Supplemental Digital Content is available in the text
Keywords: amplification, breast cancer, deletion, fluorescent in situ hybridization, HER2, tissue microarray, TOP2A
Abstract
Background:
The prognostic relevance of topoisomerase II alpha (TOP2A) copy number change remains not well established. This study is aimed to investigate the frequency and pattern of TOP2A aberrations; to correlate TOP2A alterations with human epidermal growth factor receptor 2 (HER2) status and clinicopathological parameters, and further to explore prognostic value of TOP2A and HER2 status in breast cancer in Taiwan.
Methods:
We analyzed tissue samples from 311 invasive carcinomas in tissue microarrays for TOP2A and HER2 status by fluorescent in situ hybridization.
Results:
TOP2A copy number change is an infrequent genetic event (9.8% amplification and 2.7% deletion) and is present in both HER2-amplified and nonamplified tumors. TOP2A amplification is statistically associated with age >50 at diagnosis (P = 0.016) and HER2 amplification (P < 0.001). HER2 amplification, but not TOP2A amplification, is a predictor of unfavorable prognosis (P = 0.002). Univariate and multivariate analysis showed that higher histologic grading, positive nodal involvement, and HER2 positivity were associated with poorer overall survival. Cytogenetically, double minutes-type amplification is the predominant pattern for both genes (HER2: 64% and TOP2A: 93.1%). Homogeneous staining region-type signals of both genes are resistant to RNase digestion, supporting that these were not nuclear accumulation of mRNA transcripts.
Conclusion:
Our results demonstrate the prognostic value of tumor grading, nodal involvement, and HER2 status in Taiwanese breast cancer. TOP2A aberrations are an infrequent event independent of HER2 status, and TOP2A amplification carries no prognostic value. The predictive value of TOP2A aberrations in patients of breast cancer taking athracycline-containing treatment in Taiwan remains to be determined in prospectively well-designed clinical trials.
1. Introduction
Breast cancer is a genetically heterogeneous disease with variable clinical outcomes.[1] Clinically, breast cancer is categorized into 3 therapeutic groups: estrogen receptor (ER) positive tumors; HER2-amplified tumors; and triple-negative (basal-like) tumors, defined as ER and progesterone receptor (PR) negative and HER2-non-amplified.[2] Genetically, genomic profiling has identified 3 distinct patterns of DNA copy number alteration: simple, amplifier, and complex.[3] An “amplifier” or “firestorm” type of genomic aberration is characterized by focal high-level DNA amplification, clustered on 1 or more chromosome arm, set in a background of simple to moderately complex gains/losses. This pattern is characteristic of HER2-amplified tumors and luminal B subtype of ER-positive tumors.[4] Common amplified sites found in HER2-amplified tumors include 8p12, 8q24, 11q13, 17q12, and 20q13.[3,4] Genomic analysis of 17q12-q21 amplicon has identified recurrent amplification of many genes including HER2. HER2 amplification is one of the most frequent genetic alterations in breast cancer, occurring in approximately 15% to 20% of cases.[5] HER2 amplification is an adverse prognostic factor and is a predictive biomarker of response to HER2-targeted therapy.[5] In addition, HER2 amplification is functionally implicated as a driver of genomic instability and hence can simultaneously lead to amplification and activation of other genes.[3] Coamplified genes located in the smallest region of amplification of HER2 amplicon include MED1, STARD3, GRB7, THRA, and RARA.[3,4] TOP2A (topoisomerase II alpha), located in a separate amplicon downstream to HER2 amplicon, is frequently altered in HER2-amplified tumors.[3,6,7] Topoisomerase II α, a 170-kDa protein product of TOP2A, is the primary molecular target of anthracyclines.[7] Although anthracyclines are widely used in the treatment of many cancers, their exact mechanisms of action remain uncertain due to complex interactions with different molecules or pathways.[7] Data of 2 meta-analyses indicated that only patients with HER2-positive tumors showed survival benefit from anthracyclines,[8,9] but contradictory results were also reported.[10] It is postulated that response of HER2-positve patients to anthracyclines is not a direct effect of HER2 amplification, but the result of coamplification of TOP2A. However, reported data on the predictive value of TOP2A are even more controversial. A recent meta-analysis of individual patient data indicated that the benefits of anthracycline-based treatment are not dependent on the status of either TOP2A or HER2.[11] In contrast, a prospectively planned, individual-patient pooled analysis demonstrated that duplication of chromosome 17 and/or TOP2A aberration was independently predictive of adjuvant anthracycline chemotherapy.[12] In comparison, the prognostic value of TOP2A alteration has been reported in a limited number of studies.[13,14]
Gene amplification in mammalian cells is present in 2 distinct structures: intrachromosomal homogenously staining regions (HSRs) and extrachromosomal double minutes (DMs).[15] Fluorescent in situ hybridization (FISH) is regarded as the assay of choice for detecting amplification, as it is unaffected by the problem of tumor cell heterogeneity and contaminating normal cells.[16] One of the key challenges of interphase FISH is enumeration of “large clustered signals,” which cytogenetically correspond HSRs. Recently Ooi et al[17] reported that some of HSR-type ESR1 (ER 1) signals were actually concentrated intranuclear ESR1 mRNA, indicating that it is pertinent to perform RNase sensitivity assay for HSR-type signals.
The incidence of estrogen-related tumors, including female breast, uterine, and ovarian cancers, has rapidly increased in young women in Taiwan since 1997.[18] Patients of female breast cancer in Taiwan tend to be younger than those in Western countries, with a peak incidence between the ages of 40 and 50 years.[18] We have previously reported that HER2 amplification is a frequent genetic aberration and is an unfavorable prognostic factor in a series of breast cancer in Taiwan.[19] This study was initiated to investigate the prevalence and pattern of TOP2A copy number changes and to correlate TOP2A aberration with HER2 status, hormone receptor (HR) status, and clinicopathological features. A further objective was to explore the prognostic significance of TOP2A and HER2 status. We also performed RNase sensitivity assay to examine whether large clustered HER2 and TOP2A signals were concentrated accumulation of mRNA transcripts.
2. Methods
2.1. Ethical considerations
This study was approved by the Institutional Review Board (IRB) of Chang Gung Memorial Hospital, Tao-Yuan, Taiwan, which permitted using archival formalin-fixed, paraffin-embedded tissues anonymously for research purpose without informed consent.
2.2. Specimens and patient characteristics
This study included a series of 311 patients with invasive breast cancer, newly diagnosed between January 1, 1992, and December 31, 2002, at Chang Gung Memorial Hospital, Keelung, Taiwan. Clinicopathological details of all patients including histopathological type and grade, tumor staging, HER2 status, and ER positivity have been reported elsewhere.[19] Histopathological classification was performed on the basis of the current diagnostic criteria of the World Health Organization classification.[1] Clinical information and survival data were obtained from the cancer registry of the Cancer Center, Keelung Chang Gung Memorial Hospital. Mean follow-up was 8 years (range: 0–20 years). Expression of PR and TOP2A was analyzed by immunohistochemistry.
2.3. Tissue microarray (TMA) construction
We analyzed existing TMAs containing 311 breast carcinomas of different histologic types, including invasive carcinoma of no special type (NST) (n = 273), invasive lobular carcinoma (n = 30), medullary carcinoma (n = 5), and invasive papillary carcinoma (n = 3).[19] The design and construction of TMA have been described previously in detail.[19,20] Briefly, each 81-core TMA block contained 1.5 mm triplicate cores of 26 breast tumor issues and 1 tonsil. Tonsil was inserted at different locations to serve as the landmark. Four-micrometer thick sections were cut and transferred onto polyline-coated slides for studies. Hematoxylin- and eosin-stained TMA sections were used as reference histology.
2.4. Immunohistochemistry and scoring
Mouse monoclonal antibodies against human PR (Novocastra clone 16; Leica Microsystems, Melbourne, Australia; 1 : 500 dilution) and TOP2A (NCL-TOPOIIA, Leica Microsystems, 1 : 40 dilution) were used in this study. Pre-treatment of sections (deparaffinization, rehydration, and antigen retrieval) and automated immunostaining were carried out on a BOND-MAX automated autostainer (Leica Microsystems, Melbourne, Australia) as per the manufacturer's instructions. Scoring of PR immunostaining was performed according to the Allred score.[21] Tumors with Allred score ≥3 were rated as PR-positive. Quantitative analysis of TOP2A-labeled carcinoma cells was performed using a web-based automated image analysis application: ImmunoRatio.[22] Using 20x microscope objective, 3 images per core were taken from triplicate cores of tumor. Images were then uploaded and analyzed as recommended by ImmunoRatio application. The final percentage of positive invasive carcinoma cells, as a percentage of total tumor, represented mean value of 9 independent measurements. Interpretation was performed blinded to the results of TOP2A FISH analysis and the clinicopathological information. Because no cutoff for positivity has been validated to define TOP2A overexpression, we analyzed this variable on the basis of a cutoff of 15% or greater.
2.5. Fluorescent in situ hybridization (FISH)
HER2 and TOP2A FISH were performed using Vysis LSI HER-2/neu Spectrum Orange and CEP 17 Spectrum Green probe (Abbott Laboratories, Abbott Park, IL) and Vysis LSI TOP2A Spectrum Orange/CEP17 Spectrum Green Probe (Abbott Laboratories), respectively. The TOP2A probe covers a 160 kb segment containing the TOP2A gene and two flanking genes on each side. The HER2 probe encompasses a 226 kb segment containing 5 flanking genes. TMA sections were deparaffinized for 10 minutes at 70°C and twice for 10 minutes in xylene. After rehydration in graded ethanol and rinsing in distilled water, FISH was subsequently performed as per manufacturer's standardized protocols with minor modification. Thirty microliters dual-color FISH probes were applied to the center of TMA area, sections were cover slipped, and the edges were sealed with rubber cement. Slides were placed in a Dako Hybridizer (Dako), denatured at 75°C for 10 minutes, and hybridized at 37°C for 16 to 18 hours. After hybridization, slides were washed in post-hybridization buffer (2X SSC/0.3% NP-40, pH 7.0–7.5) at 72°C for 2 minutes, dehydrated in graded ethanol and air-dried. Sections were covered with anti-fading DAPI (4’,6-diamidino-2-phenylindole) solution and incubated in the dark for 15 minutes before analyzing using an epifluorescence microscope. FISH scoring was performed by counting fluorescence signals in at least 60 malignant, nonoverlapping cell nuclei at X1000 oil-immersion magnification. The interpretation of HER2 results followed the current guidelines by the American Society of Clinical Oncology/College of American Pathologists.[5] Samples with HER2/CEP17 ratio ≥2.0 were rated as amplification, whereas tumors with HER2/CEP 17 ratio < 0.8 were rated as HER2 deletion.[11] TOP2A status was scored as the guideline of HER2. Images were taken using Nikon fluorescent microscope (Nikon, Tokyo, Japan) equipped with DP71 CCD camera image capture system (Olympus, Tokyo, Japan).
2.6. RNase sensitivity assay
In order to examine whether HER2 and TOP2A amplifications with HSRs-type signals represented concentrated mRNA transcripts sensitive to RNase A digestion, we performed RNase treatments on HER2- and TOP2A-amplified tumors as previously described.[17] After the standard pre-treatment of FISH, slides were incubated with RNase A (Roche Applied Science, Taipei, Taiwan; 100 μg/mL in 2X SSC) at 37°C for 10 minutes before hybridization. After thorough washing in 2X SSC, slides were processed for hybridization as described above.
2.7. Statistics
Statistical analysis was performed using SPSS version 18.0 for Windows (SPSS Inc., Chicago, IL). Contingency table analysis and Fisher-exact tests were used to study the relationship between clinicopathological characteristics and the TOP2A protein expression or copy number change of HER2 and TOP2A. Kalpan–Meier curves were used to assess survival and the log-rank test was used to compare the difference between individual curves. For survival analysis, patients were censored at the time of their last clinical follow-up or at the time of death. Multivariate analyses were carried out using Cox proportional hazard model to examine the hazard ratio of death in TOP2A+ versus TOP2A-, HER2+ versus HER2-patients, TOP2A index >15% versus TOP2A index ≤15%, and HER2 amplification with or without TOP2A amplification. Univariate predictors with a P value ≤0.1 were entered into a step-wise multivariate model to identify factors that independently predict overall survival (OS) probabilities. A P value <0.05 was considered statistically significant. Correlation between HER2/CEP17 and TOP2A/CEP17 ratio was plotted using R statistical package (R Foundation for Statistical Computing, Vienna, Austria).
3. Results
3.1. Immunohistochemical analysis of PR and TOP2A
All cases had well-preserved tissues for reliable evaluation of staining despite variable degrees of loss of tissue. All 311 cases were available for immunohistochemical analysis of PR and TOP2A. About 42.8% (133/311) of patients were PR-positive (Allred score ≥3). The proportion of Allred score of 0 to 2, 3 to 4, 5 to 6, and 7 to 8 was 57.2% (178/311), 9.0% (28/311), 14.8% (46/311), and 19.0% (59/311), respectively (supplementary Table 1). The actual Allred values of PR TOP2A index as measured using ImmunoRatio ranged from 0.5% to 62.3% (mean 8.97%, median 5.18%, range: 0.2–62.3%). About 82.0% (255/311) and 18% (56/311) of tumors had TOP2A expression ≤15% and >15%, respectively. One representative figure of TOP2A expression analyzed by ImmunoRatio is shown in Fig. 1.
Figure 1.

Representative micrographs of TOP2A-immunostained images analyzed by ImmunoRatio. The upper panel (A, B, and C) showed 3 different fields of TOP2A-immunostained images taken from a core of tissue microarray. Corresponding images analyzed using ImmunoRatio were shown in the lower panel (D, E, and F). (x200 original magnification).
3.1.1. HER2 FISH
We have previously reported that 24.1% (75/311) of breast cancers harbored HER2 amplification using Zytovision probes (Zytovision, Bremerhaven, Germany) and a cut-off of HER2/CEP 17 ratio ≥2.2.19 Although pre-treatment and post-hybridization procedures of these 2 probes differed greatly, results of HER2 FISH using Vysis probes and a HER2/CEP 17 ratio ≥2.0 yielded identical results. Among these 75 HER2-amplified tumors, 48 (64.0%) and 27 (36%) tumors showed double minutes (DM)- (Fig. 2A) and HRS-type (Fig. 2B) amplifications, respectively. HER2/CEP17 ratio of DM-type tumors ranged from 13.2 to 2.2 (mean and median 5.94 and 5.65, respectively). RNase A sensitivity assays showed that “HSR-like” signals (Fig. 2C) were completely unaffected by RNase A pre-treatment, indicating that they were not concentrated HER2 mRNA transcripts. No tumors showed HER2 deletion.
Figure 2.
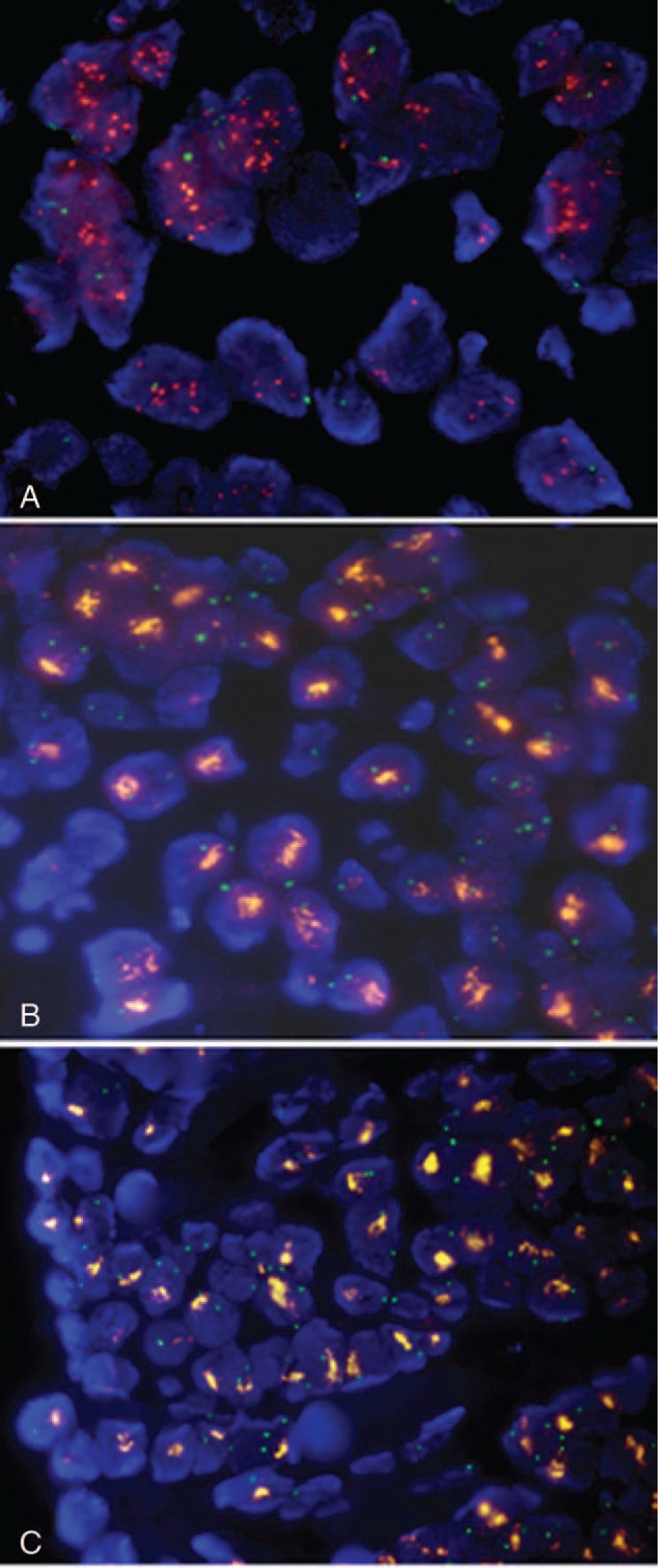
Representative micrographs of HER2 FISH. (A) A tumor with DM-type amplification displayed rather homogeneous population of cells having multiple individual HER2 signals (orange). (B) Tumor with HSR-type showed characteristic clustered HER2 signals (orange) in most tumor cells. (C) Clustered HER2 signals (orange) remained unchanged following RNase A pretreatment. (1000x magnification).
3.1.2. TOP2A FISH
TOP2A FISH analysis was successful in 95.2% (296/311) of tumors after 3 rounds of experiments. Missing results were either due to lack of interpretable FISH signals (n = 13) or loss of tissue samples (n = 2). All 15 with missing results were HER2-non-amplified. About 9.8% (29/296) of analyzable tumors showed TOP2A amplification, and 86.2% (25/29) of these tumors simultaneously harbored HER2 amplification. Overall, 8.4% (25/296) of tumor showed simultaneous HER2 and TOP2A amplifications. About 93.1% (27/29) of these tumors showed DM (Fig. 3A), and only 6.9% (2/29) had HRS-type (Fig. 3B) amplification. HSR-type TOP2A signals were also not affected by RNase A treatment (data not shown).
Figure 3.

Representative micrographs of TOP2A FISH. (A) One tumor with DM-type amplification typically displayed multiple individual TOP2A signals (orange). (B) One tumor with HSR-type amplification showed clustered TOP2A signals, resistant to RNase pre-treatment. (C) A TOP2A-deleted tumor demonstrated absence or 1 copy of TOP2A signals and 2–4 CEP 17 signals (green). (1000x magnification).
About 2.7% (8/296) of analyzable tumors showed TOP2A deletion (Fig. 3C). Seventy-five percent (6/8) of tumors showed simultaneous HER2 amplification. TOP2A/CEP17 ratio ranged from 0.15 to 0.7 (mean and median ratio; 0.43 and 0.46, respectively). HER2/CEP17 ratio plotted against TOP2A/CEP17 ratio is shown in Fig. 4.
Figure 4.

HER2/CEP17 ratio plotted against TOP2A/CEP17 ratio in 296 tumors with paired ratios.
3.2. Association of HER2 and TOP2A status with clinicopathological features
3.2.1. HER2 and TOP2A status
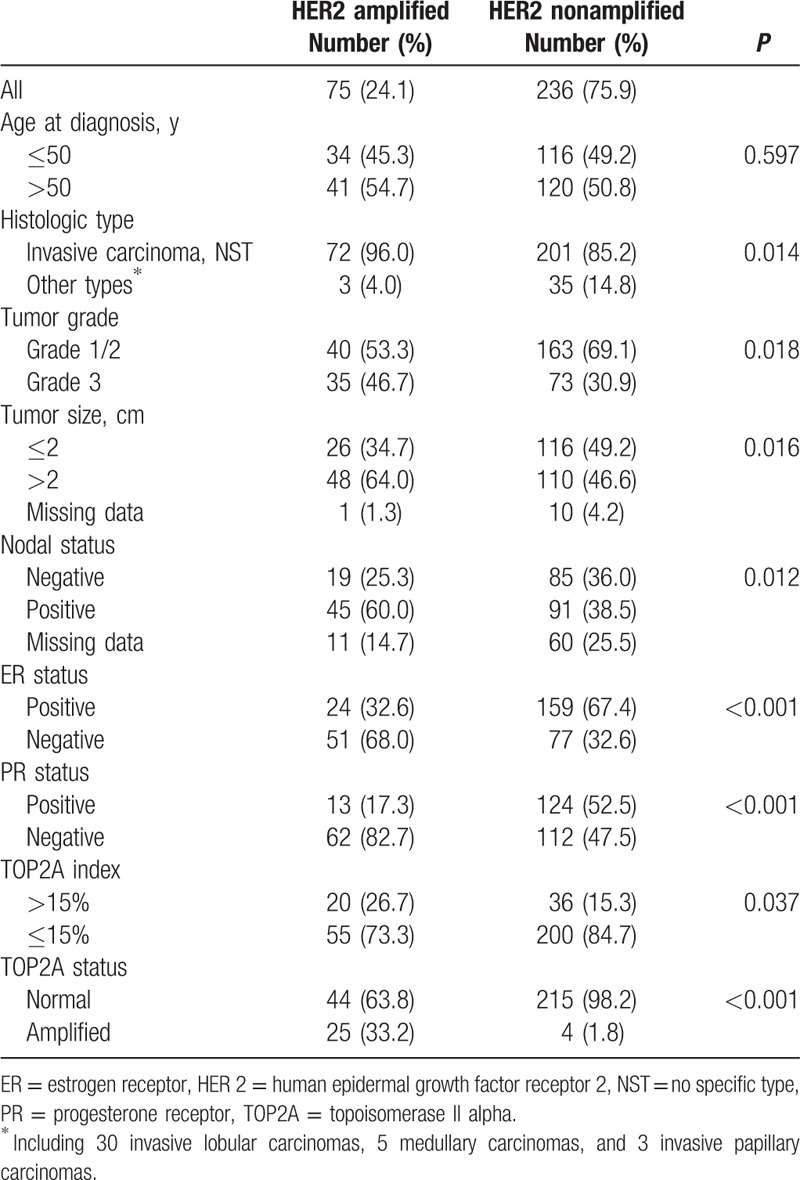
The clinical and tumor characteristics of the analytic cohort according to HER2 status were shown in Table 1. HER2 amplification is significantly associated with histologic type (P = 0.014), a higher histologic grade (P = 0.018), a large tumor size (P = 0.016), positive nodal involvement (P = 0.012), ER negativity (P < 0.001), and PR negativity (P < 0.001). HER2 amplification is not associated with age at diagnosis (P = 0.597). Normal HER2 status is significantly related to TOP2A index ≤15% (P = 0.037).
Table 1.
Patients and tumors characteristics according to HER2 status.
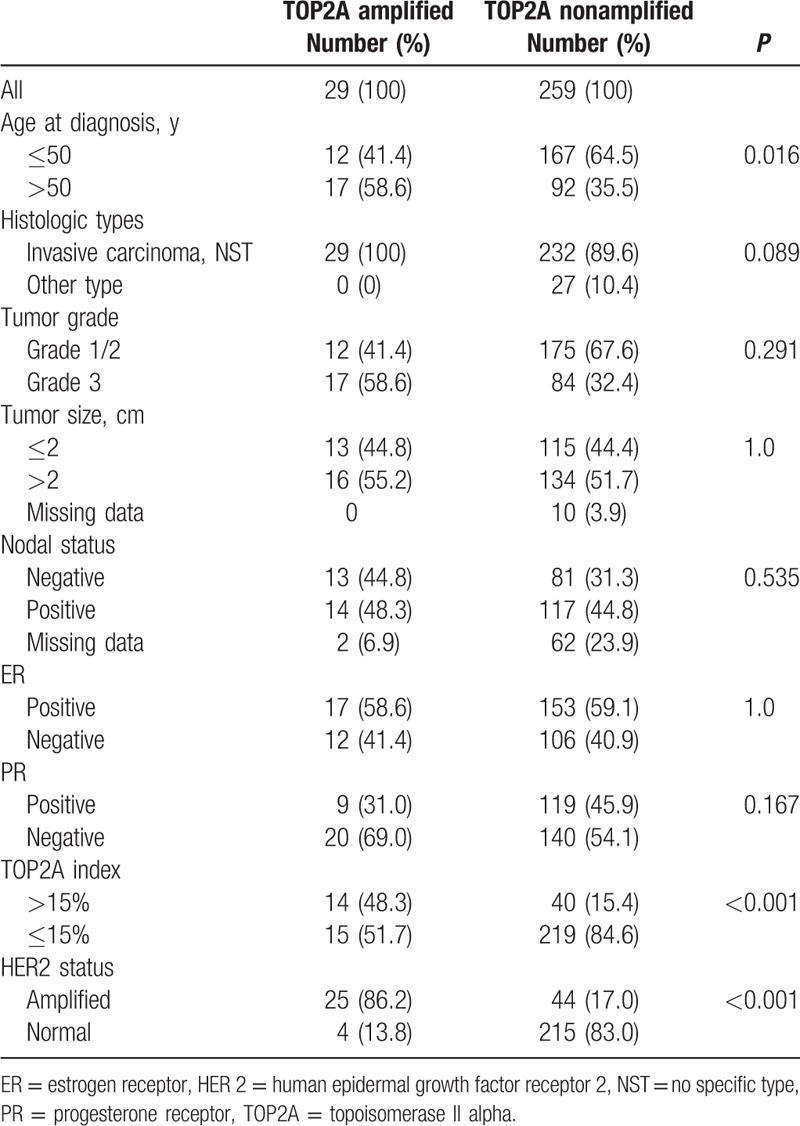
TOP2A amplification is statistically associated with age >50 at diagnosis (P = 0.016) and HER2 amplification (P < 0.001) (Table 2). Normal TOP2A status is also associated with TOP2A index < 15% (P < 0.001). TOP2A amplification is not related to all other clinicopathological parameters (Table 2). The number of TOP2A deletion is too few for significant statistical analyses.
Table 2.
Patients and tumors characteristics according to TOP2A status.
3.3. Survival analysis
Patients with HER2-amplifed tumors had a significantly poorer OS than those nonamplified (P = 0.002). During the first 5 years, risk of death from breast cancer appears to be significantly higher in patients with HER2 amplification (Fig. 5A). Patients with tumors showing either TOP2A amplification (Fig. 5B) or TOP2A index >15% (Fig. 5C) were not significantly related to better survival (P = 0.867 and P = 0.267, respectively). There was no statistical significance in survival between patients with both HER2 and TOP2A amplification and these with HER2 amplification only (P = 0.258) (Fig. 5D).
Figure 5.
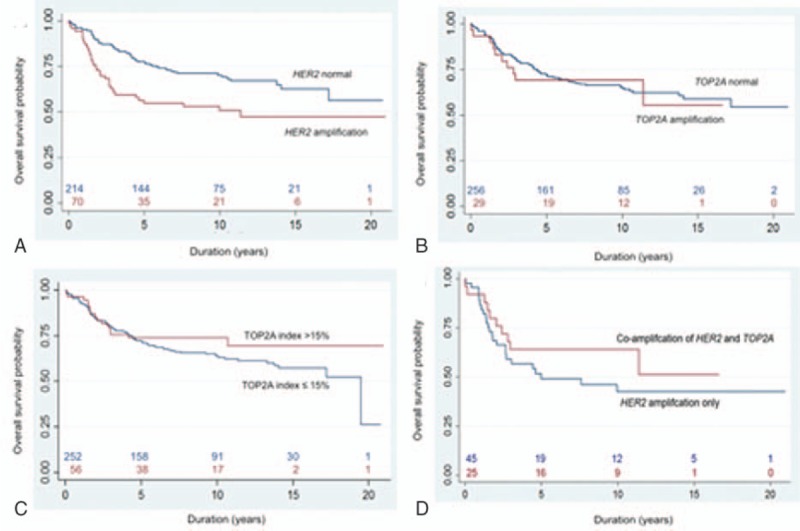
Kaplan–Meier survival curve. (A) HER2 status (P = 0.002), (B) TOP2A status (P = 0.867), (C) TOP2A index (P = 0.267), and (D) HER2 amplification with or without simultaneous amplification of TOP2A (P = 0.258).
In the univariate analysis (Table 3), factors predicting a shorter OS in all patients included older age (P = 0.007), larger tumor size (P = 0.018), positive nodal involvement (P < 0.001), higher histologic grade (P < 0.001), negativity of ER (P = 0.022), and PR (P = 0.048) and HER2 positivity (P = 0.001). In HER2-negative subgroup, older age (P = 0.031), positive nodal involvement (P < 0.001), and higher histologic grade (P < 0.001) were associated with a poorer OS, whereas positive nodal involvement (P = 0.036) is the only adverse factor in HER2-positive subgroup. TOP2A status determined by either FISH or IHC showed no prognostic value in all groups. In the stepwise multivariate analysis comprising all patients, positive nodal involvement (P = 0.001), higher histological grade (P < 0.001), and HER2-positvity (P = 0.013) are associated with a worse OS outcome (Table 4). Positive nodal involvement and higher grade are associated with a poorer OS in HER2-negative subgroup, whereas positive nodal involvement is the only risk factor (P = 0.025) in HER2-positive groups.
Table 3.
Univariate analysis of factors associated with overall survival.
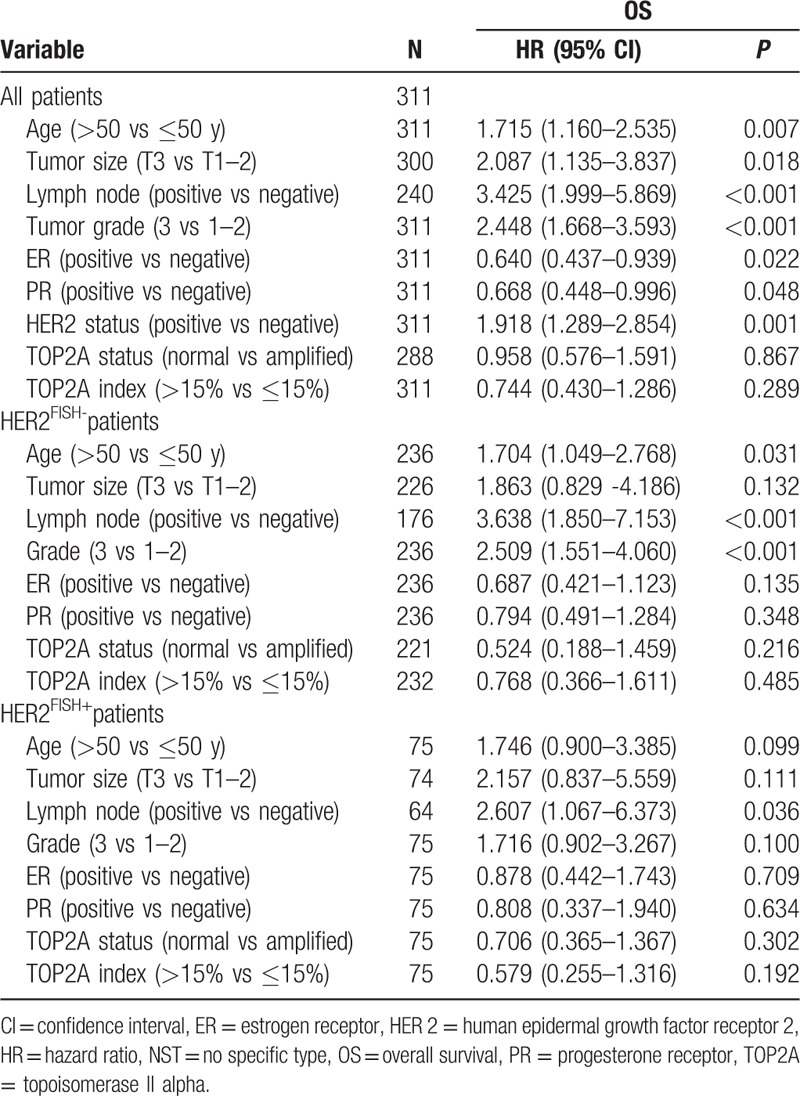
Table 4.
Multivariate Cox regression analysis of factors associated with overall survival.

4. Discussion
Results of our study of a single-institute series of breast cancers in Taiwan show TOP2A copy number change is an infrequent genetic event (9.8% amplification and 2.7% deletion). TOP2A amplification is significantly prevalent in HER2-amplified tumors (P = 0). HER2 amplification is statistically associated with the histologic subtype, higher histologic grade, positive nodal status, and negativity of HR. In contrast, TOP2A amplification is only related with age >50 at diagnosis (P = 0.016) and HER2 amplification (P < 0.001), but not with other clinicopathological parameters. HER2 amplification, but not TOP2A amplification, is a predictor of worse prognosis (P < 0.001). Double minutes (DM)-type amplification is the predominant pattern for both genes (HER2: 64.0% and TOP2A: 91.0%). HSR-type signals of both genes are resistant to RNase digestion, supporting that they were not nuclear accumulation of mRNA transcripts.
Our data showed that HER2 amplification is a negative prognostic factor, which is consistent with the results of a meta-analysis showing that in 90% of studies (81 studies with 27,161 patients), HER2 amplification or protein overexpression was correlated with worse outcome of the patients.[23] The overwhelming majority of these studies also indicate that HER2 abnormalities are strongly associated with resistance to tamoxifen therapy as well as greater benefit from anthracycline.[23] Although our data show that TOP2A amplification is strongly associated with HER2 amplification, only HER2 amplification is significantly related to poor survival. This could be explained by a crosstalk between HER2 and ER pathways or coamplification of other genes in HER2 amplicon.[7,23] Precise identification of predictive biomarkers of anthracycline benefit is complicated by several parameters, including variation between studies in anthracycline choice, dose-intensity, and patients population.[7] Also, most trials are inadequately powered for reliable biomarker subgroup analyses. Indeed, TCGA data show that all 4 major biological subtypes of breast cancer demonstrate much of the clinically observable plasticity and heterogeneity within the same subtype.[2] For instance, at least 2 types of clinically defined HER2-positive tumor were identified,[2] which might explain the variable treatment response to Herceptin therapy in HER2-positive tumors.
We identified 33.3% TOP2A amplification and 8.0% TOP2A deletion among 75 HER2-amplified tumors. Among HER2-nonamplified tumors, 5.3% TOP2A amplification and 2.6% TOP2A deletion were identified. The overall frequency of TOP2A amplification and deletion is 9.8% and 2.7%, respectively, in our study. The reported frequencies of TOP2A amplification and deletion have been variable, particularly in HER2-negative patients. The reported rate of TOP2A amplification and deletion in studies limited to HER2-positive cases is 35% to 50% and 13% to 42%, respectively.[24–26] The frequency of TOP2A deletion in studies including both HER2-positive and HER2-negative tumors is 3.7% to 17.0%,[6,10–14,27,28] and a similar frequency is reported for amplification. In the present study, 9.8% of TOP2A amplification is in line with the reported frequency by other studies, but 2.8% of TOP2A deletion is lower than that in other studies. It is noteworthy that the rate of TOP2A amplification can be influenced by the length of the hybridized probe for TOP2A locus.[29] Specifically, longer TOP2A probe of DAKO pharmDX, but not shorter PathVysion probes as used in this study could result in false-positive TOP2A amplification.[29] Deletion is far more difficult to assess than amplification due to divergent cut-off values and lack of standardized scoring systems of signals.[6,30,31] In addition, nuclear truncation in histological sections may easily lead to a falsely low estimation of copy number. Some studies have reported that TOP2A deletion is equally important for the clinical outcome as TOP2A amplification.[12,13,30] One study even reported that deleted cases had worse prognosis than amplified cases.[30] Our study demonstrates that TOP2A amplification is not a prognostic factor, which is consistent with the data of Engstrøm et al.[14] The percentage of TOP2A deletion in this study is too low for analyzing the association with the clinical outcome.
In contrast to the well-established incidence of HER2 amplification in breast cancer, the number of reports focusing on HER2 deletion is small. Tubbs et al[31] reported HER2 deletion (HER2/CEP17 ≤0.7) in 1.6% (12/742) of breast carcinomas. Nielsen et al[6] showed 2.5% (16/649) of breast carcinomas with HER2 deletion, mostly associated with TOP2A deletion (HER2/CEP17 ≤0.8). We did not detect any tumors with HER2 deletion. Collectively, current data indicate that the frequency of HER2 deletion by FISH is low. Six (75%) TOP2A-deleted tumors are HER2-amplified in this study. The molecular mechanisms underlying the neighboring HER2 amplification and TOP2A deletion are currently unknown.
In the present study, DM is the principal amplification pattern for HER2 and TOP2A, especially for the latter. Interestingly, colon cancers display predominantly HSR-type HER2 amplification, but DM-type signals for EGFR.[32] HSR is also the predominant pattern for ESR1 in breast cancer.[33] It has been shown that in several cell lines, amplified genes in the DM chromosomes were integrated into HSRs, whereas the converse breakdown of HSR to generate DM chromosomes has not been observed.[32] At present, it is not clear whether clinical significances exist between these 2 types of amplification. Technically, accurate measurement of the copy number in HSR-type signals presents a major challenge of FISH. Ooi et al[17] showed that some clustered ESR1 signals changed to small compact signals following RNase treatment, indicating that these HSR signals actually represented concentrated intranuclear ESR1 mRNA. In contrast, clustered HER2 signals were not affected by RNase treatment.[17] The epic of ESR1 amplification indicates that it is appropriate to test new FISH assays for RNase sensitivity, especially for HSR-type signals. Our results show that HSR signals of HER2 and TOP2A were not affected by RNase treatment, thus confirming they truly represented intrachromosomal amplification.
Few studies have designed to study the prognostic value of TOP2A copy number change and its relationship with clinicopathological data. Our results showed that TOP2A amplification is significantly correlated with age >50 at diagnosis and HER2 amplification, but lacks association with all other clinicopathological factors. Knoop et al[13] also reported patients with TOP2A-amplified and TOP2A-deleted tumors were more postmenopausal, and correlated with increasing old age, tumor size, nodal involvement, and ER positivity. TOP2A amplification and deletion were associated with worse relapse-free survival and OS outcomes in a prospectively designed trial.[6] Results of large-scale analysis of Affymetrix microarray data also showed that elevated TOP2A expression is an independent prognostic factor in ER-positive breast cancer and shows a strong correlation with tumor size, grading, HER2 status, Ki67 expression, and nodal status.[34] It is well recognized that TOP2A protein expression determined by immunohistochemistry does not correlate with TOP2A amplification but rather with cellular proliferation.[7] Increased TOP2A mRNA expression has been associated with cell proliferation and with highly proliferative intrinsic subtypes—both HER2+ (luminal B and HER2-enriched) and HER2- (basal-like) tumors.[35] The value of TOP2A gene expression in predicting benefit from anthracyclines remains to be determined.
In conclusion, our data show well-established clinicopathological parameters, including higher histologic grading, positive nodal involvement, and HER2 positivity, and remain robust prognostic factors in Taiwanese female breast cancer. TOP2A amplification and deletion is significantly associated with, but not limited to, tumors with HER2 amplification. TOP2A amplification carries no prognostic value. The predictive value of TOP2A aberration in patients taking anthracycline-containing treatment in Taiwan remains to be determined in prospectively well-designed clinical trials.
Acknowledgments
We thank the Cancer Center of Keelung Chang Gung Memorial Hospital and Biobank/Tissue Bank of Keelung Chang Gung Memorial Hospital for the data of cancer registry and provision of specimens, respectively. This study was supported by grant CMRPG290071 to J-R Chen from Keelung Chang Gung Memorial Hospital, Keelung, Taiwan. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Supplementary Material
Footnotes
Abbreviations: DM = double minutes, ER = estrogen receptor, FISH = fluorescent in situ hybridization, HER2 = human epidermal growth factor receptor 2, HSR = homogeneous staining regions, PR = progesterone receptor, SSC = sodium chloride-sodium citrate buffer, TMA = tissue microarray, TOP2A = topoisomerase II alpha.
Both J-R C and H-P C contributed equally to this work.
The authors have declared that no conflicting interests exist.
Supplemental Digital Content is available for this article.
References
- [1].Lakhani SR, Ellis IO, Schnitt SJ, et al. WHO Classification of Tumours of the Breast, Fourth Edition. Lyon:IARC; 2012. [Google Scholar]
- [2].TCGA Network Comprehensive molecular portraits of human breast tumours. Nature 2012;490:61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Kwei KA, Kung Y, Salari K, et al. Genomic instability in breast cancer: pathogenesis and clinical implications. Mol Oncol 2010;4:255–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Arriola E, Marchio C, Tan DS, et al. Genomic analysis of the HER2/TOP2A amplicon in breast cancer and breast cancer cell lines. Lab Invest 2008;88:491–503. [DOI] [PubMed] [Google Scholar]
- [5].Wolff AC, Hammond ME, Hicks DG, et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. J Clin Oncol 2013;31:3997–4013. [DOI] [PubMed] [Google Scholar]
- [6].Nielsen KV, Muller S, Moller S, et al. Aberrations of ERBB2 and TOP2A genes in breast cancer. Mol Oncol 2010;4:161–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Jacot W, Fiche M, Zaman K, et al. The HER2 amplicon in breast cancer: topoisomerase IIA and beyond. Biochim Biophys Acta 2013;1836:146–57. [DOI] [PubMed] [Google Scholar]
- [8].Dhesy-Thind B, Pritchard KI, Messersmith H, et al. HER2/neu in systemic therapy for women with breast cancer: a systematic review. Breast Cancer Res Treat 2008;109:209–29. [DOI] [PubMed] [Google Scholar]
- [9].Gennari A, Sormani MP, Pronzato P, et al. HER2 status and efficacy of adjuvant anthracyclines in early breast cancer: a pooled analysis of randomized trials. J Natl Cancer Inst 2008;100:14–20. [DOI] [PubMed] [Google Scholar]
- [10].Bartlett JMS, Munro A, Cameron DA, et al. Type 1 receptor tyrosine kinase profiles identify patients with enhanced benefit from anthracyclines in the BR9601 Adjuvant Breast Cancer Chemotherapy Trial. J Clin Oncol 2008;26:5027–35. [DOI] [PubMed] [Google Scholar]
- [11].Di Leo A, Desmedt C, Bartlett JM, et al. HER2 and TOP2A as predictive markers for anthracycline-containing chemotherapy regimens as adjuvant treatment of breast cancer: a meta-analysis of individual patient data. Lancet Oncol 2011;12:1134–42. [DOI] [PubMed] [Google Scholar]
- [12].Bartlett JM, McConkey CC, Munro AF, et al. Predicting anthracycline benefit: TOP2A and CEP17-not only but also. J Clin Oncol 2015;33:1680–7. [DOI] [PubMed] [Google Scholar]
- [13].Knoop AS, Knudsen H, Balslev E, et al. Retrospective analysis of topoisomerase IIa amplifications and deletions as predictive markers in primary breast cancer patients randomly assigned to cyclophosphamide, methotrexate, and fluorouracil or cyclophosphamide, epirubicin, and fluorouracil: Danish Breast Cancer Cooperative Group. J Clin Oncol 2005;23:7483–90. [DOI] [PubMed] [Google Scholar]
- [14].Engstrøm MJ, Ytterhus B, Vatten LJ, et al. TOP2A gene copy number change in breast cancer. J Clin Pathol 2014;67:420–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Storlazzi CT, Lonoce A, Guastadisegni MC, et al. Gene amplification as double minutes or homogeneously staining regions in solid tumors: origin and structure. Genome Res 2010;20:1198–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Albertson DG. Conflicting evidence on the frequency of ESR1 amplification in breast cancer. Nat Genet 2008;40:821–2. [DOI] [PubMed] [Google Scholar]
- [17].Ooi A, Inokuchi M, Harada S, et al. Gene amplification of ESR1 in breast cancers–fact or fiction? A fluorescence in situ hybridization and multiplex ligation-dependent probe amplification study. J Pathol 2012;227:8–16. [DOI] [PubMed] [Google Scholar]
- [18].Lin CH, Chen YC, Chiang CJ, et al. The emerging epidemic of estrogen-related cancers in young women in a developing Asian country. Int J Cancer 2012;130:2629–37. [DOI] [PubMed] [Google Scholar]
- [19].Chen JR, Hsieh TY, Chen HY, et al. Absence of estrogen receptor alpha (ESR1) gene amplification in a series of breast cancers in Taiwan. Virchows Arch 2014;464:689–99. [DOI] [PubMed] [Google Scholar]
- [20].Hwang CC, Pintye M, Chang LC, et al. Dual-colour chromogenic in-situ hybridization is a potential alternative to fluorescence in-situ hybridization in HER2 testing. Histopathology 2011;59:984–92. [DOI] [PubMed] [Google Scholar]
- [21].Harvey JM, Clark GM, Osborne CK, et al. Estrogen receptor status by immunohistochemistry is superior to the ligand-binding assay for predicting response to adjuvant endocrine therapy in breast cancer. J Clin Oncol 1999;17:1474–81. [DOI] [PubMed] [Google Scholar]
- [22].Tuominen VJ, Ruotoistenmaki S, Viitanen A, et al. ImmunoRatio: a publicly available web application for quantitative image analysis of estrogen receptor (ER), progesterone receptor (PR), and Ki-67. Breast Cancer Res 2010;12:R56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Ross JS, Slodkowska EA, Symmans WF, et al. The HER-2 receptor and breast cancer: ten years of targeted anti–HER-2 therapy and personalized medicine. Oncologist 2009;14:320–68. [DOI] [PubMed] [Google Scholar]
- [24].Di Leo A, Gancberg D, Larsimont D, et al. HER-2 amplification and topoisomerase IIalpha gene aberrations as predictive markers in node-positive breast cancer patients randomly treated either with an anthracycline-based therapy or with cyclophosphamide, methotrexate, and 5-fluorouracil. Clin Cancer Res 2002;8:1107–16. [PubMed] [Google Scholar]
- [25].Hicks DG, Yoder BJ, Pettay J, et al. The incidence of topoisomerase II-alpha genomic alterations in adenocarcinoma of the breast and their relationship to human epidermal growth factor receptor-2 gene amplification: a fluorescence in situ hybridization study. Hum Pathol 2005;36:348–56. [DOI] [PubMed] [Google Scholar]
- [26].Jarvinen TA, Tanner M, Rantanen V, et al. Amplification and deletion of topoisomerase IIalpha associate with ErbB-2 amplification and affect sensitivity to topoisomerase II inhibitor doxorubicin in breast cancer. Am J Pathol 2000;156:839–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Olsen KE, Knudsen H, Rasmussen BB, et al. Amplification of HER2 and TOP2A and deletion of TOP2A genes in breast cancer investigated by new FISH probes. Acta Oncol 2004;43:35–42. [DOI] [PubMed] [Google Scholar]
- [28].O’Malley FP, Chia S, Tu D, et al. Topoisomerase II alpha and responsiveness of breast cancer to adjuvant chemotherapy. J Natl Cancer Inst 2009;101:644–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Varga Z, Moelans C, Zuerrer-Hardi U, et al. Topoisomerase 2A gene amplification in breast cancer. Critical evaluation of different FISH probes. Breast Cancer Res Treat 2012;133:929–35. [DOI] [PubMed] [Google Scholar]
- [30].Nielsen KV, Ejlertsen B, Moller S, et al. The value of TOP2A gene copy number variation as a biomarker in breast cancer: update of DBCG trial 89D. Acta Oncol 2008;47:725–34. [DOI] [PubMed] [Google Scholar]
- [31].Tubbs RR, Hicks DG, Cook J, et al. Fluorescence in situ hybridization (FISH) as primary methodology for the assessment of HER2 Status in adenocarcinoma of the breast: a single institution experience. Diagn Mol Pathol 2007;16:207–10. [DOI] [PubMed] [Google Scholar]
- [32].Ooi A, Takehana T, Li X, et al. Protein overexpression and gene amplification of HER-2 and EGFR in colorectal cancers: an immunohistochemical and fluorescent in situ hybridization study. Mod Pathol 2004;17:895–904. [DOI] [PubMed] [Google Scholar]
- [33].Holst F, Stahl PR, Ruiz C, et al. Estrogen receptor alpha (ESR1) gene amplification is frequent in breast cancer. Nat Genet 2007;39:655–60. [DOI] [PubMed] [Google Scholar]
- [34].Rody A, Karn T, Ruckhaberle E, et al. Gene expression of topoisomerase II alpha (TOP2A) by microarray analysis is highly prognostic in estrogen receptor (ER) positive breast cancer. Breast Cancer Res Treat 2009;113:457–66. [DOI] [PubMed] [Google Scholar]
- [35].Romero A, Martin M, Cheang MC, et al. Assessment of Topoisomerase II alpha status in breast cancer by quantitative PCR, gene expression microarrays, immunohistochemistry, and fluorescence in situ hybridization. Am J Pathol 2011;178:1453–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


