Abstract
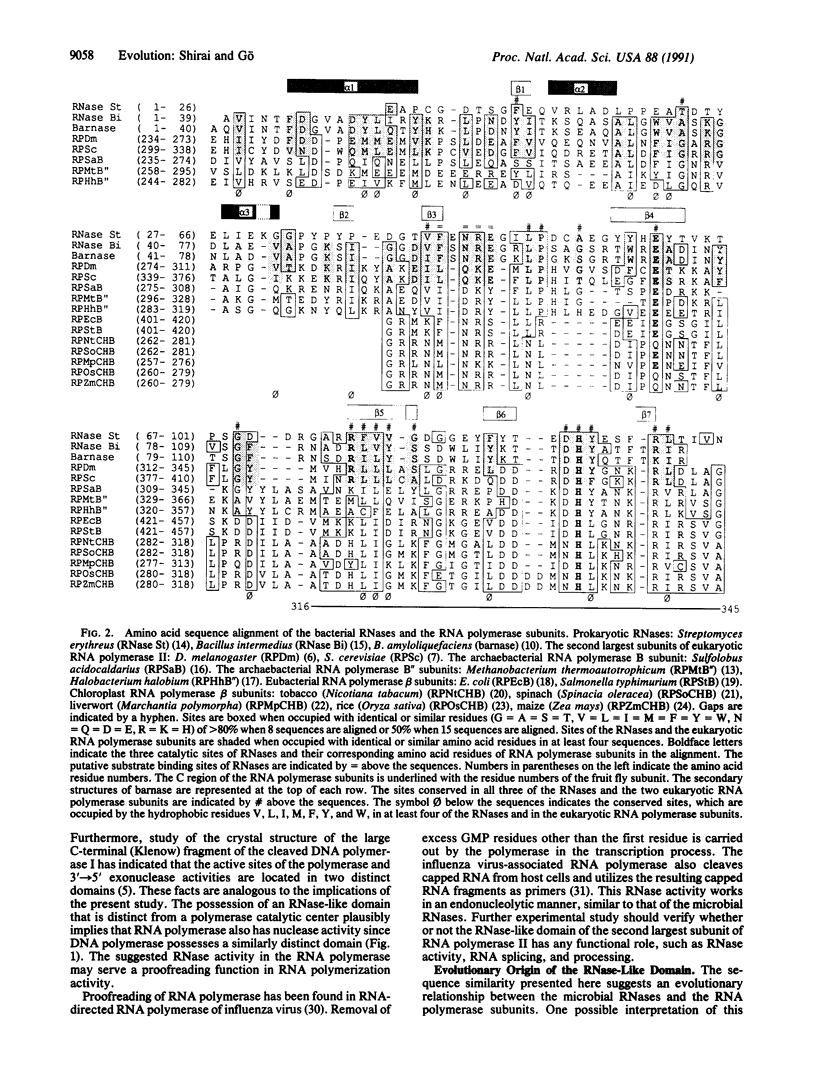
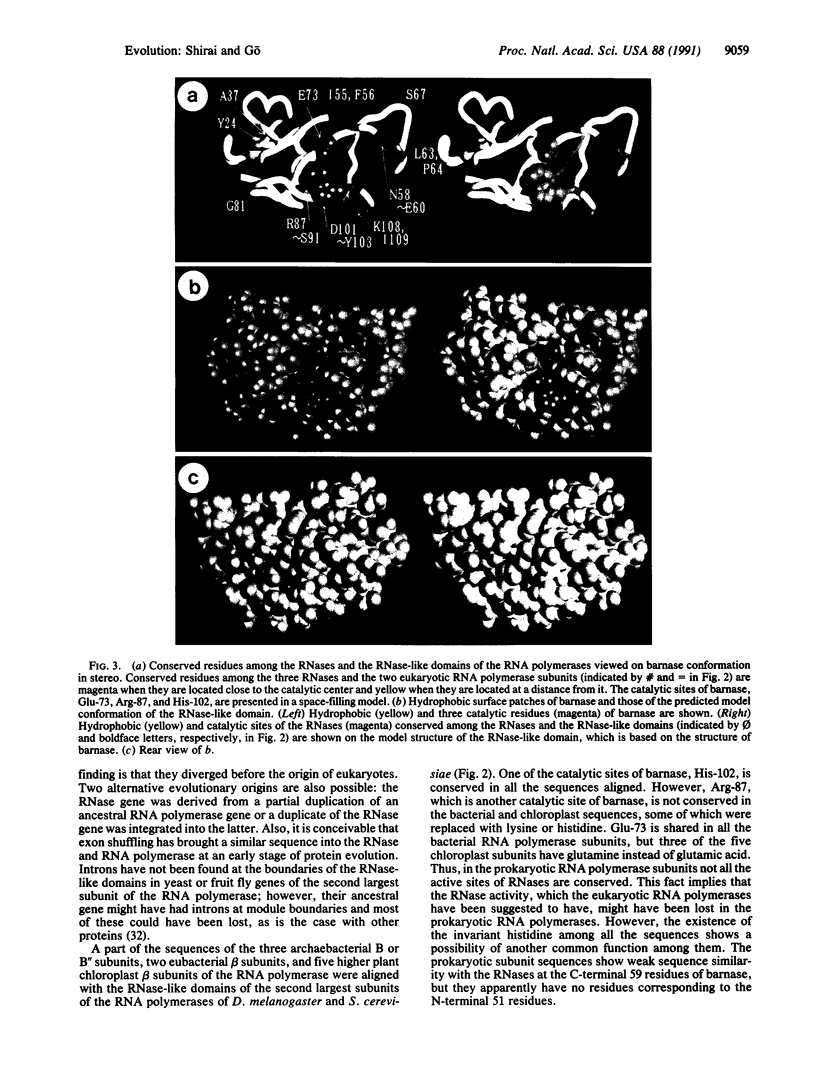
DNA-directed RNA polymerase is responsible for gene expression. Despite its importance, many details of its function and higher-order structure still remain unknown. We report here a local sequence similarity between the second largest subunit of RNA polymerase II and bacterial RNases Ba (barnase), Bi, and St. The most remarkable similarity is that the catalytic sites of the RNases are shared with the eukaryotic RNA polymerase II subunits of Drosophila melanogaster and Saccharomyces cerevisiae. Several amino acids conserved among the RNases and the RNase-like domains of the RNA polymerase subunits are located in the neighborhood of the catalytic sites of barnase, whose three-dimensional structure has been resolved. This observation suggests the functional importance of the RNase-like domain of the RNA polymerase subunits and indicates that the RNase-like domain may have RNase activity. The location of the RNase-like domain relative to the region necessary for RNA polymerization is similar to the relative proximity of 5'----3' or 3'----5' exonuclease and the region of polymerase activity of DNA polymerase I. The RNase-like domain might work in proofreading, as in RNA-directed RNA polymerase of influenza virus, or it may contribute to RNA binding through an unknown function.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allison L. A., Moyle M., Shales M., Ingles C. J. Extensive homology among the largest subunits of eukaryotic and prokaryotic RNA polymerases. Cell. 1985 Sep;42(2):599–610. doi: 10.1016/0092-8674(85)90117-5. [DOI] [PubMed] [Google Scholar]
- Aphanasenko G. A., Dudkin S. M., Kaminir L. B., Leshchinskaya I. B., Severin E. S. Primary structure of ribonuclease from Bacillus intermedius 7P. FEBS Lett. 1979 Jan 1;97(1):77–80. doi: 10.1016/0014-5793(79)80056-3. [DOI] [PubMed] [Google Scholar]
- Berghöfer B., Kröckel L., Körtner C., Truss M., Schallenberg J., Klein A. Relatedness of archaebacterial RNA polymerase core subunits to their eubacterial and eukaryotic equivalents. Nucleic Acids Res. 1988 Aug 25;16(16):8113–8128. doi: 10.1093/nar/16.16.8113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambon P. Eukaryotic nuclear RNA polymerases. Annu Rev Biochem. 1975;44:613–638. doi: 10.1146/annurev.bi.44.070175.003145. [DOI] [PubMed] [Google Scholar]
- Chothia C., Lesk A. M. The evolution of protein structures. Cold Spring Harb Symp Quant Biol. 1987;52:399–405. doi: 10.1101/sqb.1987.052.01.046. [DOI] [PubMed] [Google Scholar]
- Darnell J. E., Doolittle W. F. Speculations on the early course of evolution. Proc Natl Acad Sci U S A. 1986 Mar;83(5):1271–1275. doi: 10.1073/pnas.83.5.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkenburg D., Dworniczak B., Faust D. M., Bautz E. K. RNA polymerase II of Drosophila. Relation of its 140,000 Mr subunit to the beta subunit of Escherichia coli RNA polymerase. J Mol Biol. 1987 Jun 20;195(4):929–937. doi: 10.1016/0022-2836(87)90496-7. [DOI] [PubMed] [Google Scholar]
- Go M., Nosaka M. Protein architecture and the origin of introns. Cold Spring Harb Symp Quant Biol. 1987;52:915–924. doi: 10.1101/sqb.1987.052.01.100. [DOI] [PubMed] [Google Scholar]
- Grachev M. A., Lukhtanov E. A., Mustaev A. A., Zaychikov E. F., Abdukayumov M. N., Rabinov I. V., Richter V. I., Skoblov Y. S., Chistyakov P. G. Studies of the functional topography of Escherichia coli RNA polymerase. A method for localization of the sites of affinity labelling. Eur J Biochem. 1989 Apr 1;180(3):577–585. doi: 10.1111/j.1432-1033.1989.tb14684.x. [DOI] [PubMed] [Google Scholar]
- Hartley R. W., Barker E. A. Amino-acid sequence of extracellular ribonuclease (barnase) of Bacillus amyloliquefaciens. Nat New Biol. 1972 Jan 5;235(53):15–16. doi: 10.1038/newbio235015a0. [DOI] [PubMed] [Google Scholar]
- Hiratsuka J., Shimada H., Whittier R., Ishibashi T., Sakamoto M., Mori M., Kondo C., Honji Y., Sun C. R., Meng B. Y. The complete sequence of the rice (Oryza sativa) chloroplast genome: intermolecular recombination between distinct tRNA genes accounts for a major plastid DNA inversion during the evolution of the cereals. Mol Gen Genet. 1989 Jun;217(2-3):185–194. doi: 10.1007/BF02464880. [DOI] [PubMed] [Google Scholar]
- Hu J., Bogorad L. Maize chloroplast RNA polymerase: the 180-, 120-, and 38-kilodalton polypeptides are encoded in chloroplast genes. Proc Natl Acad Sci U S A. 1990 Feb;87(4):1531–1535. doi: 10.1073/pnas.87.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson G. S., Holton T. A., Whitfield P. R., Bottomley W. Spinach chloroplast rpoBC genes encode three subunits of the chloroplast RNA polymerase. J Mol Biol. 1988 Apr 20;200(4):639–654. doi: 10.1016/0022-2836(88)90477-9. [DOI] [PubMed] [Google Scholar]
- Ishihama A., Mizumoto K., Kawakami K., Kato A., Honda A. Proofreading function associated with the RNA-dependent RNA polymerase from influenza virus. J Biol Chem. 1986 Aug 5;261(22):10417–10421. [PubMed] [Google Scholar]
- Kawata Y., Sakiyama F., Hayashi F., Kyogoku Y. Identification of two essential histidine residues of ribonuclease T2 from Aspergillus oryzae. Eur J Biochem. 1990 Jan 12;187(1):255–262. doi: 10.1111/j.1432-1033.1990.tb15303.x. [DOI] [PubMed] [Google Scholar]
- Leffers H., Gropp F., Lottspeich F., Zillig W., Garrett R. A. Sequence, organization, transcription and evolution of RNA polymerase subunit genes from the archaebacterial extreme halophiles Halobacterium halobium and Halococcus morrhuae. J Mol Biol. 1989 Mar 5;206(1):1–17. doi: 10.1016/0022-2836(89)90519-6. [DOI] [PubMed] [Google Scholar]
- Lisitsyn N. A., Monastyrskaya G. S., Sverdlov E. D. Genes coding for RNA polymerase beta subunit in bacteria. Structure/function analysis. Eur J Biochem. 1988 Nov 1;177(2):363–369. doi: 10.1111/j.1432-1033.1988.tb14385.x. [DOI] [PubMed] [Google Scholar]
- Mauguen Y., Hartley R. W., Dodson E. J., Dodson G. G., Bricogne G., Chothia C., Jack A. Molecular structure of a new family of ribonucleases. Nature. 1982 May 13;297(5862):162–164. doi: 10.1038/297162a0. [DOI] [PubMed] [Google Scholar]
- McClure B. A., Haring V., Ebert P. R., Anderson M. A., Simpson R. J., Sakiyama F., Clarke A. E. Style self-incompatibility gene products of Nicotiana alata are ribonucleases. Nature. 1989 Dec 21;342(6252):955–957. doi: 10.1038/342955a0. [DOI] [PubMed] [Google Scholar]
- Ohme M., Tanaka M., Chunwongse J., Shinozaki K., Sugiura M. A tobacco chloroplast DNA sequence possibly coding for a polypeptide similar to E. coli RNA polymerase beta-subunit. FEBS Lett. 1986 May 5;200(1):87–90. doi: 10.1016/0014-5793(86)80516-6. [DOI] [PubMed] [Google Scholar]
- Ollis D. L., Brick P., Hamlin R., Xuong N. G., Steitz T. A. Structure of large fragment of Escherichia coli DNA polymerase I complexed with dTMP. 1985 Feb 28-Mar 6Nature. 313(6005):762–766. doi: 10.1038/313762a0. [DOI] [PubMed] [Google Scholar]
- Ovchinnikov Y. A., Monastyrskaya G. S., Gubanov V. V., Guryev S. O., Chertov OYu, Modyanov N. N., Grinkevich V. A., Makarova I. A., Marchenko T. V., Polovnikova I. N. The primary structure of Escherichia coli RNA polymerase. Nucleotide sequence of the rpoB gene and amino-acid sequence of the beta-subunit. Eur J Biochem. 1981 Jun 1;116(3):621–629. doi: 10.1111/j.1432-1033.1981.tb05381.x. [DOI] [PubMed] [Google Scholar]
- Plotch S. J., Bouloy M., Ulmanen I., Krug R. M. A unique cap(m7GpppXm)-dependent influenza virion endonuclease cleaves capped RNAs to generate the primers that initiate viral RNA transcription. Cell. 1981 Mar;23(3):847–858. doi: 10.1016/0092-8674(81)90449-9. [DOI] [PubMed] [Google Scholar]
- Pühler G., Lottspeich F., Zillig W. Organization and nucleotide sequence of the genes encoding the large subunits A, B and C of the DNA-dependent RNA polymerase of the archaebacterium Sulfolobus acidocaldarius. Nucleic Acids Res. 1989 Jun 26;17(12):4517–4534. doi: 10.1093/nar/17.12.4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riva M., Carles C., Sentenac A., Grachev M. A., Mustaev A. A., Zaychikov E. F. Mapping the active site of yeast RNA polymerase B (II). J Biol Chem. 1990 Sep 25;265(27):16498–16503. [PubMed] [Google Scholar]
- Sweetser D., Nonet M., Young R. A. Prokaryotic and eukaryotic RNA polymerases have homologous core subunits. Proc Natl Acad Sci U S A. 1987 Mar;84(5):1192–1196. doi: 10.1073/pnas.84.5.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umesono K., Inokuchi H., Shiki Y., Takeuchi M., Chang Z., Fukuzawa H., Kohchi T., Shirai H., Ohyama K., Ozeki H. Structure and organization of Marchantia polymorpha chloroplast genome. II. Gene organization of the large single copy region from rps'12 to atpB. J Mol Biol. 1988 Sep 20;203(2):299–331. doi: 10.1016/0022-2836(88)90002-2. [DOI] [PubMed] [Google Scholar]
- Varmus H. E. Form and function of retroviral proviruses. Science. 1982 May 21;216(4548):812–820. doi: 10.1126/science.6177038. [DOI] [PubMed] [Google Scholar]
- Wilmot C. M., Thornton J. M. Analysis and prediction of the different types of beta-turn in proteins. J Mol Biol. 1988 Sep 5;203(1):221–232. doi: 10.1016/0022-2836(88)90103-9. [DOI] [PubMed] [Google Scholar]
- Yoshida N., Sasaki A., Rashid M. A., Otsuka H. The amino acid sequence of ribonuclease St. FEBS Lett. 1976 Apr 15;64(1):122–125. doi: 10.1016/0014-5793(76)80264-5. [DOI] [PubMed] [Google Scholar]
- Yura T., Ishihama A. Genetics of bacterial RNA polymerases. Annu Rev Genet. 1979;13:59–97. doi: 10.1146/annurev.ge.13.120179.000423. [DOI] [PubMed] [Google Scholar]