Abstract
The association of HFE (High Iron FE) major variants with breast cancer risk and behavior has been a matter of discussion for a long time. However, their impact on the expression of iron-related proteins in the breast cancer tissue has never been addressed. In the present study, hepcidin, ferroportin 1, transferrin receptor 1 (TfR1), and ferritin expressions, as well as tissue iron deposition were evaluated in a collection of samples from breast cancers patients and analyzed according to the patients’ HFE genotype. Within the group of patients with invasive carcinoma, those carrying the p.Cys282Tyr variant in heterozygosity presented a higher expression of hepcidin in lymphocytes and macrophages than wild-type or p.His63Asp carriers. An increased expression of TfR1 was also observed in all the cell types analyzed but only in p.Cys282Tyr/p.His63Asp compound heterozygous patients. A differential impact of the two HFE variants was further noticed with the observation of a significantly higher percentage of p.Cys282Tyr heterozygous patients presenting tissue iron deposition in comparison to p.His63Asp heterozygous. In the present cohort, no significant associations were found between HFE variants and classical clinicopathological markers of breast cancer behavior and prognosis. Although limited by a low sampling size, our results provide a new possible explanation for the previously reported impact of HFE major variants on breast cancer progression, i.e., not by influencing systemic iron homeostasis but rather by differentially modulating the local cellular expression of iron-related proteins and tissue iron deposition.
Keywords: Breast cancer, Iron, HFE, p.Cys282Tyr
Introduction
Iron is an essential trace element for the human body, as a critical component of several signaling, transporter and storage molecules involved in energy production and intermediate metabolism [1, 2]. However, its characteristic chemistry contributes to the formation of hazardous molecules, such as hydroxyl radicals and hydrogen peroxide, when in excess [3, 4]. Although most organisms have their proper mechanisms to regulate iron homeostasis and avoid free iron toxicity, current knowledge suggests that the deregulation of these mechanisms may contribute to a number of chronic diseases [5]. Iron is thought to promote carcinogenesis through iron-induced oxidative stress, modulation of signaling networks associated with malignancy and by providing selective advantage to rapidly growing tumor cells [6, 7].
HFE (High Iron Fe) is a MHC class-I like protein that acts as a gatekeeper of systemic iron homeostasis by controlling hepatic hepcidin levels [8, 9]. Hepcidin, in turn, maintains normal plasma iron levels by regulating iron release from cells through the binding to its receptor, the iron exporter ferroportin 1 [10, 11]. A proposed molecular mechanism places HFE and Transferrin Receptor 1 (TfR1) in an iron-sensing complex which is disrupted by binding of circulating holotransferrin with a higher affinity for TfR1 [12]. Upon TfR1 dissociation, HFE is able to relocate to TfR2 and interact with the bone morphogenetic protein (Bmp) co-receptor Hemojuvelin [12, 13], involved in signal communication upon binding of the Bmp ligands, and whose interaction leads to the activation of hepcidin transcription [14–16]. However, previous evidences from others suggest that HFE may also act as a regulator of iron uptake through its direct interaction with the TfR1 [17–19]. HFE gene variants p.Cys282Tyr and p.His63Asp are very common in European derived populations. The p.Cys282Tyr variant disrupts the association of HFE with β-2 microglobulin, reducing the cellular surface expression of HFE [19–21]. This alteration is responsible for the large majority of hereditary hemochromatosis cases [19]. The p.His63Asp variant is believed to lower the HFE protein affinity for TfR1 [22], but its association with iron overload is controversial [23–25]. Although epidemiological studies have been inconsistent in supporting an association between HFE major variants and an increased risk for breast cancer [7], it is plausible to assume that, by interfering with the cellular and tissue iron homeostasis, they may affect the cancer cell phenotype.
We have previously shown that the deregulation of iron-related proteins in breast cancer, more specifically hepcidin, ferroportin 1 (FPN1), TfR1 and ferritin (FT), is not restricted to epithelial cells, but also extends to cells of the tumor microenvironment [26]. To our knowledge, no other group has attempted to verify if the HFE major variants had an impact on the expression of iron-related proteins in the neoplastic context.
Materials and Methods
Sample Characterization
A previously characterized group of human breast tissue samples, archived at the Pathology Service at Centro Hospitalar do Porto, was used in this study. This cohort consisted of 119 samples, including 56 cases of invasive ductal carcinomas (IDC), 14 cases of ductal carcinomas in situ (DCIS) and 49 samples without evidence of breast disease obtained from breast reduction aesthetic surgery. The study has been previously approved by the local Research Ethics Committees, as part of a more extended study (see [26]). Clinicopathological features, such as histological diagnosis, estrogen receptor (ER), progesterone receptor (PR) and human epidermal growth factor 2 (HER2) status were retrieved from interin pathology reports. ER, PR and HER-2 status were assessed by immunohistochemistry. HER2 ambiguous results were confirmed by FISH (Fluorescence in situ hybridization).
Tissue Microarray Construction and Immunohistochemistry
Tissue microarray construction and immunohistochemistry for hepcidin, FPN1, TfR1 and FT for this cohort have been extensively described before [26]. Immunostaining for hepcidin, FPN1, TfR1 and FT was evaluated in epithelial cells, lymphocytes and macrophages using the same semi-quantitative evaluation method as before [26]. Briefly, the score from the percentage of positive cells (scored from 0 to 5) was multiplied by the score of the staining intensity (scored from 0 to 3), resulting in a scale from 0 to 15. Cores from the same donor tissue were grouped and their mean score for each variable calculated.
DAB-Enhanced Perls’ Prussian Blue Staining
To evaluate the presence of iron deposition in breast samples, DAB-enhanced Perls’ Prussian Blue was performed, adapted to the Van Duijn protocol [27]. Samples’ epithelial cells and leukocytic infiltrate were considered positive for iron deposition when at least 10% of the respective cells presented the characteristic brown to dark staining.
DNA Extraction and HFE Genotyping
Genomic DNA was extracted from formalin-fixed paraffin embedded breast sections according to the Ultraprep Tissue DNA kit (AHN Biotechnologie, Germany) recommended procedures. PCR was carried out in 15.5 μL reaction volumes, containing 2 μL of the genomic DNA template, 7.5 μL of MasterMix DNA polymerase, 1 μL of Q-solution (Qiagen Multiplex PCR kit, USA) and 1 μL of each of sense and antisense primers. For the detection of the p.Cys282Tyr variant the following primers were used: 5′-CAAGTGCCTCCTTTGGTGAAGGTGACACAT-3′ as the forward primer and 5′-CTCAGGCACTCCTCTCAACC-3′ as the reverse primer. Following the verification of the 343 bp fragment amplification, RsaI was used for restriction. For the HFE p.His63Asp variant, the following forward and reverse primers’ sequences were used: 5′-ACA TGG TTA AGG CCT GTT GC-3′ and 5′-GCC ACA TCT GGC TTA AAA TT-3′ (primers from Metabion, Germany). These primers amplify a fragment with 294 bp that was then restricted by MboI (enzymes from New England Biolabs, Germany). Primers for detection of variants in the HFE gene were chosen according to the work of Feder et al. [28]. The PCR program included a step of 95 °C, for 15 min followed by 36 cycles of denaturation at 94 °C for 30 s, annealing at 58 °C for 90 s and extension at 72 °C for 90 s. Reaction was extended in the final for 10 min at 72 °C.
Statistical Analysis
Data was analyzed with IBM SPSS Statistics Version 18.0 (SPSS Inc., IBM, USA). Sample distributions were compared by the Kruskal-Wallis test followed by post-hoc testing whenever the omnibus testing was significant. Pearson’s Chi-Square was used to evaluate the differences between categorical variables. In figures, experimental errors are shown as 95% Confidence Intervals (CI). Statistical significance was accepted at p < 0.05.
Once no p.Cys282Tyr heterozygotes were found within the aesthetic breast reduction population, comparisons for this variant were restricted to breast cancer cases.
Results
Expression of Iron-Related Proteins
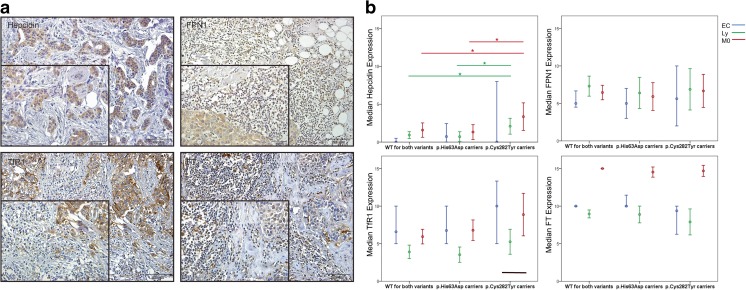
No significant differences were found in the expression of the analyzed iron-related proteins, in all cell types, between wild-type individuals and p.His63Asp carriers included in the aesthetic breast reduction population. Similarly, no significant differences were found for the expression of these proteins in DCIS samples among all the genotypes considered. In IDC cases, however, the expression of hepcidin in lymphocytes and macrophages was significantly higher in patients carrying the p.Cys282Tyr variant relative to both wild type (p = 0.049 and p = 0.050, respectively) or p.His63Asp carriers (p = 0.016 and p = 0.029, respectively) (Fig. 1a). No further differences were found regarding the expression of FPN1, TfR1 and FT in IDC cases (Fig. 1b-d).
Fig. 1.
Iron-related proteins in invasive breast carcinomas according to HFE genotype. a Representative images of Hepcidin, FPN1, TfR1 and FT immunostaining in invasive breast carcinomas. b Median hepcidin, FPN1, TfR1 and FT expression, in epithelial cells, lymphocytes and macrophages from invasive breast carcinomas, in relation to the presence or absence (WT) of the p.His63Asp and p.His63Asp variants in IDC patients. Cell types were discriminated based on their morphology. Scores ranged from 0 to 15 and errors bars present 95% CI. Abbreviations: WT Wild-Type, FPN1 ferroportin 1, TfR1 transferrin receptor 1, FT ferritin, IDC Invasive Ductal Carcinoma
To further clarify the relative impact of the two variants on the results observed, we focused the analysis on a sub-sample of p.Cys282Tyr/p.His63Asp compound heterozygotes. Remarkably, from the 9 p.Cys282Tyr carriers with IDC, 5 also carried p.His63Asp. Although these did not differ in general from the other p.Cys282Tyr/WT IDC pa-tients in terms of the expression of the iron-related proteins, they differed significantly from non-p.Cys282Tyr carriers not only by a higher expression of hepcidin in lymphocytes and macrophages (as described in the whole p.Cys282Tyr carrier population) but they showed, in addition, an increased expression of TfR1 in all the cell types analyzed (Table 1).
Table 1.
Expression of iron-related proteins in HFE p.Cys282Tyr/p.His63Asp compound heterozygous IDC patients is increased in comparison with patients without the p.Cys282Tyr variant
| Iron-Related Proteins (Mean ± SEM) | Non-p.Cys282Tyr carriers (n = 47) | p.Cys282Tyr/p.His63Asp heterozygous compounds (n = 5) | p.Cys282Tyr/WT heterozygous (n = 4) |
|---|---|---|---|
| Hepcidin | |||
| EC | 1.06 ± 0.28 | 3.4 ± 2.93 | 6.38 ± 3.33 |
| Ly | 0.80 ± 0.16 | 2.17 ± 0.69* | 2.00 ± 0.58 |
| M0 | 1.56 ± 0.32 | 3.60 ± 0.98* | 3.83 ± 1.30 |
| FPN1 | |||
| EC | 5.54 ± 0.39 | 6.11 ± 1.02 | 6.13 ± 1.71 |
| Ly | 6.67 ± 0.54 | 8.43 ± 1.06 | 4.28 ± 1.88 |
| M0 | 6.17 ± 0.43 | 7.90 ± 0.75 | 4.61 ± 1.69 |
| TfR1 | |||
| EC | 7.25 ± 0.51 | 11.00 ± 1.72* | 7.65 ± 1.98 |
| Ly | 3.70 ± 0.32 | 5.80 ± 0.97* | 4.33 ± 0.88 |
| M0 | 6.14 ± 0.38 | 9.68 ± 1.42* | 7.50 ± 2.25 |
| FT | |||
| EC | 10.09 ± 0.35 | 8.92 ± 0.74 | 9.06 ± 0.60 |
| Ly | 8.89 ± 0.24 | 8.50 ± 1.07 | 6.88 ± 0.59* |
| M0 | 14.83 ± 0.12 | 15.00 ± 0.00 | 14.17 ± 0.83 |
Abbreviations: IDC invasive ductal carcinoma, SEM Standard Error of the Mean, EC Epithelial Cells, Ly Lymphocytes, M0 Macrophages, NS Non-Significant
*represents significant differences (p < 0.05) for comparison with the non-p.Cys282Tyr carriers group
Tissue Iron Deposition
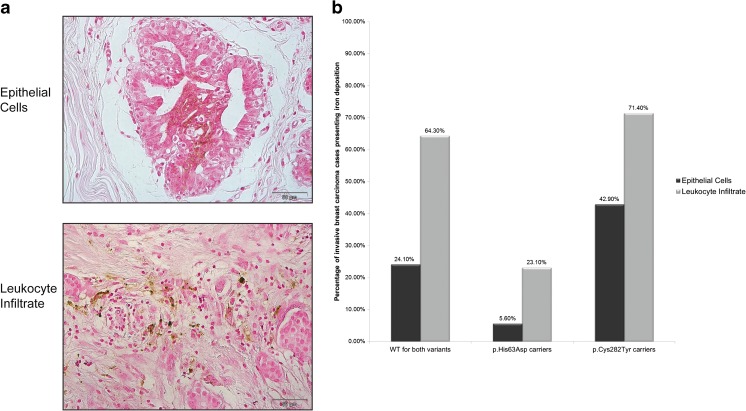
Regarding tissue iron deposition in invasive breast carcinomas, a significantly lower percentage of p.His63Asp carrier IDC patients presented iron deposits in epithelial (vs. p.Cys282Tyr patients – p = 0.024; vs. WT patients – p = 0.036) and stromal inflammatory cells (vs. p.Cys282Tyr patients – p = 0.020; vs. WT patients – p = 0.003), compared with the two other genotypes considered (Fig. 2). Differences between wild-type and p.Cys282Tyr heterozygotes were not statistically significant. All p.Cys282Tyr carrier IDC patients displaying iron deposits in epithelial cells were compound heterozygotes and 4 out of 5 patients with iron deposition in stromal inflammatory cells were also compound heterozygotes.
Fig. 2.
Iron deposition in invasive breast carcinomas according to the HFE genotype. a Representative images of iron deposition in epithelial cells and leukocyte infiltrate from invasive breast carcinomas, as detected with DAB-enhanced Perls’ Prussian Blue staining. b Percentage of invasive breast cancer cases presenting iron deposition, as assessed by DAB-enhanced Perls’ Prussian Blue staining, in epithelial cells (black, n = 51) and in the leukocyte infiltrate (grey, n = 48), in relation to the HFE genotype. Cell types were discriminated based on their morphology. Abbreviations: WT Wild-Type
Clinicopathological Data
Hormone receptor and HER2 status, lymph node involvement and tumor size were available from the interin records and were also analyzed regarding the HFE genotype. None of the different genotypes were associated with any of the considered variables of poor outcome (Table 2).
Table 2.
Clinicopathological features of breast cancer patients according to their HFE genotype
| DCIS | IDC | |||||||
|---|---|---|---|---|---|---|---|---|
| Factors | WT | p.Cys282Tyr | p.Cys282Tyr carriers | p | WT | p.His63Asp carriers | p.Cys282Tyr carriers | p |
| n (%) | n (%) | n (%) | n (%) | n (%) | n (%) | |||
| ER status, n (%)a | ||||||||
| negative | 2 (28.6%) | 1 (25.0%) | 2 (66.7%) | NS | 7 (22.6%) | 3 (16.7%) | 3 (37.5%) | NS |
| positive | 5 (71.4%) | 3 (75.0%) | 1 (33.3%) | 24 (77.4%) | 15 (83.3%) | 5 (62.5%) | ||
| PR status, n (%)a | ||||||||
| negative | 3 (42.9%) | 2 (50.0%) | 2 (66.7%) | NS | 8 (25.8%) | 5 (27.8%) | 4 (50.0%) | NS |
| positive | 4 (57.1%) | 2 (50.0%) | 1 (33.3%) | 23 (74.2%) | 13 (72.2%) | 4 (50.0%) | ||
| HER2 status, n (%)a | ||||||||
| negative | 4 (57.1%) | 1 (33.3%) | 2 (66.7%) | NS | 24 (77.7%) | 13 (76.5%) | 4 (50.00%) | NS |
| positive | 3 (42.9%) | 2 (66.7%) | 1 (33.3%) | 7 (22.6%) | 4 (23.5%) | 4 (50.00%) | ||
| LN metastasis, n (%)a | ||||||||
| non-metastized | 17 (56.7%) | 4 (22.2%) | 4 (50.0%) | NS | ||||
| metastized | 13 (43.3%) | 14 (77.8%) | 4 (50.0%) | |||||
| Median tumor size (IQR)b | 1.00 (1.00–2.00) | 1.00 (1.00–2.00) | 1.00 (1.00–1.50) | NS | ||||
Abbreviations: DCIS ductal carcinoma in situ, IDC invasive ductal carcinoma, ER estrogen receptor, PR progesterone receptor, HER2 Human Epidermal growth factor Receptor 2, LN lymph-node, IQR interquartile range, NS non-significant
aPearson Chi-Square
bKruskal-Wallis Test
Discussion
The knowledge of a remarkable high frequency but low penetrance of the HFE variants [29] contributed to an increasing general interest in the HFE gene as a risk factor or disease modifier in various chronic diseases, including cancer. However, genetic association studies have not been successful in demonstrating a clear relationship between HFE major variants and increased breast cancer risk [7].
In this study we approached the question of the impact of HFE variants in breast cancer by looking at their influence on the local tissue iron homeostasis. While there is no consistent evidence that HFE variants, in the heterozygous form, somehow influence the systemic levels of hepcidin, one cannot ignore the local iron homeostatic regulation and the possibility that an association between HFE variants and other proteins, such as p16, may be cell-specific or even restricted to the neoplastic context [30].
We describe a significantly higher expression of hepcidin in breast tissue lymphocytes and macrophages from patients with invasive ductal carcinomas (IDC) who carry the p.Cys282Tyr variant (Fig. 1a, Table 1) Furthermore, p.Cys282Tyr/p.His63Asp compound heterozygous IDC patients also display enhanced expression of TfR1 in all the cell types analyzed. The presence of the p.His63Asp and the p.Cys282Tyr variants in compound heterozygosity has been already associated with other conditions of altered iron handling at the local level namely increased hepatic iron concentrations in the context of chronic liver diseases [31]. The increased iron deposition in epithelial and stromal inflammatory cells of IDC patients with the p.Cys282Tyr variant may, thus, be a consequence of not only increased iron retention due to the local hepcidin effect but also due to increased TfR1 expression as a result of neoplastic epithelial cells’ ‘iron-deficient’ phenotype [32] and leukocyte activation [33, 34]. The biological consequence of this altered expression of hepcidin and TfR1, and consequently of breast tissue iron content, in p.Cys282Tyr carriers could be a more aggressive course of disease by favoring tumor nutrition [26]. This interpretation may thus support other studies proposing a role for HFE major variants in the regulation of breast cancer behavior [30, 35].
The fact that differences in local iron homeostasis were found only in IDC patients may partially explain the inconsistent results trying to relate the p.Cys282Tyr variant and breast cancer risk, independently of the type of tumor. While others have observed an increased prevalence of the p.Cys282Tyr variant with a higher num-ber of lymph-nodes affected [36] we have not found any association between classic clinicopathological markers in breast cancer and any of the HFE genotypes considered. A slightly higher prevalence of the p.Cys282Tyr variant was observed in patients with markers of poor outcome (ER-, PR-, HER2+) but the insufficient sampling size does not allow us to draw further conclusions.
In conclusion, in spite of the limitation of this study due to low population numbers, the results give further support to the concept of an alternative role for HFE in chronic diseases through modulation of local iron homeostasis, and highlight the need for a more insightful knowledge of the role of HFE in cancer.
Acknowledgements
OM is a recipient of the PhD grant SFRH/BD/2011/78184 from Fundação para a Ciência e Tecnologia (FCT). The authors also acknowledge financial support from ICBAS/AI&NS-UMIB and by national funds through FCT and Ministério da Educação e Ciência (MEC) and when applicable co-funded by FEDER funds within the partnership agreement PT2020 related with the research unit number 4293. The authors would like to thank Drs. Bárbara Leal, Cláudia Carvalho and Luis Cirnes for critical advice in technical procedures.
Compliance with Ethical Standards
This study was approved by Centro Hospitalar do Porto Research Ethics Health Committee (228-CES) and by Centro Hospitalar do Porto Department of Education, Development and Research (152-DEFI) ethical boards. For this type of study, informed consent is not required.
Conflict of Interest
The authors declare they have no competing interests.
References
- 1.Andrews NC. Disorders of iron metabolism. N Engl J Med. 1999;341:1986–1995. doi: 10.1056/NEJM199912233412607. [DOI] [PubMed] [Google Scholar]
- 2.Ganz T, Nemeth E. Hepcidin and disorders of iron metabolism. Annu Rev Med. 2011;62:347–360. doi: 10.1146/annurev-med-050109-142444. [DOI] [PubMed] [Google Scholar]
- 3.Fenton HJH. LXXIII.-oxidation of tartaric acid in presence of iron. J Chem Soc. 1894;65:899–910. doi: 10.1039/CT8946500899. [DOI] [Google Scholar]
- 4.Halliwell B, Gutteridge JM. Biologically relevant metal ion-dependent hydroxyl radical generation. An update. FEBS Lett. 1992;307:108–112. doi: 10.1016/0014-5793(92)80911-Y. [DOI] [PubMed] [Google Scholar]
- 5.Kell DB. Iron behaving badly: inappropriate iron chelation as a major contributor to the aetiology of vascular and other progressive inflammatory and degenerative diseases. BMC Med Genet. 2009;2:2. doi: 10.1186/1755-8794-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eaton JW, Qian M. Molecular bases of cellular iron toxicity. Free Radic Biol Med. 2002;32:833–840. doi: 10.1016/S0891-5849(02)00772-4. [DOI] [PubMed] [Google Scholar]
- 7.Marques O, da Silva BM, Porto G, Lopes C. Iron homeostasis in breast cancer. Cancer Lett. 2014;347:1–14. doi: 10.1016/j.canlet.2014.01.029. [DOI] [PubMed] [Google Scholar]
- 8.Vujic Spasic M, Kiss J, Herrmann T, Galy B, Martinache S, Stolte J, Grone HJ, Stremmel W, Hentze MW, Muckenthaler MU. Hfe acts in hepatocytes to prevent hemochromatosis. Cell Metab. 2008;7:173–178. doi: 10.1016/j.cmet.2007.11.014. [DOI] [PubMed] [Google Scholar]
- 9.Schmidt PJ, Toran PT, Giannetti AM, Bjorkman PJ, Andrews NC. The transferrin receptor modulates Hfe-dependent regulation of hepcidin expression. Cell Metab. 2008;7:205–214. doi: 10.1016/j.cmet.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nemeth E, Tuttle MS, Powelson J, Vaughn MB, Donovan A, Ward DM, Ganz T, Kaplan J. Hepcidin regulates cellular iron efflux by binding to ferroportin and inducing its internalization. Science. 2004;306:2090–2093. doi: 10.1126/science.1104742. [DOI] [PubMed] [Google Scholar]
- 11.Nemeth E, Ganz T. Regulation of iron metabolism by hepcidin. Annu Rev Nutr. 2006;26:323–342. doi: 10.1146/annurev.nutr.26.061505.111303. [DOI] [PubMed] [Google Scholar]
- 12.Goswami T, Andrews NC. Hereditary hemochromatosis protein, HFE, interaction with transferrin receptor 2 suggests a molecular mechanism for mammalian iron sensing. J Biol Chem. 2006;281:28494–28498. doi: 10.1074/jbc.C600197200. [DOI] [PubMed] [Google Scholar]
- 13.D’Alessio F, Hentze MW, Muckenthaler MU. The hemochromatosis proteins HFE, TfR2, and HJV form a membrane-associated protein complex for hepcidin regulation. J Hepatol. 2012;57:1052–1060. doi: 10.1016/j.jhep.2012.06.015. [DOI] [PubMed] [Google Scholar]
- 14.Babitt JL, Huang FW, Wrighting DM, Xia Y, Sidis Y, Samad TA, Campagna JA, Chung RT, Schneyer AL, Woolf CJ, Andrews NC, Lin HY. Bone morphogenetic protein signaling by hemojuvelin regulates hepcidin expression. Nat Genet. 2006;38:531–539. doi: 10.1038/ng1777. [DOI] [PubMed] [Google Scholar]
- 15.Babitt JL, Huang FW, Xia Y, Sidis Y, Andrews NC, Lin HY. Modulation of bone morphogenetic protein signaling in vivo regulates systemic iron balance. J Clin Invest. 2007;117:1933–1939. doi: 10.1172/JCI31342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miyazono K, Maeda S, Imamura T. BMP receptor signaling: transcriptional targets, regulation of signals, and signaling cross-talk. Cytokine Growth Factor Rev. 2005;16:251–263. doi: 10.1016/j.cytogfr.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 17.Lebron JA, Bennett MJ, Vaughn DE, Chirino AJ, Snow PM, Mintier GA, Feder JN, Bjorkman PJ. Crystal structure of the hemochromatosis protein hfe and characterization of its interaction with transferrin receptor. Cell. 1998;93:111–123. doi: 10.1016/S0092-8674(00)81151-4. [DOI] [PubMed] [Google Scholar]
- 18.Waheed A, Grubb JH, Zhou XY, Tomatsu S, Fleming RE, Costaldi ME, Britton RS, Bacon BR, Sly WS. Regulation of transferrin-mediated iron uptake by HFE, the protein defective in hereditary hemochromatosis. Proc Natl Acad Sci U S A. 2002;99:3117–3122. doi: 10.1073/pnas.042701499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feder JN, Gnirke A, Thomas W, Tsuchihashi Z, Ruddy DA, Basava A, Dormishian F, Domingo R, Jr, Ellis MC, Fullan A, Hinton LM, Jones NL, Kimmel BE, Kronmal GS, Lauer P, Lee VK, Loeb DB, Mapa FA, McClelland E, Meyer NC, Mintier GA, Moeller N, Moore T, Morikang E, Prass CE, Quintana L, Starnes SM, Schatzman RC, Brunke KJ, Drayna DT, Risch NJ, Bacon BR, Wolff RK. A novel mhc class i-like gene is mutated in patients with hereditary haemochromatosis. Nat Genet. 1996;13:399–408. doi: 10.1038/ng0896-399. [DOI] [PubMed] [Google Scholar]
- 20.Waheed A, Parkkila S, Zhou XY, Tomatsu S, Tsuchihashi Z, Feder JN, Schatzman RC, Britton RS, Bacon BR, Sly WS. Hereditary hemochromatosis: effects of C282Y and H63D mutations on association with beta2-microglobulin, intracellular processing, and cell surface expression of the HFE protein in COS-7 cells. Proc Natl Acad Sci U S A. 1997;94:12384–12389. doi: 10.1073/pnas.94.23.12384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feder JN, Tsuchihashi Z, Irrinki A, Lee VK, Mapa FA, Morikang E, Prass CE, Starnes SM, Wolff RK, Parkkila S, Sly WS, Schatzman RC. The hemochromatosis founder mutation in HLA-H disrupts beta2-microglobulin interaction and cell surface expression. J Biol Chem. 1997;272:14025–14028. doi: 10.1074/jbc.272.22.14025. [DOI] [PubMed] [Google Scholar]
- 22.Gray SG, Crowe J, Lawless MW. Hemochromatosis: as a conformational disorder. Int J Biochem Cell Biol. 2009;41:2094–2097. doi: 10.1016/j.biocel.2009.04.006. [DOI] [PubMed] [Google Scholar]
- 23.Kelley M, Joshi N, Xie Y, Borgaonkar M. Iron overload is rare in patients homozygous for the H63D mutation. Can J Gastroenterol Hepatol. 2014;28:198–202. doi: 10.1155/2014/468521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aguilar-Martinez P, Bismuth M, Picot MC, Thelcide C, Pageaux GP, Blanc F, Blanc P, Schved JF, Larrey D. Variable phenotypic presentation of iron overload in H63D homozygotes: are genetic modifiers the cause? Gut. 2001;48:836–842. doi: 10.1136/gut.48.6.836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Porto G, Brissot P, Swinkels DW, Zoller H, Kamarainen O, Patton S, Alonso I, Morris M, Keeney S. EMQN best practice guidelines for the molecular genetic diagnosis of hereditary hemochromatosis (HH) Eur J Hum Genet. 2015;24:479–495. doi: 10.1038/ejhg.2015.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marques O, Porto G, Rema A, Faria F, Cruz Paula A, Gomez-Lazaro M, Silva P, Martins da Silva B, Lopes C. Local iron homeostasis in the breast ductal carcinoma microenvironment. BMC Cancer. 2016;16:187. doi: 10.1186/s12885-016-2228-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Duijn S, Nabuurs RJ, van Duinen SG, Natte R. Comparison of histological techniques to visualize iron in paraffin-embedded brain tissue of patients with Alzheimer’s disease. J Histochem Cytochem. 2013;61:785–792. doi: 10.1369/0022155413501325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feder JN, Gnirke A, Thomas W, Tsuchihashi Z, Ruddy DA, Basava A, Dormishian F, Domingo R, Ellis MC, Fullan A, Hinton LM, Jones NL, Kimmel BE, Kronmal GS, Lauer P, Lee VK, Loeb DB, Mapa FA, McClelland E, Meyer NC, Mintier GA, Moeller N, Moore T, Morikang E, Prass CE, Quintana L, Starnes SM, Schatzman RC, Brunke KJ, Drayna DT, Risch NJ, Bacon BR, Wolff RK. A novel MHC class I-like gene is mutated in patients with hereditary haemochromatosis. Nat Genet. 1996;13:399–408. doi: 10.1038/ng0896-399. [DOI] [PubMed] [Google Scholar]
- 29.Waalen J, Nordestgaard BG, Beutler E. The penetrance of hereditary hemochromatosis. Best Pract Res Clin Haematol. 2005;18:203–220. doi: 10.1016/j.beha.2004.08.023. [DOI] [PubMed] [Google Scholar]
- 30.Lee SY, Liu S, Mitchell RM, Slagle-Webb B, Hong YS, Sheehan JM, Connor JR. HFE polymorphisms influence the response to chemotherapeutic agents via induction of p16INK4A. Int J Cancer. 2011;129:2104–2114. doi: 10.1002/ijc.25888. [DOI] [PubMed] [Google Scholar]
- 31.Walsh A, Dixon JL, Ramm GA, Hewett DG, Lincoln DJ, Anderson GJ, Subramaniam VN, Dodemaide J, Cavanaugh JA, Bassett ML, Powell LW. The clinical relevance of compound heterozygosity for the C282Y and H63D substitutions in hemochromatosis. Clin Gastroenterol Hepatol. 2006;4:1403–1410. doi: 10.1016/j.cgh.2006.07.009. [DOI] [PubMed] [Google Scholar]
- 32.Pinnix ZK, Miller LD, Wang W, D’Agostino R Jr., Kute T, Willingham MC, Hatcher H, Tesfay L, Sui G, Di X, Torti SV, Torti FM (2010) Ferroportin and iron regulation in breast cancer progression and prognosis. Sci Transl Med 2:43ra56 [DOI] [PMC free article] [PubMed]
- 33.Manger B, Weiss A, Hardy KJ, Stobo JD. A transferrin receptor antibody represents one signal for the induction of IL 2 production by a human T cell line. J Immunol. 1986;136:532–538. [PubMed] [Google Scholar]
- 34.Paulnock DM, Lambert LE. Identification and characterization of monoclonal antibodies specific for macrophages at intermediate stages in the tumoricidal activation pathway. J Immunol. 1990;144:765–773. [PubMed] [Google Scholar]
- 35.Lee SY, Patton SM, Henderson RJ, Connor JR. Consequences of expressing mutants of the hemochromatosis gene (hfe) into a human neuronal cell line lacking endogenous HFE. FASEB J. 2007;21:564–576. doi: 10.1096/fj.06-6397com. [DOI] [PubMed] [Google Scholar]
- 36.Abraham BK, Justenhoven C, Pesch B, Harth V, Weirich G, Baisch C, Rabstein S, Ko YD, Bruning T, Fischer HP, Haas S, Brod S, Oberkanins C, Hamann U, Brauch H, Network G. Investigation of genetic variants of genes of the hemochromatosis pathway and their role in breast cancer. Cancer Epidemiol Biomark Prev. 2005;14:1102–1107. doi: 10.1158/1055-9965.EPI-05-0013. [DOI] [PubMed] [Google Scholar]