Supplemental Digital Content is available in the text
Keywords: association studies, genetics, heart arrest, ion channel, risk prediction, ventricular arrhythmia
Abstract
Potassium calcium-activated channel subfamily N member 2 (KCNN2) encodes an integral membrane protein that forms small-conductance calcium-activated potassium (SK) channels. Recent studies in animal models show that SK channels are important in atrial and ventricular repolarization and arrhythmogenesis. However, the importance of SK channels in human arrhythmia remains unclear. The purpose of the present study was to test the association between genetic polymorphism of the SK2 channel and the occurrence of cardiac tachyarrhythmias in humans. We enrolled 327 Han Chinese, including 72 with clinically significant ventricular tachyarrhythmias (VTa) who had a history of aborted sudden cardiac death (SCD) or unexplained syncope, 98 with a history of atrial fibrillation (AF), and 144 normal controls. We genotyped 12 representative tag single nucleotide polymorphisms (SNPs) across a 141-kb genetic region containing the KCNN2 gene; these captured the full haplotype information. The rs13184658 and rs10076582 variants of KCNN2 were associated with VTa in both the additive and dominant models (odds ratio [OR] 2.89, 95% confidence interval [CI] = 1.505–5.545, P = 0.001; and OR 2.55, 95% CI = 1.428–4.566, P = 0.002, respectively). After adjustment for potential risk factors, the association remained significant. The population attributable risks of these 2 variants of VTa were 17.3% and 10.6%, respectively. One variant (rs13184658) showed weak but significant association with AF in a dominant model (OR 1.91, CI = 1.025–3.570], P = 0.042). There was a significant association between the KCNN2 variants and clinically significant VTa. These findings suggest an association between KCNN2 and VTa; it also appears that KCNN2 variants may be adjunctive markers for risk stratification in patients susceptible to SCD.
1. Introduction
The genetic basis of ventricular tachyarrhythmias (VTa) and sudden cardiac death (SCD) remains unclear. Even when exposed to the same risk factors, such as ischemic or nonischemic cardiomyopathy, inherited channelopathies, or metabolic syndrome, some patients experience lethal arrhythmias while others have minimal symptoms.[1–4] These findings suggest that unknown disease modifiers play a role in the differential outcomes among these patients.
Small-conductance calcium-activated potassium (SK) currents are repolarization currents responsible for afterhyperpolarization of the neurons in the central nervous system.[5,6] Recent studies have demonstrated the existence of SK currents in the atrial myocytes that are responsible for atrial repolarization.[7–9] The SK current is downregulated in diabetic mouse atria and in human chronic atrial fibrillation (AF).[10,11] Inhibition of SK current is proarrhythmic in healthy atria but antiarrhythmic in atrial tachypacing-induced remodeled atria due to the associated increase in action potential duration.[12–15] Although SK current is either very small or undetectable in normal ventricular myocytes paced at normal rates,[16–18] it is important for ventricular repolarization in normal hearts with bradycardia and/or hypokalemia, as well as in ischemic and failing ventricles.[18–25] Upregulation of SK current in failing hearts is responsible for postshock action potential duration shortening and recurrent spontaneous ventricular fibrillation (VF).[18] Although blocking SK current in failing rabbit hearts may act as an antiarrhythmic by suppressing electrical storm, it may also reduce repolarization reserve and induce early afterdepolarization and torsades de pointes ventricular arrhythmia.[24] Therefore, the role of SK current upregulation or downregulation in ventricular arrhythmia is complex, depending on the clinical scenario.[26,27] Nevertheless, these findings suggest that SK channels may play an important role in the pathogenesis of ventricular arrhythmia.
SK channels have 3 subtypes, encoded as potassium calcium-activated channel subfamily N member 1, 2, and 3 (KCNN1, KCNN2, and KCNN3). Among these, variants of KCNN3 are known to be associated with AF,[28] and SK channel subtype 2 (SK2) is known to be upregulated in failing human hearts and plays an important role in ventricular repolarization.[19,20] Although genetic variants are known to be associated with human cardiac arrhythmogenesis,[29,30] there are no reported associations between genetic variants of the KCNN2 gene and the risk of cardiac arrhythmias. The purpose of the present study was to test the association between KCNN2 common genetic variants and cardiac arrhythmias, including both VTa and AF.
2. Methods
The study protocol was in accordance with the Declaration of Helsinki and was approved by the Institutional Ethical Committee of National Taiwan University Hospital. All study subjects signed informed consent before participation.
2.1. Study populations
In a single tertiary referring medical center (National Taiwan University Hospital), 2 groups of patients were consecutively and prospectively enrolled. Group 1 was a defined VTa population that included the following: patients (N = 69) who received implantable cardioverter-defibrillator (ICD) implantation for secondary prevention, including survivors of cardiac arrest due to VF or hemodynamically unstable sustained ventricular tachycardia (VT) after evaluation to define the cause of the event and to exclude any completely reversible cause; and patients (N = 3) with unexplained syncope with clinically relevant, hemodynamically significant sustained VT or VF induced at electrophysiological study. Group 2 (N = 98) included patients with drug-refractory paroxysmal or persistent AF who underwent pulmonary vein isolation by radiofrequency catheter ablation. The control population (N = 144) comprised patients admitted to the Health Management Center of National Taiwan University Hospital. The latter patients did not have diabetes, hypertension, dyslipidemia, or coronary artery disease based on history, physical examination, routine laboratory testing, electrocardiogram, and chest X-ray. Due to wide range of minor allele frequencies (MAF) over different populations, only Han Chinese were enrolled.
2.2. Selection of SNPs and genotyping
Using the HapMap CHB databank (public data release 27 phase II + III, February 2009), 78 single nucleotide polymorphisms (SNPs) were identified in a 141-kb region containing KCNN2, 5 kb upstream, and 1 kb downstream using Haploview software (version 4.2).[31] Twelve tag SNPs (rs6884289, rs7710366, rs2416371, rs11738819, rs10076582, rs338625, rs13181189, rs163305, rs12516818, rs1599175, rs13184658, and rs12652782) were selected, capturing 100% of haplotype variance for all SNPs on KCNN2 with a minimum r[2] value of 0.8 and MAF ≥ 5%. All SNP markers were genotyped by matrix-assisted laser desorption/ionization-time of flight mass spectrometry at the National Center for Genome Medicine, and the experimental protocol can be summarized as follows. A DNA fragment (100–300 bp) encompassing the SNP site was amplified using a polymerase chain reaction GeneAmp 9700 thermal cycler (Applied Biosystems, Foster City, CA) according to the manufacturer's instructions. After polymerase chain reaction, amplification, and neutralization of the deoxynucleotide triphosphates, primer extension was performed by adding the probe, Thermo Sequenase (Amersham Pharmacia, Piscataway, NJ), and an appropriate dideoxynucleotide triphosphate/deoxynucleotide triphosphates mixture. Extension products were differentiated by matrix-assisted laser desorption/ionization-time of flight. We then compared the reference allele frequency and MAF of our population with other populations recorded in the HapMap databank (Supplemental Table S1).
2.3. Statistical analysis
Baseline characteristics of the groups were compared using Student unpaired t test (continuous data) or chi-square test (categorical data). For each SNP, the more common allele in the controls was assigned as the reference category. A Hardy–Weinberg equilibrium (HWE) test was performed for each sequence variant of the control group before marker-trait association analysis. The association of each SNP allele with VTa or AF and control was assessed using logistic regression in models of codominant, dominant, and recessive inheritance. In the model of genotyping analysis, age and sex were adjusted by multivariant logistic regression. For further robust significant association, nominal 2-sided P values were corrected for multiple tests by 10,000 permutations with Haploview software.[31] For haplotype construction, genotype data from the case and control groups were used to estimate intermarker linkage disequilibrium (LD) by measuring pairwise D′ and r2 and defining LD blocks. We used the confidence interval (CI) method component of the Haploview software to define an LD block with an extended spine if D′ was >0.8.[32] The population attributable risk (PAR) was estimated from the control group data as follows: p × (OR − 1)/[p × (OR − 1) + 1], in which p is the prevalence of risk allele among control subjects and OR is the odds ratio of the risk allele.[33] All statistical analyses were performed in IBM SPSS Statistics V20. A 2-sided P ≤ 0.05 was considered statistically significant.
3. Results
3.1. Baseline characteristics
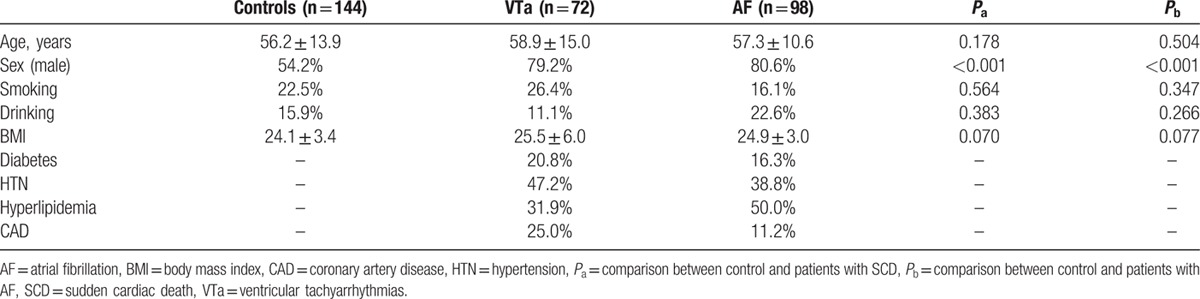
In total, there were 72 patients with history of VTa and 98 patients with AF. The underlying etiologies of the VTa were dilated cardiomyopathy in 31 (43.1%), ischemic cardiomyopathy in 20 (27.8%), and non-heart failure in 21 (29.1%), including 14 (19.4%) with idiopathicVF, 3 (4.2%) with Brugada syndrome, 2 (2.8%) with arrhythmogenic right ventricular cardiomyopathy, 1 (1.4%) with hypertrophic cardiomyopathy, and 1 (1.4%) with long QT syndrome. All the other clinical characteristics are listed in Table 1. VTa is the most disastrous and common end-stage presentation of patients with various cardiovascular diseases, especially those with congestive heart failure of different etiologies. There may be a common factor that predisposes these patients to VTa. Therefore, in the present study, we enrolled a group with various underlying diseases to investigate the association of KCNN2 SNPs with VTa.
Table 1.
Clinical characteristics.
3.2. Association between KCNN2 SNPs and VTa
There are up to 78 common SNPs in the human KCNN2 gene. To decrease genotyping effort, we only genotyped representative tag SNPs. We selected tags that captured most of haplotype variance for all SNPs on the KCNN2 gene. Ten tag SNPs, not including rs13181189 and rs12516818, were successfully genotyped, capturing 89% of haplotype variance. For these 10 SNPs, the success rate of genotyping was 99.7% (range: 99.1%–100%). A graphic representation of the SNPs in relation to the exon-intron structure (according to the National Center for Biotechnology Information) is shown in Fig. 1, middle panel. All the SNPs were in the introns. The genomic position, nucleic acid composition, MAF, HWE test P values, OR, and nominal and permuted P-values of the 10 genotyped SNPs are presented in Table 2. One SNP deviated from the expected count by HWE (rs12657682, P = 0.049).
Figure 1.
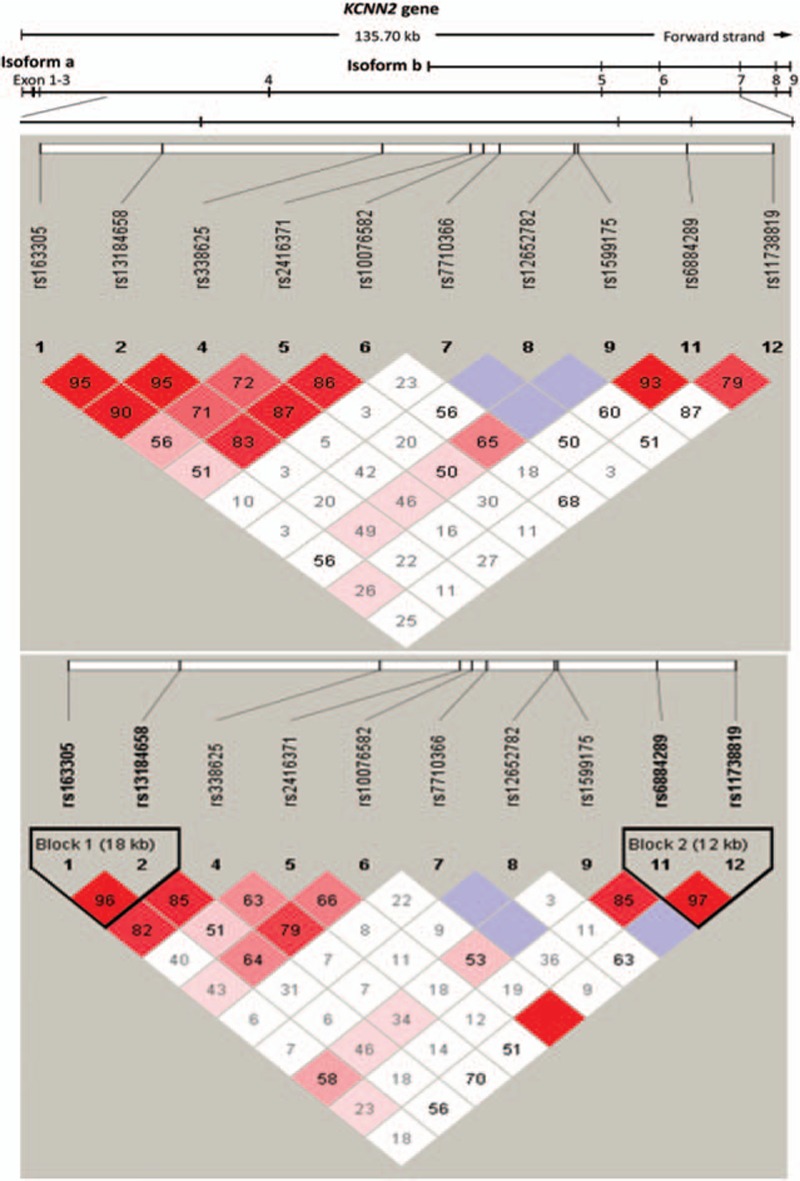
Graphical representation of single nucleotide polymorphisms in relation to the exon-intron structure (upper line) and Haploview LD graph of the KCNN2 gene (middle and lower panels). The exon regions are shown as filled rectangles and are numbered in order. Pairwise LD coefficients D′ × 100 are shown in each cell (D′ values of 1.0 are not shown). A standard Haploview color scheme was applied to the LD color display (LOD score ≥ 2 and D′ = 1 shown in bright red; LOD score ≥ 2 and D′ < 1 shown in pink; LOD score < 2 and D′ = 1 shown in blue; LOD score < 2 and D′ < 1 shown in white). The middle panel shows the association with ventricular tachyarrhythmias; lower panel shows the association with atrial fibrillation. KCNN2 = potassium calcium-activated channel subfamily N member 2, LD = linkage disequilibrium, LOD = logarithm of odds.
Table 2.
Allele association of the 10 tag SNPs in the KCNN2 gene with the risk of sudden cardiac death.
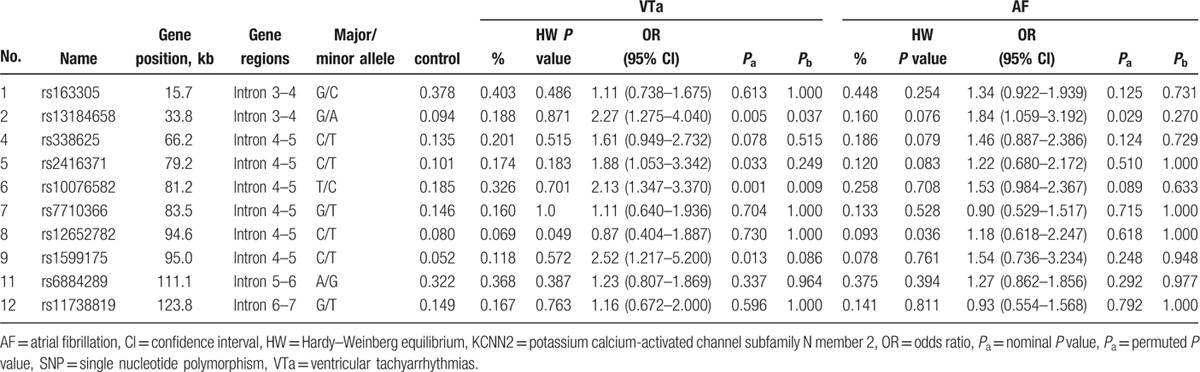
We first compared the allele frequencies of all the SNPs of case and control patients. Interestingly, 4 variants (rs13184658, rs2416371, rs10076582, and rs1599175) showed significant association with VTa. Among these, 2 SNPs (the A variant of rs13184658 and C variant of rs10076582) remained significant after 10,000 permutations (Table 2) and Bonferroni correction. The OR of rs13184658 was 2.27 (95% CI = 1.275–4.040; permutated P = 0.037; P = 0.046 with Bonferroni correction) and that of rs10076582 was 2.13 (95% CI = 1.347–3.370, permuted P = 0.009; P = 0.011 with Bonferroni correction). The estimated PARs were 17.3% and 10.6%, respectively. There was no LD detected.
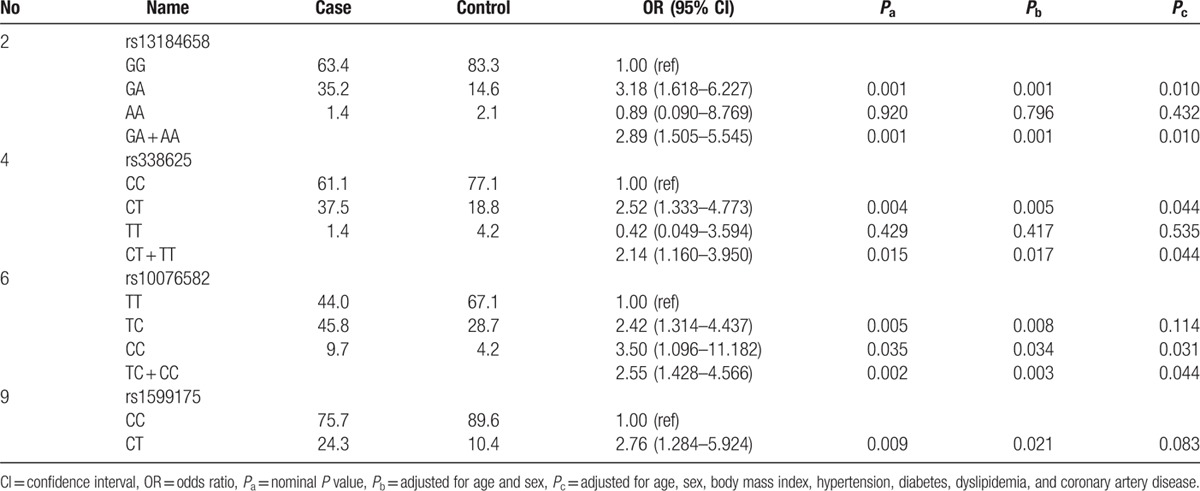
Genotypic analysis (Table 3) showed significant association of 4 SNPs (rs13184658, rs338625, rs10076582, and rs1599175) with VTa. All except rs1599175 revealed significant association in a dominant model. There was no homozygous risk allele for rs1599175. After adjustment for age and sex, all the associations remained significant. After further adjustment for other risk factors, including smoking, drinking, and body mass index, rs13184658, rs338625, and rs10076582 still showed significant association with VTa.
Table 3.
Genotype association analysis.
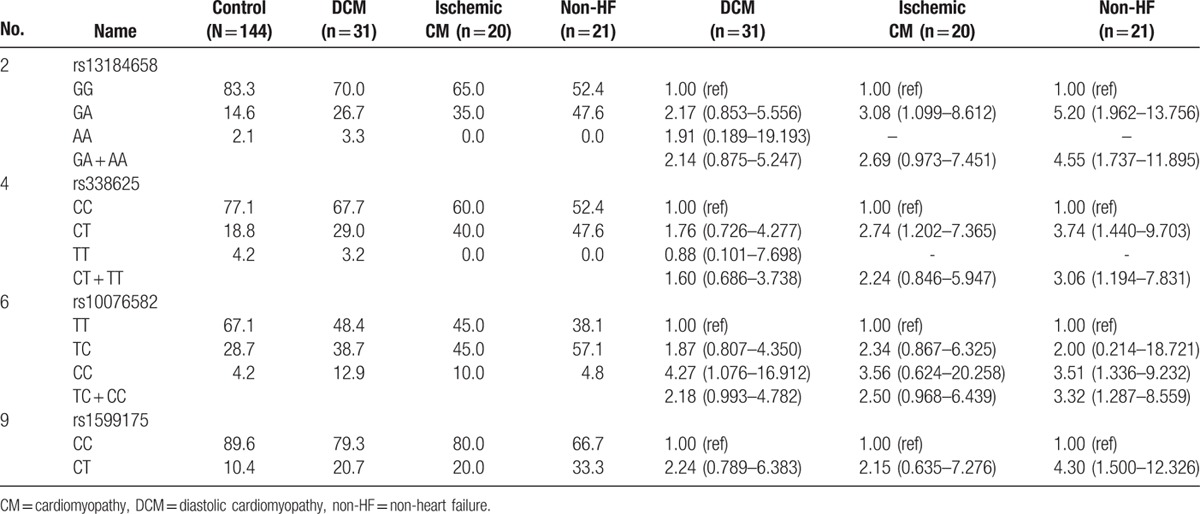
Further analysis was attempted to elucidate the SNP effect among subgroups relative to underlying disease by grouping them as dilated cardiomyopathy, ischemic cardiomyopathy, and non-heart failure (Table 4). However, the power of the analysis was limited due to the relatively small number of cases. Nonetheless, similar trends of positive association were still observed among all three subgroups, implicating the KCNN2 genetic variant as a common factor predisposing patients to VTa.
Table 4.
Subgroup analysis.
3.3. Association between SNPs and AF
Based on the standardized pairwise LD coefficient D′ and estimated r2 of the markers, 2 LD blocks were identified across the gene (Fig. 1, lower panel). One SNP (A allele of rs13184658) showed significant association with AF (OR 1.84, 95% CI = 1.059–3.192, P = 0.029). This SNP was located in LD block 1 in intron 3. The haplotypes within this LD block were significantly associated with AF. Among these haplotypes, the haplotype C-A was associated with an increased risk of AF (P = 0.02). However, both associations became insignificant after 10,000 permutations and Bonferroni correction. In a genotyping test, this variant, rs13184658, showed significant association in a dominant model (OR 1.91, 95% CI = 1.025–3.570, P = 0.042). After adjustment for age and sex, the associations remained significant (OR 3.20, 95% CI = 1.020–3.809, P = 0.044).
4. Discussion
We found a significant association between common KCNN2 variants and sustained VTa in a group of consecutively enrolled patients undergoing ICD implantation for secondary prevention of SCD. Two SNPs (rs13184658 and rs10076582) showed significant association with VTa in a dominant model, carrying 2.55- to 2.89-fold increased risk. The association was robust, as it remained significant after adjustment for multiple potential risk factors. These results suggested that SK2 current may play a role in the mechanism of human VTa.
4.1. Previous genetic association studies with the intermediate phenotype of SCD
Earlier genome-wide association studies (GWAS) and candidate gene analyses identified many rare or common variants that are associated with intermediate phenotypes predictive of SCD risk, including quantitative ECG traits, risk of coronary artery disease, and autonomic function.[34–37] However, the effects of variants associated with intermediate phenotypes were not consistent in later studies,[38] suggesting heterogeneity of underlying genetic causes for those phenotypes.[39] Although conducting a GWAS study to identify SCD genes directly might be a more comprehensive and attractive approach, not all risk variants can be detected by this approach due to its limited power, particularly in the regions that are not well covered by this technique. Focusing on specific candidate genes based on accumulated knowledge from in vivo and in vitro studies and directly using the targeted phenotype, as we have done in this study, is an efficient method of identifying potentially important risk allele(s).
4.2. Second hit theory: KCNN2 variants predispose patients to a greater risk of SCD
Previous studies also showed that while certain rare or common variants (AT1R, ADRB2, SCN5A, KCNH2, and NOS1AP) might not be directly associated with significant clinical arrhythmia syndromes, they affected carriers and predisposed them to malignant arrhythmia when exposed to a 2nd hit, like heart failure or myocardial ischemia.[40–43] Similar scenarios could also be seen in cases of acquired long QT syndrome and ischemic cardiomyopathy, in which rare variants (KCNH2, KCNE1, KCNE2, KCNQ2, SCN5A, and RyR2 in drug-induced long QT syndrome and CASQ2, RAB3GAP1, ZNF365, CXADR, GPC5, GPD1L, NOS1AP, and SCN5A in ischemic cardiomyopathy) were associated with higher risk of malignant arrhythmia and SCD.[44–49] In Brugada syndrome, the variants at SCN5A–SCN10A have a strong impact on susceptibility to SCD.[50] Population-based studies have also found associations between SCD and variants in AGTR1, AGTR2, NOS1AP, SCN5A, and ADRB2.[51–54] In the present study, although the underlying disease etiologies were diverse, a shared trait linked patients to VTa. The association of KCNN2 variants with VTa in patients with a heterogeneous background implied a role of KCNN2 in susceptibility to SCD when exposed to a 2nd hit (an environmental or nongenetic factor) regardless of the underlying cardiac pathology. This was also supported by the finding of our subgroup analysis that all 3 groups have similar trends of positive association.
4.3. Trend of association of KCNN2 genetic polymorphisms with AF
We also found a weak association between KCNN2 (rs13184658) and AF. An earlier GWAS study of lone AF patients showed an association with variants of KCNN3, another subtype of the SK family, but not with KCNN2.[28,55–57] However, downregulation of both SK2 and SK3 currents was observed in human chronic AF,[8] and ablation of SK2 channels resulted in a delay in cardiac repolarization and atrial arrhythmias.[7] Overexpression of the SK3 channels in transgenic mice led to bradyarrhythmias and heart block but not to ventricular arrhythmias.[58] Our finding of an association between AF and KCNN2 further confirms that SK currents are important in the generation or maintenance of AF.
4.4. Potential mechanisms of the association between KCNN2 variants and SCD
The pathophysiological roles of the associated KCNN2 polymorphisms remain unknown. All the polymorphisms are in intron regions and are not directly transcribed to the structure of the SK2 protein. Recent studies have shown that noncoding microRNAs (miRs), which are mainly produced in the intergenic or intron regions, play an important regulating role in gene expression, and may possibly be involved in the pathophysiology of numerous cardiovascular diseases. A single miR can regulate multiple genes, and a single gene can be regulated by multiple miRs.[59–61] It is possible that some of the polymorphisms in the LD block that is associated with our tag SNPs (rs13184658 and rs10076582) are true disease-associated SNPs and are involved in the mechanism of SCD at the epigenetic level mediated by miRs, affecting either KCNN2 or other related genes’ activities. Because SK currents may serve as rescue currents that maintain repolarization reserve in disease conditions,[26] alteration of SK current activity may contribute to cardiac arrhythmogenesis. However, further studies are needed to confirm or dismiss this possibility.
4.5. Limitations
There are several limitations to this study. First, we used the tag SNP association approach to search for common variants associated with common phenotypes among sporadic cases. This approach depends on LD to identify associated SNPs. Therefore, the true responsible variants may not be identified. Direct sequencing for all exons and introns to identify possible rare variants might be another approach, but this would dramatically increase the study cost without increasing the statistical power significantly. Family aggregation analysis would be another approach. However, we did not pursue detailed family histories or acquire blood samples from other family members. Most of the patients were sporadic cases, and we made sure that only 1 patient in each family was indexed in this study. Without knowing the exact responsible variant, further in-depth functional study was not possible. Second, the number of cases in this study is relatively small. Expanding the case numbers for this study or repeating the study results in another independent group of patients is another attractive option. Because it is not easy to clarify the association between SK2 and ventricular arrhythmias, we decided to recruit only patients with the most extreme phenotypes (lethal ventricular arrhythmias) who received ICD implantation for secondary prevention. Expanding the patient group to hemodynamically stable VTs or other intermediate phenotypes might increase the probability of false negative results. Third, the frequency of the variants in a population can affect the PAR and may vary significantly among different populations (Supplemental Table S1). It appears that most KCNN2 gene variants have much lower frequency in Han Chinese than in Caucasians. It is possible that the KCNN2 variants reported in this cohort are not associated with arrhythmias in other populations. Validation of our findings in other populations, particularly among the Caucasians, is warranted. Last, while we attempted to capture 100% of the haplotype variance using 12 tagging SNPs, we failed to genotype 2 of these. Therefore, we only analyzed 10 SNPs, capturing 89% of the haplotype variance. Among the 10 variants, 1 (rs12652782) deviated from HWE. This may be due to the low frequency of the minor allele (as shown in Table 2 and the Supplemental Table S1), a small sampling size, or both.
5. Conclusion
Variants of the KCNN2 gene are associated with the occurrence of lethal ventricular arrhythmias in patients with underlying heart diseases. The clinical impact of these results remains unclear and deserves further study.
Supplementary Material
Acknowledgements
The authors thank the staff of the Second and Eighth Core Lab, Department of Medical Research, National Taiwan University Hospital for their technical support during the study.
Footnotes
Abbreviations: AF = atrial fibrillation, CI = confidence interval, GWAS = genome-wide association studies, HWE = Hardy–Weinberg equilibrium, ICD = implantable cardioverter-defibrillator, KCNN2 = potassium calcium-activated channel subfamily N member 2, KCNN3 = potassium calcium-activated channel subfamily N member 3, LD = linkage disequilibrium, MAF = minor allele frequencies, OR = odds ratio, PAR = population attributable risk, SCD = sudden cardiac death, SK = small-conductance calcium-activated potassium, SK2 = small-conductance calcium-activated potassium channel subtype 2, SNP = single nucleotide polymorphism, VF = ventricular fibrillation, VT = ventricular tachycardia, VTa = ventricular tachyarrhythmias.
C-CY and C-TT contributed equally to this work.
Authorship: C-CY, C-TT, P-LC, W-SY, and J-LL conceived and designed the experiments; C-TT, C-KW, F-CC, F-TC, L-PL, and J-LL contributed the reagents and materials; and C-CY, P-LC, and C-LC analyzed the data; C-CY, C-TT, P-LC, and P-SC drafted the paper. All authors reviewed the manuscript.
Funding/support: This study is funded from NSC 104–2314-B-002–201 and 103–2314-B-002–145 to C-KW; MOST 104–2325-B-002–036 to F-TC; NIH/NHLBI grants R01 HL71140, P01 HL78931, a Medtronic-Zipes Endowment, and the Indiana University Health-Indiana University School of Medicine Strategic Research Initiative of Indiana University to P-SC, He also has equity interest in Arrhythmotech, LLC; NSC 102–2314-B-002–071-MY3 to J-LL.
The remaining authors have no conflicts of interest to disclose.
Supplemental Digital Content is available for this article.
References
- 1.Santulli G, Pagano G, Sardu C, et al. Calcium release channel RyR2 regulates insulin release and glucose homeostasis. J Clin Invest 125:1968–1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sardu C, Carreras G, Katsanos S, et al. Metabolic syndrome is associated with a poor outcome in patients affected by outflow tract premature ventricular contractions treated by catheter ablation. BMC Cardiovasc Disord 2014; 14:176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Santulli G, Marks AR. Essential roles of intracellular calcium release channels in muscle, brain, metabolism, and aging. Curr Mol Pharmacol 2015; 8:206–222. [DOI] [PubMed] [Google Scholar]
- 4.Rizzo MR, Sasso FC, Marfella R, et al. Autonomic dysfunction is associated with brief episodes of atrial fibrillation in type 2 diabetes. J Diabetes Complications 2015; 29:88–92. [DOI] [PubMed] [Google Scholar]
- 5.Kohler M, Hirschberg B, Bond CT, et al. Small-conductance, calcium-activated potassium channels from mammalian brain. Science 1996; 273:1709–1714. [DOI] [PubMed] [Google Scholar]
- 6.Adelman JP, Maylie J, Sah P. Small-conductance Ca2 + -activated K+ channels: form and function. Annu Rev Physiol 2012; 74:245–269. [DOI] [PubMed] [Google Scholar]
- 7.Li N, Timofeyev V, Tuteja D, et al. Ablation of a Ca2+-activated K+ channel (SK2 channel) results in action potential prolongation in atrial myocytes and atrial fibrillation. J Physiol 2009; 587 (Pt 5):1087–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Skibsbye L, Poulet C, Diness JG, et al. Small-conductance calcium-activated potassium (SK) channels contribute to action potential repolarization in human atria. Cardiovasc Res 2014; 103:156–167. [DOI] [PubMed] [Google Scholar]
- 9.Zhang XD, Timofeyev V, Li N, et al. Critical roles of a small conductance Ca(2)(+)-activated K(+) channel (SK3) in the repolarization process of atrial myocytes. Cardiovasc Res 2014; 101:317–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yi F, Ling TY, Lu T, et al. Down-regulation of the small conductance calcium-activated potassium channels in diabetic mouse atria. J Biol Chem 2015; 290:7016–7026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu T, Deng C, Wu R, et al. Decreased expression of small-conductance Ca2+-activated K+ channels SK1 and SK2 in human chronic atrial fibrillation. Life Sci 2012; 90:219–227. [DOI] [PubMed] [Google Scholar]
- 12.Hsueh CH, Chang PC, Hsieh YC, et al. Proarrhythmic effect of blocking the small conductance calcium activated potassium channel in isolated canine left atrium. Heart Rhythm 2013; 10:891–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diness JG, Sorensen US, Nissen JD, et al. Inhibition of small-conductance Ca2+-activated K+ channels terminates and protects against atrial fibrillation. Circ Arrhythm Electrophysiol 2010; 3:380–390. [DOI] [PubMed] [Google Scholar]
- 14.Qi XY, Diness JG, Brundel BJ, et al. Role of small-conductance calcium-activated potassium channels in atrial electrophysiology and fibrillation in the dog. Circulation 2014; 129:430–440. [DOI] [PubMed] [Google Scholar]
- 15.Haugaard MM, Hesselkilde EZ, Pehrson S, et al. Pharmacologic inhibition of small-conductance calcium-activated potassium (SK) channels by NS8593 reveals atrial antiarrhythmic potential in horses. Heart Rhythm 2015; 12:825–835. [DOI] [PubMed] [Google Scholar]
- 16.Nagy N, Szuts V, Horvath Z, et al. Does small-conductance calcium-activated potassium channel contribute to cardiac repolarization? J Mol Cell Cardiol 2009; 47:656–663. [DOI] [PubMed] [Google Scholar]
- 17.Xu Y, Tuteja D, Zhang Z, et al. Molecular identification and functional roles of a Ca(2+)-activated K+ channel in human and mouse hearts. J Biol Chem 2003; 278:49085–49094. [DOI] [PubMed] [Google Scholar]
- 18.Chua SK, Chang PC, Maruyama M, et al. Small-conductance calcium-activated potassium channel and recurrent ventricular fibrillation in failing rabbit ventricles. Circ Res 2011; 108:971–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu CC, Corr C, Shen C, et al. Small conductance calcium-activated potassium current is important in transmural repolarization of failing human ventricles. Circ Arrhythm Electrophysiol 2015; 8:667–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang PC, Turker I, Lopshire JC, et al. Heterogeneous upregulation of apamin-sensitive potassium currents in failing human ventricles. J Am Heart Assoc 2013; 2:e004713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stowe DF, Gadicherla AK, Zhou Y, et al. Protection against cardiac injury by small Ca(2+)-sensitive K(+) channels identified in guinea pig cardiac inner mitochondrial membrane. Biochim Biophys Acta 2013; 1828:427–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee YS, Chang PC, Hsueh CH, et al. Apamin-sensitive calcium-activated potassium currents in rabbit ventricles with chronic myocardial infarction. J Cardiovasc Electrophysiol 2013; 24:1144–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gui L, Bao Z, Jia Y, et al. Ventricular tachyarrhythmias in rats with acute myocardial infarction involves activation of small-conductance Ca2+-activated K+ channels. Am J Physiol Heart Circ Physiol 2013; 304:H118–130. [DOI] [PubMed] [Google Scholar]
- 24.Chang PC, Hsieh YC, Hsueh CH, et al. Apamin induces early afterdepolarizations and torsades de pointes ventricular arrhythmia from failing rabbit ventricles exhibiting secondary rises in intracellular calcium. Heart Rhythm 2013; 10:1516–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chan YH, Tsai WC, Ko JS, et al. Small-conductance calcium-activated potassium current is activated during hypokalemia and masks short-term cardiac memory induced by ventricular pacing. Circulation 2015; 132:1377–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chang PC, Chen PS. SK channels and ventricular arrhythmias in heart failure. Trends Cardiovasc Med 2015; 25:508–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu CC, Ko JS, Ai T, et al. Arrhythmogenic calmodulin mutations impede activation of small-conductance calcium-activated potassium current. Heart Rhythm 2016; pii: S1547-5271(16)30271-5. doi: 10.1016/j.hrthm.2016.05.009. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ellinor PT, Lunetta KL, Glazer NL, et al. Common variants in KCNN3 are associated with lone atrial fibrillation. Nat Genet 2010; 42:240–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arking DE, Junttila MJ, Goyette P, et al. Identification of a sudden cardiac death susceptibility locus at 2q24.2 through genome-wide association in European ancestry individuals. PLoS Genet 2011; 7:e1002158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsai CT, Hsieh CS, Chang SN, et al. Genome-wide screening identifies a KCNIP1 copy number variant as a genetic predictor for atrial fibrillation. Nat Commun 2016; 7:10190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barrett JC, Fry B, Maller J, et al. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 2005; 21:263–265. [DOI] [PubMed] [Google Scholar]
- 32.Jurinke C, van den Boom D, Cantor CR, et al. The use of MassARRAY technology for high throughput genotyping. Adv Biochem Eng Biotechnol 2002; 77:57–74. [DOI] [PubMed] [Google Scholar]
- 33.Rockhill B, Newman B, Weinberg C. Use and misuse of population attributable fractions. Am J Public Health 1998; 88:15–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.den Hoed M, Eijgelsheim M, Esko T, et al. Identification of heart rate-associated loci and their effects on cardiac conduction and rhythm disorders. Nat Genet 2013; 45:621–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arking DE, Pulit SL, Crotti L, et al. Genetic association study of QT interval highlights role for calcium signaling pathways in myocardial repolarization. Nat Genet 2014; 46:826–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Earle N, Yeo Han D, Pilbrow A, et al. Single nucleotide polymorphisms in arrhythmia genes modify the risk of cardiac events and sudden death in long QT syndrome. Heart Rhythm 2014; 11:76–82. [DOI] [PubMed] [Google Scholar]
- 37.Hong KW, Lim JE, Kim JW, et al. Identification of three novel genetic variations associated with electrocardiographic traits (QRS duration and PR interval) in East Asians. Hum Mol Genet 2014; 23:6659–6667. [DOI] [PubMed] [Google Scholar]
- 38.Huertas-Vazquez A, Nelson CP, Sinsheimer JS, et al. Cumulative effects of common genetic variants on risk of sudden cardiac death. IJC Heart Vasc 2015; 7:88–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seyerle AA, Young AM, Jeff JM, et al. Evidence of heterogeneity by race/ethnicity in genetic determinants of QT interval. Epidemiology 2014; 25:790–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blanco RR, Austin H, Vest RN, et al. Angiotensin receptor type 1 single nucleotide polymorphism 1166A/C is associated with malignant arrhythmias and altered circulating miR-155 levels in patients with chronic heart failure. J Card Fail 2012; 18:717–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Frangiskakis JM, London B. Targeting device therapy: genomics of sudden death. Heart Fail Clin 2010; 6:93–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shin J, Lobmeyer MT, Gong Y, et al. Relation of beta(2)-adrenoceptor haplotype to risk of death and heart transplantation in patients with heart failure. Am J Cardiol 2007; 99:250–255. [DOI] [PubMed] [Google Scholar]
- 43.Liu X, Pei J, Hou C, et al. A common NOS1AP genetic polymorphism, rs12567209 G>A, is associated with sudden cardiac death in patients with chronic heart failure in the Chinese Han population. J Card Fail 2014; 20:244–251. [DOI] [PubMed] [Google Scholar]
- 44.Refaat MM, Aouizerat BE, Pullinger CR, et al. Association of CASQ2 polymorphisms with sudden cardiac arrest and heart failure in patients with coronary artery disease. Heart Rhythm 2014; 11:646–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huertas-Vazquez A, Nelson CP, Guo X, et al. Novel loci associated with increased risk of sudden cardiac death in the context of coronary artery disease. PLoS One 2013; 8:e59905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marsman RF, Wilde AA, Bezzina CR. Genetic predisposition for sudden cardiac death in myocardial ischaemia: the Arrhythmia Genetics in the NEtherlandS study. Neth Heart J 2011; 19:96–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Arking DE, Reinier K, Post W, et al. Genome-wide association study identifies GPC5 as a novel genetic locus protective against sudden cardiac arrest. PLoS One 2010; 5:e9879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Westaway SK, Reinier K, Huertas-Vazquez A, et al. Common variants in CASQ2, GPD1L, and NOS1AP are significantly associated with risk of sudden death in patients with coronary artery disease. Circ Cardiovasc Genet 2011; 4:397–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marcsa B, Denes R, Voros K, et al. A common polymorphism of the human cardiac sodium channel alpha subunit (SCN5A) gene is associated with sudden cardiac death in chronic ischemic heart disease. PLoS One 2015; 10:e0132137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bezzina CR, Barc J, Mizusawa Y, et al. Common variants at SCN5A-SCN10A and HEY2 are associated with Brugada syndrome, a rare disease with high risk of sudden cardiac death. Nat Genet 2013; 45:1044–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sotoodehnia N, Li G, Johnson CO, et al. Genetic variation in angiotensin-converting enzyme-related pathways associated with sudden cardiac arrest risk. Heart Rhythm 2009; 6:1306–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Eijgelsheim M, Newton-Cheh C, Aarnoudse AL, et al. Genetic variation in NOS1AP is associated with sudden cardiac death: evidence from the Rotterdam Study. Hum Mol Genet 2009; 18:4213–4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lahtinen AM, Noseworthy PA, Havulinna AS, et al. Common genetic variants associated with sudden cardiac death: the FinSCDgen study. PLoS One 2012; 7:e41675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gavin MC, Newton-Cheh C, Gaziano JM, et al. A common variant in the beta2-adrenergic receptor and risk of sudden cardiac death. Heart Rhythm 2011; 8:704–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Olesen MS, Jabbari J, Holst AG, et al. Screening of KCNN3 in patients with early-onset lone atrial fibrillation. Europace 2011; 13:963–967. [DOI] [PubMed] [Google Scholar]
- 56.Chang SH, Chang SN, Hwang JJ, et al. Significant association of rs13376333 in KCNN3 on chromosome 1q21 with atrial fibrillation in a Taiwanese population. Circ J 2012; 76:184–188. [DOI] [PubMed] [Google Scholar]
- 57.Luo Z, Yan C, Zhang W, et al. Association between SNP rs13376333 and rs1131820 in the KCNN3 gene and atrial fibrillation in the Chinese Han population. Clin Chem Lab Med 2014; 52:1867–1873. [DOI] [PubMed] [Google Scholar]
- 58.Mahida S, Mills RW, Tucker NR, et al. Overexpression of KCNN3 results in sudden cardiac death. Cardiovasc Res 2014; 101:326–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Santulli G, Iaccarino G, De Luca N, et al. Atrial fibrillation and microRNAs. Front Physiol 2014; 5:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wronska A, Kurkowska-Jastrzebska I, Santulli G. Application of microRNAs in diagnosis and treatment of cardiovascular disease. Acta Physiol 2015; 213:60–83. [DOI] [PubMed] [Google Scholar]
- 61.Sardu C, Santamaria M, Paolisso G, et al. microRNA expression changes after atrial fibrillation catheter ablation. Pharmacogenomics 2015; 16:1863–1877. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


