and the greater effect of additional phosphorylation show that phosphorylation can fine-tune the network interactions. Thus, the endocytic apparatus is primed following exocytosis, where calcium triggers a protein phosphatase activity that dephosphorylates many endocytic proteins.
Temperature-dependent trapping of assembly-zones
At 37°C, there are fewer adaptor puncta on cell surfaces than at lower temperatures, and thus many colocalisation studies with AP2 adaptors use low-temperature fixation. At lower temperatures, the affinity of protein interactions will increase and transient complexes will be stabilised. We previously noted that overexpression of full-length epsin1 in COS cells leads to an accumulation of puncta on the plasma membrane (Ford et al, 2002). We now show that the epsin1 puncta appearance is temperature dependent and by TIRF microscopy we show that they are plasma membrane associated (Figure 8A and Supplementary Figures 5 and 6). The puncta begin to appear at temperatures below 18°C and colocalise with endogenous AP2. These puncta have predicted hallmarks of arrested network assembly-zones that cannot proceed to functional CCVs because of the perturbations. These hallmarks include (a) the appearance of puncta as the temperature is lowered and the disappearance as the temperature is raised again (data not shown), (b) the accumulation of accessory proteins like eps15 and dynamin not normally concentrated in CCVs or in clathrin-puncta at normal temperatures (see Figure 6B and Supplementary material in Ford et al, 2002) and (c) the accumulation of MDs (Supplementary Figure 5) or wild-type α-appendage when these are overexpressed. At 6°C, there are frequently cells that have no puncta, but these cells always have a much lower level of AP2 (Figure 6A). We could also abolish the epsin1 puncta by RNAi against the μ2 subunit of AP2 complexes (data not shown). This shows that an AP2 hub is needed for the puncta formation. Transfection of the epsin1 lipid binding mutant (R63L+H73L) does not result in puncta formation at 6°C and AP2 is no longer on the membrane. In these cells, AP2s frequently form aggregates inside the cell, but the epsin mutant does not colocalise. Thus, it appears that membrane-associated AP2 is necessary to get these arrested complexes.
Figure 8.
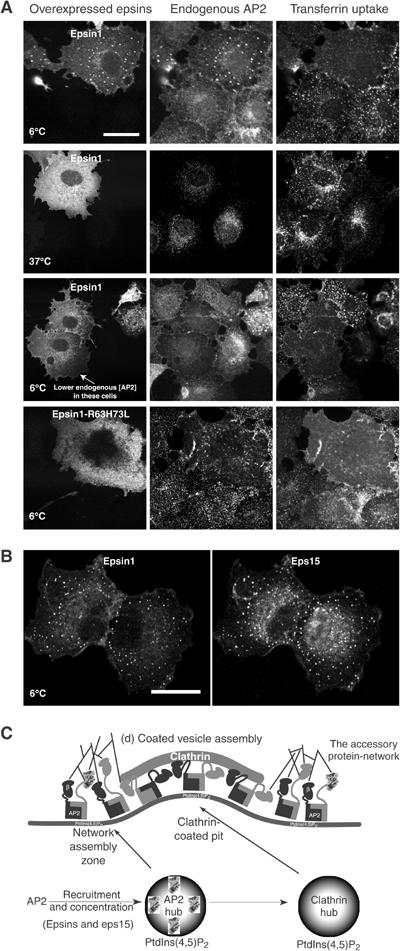
Adaptor protein-network assembly-zones. (A) Overexpression of epsin1 in COS cells leads to a temperature-dependent accumulation of puncta. This results in a redistribution of endogenous AP2 into these same puncta. Their presence is absent in cells expressing low levels of endogenous AP2 (50 out of 50 cells). The epsin1 lipid binding mutant R63L+H73L, which we previously showed inhibits transferrin at 37°C, does not form these puncta at 6°C and indeed AP2 becomes aggregated in the cytosol, but does not colocalise with other endocytic markers (not shown). Based on these experiments and the temperature-dependent appearance and our previous observations of colocalisation with eps15 and dynamin, we call these puncta ‘adaptor protein-network assembly-zones'. Scale bar, 20 μm. (B) Colocalisation of endogenous eps15 (detected with Ra15) with epsin1 in puncta. Scale bar, 20 μm. (C) Model of clathrin-coated pit assembly illustrating the adaptor protein-network assembly-zones at the edges of the pit. In these zones, proteins like epsins and eps15 are capturing more adaptors, so the vesicles are filled with cargo and shaping the membrane in preparation for clathrin assembly. Proteins with α-appendage side site interaction motifs will be able to bind simultaneously to the α-appendage along with epsins and eps15, which cluster primarily by using the top site. Thus, synaptojanin may well be recruited at this point and with its lipid phosphatase activity it will be able to destabilise the membrane attachments unless there is prior clathrin polymerisation. Amphiphysins are also found here and recruit dynamin to the edges of coated pits in preparation for the vesicle scission process. Assembly-zones may be too small and too unstable to visualise at 37°C by microscopy. A stable network will naturally progress into a coated pit due to the presence of clathrin-polymerising molecules like amphiphysin, epsins and AP180. This means that the network self-destructs as clathrin is polymerised and thus the network does not grow indefinitely. The initial AP2 recruitment to membranes is likely aided by high-affinity single appendage interactions like those for epsin1 and eps15. The affinity of both AP2 and epsin1 for membranes and cargo will help stabilise the membrane interactions.
Discussion
To understand the dynamics of α-hub assembly, we first examined the molecular basis and affinities of α-appendage interactions. Our structural information provides clear evidence for two motif interaction sites on the α-appendage (Figure 3). A side site binds WVxF motifs and at least one other unidentified motif (present in proteins like eps15). A top site binds to DxF/W, FxDxF and to peptides containing FxxFxxL (Figure 5). We showed the contribution that both these sites make to α-appendage interactions (Figure 6). These data led us to examine the binding properties of AP2 appendages in solution and when clustered, and from these data we discuss a model for the dynamics of the α-hub in coated vesicle assembly.
Motif-domains
Adaptor appendages interact with short unstructured sequence motifs normally concentrated into domains. The concept of MDs is not unique to AP2 accessory proteins. Other examples are proline-rich domains that contain SH3 binding motifs and proteins lining nuclear pores with their FG repeats. We make the following observations on the basis of our affinity measurements.
Only a limited number of possible interaction motifs can be occupied simultaneously and the presentation of many juxtapositioned motifs leads to an increased affinity for the α-appendage. Another way to increase affinity is to use two different motifs on the same polypeptide chain to interact with the top and side sites of the α-appendage. The major advantage of this mode of interaction (multiple motifs joined by a flexible linker interacting with a three-dimensional surface) is the readily reversible nature of the interactions despite the high avidity (apparent affinity). Individual ‘off rates' for motifs in the network should not be significantly altered and therefore protein partners can readily change and secondary modifications and changes in local environments can dramatically alter the protein network interactions.
For all of these MDs that we have tested in circular dichroism we have not detected any significant structure (see also Kalthoff et al, 2002). However, in Figure 6, we observe that the addition of another motif onto the end of an epsin1-MD construct affects the apparent affinities of all interactions. Previously, we also noted that mutagenesis of an AP1 γ-appendage motif in epsinR affected the affinity of a distant motif (and vice versa) (Mills et al, 2003). Thus, nature has a way of packaging many motifs in a small domain, and when ligands are presented and a motif interacts this may well lead to local changes in the conformation of the domain that either favours or disfavours additional ligand interactions (by unzipping or zipping).
Maturation of the α-hub
In solution, AP2 appendages will interact predominantly with proteins that show a high affinity for isolated AP2 complexes. The eps15:epsin partnership is a prime candidate for an AP2 binding partner at this stage and thus is likely to play a role in adaptor recruitment to membranes. The sensitivity of eps15 network interactions to phosphorylation may well help to regulate this. In assembly-zones, avidity plays a much greater role and therefore proteins with multiple motifs can interact more strongly. However, side site interaction motifs were less abundant in the MDs that we studied and are predicted to be generally less abundant in most accessory proteins with the exception of stonin2 and NECAP. Thus, there will be less competition for side sites on clustered adaptors and consequently WVxF-like motif proteins are likely to play a much more significant role in assembly-zones. From the crystal structure with synaptojanin motifs, we also noted that the presence of top and side binding motifs does not mean that they can interact with the same appendage; however, this depends on the relative orientation of the motifs and the distance apart. It is interesting to note that the motifs in amphiphysin and in the C-terminus of intersectin2 are in a similar orientation and thus these proteins are also likely to be recruited to clustered adaptors. While we believe that the α-appendage samples its environment for affinity and abundance of binding partners, there is a maturation in the appendage hub from being a free α-hub to a clustered α-hub with resultant different affinities for binding partners.
How the α-hub fits into coated vesicle dynamics
Networks of protein interactions provide high avidity and specificity, and yet the reversibility necessary in biological processes. In assembly-zones, AP2 appendages clustered on a two-dimensional surface orchestrate dynamic assemblies of endocytic components. These webs of interactions give stability to many weak protein:protein and protein:lipid interactions. These adaptor protein-network assembly-zones are likely to be transient in nature, but given enough cargo molecules in the membrane to bind to adaptors and a critical PtdIns(4,5)P2 concentration to interact with epsins and other adaptor molecules, then the network is stabilised, clathrin is recruited and the pathway is biased towards coated pit assembly.
While α-appendages may work as protein recruitment hubs, clathrin terminal domain interactions are likely to be organising hubs, given the very weak interactions with binding partners (Figure 4A) and the self-assembly of clathrin. These two hubs are used sequentially during coated vesicle maturation. If clathrin terminal domains were the prime protein recruitment hubs, then it would be difficult to ensure against the formation of empty coated pits without cargo adaptors and membrane. Thus, the cargo adaptor is central in the recruitment of accessory proteins for vesicle formation. This central role of AP2 adaptors in plasma membrane coated pit formation is not surprising and is reflected in the vast reduction of coated pits formed after depletion of AP2 by RNAi (Motley et al, 2003) and in the absence of membrane puncta containing epsin1 when AP2 expression levels are low (Figure 8A). A question arises as to why AP2 adaptors do not self-assemble like clathrin to make the clustered α-hub. This is likely to ensure that AP2 recruitment is dependent on cargo binding, and thus coated pit assembly is dependent on the same. It also gives a greater dynamic instability to the network in contrast to more stable protein oligomerisation. This clustering of AP2 by MD proteins is a novel way to make a ‘sensitive' protein recruitment hub.
In CCVs where AP2 adaptors are enriched, there is no coenrichment of most accessory proteins (Mills et al, 2003), showing that clathrin polymerisation dramatically changes the dynamics of the assembly-zone and of clustered α-hubs. Therefore, adaptor protein-network assembly-zones will be restricted to the edges of forming coated vesicles as supported by the immunolocalisation of eps15 to the edges of coated pits (Tebar et al, 1996), as illustrated in Figure 8C. These assembly-zones mature into coated pits when clathrin begins its lattice assembly or when adjacent preformed clathrin lattices capture assembly-zones. From this point onwards, clathrin interacting proteins will be guiding the fate of the nascent vesicle, whereas the adaptor protein-network assembly-zones will be pushed towards the neck of the nascent vesicle where dynamin is to be finally deposited.
In the process of review, two other papers have appeared in online versions that verify the α-appendage side interaction site that we also find in this paper (Mishra et al, 2004; Ritter et al, 2004).
Materials and methods
Brain protein interaction with GST-appendages
Rat brain extract was prepared by homogenising one brain in 10 ml of 150 mM NaCl, 10 mM HEPES pH 7.4 with 2 mM DTT and protease inhibitors. For interaction experiments, 0.05% Triton X-100 was added to 0.6 ml of this extract+30–100 μg of GST fusion protein+glutathione-Sepharose beads, incubated for 1 h and then the bead-bound proteins were washed three to four times with the same buffer with Triton X-100. The specificity of these experiments was established by looking for an absence of amphiphysin interaction with a parallel GST-β2-appendage pull-down. This may be an arbitrary stringency setting for this experiment (because amphiphysin does interact weakly with β2), but because of the network of interactions that can be formed by endocytic proteins we took this approach to exclude indirect protein interactions. For the α-mutant analysis in Figure 4A, we washed our beads multiple times over a period of 45 min at room temperature. With these conditions, we lose synaptojanin-145 and dynamin interactions but a surprising number of proteins remain attached and thus must be binding extremely tightly. For peptide competition, the peptides were included in both the brain extract and in the first wash at 100 μM unless otherwise stated. To analyse the protein interaction partners of free appendages, we have incubated GST-appendages in brain extract for 1 h and then captured these by centrifugation through a layer of GSH beads on a filter and subsequently washed these beads three times with wash buffer. This was compared with bead-bound appendages incubated with extract for 1 h before capturing the beads+GST-appendages on a filter. The assumption in this experiment is that the assembly of a protein interaction network should not take place efficiently on the moment capture and centrifugation. Brain extract was phosphorylated by addition of 2 mM MgATP+2.5 mM Na-orthovanadate, 0.5 μM cyclosporine A and 0.5 μM okadaic acid followed by 30 min incubation at 37°C. For dephosphorylated extract, 10 U of alkaline phosphatase was added per ml.
For constructs, affinity measurements, crystallography and immunofluorescence, see Supplementary methods.
Mass spectrometry analysis
Peptides of in-gel trypsin-digested protein bands were separated by liquid chromatography on a reverse-phase C18 column (150 × 0.075 mm i.d., flow rate 0.15 μl/min). The eluate was introduced directly into a Q-STAR hybrid tandem mass spectrometer (MDS Sciex, Concord, Ontario, Canada). The spectra were searched against an NCBI non-redundant database with MASCOT MS/MS Ions search (www.matrixscience.com). For proteins with a low number of peptides, we have confirmed their identity by searching the PeptideSearch nrdb database using sequence tags from our data.
Supplementary Material
Supplemental Materials
Supplementary Data
Acknowledgments
We thank Peter McPherson for NECAP and synaptojanin2 antibodies, Volker Haucke for stonin2 antibody, Tom Südhof for synaptojanin antibodies, Phil Evans and our lab members for fruitful discussions and Roger Williams for assistance with data collection. This work was supported by a Marie Curie Fellowship of the EU (contract no. HPMF-CT-2000-01086) to GJKP and by a Research Fellowship from Downing College, Cambridge, to MGJF.
References
- Brett TJ, Traub LM, Fremont DH (2002) Accessory protein recruitment motifs in clathrin-mediated endocytosis. Structure (Camb) 10: 797–809 [DOI] [PubMed] [Google Scholar]
- Brodsky FM, Chen CY, Knuehl C, Towler MC, Wakeham DE (2001) Biological basket weaving: formation and function of clathrin-coated vesicles. Annu Rev Cell Dev Biol 17: 517–568 [DOI] [PubMed] [Google Scholar]
- Chen H, Slepnev VI, Di Fiore PP, De Camilli P (1999) The interaction of epsin and Eps15 with the clathrin adaptor AP-2 is inhibited by mitotic phosphorylation and enhanced by stimulation-dependent dephosphorylation in nerve terminals. J Biol Chem 274: 3257–3260 [DOI] [PubMed] [Google Scholar]
- Collins BM, Praefcke GJ, Robinson MS, Owen DJ (2003) Structural basis for binding of accessory proteins by the appendage domain of GGAs. Nat Struct Biol 10: 607–613 [DOI] [PubMed] [Google Scholar]
- Conner SD, Schmid SL (2003) Regulated portals of entry into the cell. Nature 422: 37–44 [DOI] [PubMed] [Google Scholar]
- Evans PR, Owen DJ (2002) Endocytosis and vesicle trafficking. Curr Opin Struct Biol 12: 814–821 [DOI] [PubMed] [Google Scholar]
- Ford MG, Mills IG, Peter BJ, Vallis Y, Praefcke GJ, Evans PR, McMahon HT (2002) Curvature of clathrin-coated pits driven by epsin. Nature 419: 361–366 [DOI] [PubMed] [Google Scholar]
- Fritz CC, Zapp ML, Green MR (1995) A human nucleoporin-like protein that specifically interacts with HIV Rev. Nature 376: 530–533 [DOI] [PubMed] [Google Scholar]
- Heuser JE, Keen J (1988) Deep-etch visualization of proteins involved in clathrin assembly. J Cell Biol 107: 877–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha A, Agostinelli NR, Mishra SK, Keyel PA, Hawryluk MJ, Traub LM (2004) A novel AP-2 adaptor interaction motif initially identified in the long-splice isoform of synaptojanin 1, SJ170. J Biol Chem 279: 2281–2290 [DOI] [PubMed] [Google Scholar]
- Kalthoff C, Alves J, Urbanke C, Knorr R, Ungewickell EJ (2002) Unusual structural organization of the endocytic proteins AP180 and epsin 1. J Biol Chem 277: 8209–8216 [DOI] [PubMed] [Google Scholar]
- Kim S, Cullis DN, Feig LA, Baleja JD (2001) Solution structure of the Reps1 EH domain and characterization of its binding to NPF target sequences. Biochemistry 40: 6776–6785 [DOI] [PubMed] [Google Scholar]
- Kirchhausen T (2000) Clathrin. Annu Rev Biochem 69: 699–727 [DOI] [PubMed] [Google Scholar]
- Lui WW, Collins BM, Hirst J, Motley A, Millar C, Schu P, Owen DJ, Robinson MS (2003) Binding partners for the COOH-terminal appendage domains of the GGAs and gamma-adaptin. Mol Biol Cell 14: 2385–2398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundmark R, Carlsson SR (2003) Sorting nexin 9 participates in clathrin-mediated endocytosis through interactions with the core components. J Biol Chem 278: 46772–46781 [DOI] [PubMed] [Google Scholar]
- Marks B, McMahon HT (1998) Calcium triggers calcineurin-dependent synaptic vesicle recycling in mammalian nerve terminals. Curr Biol 8: 740–749 [DOI] [PubMed] [Google Scholar]
- Maycox PR, Link E, Reetz A, Morris SA, Jahn R (1992) Clathrin-coated vesicles in nervous tissue are involved primarily in synaptic vesicle recycling. J Cell Biol 118: 1379–1388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miele AE, Watson PJ, Evans PR, Traub LM, Owen DJ (2004) Two distinct interaction motifs in amphiphysin bind two independent sites on the clathrin terminal domain beta-propeller. Nat Struct Mol Biol 11: 242–248 [DOI] [PubMed] [Google Scholar]
- Miller GJ, Mattera R, Bonifacino JS, Hurley JH (2003) Recognition of accessory protein motifs by the gamma-adaptin ear domain of GGA3. Nat Struct Biol 10: 599–606 [DOI] [PubMed] [Google Scholar]
- Mills IG, Praefcke GJ, Vallis Y, Peter BJ, Olesen LE, Gallop JL, Butler PJ, Evans PR, McMahon HT (2003) EpsinR: an AP1/clathrin interacting protein involved in vesicle trafficking. J Cell Biol 160: 213–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra SK, Hawryluk MJ, Brett TJ, Keyel PA, Dupin AL, Jha A, Heuser JE, Fremont DH, Traub LM (2004) Dual-engagement regulation of protein interactions with the AP-2 adaptor alpha appendage. J Biol Chem, Advance online publication 2 August 2004: doi: 10.1074/jbc.M408095200 [DOI] [PubMed] [Google Scholar]
- Motley A, Bright NA, Seaman MN, Robinson MS (2003) Clathrin-mediated endocytosis in AP-2-depleted cells. J Cell Biol 162: 909–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen DJ, Vallis Y, Noble ME, Hunter JB, Dafforn TR, Evans PR, McMahon HT (1999) A structural explanation for the binding of multiple ligands by the alpha-adaptin appendage domain. Cell 97: 805–815 [DOI] [PubMed] [Google Scholar]
- Pearse BM, Robinson MS (1984) Purification and properties of 100-kD proteins from coated vesicles and their reconstitution with clathrin. EMBO J 3: 1951–1957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritter B, Denisov AY, Philie J, Deprez C, Tung EC, Gehring K, McPherson PS (2004) Two WXXF-based motifs in NECAPs define the specificity of accessory protein binding to AP-1 and AP-2. EMBO J, Advance online publication 9 September 2004: doi: 10.1038/sj.emboj.7600378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritter B, Philie J, Girard M, Tung EC, Blondeau F, McPherson PS (2003) Identification of a family of endocytic proteins that define a new alpha-adaptin ear-binding motif. EMBO Rep 4: 1089–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slepnev VI, De Camilli P (2000) Accessory factors in clathrin-dependent synaptic vesicle endocytosis. Nat Rev Neurosci 1: 161–172 [DOI] [PubMed] [Google Scholar]
- Slepnev VI, Ochoa GC, Butler MH, Grabs D, Camilli PD (1998) Role of phosphorylation in regulation of the assembly of endocytic coat complexes. Science 281: 821–824 [DOI] [PubMed] [Google Scholar]
- Takei K, Haucke V (2001) Clathrin-mediated endocytosis: membrane factors pull the trigger. Trends Cell Biol 11: 385–391 [DOI] [PubMed] [Google Scholar]
- Tebar F, Sorkina T, Sorkin A, Ericsson M, Kirchhausen T (1996) Eps15 is a component of clathrin-coated pits and vesicles and is located at the rim of coated pits. J Biol Chem 271: 28727–28730 [DOI] [PubMed] [Google Scholar]
- Walther K, Diril MK, Jung N, Haucke V (2004) Functional dissection of the interactions of stonin 2 with the adaptor complex AP-2 and synaptotagmin. Proc Natl Acad Sci USA 101: 964–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Zhao X, Puertollano R, Bonifacino JS, Eisenberg E, Greene LE (2003) Adaptor and clathrin exchange at the plasma membrane and trans-Golgi network. Mol Biol Cell 14: 516–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Materials
Supplementary Data


