Abstract
The outer membrane (OM) of Gram-negative bacteria is composed of lipopolysaccharide (LPS) in the outer leaflet and phospholipids in the inner leaflet. The outer membrane protein H (OprH) of Pseudomonas aeruginosa provides an increased stability to the OMs by directly interacting with LPS. Here we report the influence of various P. aeruginosa and, for comparison, Escherichia coli LPS environments on the physical properties of the OMs and OprH using all-atom molecular dynamics simulations. The simulations reveal that although the P. aeruginosa OMs are thinner hydrophobic bilayers than the E. coli OMs, which is expected from the difference in the acyl chain length of their lipid A, this effect is almost imperceptible around OprH due to a dynamically adjusted hydrophobic match between OprH and the OM. The structure and dynamics of the extracellular loops of OprH show distinct behaviors in different LPS environments. Including the O-antigen greatly reduces the flexibility of the OprH loops and increases the interactions between these loops and LPS. Furthermore, our study shows that the interactions between OprH and LPS mainly depend on the secondary structure of OprH and the chemical structure of LPS, resulting in distinctive patterns in different LPS environments.
Introduction
Cells of Gram-negative bacteria are surrounded by two lipid bilayer membranes. The inner (cytoplasmic) membrane is exclusively composed of phospholipids, mainly phosphatidylethanolamine (PE), phosphatidylglycerol (PG), and cardiolipin (CL), distributed equally among the inner and outer leaflets. In contrast, the outer membrane (OM) is highly asymmetric, with the inner leaflet having the same lipid composition as the cytoplasmic membrane, and the outer leaflet consisting almost exclusively of lipopolysaccharide (LPS) molecules (1, 2). LPS contains a hydrophobic lipid A region, a hydrophilic core oligosaccharide chain, and an O-antigen polysaccharide chain. Among Gram-negative bacteria, the basic chemical structures of lipid A and the core are more conserved than those of O-antigen polysaccharides. The latter consists of highly variable repeating units (up to ∼25) of about five sugars whose sequence is the basis for serogroup determination of bacteria. The lipid A structure contains an N- and O-acylated β-(1→6)-linked diglucosamine backbone, with chemical variation in the number of primary acyl groups and the types of fatty acids substituting the primary and secondary acyl chains, which anchor lipid A to the OM (3, 4).
The Gram-negative bacterium Pseudomonas aeruginosa is a major opportunistic pathogen and one of the most common causes of pneumonia in cystic fibrosis patients (5, 6, 7). The stability and impermeability of the OM contribute substantially to the high antibiotic resistance of this human pathogen (8, 9). LPS produced by P. aeruginosa is also the main factor in virulence as well as the innate and acquired host responses to infection, and lipid A is responsible for the majority of the cytotoxic effects caused by the molecule (10). Changes in the growth conditions of P. aeruginosa can induce extensive remodeling of lipid A, including addition or removal of phosphate groups and acyl chains (11). The length and number of lipid A acyl chains influence the efficiency of activation of inflammatory signaling through Toll-like receptor 4 (TLR-4), with penta-acylated LPS producing a weaker immune system response than hexa-acylated LPS (12). Strains isolated from chronically infected cystic fibrosis patients synthesize a hexa-acylated form of LPS and are generally unable to synthesize O-antigen polysaccharides (3, 10).
Another factor that contributes to the high stability of the P. aeruginosa OM is the presence of the outer membrane protein H (OprH). OprH is a 21-kDa, 200-residue, slightly basic protein that is integral to the P. aeruginosa OM (13). OprH is genetically linked to the PhoP-PhoQ two-component regulatory system that is upregulated in response to Mg2+-limited growth conditions (14, 15). When P. aeruginosa is cultured in a low concentration of divalent cations, OprH is upregulated and overexpressed, so that it becomes a major component of the OM. OprH acts as a surrogate for Mg2+ and Ca2+ by cross-linking LPS, thereby tightening the OM under conditions of divalent cation deficiency (13). Interestingly, it was reported that OprH is also highly upregulated during the early stages of P. aeruginosa infection (16), despite the presence of millimolar concentrations of Mg2+ that would normally suppress transcription of the oprH-phoP-phoQ operon (17). Although there is no OprH homolog in Escherichia coli, a BLAST search returns a list of hundreds of similar proteins present in other Pseudomonas species (18).
The solution NMR structure of OprH in 1,2-dihexanoyl-sn-glycero-3-phosphocholine (DHPC) micelles reveals an eight-stranded β-barrel protein with four extracellular loops of unequal size (PDB: 2LHF) (19). Fast-timescale dynamics measurements show that the extracellular loops are disordered and unstructured. NMR chemical-shift perturbations observed upon addition of P. aeruginosa LPS to OprH in lipid micelles indicate that the OprH-LPS interaction is predominantly electrostatic and localized to charged regions near the upper rim of the barrel, especially residues Lys70, Arg72, Lys103, Arg113, and Arg145 (18). In this study, we built various simulation systems of OprH in P. aeruginosa and, for comparison, E. coli OMs, and performed all-atom molecular dynamics (MD) simulations to investigate the impact of different LPS molecules on the structure and dynamics of OprH. We identified multiple OprH regions that interact with LPS and likely contribute to the significant antibiotic resistance of P. aeruginosa.
Materials and Methods
System setup
The solution NMR structure of OprH (PDB: 2LHF) (19) was inserted into bacterial OMs with different LPS compositions from P. aeruginosa (20, 21) (Fig. 1; Table 1). We used model 13 of the 20 models in the NMR structural ensemble to avoid severe bad contacts between the LPS structures and the OprH extracellular loops (which are ill defined by NMR due to the paucity of NMR restraints) during the initial system building. In addition, the molecular interactions between OprH and LPS were studied in recent NMR experiments using deep-rough LPS from E. coli strain F583 due to the high viscosity, variable chemical compositions, and high content of impurities of commercially available LPS from P. aeruginosa (18). Therefore, to determine how LPS from different species affects the structural stability of OprH and its interaction with LPS, we also built two additional systems with E. coli LPS (Fig. S1 in the Supporting Material; Table 1). The phospholipid composition of the inner leaflet of all OMs is a mixture of 1-palmitoyl(16:0)-2-palmitoleoyl(16:1 cis-9)-PE (PPPE), 1-palmitoyl(16:0)-2-vacenoyl(18:1 cis-11)-PG (PVPG), and 1,1′-palmitoyl-2,2′-vacenoyl CL, with a net charge of −2e (PVCL2) at a ratio of 15:4:1 (22, 23). Assuming that the inner-leaflet composition of the P. aeruginosa OM is similar to the E. coli OM, the difference in the OM properties of the two bacteria arises solely from the difference in LPS. We used the step-by-step protocol of Membrane Builder (24, 25, 26) in CHARMM-GUI (27) to build and assemble OM-only and protein-OM complex structures. This approach was previously applied successfully for E. coli LPS bilayer simulations (28), various bacteria lipid A bilayer simulations (29), and OmpLA (30), OmpF (31), and BamA (32) simulation studies in E. coli OM environments. The detailed system information is summarized in Table S1. We used 150 mM KCl to mimic the physiological bulk ion concentration, and initially placed neutralizing Ca2+ ions in the LPS regions. Three replicas for each system were independently built and simulated to improve sampling and to check for simulation convergence.
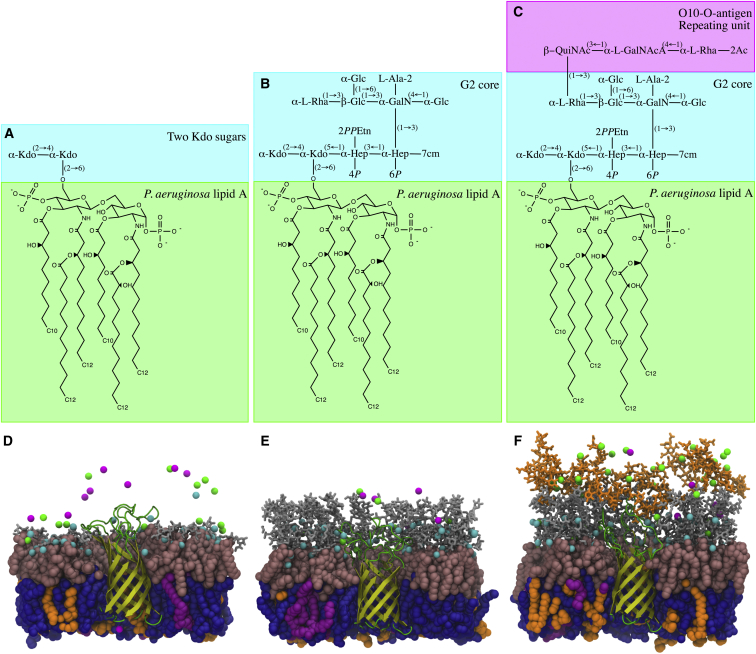
Figure 1.
(A–C) Chemical structures of lipid A and sequences of the LPS core and O-antigen of (A) Pa.Kdo, (B) Pa.G2, and (C) Pa.G2.O10 (Kdo, 2-keto-3-deoxyoctulosonate; Hep, l-glycero-d-manno heptose; Glc, d-glucose; Gal, d-galactose; GalN, d-galactosamine; Rha, l-rhamnose; QuiNAc, N-acetyl-6-deoxy-d-glucosamine; GalNAcA, 2-acetamido-2-deoxy-l-galacturonic acid; GalNAc, N-acetyl-d-galactosamine; PPEtn, pyrophosphoryl ethanolamine; Ala, l-alanine; cm, carbamoyl) (20, 21). The lipid A molecule in this study consists of two d-glucosamine (GlcN) residues joined by a β-(1→6)-linkage, two monophosphoester groups at O1 and O4′, and six amide/ester-linked fatty acids. The P. aeruginosa G2 core has two Kdo residues and two Hep residues: one has a PPEtn group at the O2 position and a monophosphoester group at the O4 position, and the other one has a monophosphoester group and cm group at the O6 and O7 positions. The outer core consists of three Glc residues, one Rha residue, and one GalN residue with the Ala group at the O2 position. They are α-linked except for the β-linkage between Glc and GalN. The O10-antigen unit consists of one QuiNAc residue, one GalNAcA residue, and one Rha residue with an acetyl group at the O2 position. QuiNAc is attached to the O3 position of the terminal Rha residue in the outer core and its linkage has the β-configuration, in contrast to the corresponding α-(1→3)-linkage between the repeating units. In this study, two units of O10-antigen are considered. (D–F) Representative snapshots of OprH embedded in (D) Pa.Kdo, (E) Pa.G2, and (F) Pa.G2.O10 OMs (pink spheres, lipid A; gray sticks, core sugars; orange sticks, O10-antigen polysaccharides; blue spheres, PPPE; orange spheres, PVPG; magenta spheres, PVCL2; small cyan spheres, Ca2+ ions; small magenta spheres, K+ ions; small green spheres, Cl− ions). For clarity, some portion of each system is truncated and water molecules are not shown. To see this figure in color, go online.
Table 1.
Different Outer Membrane Systems
| System | Composition of the Outer Leaflet |
|---|---|
| Pa.Kdo | P. aeruginosa lipid A + two Kdo sugars |
| Pa.G2 | P. aeruginosa lipid A + G2 core |
| Pa.G2.O10 | Pa.G2 + two repeating units of O10-antigen |
| Ec.Kdo | E. coli lipid A + two Kdo sugars |
| Ec.K12 | E. coli lipid A + K12 core |
MD simulations
Based on the step-by-step equilibration protocol used in CHARMM-GUI Membrane Builder, all OM systems were equilibrated for 1.45 ns using CHARMM (33) with the C36 lipid (34), carbohydrate (35, 36, 37, 38), and LPS force fields (29, 39) with the TIP3P water model (40). A gradual equilibration was performed with NVT (constant particle number, volume, and temperature) dynamics and a 1-fs time step for 100 ps, followed by 1.35-ns NPT (constant particle number, pressure, and temperature) dynamics with a 2-fs time step. During these equilibration steps, previously validated planar and dihedral harmonic restraints (24, 25, 39) were applied to the LPS molecules, phospholipids, and water molecules; these restraints were gradually reduced to zero for the production simulations. Additional dihedral angle restraints were applied to restrain all sugar rings to the pertinent chair conformation, because NMR studies as well as MD simulations show that common pyranose sugar residues are mainly present in their chair conformation on the submicrosecond timescale (41). These ring dihedral restraints were maintained during the production simulations. During NVT equilibration, constant temperature was maintained by Langevin dynamics. Temperature and pressure controls were achieved with a Hoover thermostat (42) and Langevin piston for NPT dynamics (43, 44). NPT production runs of 300 ns were performed for all systems except for the OprH-Pa.G2.O10 (550-ns production) and OM-only (100-ns production except for 250 ns for Pa.G2.O10) systems using NAMD (45). Langevin dynamics was used to maintain constant temperature with the Langevin coupling coefficient set to 1 ps−1. A Nosé-Hoover Langevin piston (46, 47) was used to maintain constant pressure with a piston period of 50 fs and a piston decay time of 25 fs. van der Waals interactions were smoothly switched off between 10 and 12 Å by a force-switching function (48), and the particle-mesh Ewald method (49) was used for long-range electrostatic interactions. The temperature and pressure were held at 310.15 K and 1 bar, respectively. A 2-fs time step with the SHAKE algorithm (50) was used.
Results and Discussion
Structural properties of P. aeruginosa and E. coli OMs
Environmental and growth conditions affect the LPS structure (i.e., rough versus smooth LPS), which may subsequently influence the molecular packing of outer membrane proteins (OMPs) and LPS via changes in the local environments of embedded OMPs and in OMP-LPS/OMP-OMP interactions. To explore the impact of various LPS structures on the molecular packing of lipids, we calculated the area per lipid (APL) for both E. coli (Ec) and P. aeruginosa (Pa) OM-only systems by utilizing a Voronoi tessellation approach (51). The time-series of APL were used to check for system equilibration and convergence of simulations, and the average APL was calculated using the last 50-ns trajectory. For example, as shown in Fig. S2, the APLs for each lipid from all three replicas of Pa.G2.O10 OM-only systems were equilibrated and well converged after 50-ns simulations. In general, the full core systems (Pa.G2 and Pa.G2.O10) have a slightly larger APL (∼190 Å2) than the Kdo-only core systems (∼180 Å2; Fig. S3). An increase in APL correlates with the increase in size of the LPS molecule. An identical trend is observed for the lipids in the inner leaflet, indicating that LPS disturbs the inner leaflet lipids, probably due to a subtle area mismatch in the asymmetric bilayers. This is consistent with previous findings for Ec.R1 rough LPS and Ec.R1.O6 smooth LPS bilayer systems, which showed that including O6-antigen repeating units in Ec.R1.O6 resulted in a larger APL compared to the Ec.R1 system (28).
We further examined the OM properties in terms of the acyl chain deuterium order parameters (SCD) for each lipid: , where is the time-dependent angle between the C-H bond vector and the bilayer normal (i.e., the z axis), and the angular bracket denotes a time and ensemble average (52). In general, an increase in SCD correlates with an increase in the chain order and tighter packing of acyl chains. In contrast to Ec systems, a change in the LPS sugar compositions in Pa systems has almost no influence on the average SCD of lipid A, probably due to its short acyl chain lengths (Fig. S4). Overall, relatively low SCD values (generally less than 0.3) indicate that all OMs are in a liquid-disordered bilayer phase at 310 K.
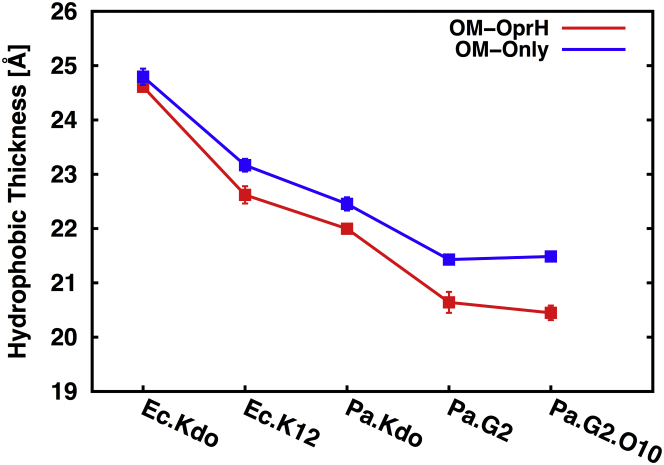
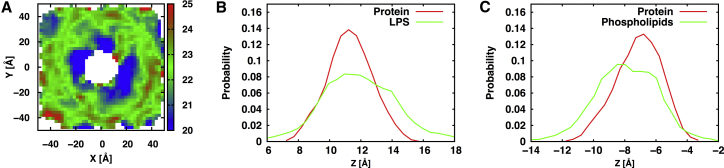
As shown in Fig. 2, the Pa OMs are thinner hydrophobic bilayers than the Ec OMs, which is expected from the difference in the acyl chain length of their lipid A (Figs. 1 and S1). This is also consistent with a recent report based on an analysis of 21 distinct lipid A molecules from 12 bacterial species, showing that different lengths and numbers of lipid A acyl chains influence the molecular packing and membrane properties of Gram-negative bacteria (29). We observe that not only the length and number of acyl chains but also the different chemical structures of LPS molecules influence membrane packing, with the Kdo-only core systems having a greater thickness and smaller APL than the full core systems. Notably, the OM-only systems are thicker than the OprH-OM systems, indicating membrane thinning by OprH (Fig. 3). A comparison of two-dimensional thickness distributions clearly shows membrane thinning of ∼2 Å for lipids surrounding OprH in the Pa systems, and ∼4 Å in Ec systems (Fig. S5), compared to their respective OM-only systems (Fig. S6). In general, such local thinning results from a mismatch between the hydrophobic regions of a membrane and membrane proteins, which is energetically costly and leads to structural adjustments to minimize the exposure of nonpolar groups to the aqueous phase (53, 54, 55). These observations are also consistent with previous studies on OmpLA-OM (30) and BamA-OM (32) systems. To determine whether the membrane thinning near OprH is the result of maximizing a hydrophobic match between OprH and asymmetric OMs, we calculated the two-dimensional z-position distributions of the C2 and C4 atoms (i.e., the carbon bonded to carbonyl group) of lipid A, the acyl chain C2 carbon atoms (i.e., the carbon bonded to carbonyl group) of phospholipids, and the center of mass (COM) of the side chain of the hydrophobic residues on the rim of each β-strand. As shown in Fig. S7, the hydrophobic residues in the OprH barrel are located in z positions similar to the C2 and C4 atoms of lipid A and the C2 atoms of phospholipids. Interestingly, as shown in Fig. 3, there is a good correlation between the distributions of the z position of the COM of protein hydrophobic residues, the C2 and C4 atoms of lipid A, and the C2 atoms of phospholipids. This analysis reveals that the components of the OM (OMPs, lipid A, and phospholipids) dynamically adjust to each other to maximally match their hydrophobic regions during simulations to yield such a time-averaged hydrophobic match (and membrane thinning).
Figure 2.
Average hydrophobic thickness of each OM system, with the standard errors over three replicas. The hydrophobic thickness of each system was calculated by measuring the average distance between acyl chain C2 atoms (i.e., the carbon bonded to carbonyl group) for the phospholipids, and C2 and C4 atoms (i.e., the carbon bonded to carbonyl group) for the lipid A molecule. To see this figure in color, go online.
Figure 3.
(A) Two-dimensional thickness distribution of the OM in OprH-Pa.Kdo. (B and C) Distributions of the z position of the COM of hydrophobic residues of OprH with (B) the C2 and C4 atoms (i.e., the carbon bonded to carbonyl group) of lipid A and (C) C2 atoms (i.e., the carbon-bonded-to-carbonyl group) of phospholipids that were used to calculate the hydrophobic thickness distribution in (A). The OM is centered at z = 0 and the OM normal is the z axis. To see this figure in color, go online.
Water molecules inside OprH in P. aeruginosa and E. coli OMs
We also investigated the lipid and protein distributions along the membrane normal (i.e., the z axis) by calculating the heavy-atom number density profiles for all systems (Fig. S8). A subtle height difference in the density profile of carbon tails is observed between the upper leaflets of Ec OMs and Pa OMs, which arises mainly from the difference in acyl chain lengths between Ec lipid A and Pa lipid A. A similar trend is observed in the core distributions, in that the Ec.K12 core (10 sugar residues in the core) has a broader density distribution than the Pa.G2 core (9 sugar residues in the core). For all OMs, water molecules penetrate deep inside the core sugar and lipid headgroup regions, whereas there is slight penetration into the hydrophobic hydrocarbon tail region. Interestingly, a continuous density of water along the membrane normal is not observed even in the OprH-OM systems. Although there are some water molecules inside the OprH barrel, they do not translocate through the pore from one leaflet to the other leaflet (Fig. S9), because there are strong electrostatic interactions between charged residues (especially Arg54, Asp81, and Glu138) that block water permeation through the OprH barrel. As shown in Fig. S9 A, water molecules in Pa.G2.O10 are clearly disconnected inside the OprH barrel. Although water molecules in Pa.Kdo (Fig. S9 B) seem to be connected inside the OprH barrel, they never passed through the pore during the simulation times in this study. However, it is possible that different protonation states (due to pH changes) could alter the electrostatic interactions and allow water permeation through the pore.
Structure and dynamics of OprH in P. aeruginosa and E. coli OMs
The solution NMR study of OprH in DHPC micelles revealed that the extracellular loops are highly disordered and undergo motions on the picosecond and nanosecond timescales, but there is no prior information about the loop dynamics in more biologically relevant membrane mimetics, including phospholipid and OM bilayers. This is important because even though detergent micelles mimic lipid bilayers well by providing both hydrophobic and hydrophilic environments for the transmembrane domain of a membrane protein, they do not provide the geometric constraints imposed by lipid bilayers, so the orientations of the long loops or intra- or extracellular domains of a membrane protein could be ill defined in micelles (56). First, we evaluated the structural stability of OprH in various LPS environments by measuring the root mean-square deviation (RMSD) of the β-barrel backbone atoms from the initial structure. Overall, the RMSD increased initially and reached a plateau around 2 Å, indicating that there was no significant difference in β-barrel structural stability among the systems (Fig. S10), which agrees well with the stability of other OMP β-barrel structures observed in previous studies (30, 31, 57, 58, 59, 60, 61, 62, 63).
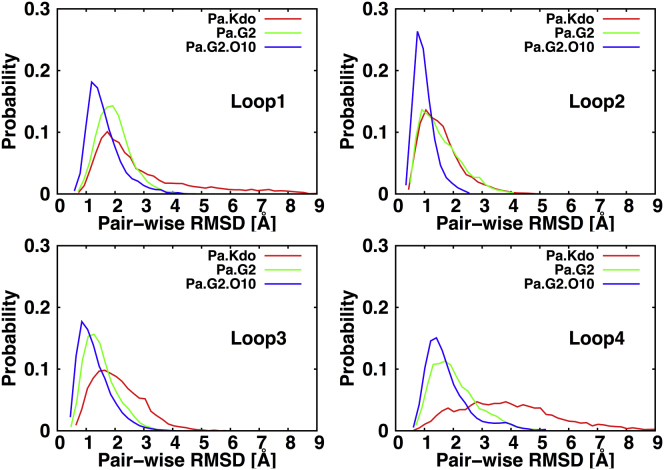
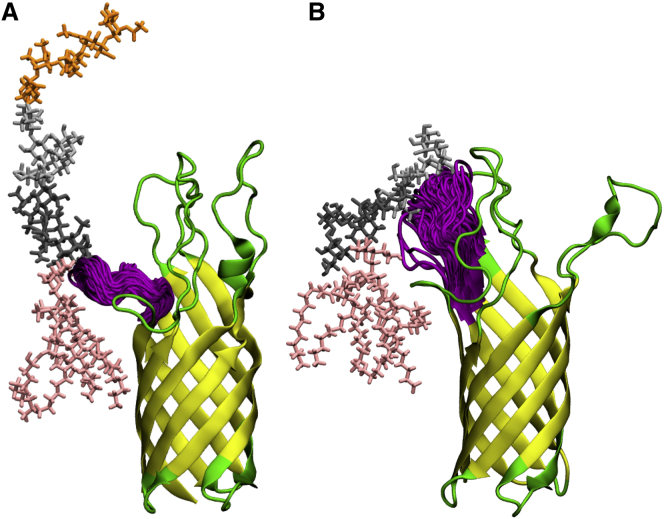
To check the conformational variability of the OprH loops, we calculated pairwise RMSD distributions by aligning each of the last 100-ns snapshot structures to the β-barrel atoms of the initial structure and then measuring the RMSD of each pair using the loop Cα atoms. As shown in Fig. 4, each loop shows various extents of conformational variability because of varied interactions with LPS molecules (see next section) as well as the available physical space (see Fig. S11 for Ec systems). For example, the loops in Pa.G2.O10 exhibit narrower RMSD distributions than those in other systems because the O10-antigen sugar residues interact with the loops and restrict their mobility. However, the Pa.Kdo system shows broader RMSD distributions, as there are only two Kdo sugar residues that can interact with the water-exposed loops. Therefore, the loop orientations (relative to the β-barrel) in the Kdo-only core systems and similarly in solution NMR structures vary widely and are not restricted like those in the full core or O10-antigen systems. This is clearly shown by a comparison of the starting NMR structure and MD-averaged structures in each system (Fig. S12). Together with these pairwise RMSD distributions, the per-residue root mean-square fluctuation (RMSF) plots in Fig. S13 support our claim that the interactions between the loops and LPS constrain the loop motions; for example, the OprH-Pa.G2.O10 system has the least dynamic loops and OprH-Pa.Kdo system shows the largest loop fluctuation.
Figure 4.
Pairwise RMSD distributions of the OprH loops for Pa systems, calculated by aligning each of the last 100-ns snapshot structures to the β-barrel Cα atoms of the initial structure and then measuring the RMSD of each pair using the loop Cα atoms. To see this figure in color, go online.
Interactions between OprH and LPS in P. aeruginosa and E. coli OMs
To understand how OprH interacts with OMs with different LPS compositions, we performed a heavy-atom contact analysis between each protein residue and surrounding molecules (see Fig. 5 for OprH-Pa.G2.O10 and Figs. S14–S17 for other systems; n.b., hereafter, for brevity, the system name does not include OprH). A cutoff distance of 4 Å was used to define a contact in these calculations, and the average of three replicas was used in the figures. In addition, the occupancy of hydrogen bonds between each loop (L1–L4) and LPS was calculated (Fig. S18). In general, the protein-LPS interaction patterns of the OprH-OM systems are similar to those of the OmpLA-OM (30) and BamA-OM (32) systems. The β-strands mainly interact with LPS or phospholipid acyl tails; the extracellular loops primarily interact with lipid A headgroups, the core, O-antigen, and water; and the turns on the periplasmic side mainly interact with phospholipids and water.
Figure 5.
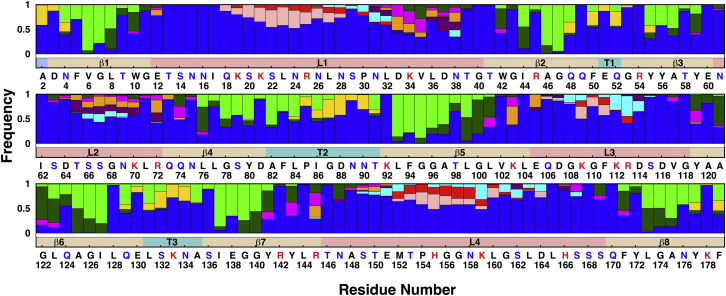
Patterns of the interactions of protein residues with their environment in Pa.G2.O10. The frequencies of various environmental entities within 4 Å of each OprH residue are shown for water (blue), phospholipid headgroups (yellow), phospholipid carbon tails (green), lipid A tails (dark green), lipid A headgroups (orange), the region between lipid A headgroups and lipid A tails (magenta), Kdo sugars in the inner core (purple), Hep sugars in the inner core (cyan) and outer core (red), and O10-antigen (pink). The bar below each set of patterns indicates the protein secondary structure: β-barrel (beige), loop (coral), turn (turquoise), and N-terminus (light blue). The red- and blue-colored characters indicate basic and polar residues, respectively. To see this figure in color, go online.
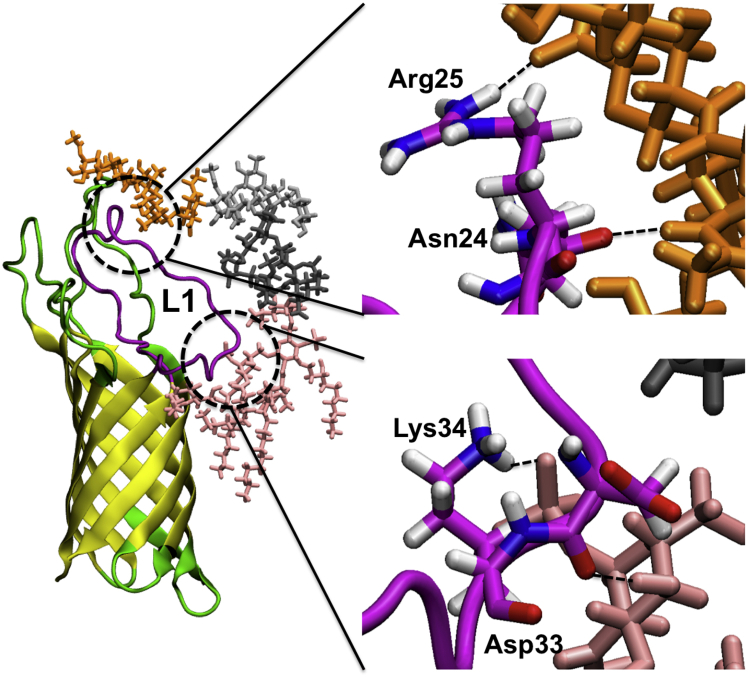
In Pa.G2.O10, three different interaction patterns of loop L1 residues are observed. Residues 21–29 preferentially interact with outer-core and O10-antigen sugars, residues 30–32 mainly interact with inner-core sugars, and residues 33–38 mainly interact with lipid A headgroups. Similar interaction patterns for L1 residues are observed in Pa.Kdo (Fig. S14) and Pa.G2 (Fig. S15), i.e., residues 33–38 interact with lipid A headgroups in Pa.Kdo and Pa.G2, and residues 30–32 interact with inner-core sugars in Pa.G2. Interactions between L1 residues 21–29 and O10-antigen appear to slightly restrict the overall dynamics of L1 (Fig. 6), leading to a higher frequency of interactions between L1 residues 33–38 and lipid A headgroups in Pa.G2.O10 compared to Pa.G2 and Pa.Kdo. OprH L1 residues show similar patterns of interaction with lipid A headgroups in Pa.Kdo (Fig. S14) and Ec.Kdo (Fig. S16). Increased frequencies of interactions between L1 residues 21–29 and core sugars, and between L1 residues 33–38 and lipid A headgroups are observed in Ec.K12 (Fig. S17) compared to Pa.G2 (Fig. S15). These different interaction patterns suggest different exposures of the residues to surrounding LPS environments due to the conformational preferences of constituent LPS sugars in the outer core in these systems (Fig. S19).
Figure 6.
Snapshot showing interactions of OprH L1 with LPS in Pa.G2.O10 (magenta, L1; pink sticks, lipid A; dark gray sticks, inner-core sugars; gray sticks, outer-core sugars; orange sticks, O10-antigen polysaccharide). Asn24, Arg25, and Lys34 in L1 mainly form hydrogen bonds with sugar residues in both lipid A and O10-antigen regions. To see this figure in color, go online.
The OprH loops are rich in positively charged residues (20.5% of all loop residues), which enhance the binding to negatively charged LPS. This is clearly reflected by the majority of interactions arising from electrostatic interactions between basic or polar residues in OprH loops and negatively charged and groups in LPS (Fig. 5). In the case of loop L1, frequent contacts are made through residues Asp33, Lys34, Asp37, and Asn38. This observation is supported by experimental data showing that addition of P. aeruginosa LPS protects this region of OprH from trypsin digestion (19). Interestingly, hydrophobic residue Val35 is also important for LPS binding, especially in Pa.G2.O10, Ec.Kdo, and Ec.K12.
Interactions of loop L2 are observed to be dominated by lipid A headgroups and Kdo and Hep in the inner core, with the exception of Ec.K12 (Fig. S17), where residues 67–69 interact with outer-core sugars. Interestingly, this different interaction pattern does not arise from variations in the overall membrane hydrophobic thickness (as the membrane thinning occurs near OprH (Fig. 3)), but rather from the conformational difference of L2 in different environments. For example, as shown in Fig. 7, the L2 residues manifest a larger fluctuation in Ec.K12 than in Pa.G2.O10, which is consistent with the results of the pairwise RMSD of L2 (Fig. 4).
Figure 7.
Fluctuations of loop L2 (magenta) and the nearest LPS molecule in (A) Pa.G2.O10 and (B) Ec.K12 from a representative replica of the systems (pink sticks, lipid A; dark gray sticks, inner-core sugars; gray sticks, outer-core sugars; orange sticks, O-antigen polysaccharide). The L2 structures were taken from the last 200-ns trajectories with a 1-ns interval. Similar L2 fluctuations were observed in other replicas of each system (data not shown). To see this figure in color, go online.
In all systems, Lys70 and Arg72 in loop L2 show similar interactions with LPS (Figs. 5 and S14–S17). Both residues interact with lipid A headgroups and Kdo sugars, indicating that these residues are important binding sites between OprH and LPS. This observation is consistent with recent experimental data showing that conserved Lys70 and Arg72 contribute significantly to interactions between OprH and LPS (18). A stretch of mostly charged residues from 64 to 69 also substantially aids LPS binding, but in the case of these residues, the interaction sites vary among lipid A headgroups, Kdo sugars, and inner- and outer-core sugars (Figs. 5 and S14–S17).
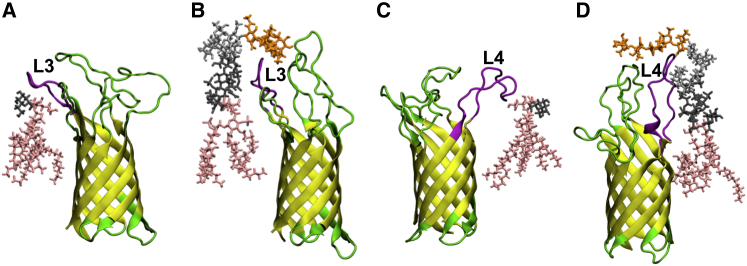
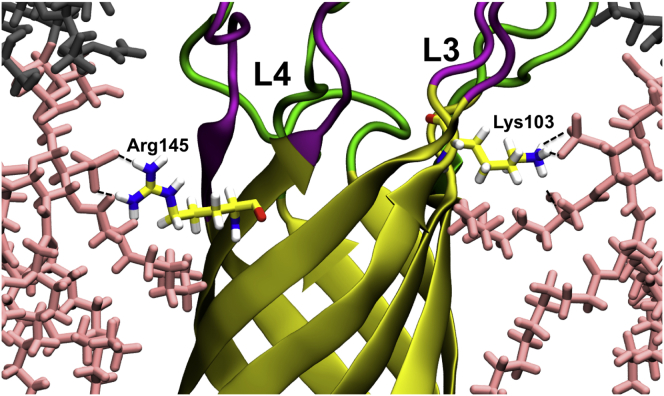
In the case of loop L3 and L4 residues, different interaction patterns are observed in Pa.G2 and Pa.Kdo compared to Pa.G2.O10. These interactions appear to affect the conformational dynamics of loops L3 and L4. For example, to establish interactions with Kdo sugars, the conformations of L3 (Fig. 8 A) and L4 (Fig. 8 C) need to be more tilted and extended in Pa.Kdo compared to relatively straight and restricted conformations in Pa.G2.O10 (Fig. 8, B and D, respectively). This is also consistent with the observed fluctuations of L3 and L4 (Fig. S20) and the broad pairwise RMSD distributions of L3 and L4 in Pa.Kdo compared to Pa.G2.O10 (Fig. 4). We observed a similar behavior in Ec systems (Fig. S21). The simulation results indicate that the conformational dynamics of the loops, especially L3 and L4, vary with the LPS composition, which may be necessary to maintain the functionality of the protein for bacterial survival. It is worth noting that although most residues in L3 and L4 significantly contribute to LPS conformational dynamics, some of the residues localized in the upper rim of the β-barrel, especially Lys103 and Arg145 (Fig. 9), play a crucial role in LPS binding (via lipid A headgroups and the region between the lipid A headgroups and tails) to OprH. This observation is consistent with experimental data (18) and the fact that these residues are evolutionarily highly conserved (11).
Figure 8.
(A–D) Representative snapshots showing interactions of loops L3 and L4 with LPS: (A) L3 in Pa.Kdo, (B) L3 in Pa.G2.O10, (C) L4 in Pa.Kdo, and (D) L4 in Pa.G2.O10. Each loop that interacts with LPS is colored magenta (pink sticks, lipid A; dark gray sticks, inner-core sugars; gray sticks, outer-core sugars; orange sticks, O-antigen polysaccharide). To see this figure in color, go online. To see this figure in color, go online.
Figure 9.
Snapshot showing interactions of Lys103 and Arg145 with lipid A headgroups in Pa.G2.O10. L3 and L4 residues are colored magenta, lipid A is represented as pink sticks, and inner-core sugars are shown as dark gray sticks. To see this figure in color, go online.
Conclusions
Since the introduction of the fluid mosaic model of biological membranes, there has been an interest in how membrane proteins interact with lipid molecules. Recently, several new protein-lipid complexes have been described by x-ray crystallography and mass spectroscopy (64, 65). Despite this recent progress, the mechanisms used by lipids to regulate protein structure and function are still largely unknown, as only a limited number of methods are capable of systematically characterizing protein-lipid interactions (66).
In this work, we used all-atom MD simulations to explore the structural properties, dynamics, and interactions of OprH in native-like asymmetric P. aeruginosa OMs and, for comparison, in E. coli OMs. Analyses of OM-only membrane simulations show that as more LPS components are added, the area per lipid A slightly increases, indicating slightly looser lipid A packing in the full core systems (Pa.G2, Pa.G2.O10, and Ec.K12) compared to Kdo-only core systems (Pa.Kdo and Ec.Kdo). As expected from their different lipid A chain lengths, the Ec systems have a larger hydrophobic thickness than the Pa systems. Nonetheless, it should be noted that all OprH-OM systems show similar hydrophobic thicknesses near OprH due to membrane thinning arising from specific OprH-OM interactions by which the hydrophobic match between OprH and the OM is dynamically adjusted.
The chemical structure of LPS can be extensively modified depending on the conditions in which the bacteria are cultured (11). However, Pseudomonas species generally have only one gene coding OprH-like protein. In that case, it may be necessary to change the internal conformational dynamics of loops with different LPS compositions to maintain the functional role of OprH as a membrane stabilizer and thus the survival of the bacteria. Interestingly, the dynamics of the OprH loops indeed depend on the LPS composition in the OMs and are greatly reduced in OMs with LPS O-antigen compared to those in Kdo-only OMs and in solution NMR structures. As it is difficult to obtain well-defined orientations of OprH loops by solution NMR experiments in detergent micelles, the OprH-OM simulations with different LPS compositions reported in this study may also provide an opportunity to refine functionally important loop conformations of OprH (and other outer membrane proteins).
Notably, in every simulation performed here, water molecules were unable to pass through the OprH pore from one leaflet to the other, due to the presence of charged residues occluding the pore. This further suggests that the functional role of this protein does not involve substrate transport. However, different protonation states of the charged residues at different pH values may change these interaction patterns, which could lead to water permeation through the pore.
Consistent with previous experimental findings (18), the calculated interaction patterns between OprH and LPS show that Lys70, Arg72, Lys103, Arg113, and Arg145 are key residues for interactions between OprH and LPS, although the interaction frequencies vary among the different LPS environments. In all LPS simulation systems, Lys70, Arg72, Lys103, and Arg145 interact mainly with the lipid A headgroups and the region between the headgroups and acyl tails of lipid A. This region of LPS is evolutionarily highly conserved (11) and plays an important role in the interactions of LPS with other outer membrane proteins (67) as well as with components of the innate immune system (68).
In summary, the results of the MD simulations presented here provide valuable insight into OprH-LPS interactions that increase the integrity of the OM of P. aeruginosa under stress conditions and during early stages of infection. Due to the recent increase in the number of P. aeruginosa strains that are capable of chemically modifying their lipid A structure to avoid detection by the immune system, OprH might also be considered a potential target for novel antimicrobial therapies (13, 69, 70).
Author Contributions
W.I. conceived the study. J.L. and W.I. performed research. J.L., D.S.P., I.K., L.K.T., and W.I. analyzed data. J.L., D.S.P., I.K., and W.I. wrote the manuscript. All authors contributed to editing the manuscript.
Acknowledgments
This work was supported in part by grants from the National Science Foundation (MCB-1516154, DBI-1145987, IIA-1359530, and XSEDE MCB070009 to W.I.) and the National Institutes of Health (R01 51329 to L.K.T.).
Editor: Francesca Marassi.
Footnotes
Twenty-one figures and one table are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(16)34281-3.
Supporting Material
References
- 1.Bos M.P., Robert V., Tommassen J. Biogenesis of the gram-negative bacterial outer membrane. Annu. Rev. Microbiol. 2007;61:191–214. doi: 10.1146/annurev.micro.61.080706.093245. [DOI] [PubMed] [Google Scholar]
- 2.Ruiz N., Kahne D., Silhavy T.J. Advances in understanding bacterial outer-membrane biogenesis. Nat. Rev. Microbiol. 2006;4:57–66. doi: 10.1038/nrmicro1322. [DOI] [PubMed] [Google Scholar]
- 3.Maldonado R.F., Sá-Correia I., Valvano M.A. Lipopolysaccharide modification in Gram-negative bacteria during chronic infection. FEMS Microbiol. Rev. 2016;40:480–493. doi: 10.1093/femsre/fuw007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang G., Meredith T.C., Kahne D. On the essentiality of lipopolysaccharide to Gram-negative bacteria. Curr. Opin. Microbiol. 2013;16:779–785. doi: 10.1016/j.mib.2013.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Govan J.R., Brown A.R., Jones A.M. Evolving epidemiology of Pseudomonas aeruginosa and the Burkholderia cepacia complex in cystic fibrosis lung infection. Future Microbiol. 2007;2:153–164. doi: 10.2217/17460913.2.2.153. [DOI] [PubMed] [Google Scholar]
- 6.Lyczak J.B., Cannon C.L., Pier G.B. Lung infections associated with cystic fibrosis. Clin. Microbiol. Rev. 2002;15:194–222. doi: 10.1128/CMR.15.2.194-222.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saiman L., Siegel J. Infection control in cystic fibrosis. Clin. Microbiol. Rev. 2004;17:57–71. doi: 10.1128/CMR.17.1.57-71.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 2003;67:593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nikaido H., Hancock R.E. Outer membrane permeability of Pseudomonas aeruginosa. In: Sokatch J.R., editor. The Bacteria: A Treatise on Structure and Function. Academic Press; London: 1986. pp. 145–193. [Google Scholar]
- 10.Pier G.B. Pseudomonas aeruginosa lipopolysaccharide: a major virulence factor, initiator of inflammation and target for effective immunity. Int. J. Med. Microbiol. 2007;297:277–295. doi: 10.1016/j.ijmm.2007.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Needham B.D., Trent M.S. Fortifying the barrier: the impact of lipid A remodelling on bacterial pathogenesis. Nat. Rev. Microbiol. 2013;11:467–481. doi: 10.1038/nrmicro3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pier G.B., Markham R.B., Eardley D. Correlation of the biologic responses of C3H/HEJ mice to endotoxin with the chemical and structural properties of the lipopolysaccharides from Pseudomonas aeruginosa and Escherichia coli. J. Immunol. 1981;127:184–191. [PubMed] [Google Scholar]
- 13.Rehm B.H., Hancock R.E. Membrane topology of the outer membrane protein OprH from Pseudomonas aeruginosa: PCR-mediated site-directed insertion and deletion mutagenesis. J. Bacteriol. 1996;178:3346–3349. doi: 10.1128/jb.178.11.3346-3349.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Macfarlane E.L., Kwasnicka A., Hancock R.E. Role of Pseudomonas aeruginosa PhoP-phoQ in resistance to antimicrobial cationic peptides and aminoglycosides. Microbiology. 2000;146:2543–2554. doi: 10.1099/00221287-146-10-2543. [DOI] [PubMed] [Google Scholar]
- 15.McPhee J.B., Bains M., Hancock R.E. Contribution of the PhoP-PhoQ and PmrA-PmrB two-component regulatory systems to Mg2+-induced gene regulation in Pseudomonas aeruginosa. J. Bacteriol. 2006;188:3995–4006. doi: 10.1128/JB.00053-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gellatly S.L., Needham B., Hancock R.E. The Pseudomonas aeruginosa PhoP-PhoQ two-component regulatory system is induced upon interaction with epithelial cells and controls cytotoxicity and inflammation. Infect. Immun. 2012;80:3122–3131. doi: 10.1128/IAI.00382-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Macfarlane E.L., Kwasnicka A., Hancock R.E. PhoP-PhoQ homologues in Pseudomonas aeruginosa regulate expression of the outer-membrane protein OprH and polymyxin B resistance. Mol. Microbiol. 1999;34:305–316. doi: 10.1046/j.1365-2958.1999.01600.x. [DOI] [PubMed] [Google Scholar]
- 18.Kucharska I., Liang B., Tamm L.K. Molecular interactions of lipopolysaccharide with an outer membrane protein from pseudomonas aeruginosa probed by solution NMR. Biochemistry. 2016;55:5061–5072. doi: 10.1021/acs.biochem.6b00630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Edrington T.C., Kintz E., Tamm L.K. Structural basis for the interaction of lipopolysaccharide with outer membrane protein H (OprH) from Pseudomonas aeruginosa. J. Biol. Chem. 2011;286:39211–39223. doi: 10.1074/jbc.M111.280933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knirel Y.A., Valvano M.A. Springer; New York: 2011. Bacterial Lipopolysaccharides. [Google Scholar]
- 21.Raetz C.R., Whitfield C. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 2002;71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raetz C.R.H. Enzymology, genetics, and regulation of membrane phospholipid synthesis in Escherichia coli. Microbiol. Rev. 1978;42:614–659. doi: 10.1128/mr.42.3.614-659.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vance D.E., Vance J.E. Elsevier; Amsterdam: 2002. Biochemistry of Lipids, Lipoproteins and Membranes. [Google Scholar]
- 24.Jo S., Kim T., Im W. Automated builder and database of protein/membrane complexes for molecular dynamics simulations. PLoS One. 2007;2:e880. doi: 10.1371/journal.pone.0000880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jo S., Lim J.B., Im W. CHARMM-GUI Membrane Builder for mixed bilayers and its application to yeast membranes. Biophys. J. 2009;97:50–58. doi: 10.1016/j.bpj.2009.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu E.L., Cheng X., Im W. CHARMM-GUI Membrane Builder toward realistic biological membrane simulations. J. Comput. Chem. 2014;35:1997–2004. doi: 10.1002/jcc.23702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jo S., Kim T., Im W. CHARMM-GUI: a web-based graphical user interface for CHARMM. J. Comput. Chem. 2008;29:1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- 28.Wu E.L., Engström O., Im W. Molecular dynamics and NMR spectroscopy studies of E. coli lipopolysaccharide structure and dynamics. Biophys. J. 2013;105:1444–1455. doi: 10.1016/j.bpj.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim S., Patel D.S., Im W. Bilayer properties of lipid A from various Gram-negative bacteria. Biophys. J. 2016;111:1750–1760. doi: 10.1016/j.bpj.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu E.L., Fleming P.J., Im W. E. coli outer membrane and interactions with OmpLA. Biophys. J. 2014;106:2493–2502. doi: 10.1016/j.bpj.2014.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Patel D.S., Re S., Im W. Dynamics and interactions of OmpF and LPS: influence on pore accessibility and ion permeability. Biophys. J. 2016;110:930–938. doi: 10.1016/j.bpj.2016.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fleming P.J., Patel D.S., Im W. BamA POTRA domain interacts with a native lipid membrane surface. Biophys. J. 2016;110:2698–2709. doi: 10.1016/j.bpj.2016.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brooks B.R., Brooks C.L., 3rd, Karplus M. CHARMM: the biomolecular simulation program. J. Comput. Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klauda J.B., Venable R.M., Pastor R.W. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J. Phys. Chem. B. 2010;114:7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guvench O., Hatcher E.R., Mackerell A.D. CHARMM additive all-atom force field for glycosidic linkages between hexopyranoses. J. Chem. Theory Comput. 2009;5:2353–2370. doi: 10.1021/ct900242e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guvench O., Mallajosyula S.S., Mackerell A.D., Jr. CHARMM additive all-atom force field for carbohydrate derivatives and its utility in polysaccharide and carbohydrate-protein modeling. J. Chem. Theory Comput. 2011;7:3162–3180. doi: 10.1021/ct200328p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guvench O., Greene S.N., Mackerell A.D., Jr. Additive empirical force field for hexopyranose monosaccharides. J. Comput. Chem. 2008;29:2543–2564. doi: 10.1002/jcc.21004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hatcher E., Guvench O., Mackerell A.D. CHARMM additive all-atom force field for aldopentofuranoses, methyl-aldopentofuranosides, and fructofuranose. J. Phys. Chem. B. 2009;113:12466–12476. doi: 10.1021/jp905496e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jo S., Wu E.L., Im W. Lipopolysaccharide membrane building and simulation. Methods Mol. Biol. 2015;1273:391–406. doi: 10.1007/978-1-4939-2343-4_24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jorgensen W.L., Chandrasekhar J., Klein M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 41.Sattelle B.M., Almond A. Assigning kinetic 3D-signatures to glycocodes. Phys. Chem. Chem. Phys. 2012;14:5843–5848. doi: 10.1039/c2cp40071e. [DOI] [PubMed] [Google Scholar]
- 42.Hoover W.G. Canonical dynamics: equilibrium phase-space distributions. Phys. Rev. A Gen. Phys. 1985;31:1695–1697. doi: 10.1103/physreva.31.1695. [DOI] [PubMed] [Google Scholar]
- 43.Andersen H.C. Molecular-dynamics simulations at constant pressure and-or temperature. J. Chem. Phys. 1980;72:2384–2393. [Google Scholar]
- 44.Nose S., Klein M.L. A study of solid and liquid carbon tetrafluoride using the constant pressure molecular-dynamics technique. J. Chem. Phys. 1983;78:6928–6939. [Google Scholar]
- 45.Phillips J.C., Braun R., Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martyna G.J., Tobias D.J., Klein M.L. Constant-pressure molecular-dynamics algorithms. J. Chem. Phys. 1994;101:4177–4189. [Google Scholar]
- 47.Feller S.E., Zhang Y.H., Brooks B.R. Constant-pressure molecular-dynamics simulation—the Langevin piston method. J. Chem. Phys. 1995;103:4613–4621. [Google Scholar]
- 48.Steinbach P.J., Brooks B.R. New spherical-cutoff methods for long-range forces in macromolecular simulation. J. Comput. Chem. 1994;15:667–683. [Google Scholar]
- 49.Essmann U., Perera L., Pedersen L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995;103:8577–8593. [Google Scholar]
- 50.Ryckaert J.P., Ciccotti G., Berendsen H.J.C. Numerical integration of Cartesian equations of motion of a system with constraints—molecular-dynamics of N-alkanes. J. Comput. Phys. 1977;23:327–341. [Google Scholar]
- 51.Pandit S.A., Vasudevan S., Scott H.L. Sphingomyelin-cholesterol domains in phospholipid membranes: atomistic simulation. Biophys. J. 2004;87:1092–1100. doi: 10.1529/biophysj.104.041939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhuang X., Makover J.R., Klauda J.B. A systematic molecular dynamics simulation study of temperature dependent bilayer structural properties. Biochim. Biophys. Acta. 2014;1838:2520–2529. doi: 10.1016/j.bbamem.2014.06.010. [DOI] [PubMed] [Google Scholar]
- 53.Rui H., Lee J., Im W. Comparative molecular dynamics simulation studies of protegrin-1 monomer and dimer in two different lipid bilayers. Biophys. J. 2009;97:787–795. doi: 10.1016/j.bpj.2009.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim T., Lee K.I., Im W. Influence of hydrophobic mismatch on structures and dynamics of gramicidin a and lipid bilayers. Biophys. J. 2012;102:1551–1560. doi: 10.1016/j.bpj.2012.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hong H., Tamm L.K. Elastic coupling of integral membrane protein stability to lipid bilayer forces. Proc. Natl. Acad. Sci. USA. 2004;101:4065–4070. doi: 10.1073/pnas.0400358101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cheng X., Jo S., Im W. CHARMM-GUI micelle builder for pure/mixed micelle and protein/micelle complex systems. J. Chem. Inf. Model. 2013;53:2171–2180. doi: 10.1021/ci4002684. [DOI] [PubMed] [Google Scholar]
- 57.Fleming P.J., Freites J.A., Fleming K.G. Outer membrane phospholipase A in phospholipid bilayers: a model system for concerted computational and experimental investigations of amino acid side chain partitioning into lipid bilayers. Biochim. Biophys. Acta. 2012;1818:126–134. doi: 10.1016/j.bbamem.2011.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Baaden M., Meier C., Sansom M.S.P. A molecular dynamics investigation of mono and dimeric states of the outer membrane enzyme OMPLA. J. Mol. Biol. 2003;331:177–189. doi: 10.1016/s0022-2836(03)00718-6. [DOI] [PubMed] [Google Scholar]
- 59.Piggot T.J., Holdbrook D.A., Khalid S. Conformational dynamics and membrane interactions of the E. coli outer membrane protein FecA: a molecular dynamics simulation study. Biochim. Biophys. Acta. 2013;1828:284–293. doi: 10.1016/j.bbamem.2012.08.021. [DOI] [PubMed] [Google Scholar]
- 60.Straatsma T.P., Soares T.A. Characterization of the outer membrane protein OprF of Pseudomonas aeruginosa in a lipopolysaccharide membrane by computer simulation. Proteins. 2009;74:475–488. doi: 10.1002/prot.22165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Im W., Roux B. Ions and counterions in a biological channel: a molecular dynamics simulation of OmpF porin from Escherichia coli in an explicit membrane with 1 M KCl aqueous salt solution. J. Mol. Biol. 2002;319:1177–1197. doi: 10.1016/S0022-2836(02)00380-7. [DOI] [PubMed] [Google Scholar]
- 62.Rui H., Lee K.I., Im W. Molecular dynamics studies of ion permeation in VDAC. Biophys. J. 2011;100:602–610. doi: 10.1016/j.bpj.2010.12.3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tamm L.K., Hong H., Liang B. Folding and assembly of beta-barrel membrane proteins. Biochim. Biophys. Acta. 2004;1666:250–263. doi: 10.1016/j.bbamem.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 64.Yeagle P.L. Non-covalent binding of membrane lipids to membrane proteins. Biochim. Biophys. Acta. 2014;1838:1548–1559. doi: 10.1016/j.bbamem.2013.11.009. [DOI] [PubMed] [Google Scholar]
- 65.Lee A.G. Lipid-protein interactions in biological membranes: a structural perspective. Biochim. Biophys. Acta. 2003;1612:1–40. doi: 10.1016/s0005-2736(03)00056-7. [DOI] [PubMed] [Google Scholar]
- 66.Saliba A.E., Vonkova I., Gavin A.C. The systematic analysis of protein-lipid interactions comes of age. Nat. Rev. Mol. Cell Biol. 2015;16:753–761. doi: 10.1038/nrm4080. [DOI] [PubMed] [Google Scholar]
- 67.Ferguson A.D., Welte W., Diederichs K. A conserved structural motif for lipopolysaccharide recognition by procaryotic and eucaryotic proteins. Structure. 2000;8:585–592. doi: 10.1016/s0969-2126(00)00143-x. [DOI] [PubMed] [Google Scholar]
- 68.Park B.S., Song D.H., Lee J.O. The structural basis of lipopolysaccharide recognition by the TLR4-MD-2 complex. Nature. 2009;458:1191–1195. doi: 10.1038/nature07830. [DOI] [PubMed] [Google Scholar]
- 69.Miller A.K., Brannon M.K., Moskowitz S.M. PhoQ mutations promote lipid A modification and polymyxin resistance of Pseudomonas aeruginosa found in colistin-treated cystic fibrosis patients. Antimicrob. Agents Chemother. 2011;55:5761–5769. doi: 10.1128/AAC.05391-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Muller C., Plésiat P., Jeannot K. A two-component regulatory system interconnects resistance to polymyxins, aminoglycosides, fluoroquinolones, and β-lactams in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2011;55:1211–1221. doi: 10.1128/AAC.01252-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.