Abstract
Recognition of the large secreted protein Slit by receptors of the Robo family provides fundamental signals in axon guidance and other developmental processes. In Drosophila, Slit–Robo signalling regulates midline crossing and the lateral position of longitudinal axon tracts. We report the functional dissection of Drosophila Slit, using structure analysis, site-directed mutagenesis and in vitro assays. The N-terminal region of Slit consists of a tandem array of four independently folded leucine-rich repeat (LRR) domains, connected by disulphide-tethered linkers. All three Drosophila Robos were found to compete for a single highly conserved site on the concave face of the second LRR domain of Slit. We also found that this domain is sufficient for biological activity in a chemotaxis assay. Other Slit activities may require Slit dimerisation mediated by the fourth LRR domain. Our results show that a small portion of Slit is able to induce Robo signalling and indicate that the distinct functions of Drosophila Robos are encoded in their divergent cytosolic domains.
Keywords: axon guidance, cell migration, leucine-rich repeat, mutagenesis, X-ray crystallography
Introduction
During development, growing axons navigate through the embryo by processing a number of attractive and repulsive signals delivered by cell contact or diffusion through the extracellular matrix. The key ligand–receptor systems involved in axon guidance have been highly conserved in evolution. Many of them have additional functions, for example, in cell migration during development and in human cancer (for reviews, see Tessier-Lavigne and Goodman, 1996; Dickson, 2002; Araujo and Tear, 2003; Guan and Rao, 2003). A particularly well-studied example involves the interaction of Slit and Roundabout (Robo) at the midline of the central nervous system of invertebrates and vertebrates (Brose et al, 1999; Kidd et al, 1999). Slit is a large protein secreted by midline glial cells, while Robo is a transmembrane receptor expressed on the axon growth cone. Drosophila has a single Slit and three Robos (Robo, Robo2, Robo3) and Slit–Robo signalling results in axon repulsion away from the midline. To allow midline crossing of commissural axons, Robo levels at the growth cone surface are regulated by the Drosophila protein Commissureless (Keleman et al, 2002; Myat et al, 2002). Differential expression of the three Robo receptors (the ‘Robo code') determines the lateral position of longitudinal axon tracts relative to the midline (Rajagopalan et al, 2000a, 2000b; Simpson et al, 2000a, 2000b). In mammals, which have three Slits and four Robos, Slit–Robo signalling plays important roles in the development of the nervous system (Jen et al, 2004; Long et al, 2004), lung and kidney formation (Xian et al, 2001; Grieshammer et al, 2004), leucocyte chemotaxis (Wu et al, 2001) and tumour angiogenesis (Wang et al, 2003). Interestingly, mammals lack a Commissureless orthologue, and the repulsion by Slit of precrossing commissural axons is suppressed by a divergent Robo receptor, Robo3/Rig-1 (Marillat et al, 2004; Sabatier et al, 2004).
Slit and Robo are both large multidomain proteins (Figure 1A). Genetic and biochemical experiments have shown that the leucine-rich repeat (LRR) region of Slit and the immunoglobulin-like (IG) domains of Robo are important for Slit–Robo signalling (Battye et al, 2001; Chen et al, 2001; Nguyen Ba-Charvet et al, 2001), but a more detailed characterisation, in particular of the unique LRR structure of Slit, is lacking. We have carried out a thorough structure–function study of the LRR domains of Drosophila Slit and found that all three Robo receptors compete for a single active site located in the second of the four LRR domains of Slit.
Figure 1.
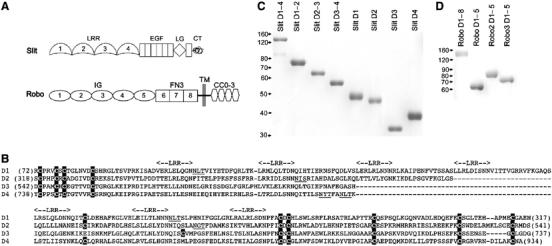
Recombinant Slit and Robo constructs. (A) Domain organisation of Drosophila Slit and Robo: LRR, leucine-rich repeat; EGF, epidermal growth factor-like; LG, laminin G-like; CT, C-terminal cystine-knot; IG, immunoglobulin-like; FN3, fibronectin type 3-like; TM, transmembrane; CC0–3, conserved cytosolic motifs. (B) Sequence alignment of Drosophila Slit LRR domains D1–4. Cysteines are shaded black, putative N-linked glycosylation sites are underlined and the positions of LRR core motifs (LX1X2LX3LX4X5N) are indicated above the alignment. (C) Coomassie blue-stained reducing SDS–PAGE gel of recombinant His-myc-tagged Slit proteins used in this study. (D) Reducing SDS–PAGE gel of recombinant Robo proteins. Robo D1–8 is a fusion protein with a dimerising Fc-tag; all other Robo proteins have a C-terminal FLAG-tag. The positions of molecular mass markers are indicated on the left.
Results
Expression and structure determination of Slit domains
Sequence analysis suggests that the LRR region of Slit consists of four distinct domains, D1–4, each consisting of an N-terminal cap, a variable number of LRRs and a C-terminal cap (Figure 1B). We made expression vectors for the entire LRR region of Drosophila Slit (D1–4), three domain pairs (D1–2, D2–3, D3–4), and all four individual domains (D1, D2, D3, D4). All constructs were produced in good yields by human embryonic kidney 293 cells, demonstrating that the Slit domains defined by sequence analysis indeed correspond to folding units (Figure 1C). On SDS–PAGE, only D3 migrates according to its calculated molecular mass, whereas all other constructs appear to bear N-linked glycans, consistent with sequence analysis (Figure 1B).
To obtain detailed structural information about the Slit LRR domains, we determined the crystal structure of D3, the only domain not modified by glycosylation (Table I and Figure 2A). Slit D3 contains only five LRRs and appears less dramatically curved than larger LRR proteins, such as the ectodomain of Nogo receptor (NogoR) (Barton et al, 2003; He et al, 2003). NogoR consists of nine rather than five LRRs, but the N- and C-terminal portions, including the cap structures, closely match the corresponding regions of Slit D3 (N-terminal 45 Cα atoms, 0.94 Å r.m.s.d.; C-terminal 128 Cα atoms, 1.9 Å r.m.s.d.). The concave face of Slit D3 is formed from seven β-strands, namely a β-hairpin from the N-terminal cap and five parallel strands contributed by the LRRs. The convex back of Slit D3 is made up of loops largely devoid of secondary structure. Compared with all other LRR proteins, the Slit domains contain an additional disulphide bridge tethering the interdomain linker to the convex backs of the LRR array (Figures 1B and 2A).
Table 1.
Crystallographic statistics for Slit D3
| Data collection and reduction | |
| Space group | C2 |
| Unit cell dimensions | a=121.21 Å, b=31.85 Å, c=49.53 Å, β=100.72° |
| Resolution range (Å) | 20–2.8 |
| Unique reflections | 4705 |
| Multiplicity | 4.1 |
| Completeness (%) | 98.5 |
| Rmerge | 0.074 |
| Refinement | |
| Reflections (working set/test set) | 4212/493 |
| Protein atoms | 1465 |
| Water molecules | 5 |
| Rcryst/Rfree | 0.199/0.269 |
| R.m.s.d. bonds (Å) | 0.007 |
| R.m.s.d. angles (deg) | 1.5 |
| R.m.s.d. B-factors (Å2) | 1.7 |
| Ramachandran plot (%)a | 71.1, 28.3, 0.6, 0.0 |
| aResidues in most favoured, additionally allowed, generously allowed and disallowed regions (Laskowski et al, 1993). | |
Figure 2.
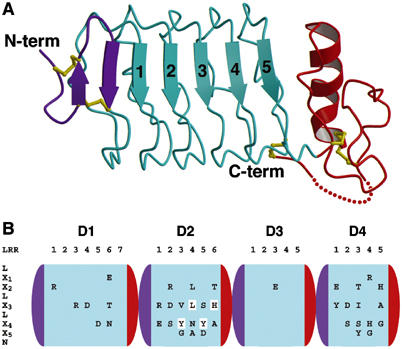
Structure of Slit LRR domains. (A) Cartoon drawing of the crystal structure of Slit D3. The N- and C-terminal caps are in violet and red, respectively, and LRRs 1–5 are in cyan. Disulphide bridges are in yellow. The N- and C-terminus are labelled. (B) Sequence conservation of the concave faces of D1–4. The domains are shown schematically in the orientation and colour scheme used in (A). The LRR consensus sequence is indicated on the left, running vertically from top to bottom. Within each domain, the LRRs are numbered horizontally and invariant residues at solvent-exposed positions X1–X5 are shown. Four residues in D2 that were mutated in this study are highlighted.
The heart of the Slit LRRs is characterised by the sequence LX1X2LX3LX4X5N: the leucine side chains are buried in the hydrophobic core, while residues X1–X5 are exposed on the concave face of the domain and participate in ligand binding in other LRR proteins (Kobe and Kajava, 2001; Huizinga et al, 2002; Schubert et al, 2002). Since cross-species experiments have shown that the Slit–Robo interaction is conserved between invertebrates and vertebrates (Brose et al, 1999), we examined the conservation of residues X1–X5 in six Slit sequences (Caenorhabditis elegans, Drosophila and Xenopus Slit; human Slit1, Slit2 and Slit3). This analysis revealed that the concave faces of D2 and D4 are markedly more conserved than those of D1 and D3 (Figure 2B), whereas residues on the convex back are not conserved in any of the domains. Thus, the concave faces of D2 and/or D4 are likely to be important for Slit function.
Robo binding site on Slit D2
Previous studies have shown that binding of the Slit LRR region to Robo results in growth cone repulsion (Battye et al, 2001; Chen et al, 2001; Nguyen Ba-Charvet et al, 2001). To locate the Robo binding site within this large Slit region (≈900 residues), we prepared soluble Robo proteins (Figure 1D) and analysed their interaction with Slit fragments in a stringent solid-phase assay. Dimeric Drosophila Robo D1–8 bound to immobilised Slit D1–4 with half-maximal saturation at 2–3 μg/ml (Figure 3A). The corresponding KD of ≈10 nM is in good agreement with previous data obtained with Slit proteins captured at cell surfaces (Brose et al, 1999; Li et al, 1999; Nguyen Ba-Charvet et al, 2001). FLAG-tagged Robo D1–5 also bound to Slit D1–4 at similar levels, demonstrating that neither the FN3 domains nor Robo dimerisation are required for Slit binding (Figure 3B). Of the smaller Slit fragments, only D1–2, D2–3 and D2 were recognised by Robo, showing that the (major) Robo binding site is contained within D2 (Figure 3A and B). The same result was obtained when we reversed the order of proteins in the assay and measured binding of Slit fragments to immobilised Robo (Figure 3C). In all experiments, Robo binding by Slit D1–4 was consistently more avid than binding by any of the smaller Slit fragments; we think that this is due to the dimeric nature of D1–4 (see below). Further experiments with Drosophila Robo2 and Robo3 showed that these receptors also have a unique binding site in Slit D2 (Figure 3D and E); competition experiments indicated that the binding of the different Robos to Slit is mutually exclusive (Figure 3F), suggesting a very similar mode of interaction. Importantly, we observed no dramatic differences in the affinities of the three Robos for Slit: they all bound to immobilised Slit D1–4 with half-maximum saturation at 5–20 μg/ml, corresponding to KD values in the low 100 nM range. In the competition experiment, which is less straightforward to quantify, Robo2 appeared to bind somewhat more tightly than Robo or Robo3.
Figure 3.
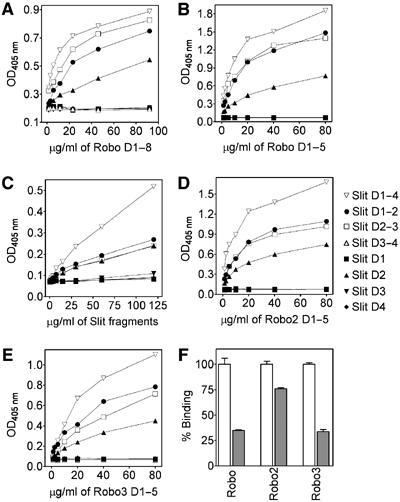
All three Drosophila Robos bind to Slit D2. (A) Binding of dimeric Fc-tagged Robo D1–8 to immobilised Slit fragments. (B) Binding of monomeric FLAG-tagged Robo D1–5 to immobilised Slit fragments. (C) Binding of His-myc-tagged Slit fragments to immobilised Robo D1–5. (D, E) Binding of FLAG-tagged Robo2 D1–5 (D) and Robo3 D1–5 (E) to immobilised Slit fragments. (F) Binding of FLAG-tagged Robos (50 μg/ml) to immobilised Slit D1–4 in the absence (open bars) and presence (grey bars) of a 10-fold excess of His-tagged Robo D1–5. The error bars indicate standard errors of the mean (n=3). Each experiment was carried out at least three times with similar results.
To analyse the role of the conserved concave face of Slit D2 in Robo binding, we mutated solvent-exposed residues in the central LRRs of D2 (Figure 4A). Because the LRR fold is very resilient to nonconservative replacements of such residues (Smits et al, 2003; Vischer et al, 2003), we introduced a large charged side chain into the centre of the presumed Robo binding site (L424R) and removed three conserved aromatic side chains (Y402S, Y450S and H472S). Correct folding of the mutants was indicated by their high expression levels and protein solubility (not shown). All three Drosophila Robos failed to bind to any of the D2 concave face mutants, whereas control mutations on the convex back of D2 (D408S) or concave face of D3 (N649R) had little effect on binding (Figure 4B–D). Thus, the concave face of Slit D2 contains a unique binding site for Robo receptors and critical interactions are provided by at least four consecutive LRRs.
Figure 4.
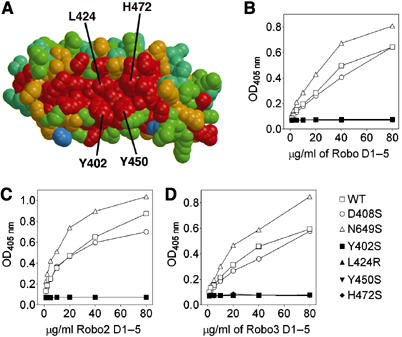
Conserved residues on the concave face of Slit D2 are important in Robo binding. (A) Sequence conservation mapped onto the concave face of a Slit D2 homology model (see text and Materials and methods). The colour scheme ranges from red (identity) to blue (no conservation). The orientation is similar to Figure 2A. Residues mutated in this study are labelled. (B–D) Binding of FLAG-tagged Robo D1–5 (B), Robo2 D1–5 (C) and Robo3 D1–5 (D) to immobilised Slit D2–3 proteins. Each experiment was carried out at least three times with similar results.
Biological activity of Slit D2
Having demonstrated that Slit D2 is sufficient for Robo binding, we wanted to determine whether D2 is also sufficient for productive Robo signalling. Because functional studies of Drosophila Slit and Robo have been limited to genetic experiments, we exploited the remarkable cross-species reactivity of these highly conserved proteins (Brose et al, 1999). A recent study has shown that tumour-derived Slit2 promotes angiogenesis and that purified human Slit2 is chemotactic for human umbilical vein endothelial cells (HUVECs), which express Robo1, a human orthologue of Drosophila Robo (Wang et al, 2003). Like human Slit2, Drosophila Slit D1–4 robustly stimulated HUVEC migration (Figure 5A). D2, the smallest Slit fragment retaining Robo binding activity, stimulated migration to the same extent as D1–4. The chemotactic activity of Drosophila Slit fragments appeared to be mediated by an HUVEC Robo, since Slit-induced, but not bFGF-induced, migration could be reduced to basal levels by an excess of soluble Drosophila Robo. To confirm that the effect of Drosophila Slit on HUVEC migration is directly correlated to Robo binding, we compared wild-type Slit D2–3 and a point mutant defective in Robo binding (L424R); indeed, only the wild-type protein significantly stimulated migration (Figure 5B). Thus, Slit D2 is sufficient to induce Robo-dependent chemotaxis of endothelial cells.
Figure 5.
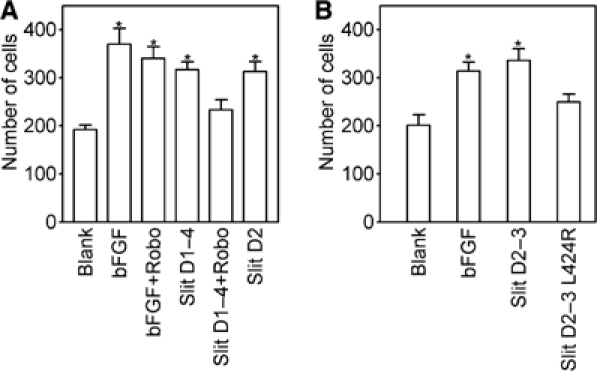
Slit D2 is sufficient to induce Robo-mediated HUVEC chemotaxis. (A) Comparison of the chemotactic activities of 0.4 nM basic fibroblast growth factor (bFGF; positive control) and 5 nM Slit proteins, as well as inhibition of Slit-induced chemotaxis by a 10-fold molar excess of Robo D1–5. (B) Comparison of chemotaxis induced by wild-type and mutant Slit D2–3 (5 nM). The number of migrated cells on the whole filter was determined by a person blinded to the experimental conditions. The bars indicate standard errors of the mean (n=5). An asterisk indicates significantly increased migration relative to unstimulated conditions (P<0.05, Student's t-test). Each experiment was carried out at least three times with similar results.
Noncovalent dimerisation of Slit D4
In the solid-phase binding assay, all Robos bound to Slit D1–4 more avidly than to D2, suggesting that Slit domains other than D2 might contribute to Robo binding. This contribution evidently is not important for Slit-induced chemotaxis, since D2 was as active as D1–4, but the same may not be true for other Slit activities. Using gel filtration chromatography, we found that Slit D4 is a stable noncovalent dimer (Figure 6); chemical crosslinking demonstrated dimerisation also of D1–4 (data not shown). Thus, the enhanced Robo binding of Slit D1–4 is most likely due to Slit dimerisation, and not to an additional (minor) Robo binding site outside of D2.
Figure 6.
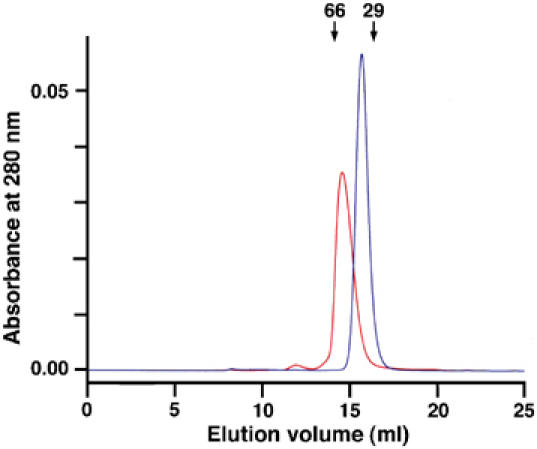
Dimerisation of Slit D4. Shown are gel filtration chromatograms of His-myc-tagged Slit D2 (blue; calculated monomer mass, 30.9 kDa; two N-linked carbohydrate chains) and Slit D4 (red; monomer mass, 27.5 kDa; one N-linked carbohydrate chain). Both proteins were injected at a concentration of 4 mg/ml. The elution volumes of two globular molecular mass standards are indicated by labelled arrows.
Discussion
Slit–Robo signalling has multiple and critical functions in the development of the nervous system and other organs, as well as in human disease, but the structural basis of Slit recognition by Robo receptors is not understood. We have carried out a molecular dissection of the large LRR region of Drosophila Slit and found that all three Drosophila Robos bind, with comparable affinities, to a common site located on the concave face of the second of four LRR domains of Slit (Figures 3 and 4). Thus, the Robo code that determines the lateral position of axon tracts in Drosophila (Rajagopalan et al, 2000a, 2000b; Simpson et al, 2000a, 2000b) is not the result of fundamental differences in Slit recognition, but must be encoded in the divergent cytosolic signalling domains of Robos and/or their expression patterns. This conclusion is consistent with the identification of distinct sets of signalling motifs in the Robo family (Bashaw et al, 2000; Wong et al, 2001; Fan et al, 2003).
Because of the extraordinary conservation of the Robo binding site in Slit D2, and our observation that Drosophila Slit D2 is chemoattractive for human endothelial cells, we believe that the mode of the Slit–Robo interaction is conserved in vertebrates. Thus, we predict that Robo1, Robo2 and Robo3/Rig-1 all bind to Slit D2 in fundamentally the same manner. As in Drosophila, the distinct Robo functions in mammals (Marillat et al, 2004; Sabatier et al, 2004) most likely result from differential expression and/or intracellular signalling, and not from differences in Slit recognition. Furthermore, the three mammalian Slit orthologues are likely to function interchangeably in many settings. Indeed, commissural axon guidance defects are only observed in mice lacking all six Slit alleles (Long et al, 2004).
Slit is unique among LRR proteins in that it contains a tandem of four LRR domains. As a result of the unusual disulphide bridges to the domain linkers, this tandem most likely assumes a complex, nonextended structure, which is further constrained by noncovalent dimerisation via the D4 domain. Given the high conservation of the concave face of D4 (Figure 2B), this surface is likely to form the dimer interface, as suggested for other dimeric LRR proteins (Le Goff et al, 2003). The functional consequences of Slit dimerisation remain to be explored. Recently, a Robo2–Slit–Robo2 sandwich has been proposed to regulate the migration of sensory neurons in Drosophila (Kraut and Zinn, 2004). Since we have shown that there is only one high-affinity Robo2 binding site on Slit, such a sandwich would require Slit to be dimeric.
C-terminal to the LRR region, Slits contain a number of conserved domains that do not appear to be critical for Slit–Robo signalling in vitro (Battye et al, 2001; Chen et al, 2001; Nguyen Ba-Charvet et al, 2001). Proteoglycan binding by the C-terminal region (Ronca et al, 2001) may control Slit diffusion in vivo or help concentrate Slit protein at the axon growth cone surface (Hu, 2001; Johnson et al, 2004; Steigemann et al, 2004). An obligate coreceptor function of proteoglycans appears unlikely, given that the LRR region is biologically active despite a low affinity for heparan sulphate (Ronca et al, 2001). The C-terminal domain of Slit is distantly related to cystine-knot domains of dimeric growth factors, such as TGF-β; whether it contributes to Slit dimerisation remains to be seen.
Our study did not address the question which of the five IG domains of Robo contain the Slit binding site, but several studies have strongly implicated the N-terminal IG domain pair: genetic deletion in mice of Robo1 IG1 results in abnormal lung development (Xian et al, 2001); antibodies against Robo1 IG1 inhibit tumour growth in mice (Wang et al, 2003) and neurite outgrowth in vitro (Hivert et al, 2002); and Robo1 IG1–2 is important for Slit binding and function in vitro (Liu et al, 2004). The Robo binding site on the concave face of Slit D2 is of similar size as a single IG domain, and a direct interaction with Robo IG1 is therefore an attractive hypothesis.
In summary, our results reveal a functional role of the second LRR domain of Slit in Slit–Robo signalling. Importantly, Slit D2 appears to be the sole recognition site for all three Drosophila Robos, and this is most likely the case for vertebrate Slit–Robo signalling as well. Our functional characterisation of the conserved Robo binding site of Slit, along with the structure determination of a Slit LRR domain, will be valuable in future studies into the mechanisms of axon guidance and the development of specific reagents for the blocking of Slit-induced tumour angiogenesis.
Materials and methods
Construction of expression vectors
All constructs were made by PCR amplification from complete cDNA clones of Drosophila Slit and Robos. We used modified pCEP-Pu expression vectors (Kohfeldt et al, 1997) for all but one construct. After cleavage of the signal sequence, the mature proteins contain N- or C-terminal His-tags, an N-terminal His-myc-tag or a C-terminal FLAG-tag. The following domain boundaries were used: Slit D1, 62–318; Slit D2, 314–542; Slit D3, 540–737; Slit D4, 735–934; Robo D1–5, 51–546; Robo2 D1–5, 24–528; Robo3 D1–5, 19–516. Mutant Slit D2–3 constructs were made by overlap extension PCR from the full-length Slit cDNA. A C-terminally Fc-tagged Robo D1–8 (residues 1–881) construct was made by PCR amplification and ligation of the PCR product into the pcFc vector (Leitinger, 2003). The insert sequences of all expression vectors were verified by DNA sequencing.
Protein expression and purification
Proteins were purified from the conditioned medium of episomally transfected 293T cells (Fc-tagged Robo D1–8) or 293-EBNA cells (all other proteins). Cells were maintained in Dulbecco's modified Eagle's medium (DMEM) (Invitrogen) plus 10% fetal bovine serum (FBS) and transfected using Fugene reagent (Roche). Cells containing the episome were selected with either 1 μg/ml of puromycin (293-EBNA cells) or 100 μg/ml of Zeocin (Invitrogen) (293T cells). Resistant cells were grown to confluence and used for the collection of serum-free conditioned medium. His-tagged and His-myc-tagged Slit proteins were purified on TALON metal affinity beads (Clontech). Fc-tagged and FLAG-tagged Robo proteins were purified on columns of protein A–Sepharose and anti-FLAG M2 agarose resin (Sigma), respectively. Purified proteins were dialysed against Tris-buffered saline (TBS); final yields were 2–10 mg of protein per litre of medium. Protein concentrations were determined by measuring absorbance at 280 nm. Gel filtration chromatography of Slit proteins was carried out in TBS using a 24 ml Superdex 200 column on an Äkta system (Amersham Biosciences).
Solid-phase protein binding assay
His-myc-tagged Slit proteins (80 μg/ml) were coated onto 96-well microtitre plates (Maxisorp, Nalge NUNC International) for 2 h. Wells were blocked with TBS/3% casein/0.05% Tween 20 (2 h), and then washed three times with TBS/0.05% Tween 20 and once with TBS. Robo D1–8 in TBS/0.5% casein/0.05% Tween 20 was added for 1 h. After four washes, alkaline phosphatase (AP)-conjugated goat anti-human Fc antibody (Sigma; 1:5000 dilution) was added for 1 h. After further washing, 4 mM of 4-nitrophenyl phosphate in 0.1 M glycine pH 9.6, 1 mM MgCl2 was added to each well, and the reaction was stopped with 5 M NaOH after 20 min. Plates were read in an ELISA reader (Tecan Sunrise) at 405 nm. All other binding assays followed the same protocol, with the exception that FLAG-tagged and myc-tagged proteins were detected, respectively, with AP-conjugated mouse anti-FLAG M2 and anti-myc monoclonal antibodies (Sigma; 1:500 dilution).
HUVEC migration assay
Assays were performed using 24-well multidishes and transwells with polycarbonate membranes (8 μm pores, NUNC). The underside of the filters was coated with 10 μg/ml fibronectin overnight at 4°C and blocked for 1 h with DMEM supplemented with 1% heat-inactivated fetal calf serum (FCS). The bottom chambers were loaded with or without bFGF (Sigma) and various Slit proteins. The upper chambers were seeded with 15 000 HUVECs (TCS Cell Work) resuspended in DMEM+1% FCS. For inhibition experiments, Slit D1–4 was incubated with excess Robo D1–5 (30 min, 37°C) before adding to the bottom chambers. Wells were incubated at 37°C for 4 h in 5% CO2. The filters were then fixed with 4% paraformaldehyde and cells stained with 1% crystal violet for counting. The person counting the migrated cells was blinded to the experimental conditions.
Crystal structure determination of Slit D3
N-terminally His-tagged Slit D3 was concentrated to 10 mg/ml in TBS and crystals were obtained by the hanging drop method at room temperature using 25% (w/v) PEG4000, 0.1 M Tris pH 8.5 and 0.3 M Na-acetate as precipitant. Attempts to flash-freeze the crystals were unsuccessful and X-ray data were collected at room temperature using a MAR image plate detector mounted on a Rigaku RU-H3R rotating anode X-ray generator equipped with OSMIC focusing mirrors (CuKα radiation; λ=1.54 Å). The diffraction data were processed with MOSFLM and programs of the CCP4 suite (CCP4-Collaborative Computing project No. 4, 1994). The crystals are in space group C2 with one Slit D3 molecule in the asymmetric unit (Table I). A search model for molecular replacement was constructed from the human NogoR structure (26% sequence identity) (He et al, 2003). To remove four LRRs not present in Slit D3, NogoR residues 26–77 were fused to residues 175–290 in a manner that maintained the curvature of the LRR region. A solution was obtained with the CCP4 program AMoRe (R=0.49 after rigid-body refinement at 4 Å resolution), and alternating cycles of refinement with CNS (Brunger et al, 1998) and model building with O (Jones et al, 1991) resulted in the final Slit D3 model. Residues 728–730 have no electron density and are presumed disordered. Due to the limited resolution of the data, only a few internal water molecules were included in the model. Crystallographic statistics are summarised in Table I. The coordinates have been deposited in the Protein Data Bank (entry 1w8a). The figures were made with BOBSCRIPT (Esnouf, 1997) and RASTER3D (Merritt and Bacon, 1997).
Homology modelling of Slit D2
A model of Slit D2 was made in O (Jones et al, 1991) using the D3 crystal structure as a template. The additional LRR was added in a similar manner as described for the molecular replacement model. A few minor changes were made to the main chain, but most steric clashes could be relieved by choosing alternative side-chain rotamers from the O database. The model was energy-minimised in CNS (Brunger et al, 1998), yielding Ramachandran statistics comparable to the D3 crystal structure and only a single bad contact according to PROCHECK (Laskowski et al, 1993).
Acknowledgments
We thank Dr Guy Tear (King's College, London) for Slit and Robo cDNAs, Dr Patrik Maurer (University of Cologne) for the pCEP-Pu vectors, Dr Birgit Leitinger (University College London) for the pcFc vector and Dr Jonathan Goldberg (Sloan-Kettering Institute) for a gift of transwell filters. This work was funded by Wellcome Trust grants 067203 and 054334. EH is a Wellcome Senior Research Fellow.
References
- Araujo SJ, Tear G (2003) Axon guidance mechanisms and molecules: lessons from invertebrates. Nat Rev Neurosci 4: 910–922 [DOI] [PubMed] [Google Scholar]
- Barton WA, Liu BP, Tzvetkova D, Jeffrey PD, Fournier AE, Sah D, Cate R, Strittmatter SM, Nikolov DB (2003) Structure and axon outgrowth inhibitor binding of the Nogo-66 receptor and related proteins. EMBO J 22: 3291–3302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashaw GJ, Kidd T, Murray D, Pawson T, Goodman CS (2000) Repulsive axon guidance: Abelson and Enabled play opposing roles downstream of the roundabout receptor. Cell 101: 703–715 [DOI] [PubMed] [Google Scholar]
- Battye R, Stevens A, Perry RL, Jacobs JR (2001) Repellent signaling by Slit requires the leucine-rich repeats. J Neurosci 21: 4290–4298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brose K, Bland KS, Wang KH, Arnott D, Henzel W, Goodman CS, Tessier-Lavigne M, Kidd T (1999) Slit proteins bind Robo receptors and have an evolutionarily conserved role in repulsive axon guidance. Cell 96: 795–806 [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D 54: 905–921 [DOI] [PubMed] [Google Scholar]
- CCP4-Collaborative Computing Project No. 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr D 50: 760–763 [DOI] [PubMed] [Google Scholar]
- Chen JH, Wen L, Dupuis S, Wu JY, Rao Y (2001) The N-terminal leucine-rich regions in Slit are sufficient to repel olfactory bulb axons and subventricular zone neurons. J Neurosci 21: 1548–1556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson BJ (2002) Molecular mechanisms of axon guidance. Science 298: 1959–1964 [DOI] [PubMed] [Google Scholar]
- Esnouf RM (1997) An extensively modified version of MolScript that includes greatly enhanced coloring capabilities. J Mol Graph Model 15: 132–134 [DOI] [PubMed] [Google Scholar]
- Fan X, Labrador JP, Hing H, Bashaw GJ (2003) Slit stimulation recruits Dock and Pak to the roundabout receptor and increases Rac activity to regulate axon repulsion at the CNS midline. Neuron 40: 113–127 [DOI] [PubMed] [Google Scholar]
- Grieshammer U, Le M, Plump AS, Wang F, Tessier-Lavigne M, Martin GR (2004) SLIT2-mediated ROBO2 signaling restricts kidney induction to a single site. Dev Cell 6: 709–717 [DOI] [PubMed] [Google Scholar]
- Guan KL, Rao Y (2003) Signalling mechanisms mediating neuronal responses to guidance cues. Nat Rev Neurosci 4: 941–956 [DOI] [PubMed] [Google Scholar]
- He XL, Bazan JF, McDermott G, Park JB, Wang K, Tessier-Lavigne M, He Z, Garcia KC (2003) Structure of the Nogo receptor ectodomain: a recognition module implicated in myelin inhibition. Neuron 38: 177–185 [DOI] [PubMed] [Google Scholar]
- Hivert B, Liu Z, Chuang CY, Doherty P, Sundaresan V (2002) Robo1 and Robo2 are homophilic binding molecules that promote axonal growth. Mol Cell Neurosci 21: 534–545 [DOI] [PubMed] [Google Scholar]
- Hu H (2001) Cell-surface heparan sulfate is involved in the repulsive guidance activities of Slit2 protein. Nat Neurosci 4: 695–701 [DOI] [PubMed] [Google Scholar]
- Huizinga EG, Tsuji S, Romijn RA, Schiphorst ME, de Groot PG, Sixma JJ, Gros P (2002) Structures of glycoprotein Ibα and its complex with von Willebrand factor A1 domain. Science 297: 1176–1179 [DOI] [PubMed] [Google Scholar]
- Jen JC, Chan WM, Bosley TM, Wan J, Carr JR, Rub U, Shattuck D, Salamon G, Kudo LC, Ou J, Lin DD, Salih MA, Kansu T, Al Dhalaan H, Al Zayed Z, MacDonald DB, Stigsby B, Plaitakis A, Dretakis EK, Gottlob I, Pieh C, Traboulsi EI, Wang Q, Wang L, Andrews C, Yamada K, Demer JL, Karim SS, Alger JR, Geschwind DH, Deller T, Sicotte NL, Nelson SF, Baloh RW, Engle EC (2004) Mutations in a human ROBO gene disrupt hindbrain axon pathway crossing and morphogenesis. Science 304: 1509–1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KG, Ghose A, Epstein E, Lincecum J, O'Connor MB, Van Vactor D (2004) Axonal heparan sulfate proteoglycans regulate the distribution and efficiency of the repellent slit during midline axon guidance. Curr Biol 14: 499–504 [DOI] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A 47: 110–119 [DOI] [PubMed] [Google Scholar]
- Keleman K, Rajagopalan S, Cleppien D, Teis D, Paiha K, Huber LA, Technau GM, Dickson BJ (2002) Comm sorts Robo to control axon guidance at the Drosophila midline. Cell 110: 415–427 [DOI] [PubMed] [Google Scholar]
- Kidd T, Bland KS, Goodman CS (1999) Slit is the midline repellent for the Robo receptor in Drosophila. Cell 96: 785–794 [DOI] [PubMed] [Google Scholar]
- Kobe B, Kajava AV (2001) The leucine-rich repeat as a protein recognition motif. Curr Opin Struct Biol 11: 725–732 [DOI] [PubMed] [Google Scholar]
- Kohfeldt E, Maurer P, Vannahme C, Timpl R (1997) Properties of the extracellular calcium binding module of the proteoglycan testican. FEBS Lett 414: 557–561 [DOI] [PubMed] [Google Scholar]
- Kraut R, Zinn K (2004) Roundabout 2 regulates migration of sensory neurons by signaling in trans. Curr Biol 14: 1319–1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr 26: 283–291 [Google Scholar]
- Le Goff MM, Hindson VJ, Jowitt TA, Scott PG, Bishop PN (2003) Characterization of opticin and evidence of stable dimerization in solution. J Biol Chem 278: 45280–45287 [DOI] [PubMed] [Google Scholar]
- Leitinger B (2003) Molecular analysis of collagen binding by the human discoidin domain receptors, DDR1 and DDR2. Identification of collagen binding sites in DDR2. J Biol Chem 278: 16761–16769 [DOI] [PubMed] [Google Scholar]
- Li HS, Chen JH, Wu W, Fagaly T, Zhou L, Yuan W, Dupuis S, Jiang ZH, Nash W, Gick C, Ornitz DM, Wu JY, Rao Y (1999) Vertebrate Slit, a secreted ligand for the transmembrane protein Roundabout, is a repellent for olfactory bulb axons. Cell 96: 807–818 [DOI] [PubMed] [Google Scholar]
- Liu Z, Patel K, Schmidt H, Andrews W, Pini A, Sundaresan V (2004) Extracellular Ig domains 1 and 2 of Robo are important for ligand (Slit) binding. Mol Cell Neurosci 26: 232–240 [DOI] [PubMed] [Google Scholar]
- Long H, Sabatier C, Ma L, Plump A, Yuan W, Ornitz DM, Tamada A, Murakami F, Goodman CS, Tessier-Lavigne M (2004) Conserved roles for Slit and Robo proteins in midline commissural axon guidance. Neuron 42: 213–223 [DOI] [PubMed] [Google Scholar]
- Marillat V, Sabatier C, Failli V, Matsunaga E, Sotelo C, Tessier-Lavigne M, Chedotal A (2004) The Slit receptor Rig-1/Robo3 controls midline crossing by hindbrain precerebellar neurons and axons. Neuron 43: 69–79 [DOI] [PubMed] [Google Scholar]
- Merritt EA, Bacon DJ (1997) Raster3D: photorealistic molecular graphics. Methods Enzymol 277: 505–524 [DOI] [PubMed] [Google Scholar]
- Myat A, Henry P, McCabe V, Flintoft L, Rotin D, Tear G (2002) Drosophila Nedd4, a ubiquitin ligase, is recruited by Commissureless to control cell surface levels of the Roundabout receptor. Neuron 35: 447–459 [DOI] [PubMed] [Google Scholar]
- Nguyen Ba-Charvet KT, Brose K, Ma L, Wang KH, Marillat V, Sotelo C, Tessier-Lavigne M, Chedotal A (2001) Diversity and specificity of actions of Slit2 proteolytic fragments in axon guidance. J Neurosci 21: 4281–4289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan S, Nicolas E, Vivancos V, Berger J, Dickson BJ (2000a) Crossing the midline: roles and regulation of Robo receptors. Neuron 28: 767–777 [DOI] [PubMed] [Google Scholar]
- Rajagopalan S, Vivancos V, Nicolas E, Dickson BJ (2000b) Selecting a longitudinal pathway: Robo receptors specify the lateral position of axons in the Drosophila CNS. Cell 103: 1033–1045 [DOI] [PubMed] [Google Scholar]
- Ronca F, Andersen JS, Paech V, Margolis RU (2001) Characterization of Slit protein interactions with glypican-1. J Biol Chem 276: 29141–29147 [DOI] [PubMed] [Google Scholar]
- Sabatier C, Plump AS, Le M, Brose K, Tamada A, Murakami F, Lee EY, Tessier-Lavigne M (2004) The divergent Robo family protein Rig-1/Robo3 is a negative regulator of slit responsiveness required for midline crossing by commissural axons. Cell 117: 157–169 [DOI] [PubMed] [Google Scholar]
- Schubert WD, Urbanke C, Ziehm T, Beier V, Machner MP, Domann E, Wehland J, Chakraborty T, Heinz DW (2002) Structure of internalin, a major invasion protein of Listeria monocytogenes, in complex with its human receptor E-cadherin. Cell 111: 825–836 [DOI] [PubMed] [Google Scholar]
- Simpson JH, Bland KS, Fetter RD, Goodman CS (2000a) Short-range and long-range guidance by Slit and its Robo receptors: a combinatorial code of Robo receptors controls lateral position. Cell 103: 1019–1032 [DOI] [PubMed] [Google Scholar]
- Simpson JH, Kidd T, Bland KS, Goodman CS (2000b) Short-range and long-range guidance by Slit and its Robo receptors. Robo and Robo2 play distinct roles in midline guidance. Neuron 28: 753–766 [DOI] [PubMed] [Google Scholar]
- Smits G, Campillo M, Govaerts C, Janssens V, Richter C, Vassart G, Pardo L, Costagliola S (2003) Glycoprotein hormone receptors: determinants in leucine-rich repeats responsible for ligand specificity. EMBO J 22: 2692–2703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steigemann P, Molitor A, Fellert S, Jackle H, Vorbruggen G (2004) Heparan sulfate proteoglycan syndecan promotes axonal and myotube guidance by Slit/Robo signaling. Curr Biol 14: 225–230 [DOI] [PubMed] [Google Scholar]
- Tessier-Lavigne M, Goodman CS (1996) The molecular biology of axon guidance. Science 274: 1123–1133 [DOI] [PubMed] [Google Scholar]
- Vischer HF, Granneman JC, Noordam MJ, Mosselman S, Bogerd J (2003) Ligand selectivity of gonadotropin receptors. Role of the β-strands of extracellular leucine-rich repeats 3 and 6 of the human luteinizing hormone receptor. J Biol Chem 278: 15505–15513 [DOI] [PubMed] [Google Scholar]
- Wang B, Xiao Y, Ding BB, Zhang N, Yuan X, Gui L, Qian KX, Duan S, Chen Z, Rao Y, Geng JG (2003) Induction of tumor angiogenesis by Slit–Robo signaling and inhibition of cancer growth by blocking Robo activity. Cancer Cell 4: 19–29 [DOI] [PubMed] [Google Scholar]
- Wong K, Ren XR, Huang YZ, Xie Y, Liu G, Saito H, Tang H, Wen L, Brady-Kalnay SM, Mei L, Wu JY, Xiong WC, Rao Y (2001) Signal transduction in neuronal migration: roles of GTPase activating proteins and the small GTPase Cdc42 in the Slit–Robo pathway. Cell 107: 209–221 [DOI] [PubMed] [Google Scholar]
- Wu JY, Feng L, Park HT, Havlioglu N, Wen L, Tang H, Bacon KB, Jiang Z, Zhang X, Rao Y (2001) The neuronal repellent Slit inhibits leukocyte chemotaxis induced by chemotactic factors. Nature 410: 948–952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xian J, Clark KJ, Fordham R, Pannell R, Rabbitts TH, Rabbitts PH (2001) Inadequate lung development and bronchial hyperplasia in mice with a targeted deletion in the Dutt1/Robo1 gene. Proc Natl Acad Sci USA 98: 15062–15066 [DOI] [PMC free article] [PubMed] [Google Scholar]


