Abstract
Candida albicans is the most frequently encountered fungal pathogen in humans, capable of causing mucocutaneous and systemic infections in immunocompromised individuals. C. albicans virulence is influenced by multiple factors. Importantly, iron acquisition and avoidance of the immune oxidative burst are two critical barriers for survival in the host. Prior studies using whole genome microarray expression data indicated that the CCAAT-binding factor is involved in the regulation of iron uptake/utilization and the oxidative stress response. This study examines directly the role of the CCAAT-binding factor in regulating the expression of oxidative stress genes in response to iron availability. The CCAAT-binding factor is a heterooligomeric transcription factor previously shown to regulate genes involved in respiration and iron uptake/utilization in C. albicans. Since these pathways directly influence the level of free radicals, it seemed plausible the CCAAT-binding factor regulates genes necessary for the oxidative stress response. In this study, we show the CCAAT-binding factor is involved in regulating some oxidative stress genes in response to iron availability, including CAT1, SOD4, GRX5, and TRX1. We also show that CAT1 expression and catalase activity correlate with the survival of C. albicans to oxidative stress, providing a connection between iron obtainability and the oxidative stress response. We further explore the role of the various CCAAT-binding factor subunits in the formation of distinct protein complexes that modulate the transcription of CAT1 in response to iron. We find that Hap31 and Hap32 can compensate for each other in the formation of an active transcriptional complex; however, they play distinct roles in the oxidative stress response during iron limitation. Moreover, Hap43 was found to be solely responsible for the repression observed under iron deprivation.
Introduction
Candida albicans exists as a commensal in healthy individuals; however, it is capable of causing infections ranging from superficial mucosal to systemic life threatening infections in immunocompromised individuals [1,2]. While the virulence of C. albicans is multifactorial, one necessary aspect of survival is the ability to survive the host immune response. By contrast, innate immune cells, such as macrophages and neutrophils, attempt to kill invading pathogens by exposing them to superoxides, peroxides, and hydroxyl radicals, collectively called Reactive Oxygen Species (ROS), through a process called the respiratory burst [3–7]. C. albicans defends against the respiratory burst by expressing an array of antioxidant enzymes such as catalase, superoxide dismutases, glutaredoxins and thioredoxins [4,8–12].
While our current understanding of the Oxidative Stress Response (OSR) developed through the study of various yeast and fungi, including Saccharomyces cerevisiae, the response in C. albicans displays distinct differences in the regulatory circuits that govern the stress response across fungal clades [13,14]. For example, C. albicans lacks the general Msn2/Msn4-mediated stress response and the cross protection to different stresses is poor to negligible, unlike the response seen in S. cerevisiae or Schizosaccharomyces pombe [14,15]. Similarly, the Stress Activated Protein Kinase (SAPK), Hog1, functions in osmotic stress in both S. cerevisiae and C. albicans; however, it is also involved in the OSR in C. albicans [16–19]. Even within the same genus, C. albicans and C. glabrata have strikingly different regulatory mechanisms for dealing with oxidative stress [4].
The genes involved in the OSR are conserved among fungal pathogens and benign model yeasts. C. albicans has a single gene encoding catalase (CAT1) that converts hydrogen peroxide to water and oxygen [20]. C. albicans encodes six distinct superoxide dismutases, with SOD2 and SOD3 being homologous to the Mn-Sod family while SOD1, SOD4, SOD5, and SOD6 are homologous to the CuZn-Sod family [21,22]. More recently it was demonstrated that Sod5 is a unique Cu-only superoxide dismutase [9]. These enzymes convert superoxide anions to hydrogen peroxide which is further processed by catalase to water and oxygen. In addition, C. albicans encodes four putative glutaredoxins, GRX1, GRX2, GRX3 and GRX5 and two thioredoxins, TRX1 and TRX2 [23,24]. With the large array of proteins involved in the OSR, it is likely that a subset of these antioxidant enzymes will be coordinately regulated to facilitate the survival of C. albicans in the dynamic micro-niches of the host.
The expression of some antioxidant enzymes appears to be regulated by iron availability [25,26]. Iron poses an interesting dilemma for cells since it is essential for the activity of many enzymes, yet an excess of intracellular iron can catalyze the formation of reactive oxygen species, via the Fenton reaction, resulting in oxidative cell damage [27]. Thus, the maintenance of intracellular iron homeostasis is essential for normal growth and minimizing the oxidative damage associated with iron overload. For an invading pathogen, the human host is essentially a low-iron environment with limited free iron [28,29]. To combat the iron sequestration by the host, C. albicans has evolved multiple mechanisms to acquire iron that involve a reductive uptake mechanism, a siderophore scavenging pathway and a hemoglobin uptake pathway [30–32].
In C. albicans, several transcription factors have been found to be involved in iron acquisition/utilization pathways [30,31,33]. One of these transcription factors, the CCAAT-binding factor, has been shown to be important for virulence [25,26,34]. Work from our lab has previously demonstrated that the CCAAT-binding factor is important for the regulation of genes involved in respiratory metabolism, namely CYC1 and COX5a, in response to carbon source availability [35]. Moreover, it has been observed that the CCAAT-binding factor is important for the regulation of genes involved in acquisition/utilization of iron [25,26,34,36,37]. Since the generation of reactive oxygen species in actively growing cells occurs via the Fenton reaction or as a byproduct of respiratory metabolism, we hypothesized that the CCAAT-binding factor may play a central role in regulating the OSR, thereby coordinately regulating iron acquisition/utilization, respiratory metabolism, and the OSR. In fact, whole genome microarray expression studies performed by Singh et al. [34] suggested that genes involved in the OSR were regulated by the CCAAT-binding factor.
The CCAAT-binding factor is an evolutionarily conserved heterooligomeric transcription factor that binds to the consensus 5’-CCAAT-3’ sequence in the promoters of target genes [38]. In S. cerevisiae, the CCAAT-binding factor is composed of three subunits, Hap2, Hap3, and Hap5, necessary for DNA-binding and a fourth subunit, Hap4, the effector subunit responsible for transcriptional activation [39–41]. It has been well-characterized in S. cerevisiae as the activator of genes involved in respiratory metabolism as well as other pathways [41–45]. In C. albicans, the CCAAT-binding factor is comprised of the Hap2 and Hap5 subunits; however, there are two distinct homologs of Hap3, termed Hap31 and Hap32 [34,35,37]. In addition, there are three putative homologs of Hap4, termed Hap41, Hap42, and Hap43 [34,35]. Previous studies have shown that deletion of either HAP2 or HAP5 leads to complete abolishment of the DNA-binding activity in both S. cerevisiae and C. albicans [35,36,40]. In C. albicans it is plausible that the Hap31 and Hap32 may individually interact with Hap2 and Hap5 to form DNA-binding complexes with differing regulatory functions via interaction with the three Hap4-like proteins [34].
In the experiments presented herein, we show that the CCAAT-binding factor is involved in the iron-dependent differential expression of CAT1 in C. albicans. We further provide evidence to support a role for the CCAAT-binding factor in the iron-dependent regulation of other genes involved in the OSR, including SODs, GRXs and TRXs. We used CAT1 as the prototype gene to ask whether the differential regulatory pattern is achieved, at least partially, through the iron-dependent recruitment of distinct CCAAT-binding factor complexes to target promoters. Lastly, we discuss a framework for the role of the CCAAT-binding factor in the iron-dependent transcriptional regulation of the OSR.
Materials and Methods
Yeast strain and growth conditions
The yeast strains used in this study are listed in S1 Table. Strains were routinely cultured in yeast extract-peptone-dextrose (YPD) medium [46]. For DNA transformations, synthetic complete (SC) medium lacking auxotrophic supplements or synthetic minimal medium (SD) augmented with the auxotrophic requirements was used [46]. To generate iron depleted growth conditions, bathophenanthroline disulfonate (BPS) (Sigma) was added to the growth medium at the indicated concentration.
Oligonucleotides and plasmid construction
The oligonucleotides used in this study are listed in S2 Table. The plasmid pDM588 containing HAP41 was generated by PCR amplification of C. albicans orf19.740 using the oligonucleotide primers oDM0343/oDM0344. The HAP41 PCR product was digested with BamHI/PstI cloned into the same sites of pSP65 (Promega Corp). Plasmid pDM588 was digested with NdeI/ClaI, the ends blunted with T4 polymerase and a BglII linker ligated to generate pDM589 containing a deletion in the HAP41 coding sequence. To generate pDM592 with the hap41::URA3 allele, URA3 was amplified from pGEM-URA3 [47] using primers oDM0382/oDM0383 that generated BamHI sites on both the 5’ and 3’ ends of the gene. Plasmid pDM589 and the URA3 PCR product were digested with BglII and BamHI, respectively, and ligated. To generate pDM598 containing the hap41::HIS1 allele, HIS1 was amplified from pGEM-HIS1 [47] with primers oDM0384/oDM0385 that generated BclI sites on both the 5’ and 3’ ends of the gene. Plasmid pDM589 and the HIS1 PCR product were digested with BglII and BclI, respectively, and ligated. The plasmid pDM571 containing HAP42 was generated by PCR amplification of the C.albicans orf19.1481 with the oligonucleotide primers oDM0345/oDM0346 that incorporated unique BamHI and HindIII restriction sites into the 5’ and 3’ ends of the gene, respectively. The plasmid YEplac181 and the HAP42 PCR product were digested with BamHI/HindIII and ligated to generate pDM571. To create hap42Δ::hisG-URA3-hisG knockout allele, pDM571 was digested with BamHI/HindIII and HAP42 was ligated into pSP65 digested with BamHI/HindIII to generate pDM800. The plasmid pDM800 was subsequently amplified by PCR using primers oDM0588/oDM0589 that yielded a plasmid product with a deletion of the HAP42 coding region, but retaining approximately 500bp of flanking sequence and a unique BglII restriction site on the 5’ and 3’ ends. The DNA was digested with BglII and ligated with the BamHI/BglII digested hisG-URA3-hisG from the plasmid p5921 [48]. The plasmid pDM602 contains HAP43 that was generated by PCR amplification of the C. albicans orf19.681 with the oligonucleotide primers oDM0394/oDM0395 that incorporated unique BamHI and SalI restriction sites. The BamHI/SalI digested PCR product was cloned into BamHI/SalI digested pSP65 to generate pDM801. The plasmid pDM801 was used as a template for PCR with primers oDM0590/oDM0591 that created a plasmid product with a deleted coding sequence, but containing approximately 1000bp of HAP43 flanking sequence and a unique BglII restriction site on the 5’ and 3’ ends. The DNA was digested with BglII, ligated with the BamHI/BglII digested hisG-URA3-hisG from the plasmid p5921 [48] and the plasmid designated pDM803. For construction of pDM802 containing the CAT1 promoter fused to Renilla luciferase (Rluc), a 1000bp region of CAT1, upstream of the start codon of orf19.13609, was amplified by PCR using the oligonucleotide primers oDM0660/oDM0661, which incorporated unique SphI and BamHI sites into the 5’ and 3’ ends of the PCR product, respectively. The CAT1 promoter fragment was digested with SphI/BamHI and cloned into the SphI/BamHI sites of pDM692.
Construction of C. albicans strains
All DNA transformation procedures were performed using the lithium acetate transformation kit (QBiogene, Inc.) per the manufacturer instructions. All of the strains described are isogenic derivatives of BWP17 except for the indicated hap allele under study. We have noted BWP17-derived strains carry a mutation in IRO1, a gene adjacent to URA3 locus that has been implicated in iron utilization ([49,50]. However, any potential effect of iro1 in our study would be the same for all strains. The construction of the hap2Δ/Δ (DMC249), hap31Δ/Δ (DMC280), hap32Δ/Δ (DMC285) and the hap31Δ/Δ hap32Δ/Δ (DMC290) mutants have be described elsewhere [51]. The hap41Δ/Δ mutant (DMC190) was generated by two consecutive rounds of transformation of the parent strain BWP17 using the hap41Δ::URA3 and hap41Δ::HIS1 disruption cassettes. The hap41Δ::URA3 was released from plasmid pDM592 by digestion with BamHI/HindIII, introduced into BWP17, and selected on SC-Ura medium. To verify the HAP41/hap41Δ::URA3 heterozygote, genomic DNA was isolated from transformants as described previously [52] and PCR was used to confirm the appropriate recombination. For PCR the oligonucleotide primers oDM0369 (anneals within the URA3 gene) and oDM0620 (anneals to HAP41 locus upstream of the recombination) were used. The HAP41/hap41Δ::URA3 heterozygote was subsequently transformed with BamHI/HindIII-digested pDM598 containing the hap41Δ::HIS1 allele and transformants were selected on SC-His medium. The transformants were subsequently tested on SC-His-Ura medium to verify knockout of both loci. Genomic DNAs were prepared from His+ Ura+ transformants and PCR was used to verify the correct recombination, using oligonucleotide primers oDM0370 (anneals within HIS1) and oDM0620. The hap42Δ/Δ mutant DMC350 was generated as follows. C. albicans BWP17 was subjected to two consecutive rounds of DNA transformation with the hap42Δ::hisG-URA3-hisG cassette after release of the knockout cassette from pDM800 by digestion with BamHI/HindIII. Transformants were selected on SC-Ura medium. To confirm deletion of the first copy of HAP42, genomic DNA was isolated and PCR was performed with oligonucleotides primers oDM0617 (anneals to HAP42 locus upstream of the recombination) and oDM0369 (anneals within URA3). Following confirmation, the HAP42/hap42Δ::hisG-URA3-hisG heterozygote was grown on 5-fluoroorotic acid (5-FOA) medium to select from Ura- recombinants. The transformation was repeated for the deletion of the second allele of HAP42 and the hap42Δ/Δ mutant confirmed by PCR with same oligonucleotide primers. The hap43Δ/Δ strain DMC351 was generated in a similar manner using the hap43Δ::hisG-URA3-hisG cassette after release of the cassette from pDM803 by digestion with BamHI and SalI. The gene disruptions were verified using oligonucleotide primers oDM0396 (anneals to the HAP43 locus upstream of recombination) and oDM369 (anneals within URA3). The hap42Δ/Δ hap43Δ/Δ double mutant (DMC352) was constructed by disrupting HAP43 using the hap43Δ::hisG-URA3-hisG in the hap42Δ/Δ strain DMC350. The hap41Δ/Δ hap42Δ/Δ (DMC353) and the hap41Δ/Δ hap43Δ/Δ (DMC354) double mutants were generated from the hap42Δ (DMC350) and hap43Δ (DMC351) strains, respectively, by transformation with the hap41Δ::URA3 and the hap41Δ::HIS1 disruption constructs sequentially and the resulting disruption was confirmed by PCR as outlined above. The hap41Δ/Δ hap42Δ/Δ hap43Δ/Δ triple mutant (DMC355) was generated using the hap42Δ/Δ hap43Δ/Δ strain DMC352 and disrupting the HAP41 with hap41Δ::URA3 and the hap41Δ::HIS1 disruption constructs sequentially as described above. All of the strains were confirmed by Southern blot as previously described [35]. C. albicans strains expressing the CAT1-Rluc reporter pDM802 were generated by linearizing pDM802 with HpaI within ARG4 and introducing the plasmid into the appropriate strains (BWP17 and DMC108) containing the arg4 auxotropy yielding the strains DMC356 and DMC357, respectively. The resulting transformants were selected on SC-Arg and at least three independent colonies were used for the luciferase assays. After strain construction, the remaining auxotrophies were rescued. For arg4, the plasmid pDM583 was linearized with HpaI within ARG4 and introduced into the appropriate strains. For the ura3 and his1, the plasmid pDM605 (containing URA3 and HIS1) was linearized with NruI within HIS1 and introduced into the appropriate strains. The final prototrophic strains were confirmed by growth on synthetic minimal medium.
Northern blot analysis
C. albicans strains were grown to saturation in YPD for iron replete conditions or YPD + 200 μM BPS for iron-limiting conditions, and subsequently reinoculated into the respective medium and grown to an OD600nm of 0.5–0.8 at 30°C. The cells were harvested by centrifugation, and total RNA was prepared by the glass bead-acid phenol method as previously described [53]. Approximately 20 μg of total RNA was loaded, separated by formaldehyde-1% agarose gel electrophoresis, and transferred to GeneScreen Plus membranes (Dupont-NEN Research products) according to manufacturer’s protocol. The membranes were hybridized and washed under standard high-stringency conditions [35]. The CAT1 and 26S rRNA probes were obtained by PCR amplification from C. albicans BWP17 genomic DNA using the following primer pair oDM0621/oDM0622 and oDM0459/oDM0460, respectively. The SOD1, SOD2, SOD3, SOD4, SOD5, and SOD6 probes were obtained by PCR amplification using the following primer pair oDM0650/oDM0651, oDM0626/oDM0627, oDM0628/oDM0629, oDM0652/oDM0653, oDM0654/oDM0655 and oDM0656/oDM0657, respectively. The GRX2, GRX3, GRX5 and TRX1 probes were obtained by PCR amplification using oligonucleotide pairs oDM0665/oDM0666, oDM0632/oDM0633, oDM0630/oDM0631 and oDM0634/oDM0635, respectively. The probes were purified by agarose gel electrophoresis and GeneClean (Qbiogene, Inc.), and radiolabeled with [α-32P] dATP (MP Biomedicals, LLC) by a random primer labelling kit (U.S. Biochemicals) according to the manufacturer’s protocol. The transcript levels were quantified on a model 9600 Typhoon imager (GE Healthcare Life Sciences, Piscataway, NJ). All Northern blotting experiments shown are representative of at least two, and most cases three, independent experiments from different total RNA preparations.
Catalase enzymatic assays
Catalase activity was determined by monitoring the decomposition of hydrogen peroxide spectrophotometrically at 240nm as previously described [54]. The cells were grown to saturation in YPD or YPD + 200 μM BPS medium, the cultures were subsequently diluted in YPD or YPD + 150 μM BPS and grown to an OD600nm of 0.5 to 0.8. The cells were harvested by centrifugation at 14,000 X g, washed with water and the cell pellets weighed. The pellet was suspended in 50 mM potassium phosphate buffer (pH 7.2) containing 0.2 mM phenylmethylsulfonylfluoride (PMSF) such that the final cell concentration was 0.25 gm of cells/ml wet weight. The cells were disrupted using the Mini-Beadbeater (Biospec products) in the presence of 0.5 mm glass beads and subsequently centrifuged at 14,000 X g to obtain cell free lysate for the catalase assay. The total protein concentration of the lysates was determined by the Bradford’s protein assay (Biorad). For the catalase activity assay, the Beckman-Coulter DU 800 spectrophotometer was zeroed using 50 mM potassium phosphate buffer containing 40 μl of the cell lysate. Following the addition of 400 μl of 30% w/w hydrogen peroxide (Sigma), the decomposition of hydrogen peroxide was determined by measuring the continuous decrease in absorbance at 240 nm and the activity was calculated from the linear range of the curve. The catalase activity of each strain was proportional to the amount of hydrogen peroxide decomposed as determined by: Δμmol H2O2/min./μg cell lysate = ΔA240 /(1.5min X 39.7m-1cm-1 X 1000 X μg cell lysate), where the Δμmol H2O2 is the change in micromoles of hydrogen peroxide per min per microgram lysate, ΔA240 is the change in absorbance at 240nm, 39.7m-1cm-1 is the molar extinction coefficient of hydrogen peroxide at 240 nm.
Hydrogen peroxide sensitivity assays
Each strain was grown to saturation in YPD or YPD + 200 μM BPS medium, the cultures were subsequently diluted in YPD or YPD + 150 μM BPS, respectively, and grown to an OD600nm of 0.5 to 0.8. The cells were harvested by centrifugation for 1 min. at 14,000 X g, washed twice with sterile deionized water, and quantified using a hemocytometer. Approximately 1 x 107 cells of each strain were suspended in YPD medium containing 0, 40, and 80 mM hydrogen peroxide and incubated for 2 h at 30°C. To assess the hydrogen peroxide sensitivity, ten-fold serial dilutions were plated on YPD medium and incubated at 30°C.
Renilla luciferase assays
Renilla Luciferase assays were performed using the Renilla Luciferase reporter assay system (Promega Corp., Madison, WI). For the luciferase measurements, all yeast strains were grown overnight in YPD medium with or without 100 μM BPS for iron replete or iron-limiting growth, respectively. The cultures were subsequently diluted and grown to mid-log phase in YPD or YPD + 150 μM BPS at 30°C. The cell density of the cultures was determined by absorbance at A600nm. A 1 ml aliquot of each culture was removed and centrifuged at 14,000 rpm for one min. The supernatant was removed and the cells were resuspended on 100 μl of 1X lysis buffer (Promega Corp.), and sterile glass beads were added. The samples were vortexed for one min, cooled on ice for 30 seconds, and vortexed for another one min. The samples were centrifuged for one minute at 14,000 rpm to clarify the lysate. For the luciferase assay, 10 μl of cell lysate was added to a luminometer tube along with 100μl of Renilla luciferase substrate and luminescence was measured using a Turner Designs TD-20/20 luminometer. The final Renilla Luciferase activity was calculated with the following formula: RLA = RLU/OD X (Va X Vc/Vb), where RLA is Relative Luciferase Activity in arbitrary units, RLU is the Renilla Luciferase luminescence determined by luminometry, OD is optical density of the cell culture at A600nm, Va is the volume of lysate used in the assay (0.01 ml), Vb is volume of lysis buffer (0.1 ml), Vc is volume taken from original culture (1 ml).
Results
The CCAAT-binding factor is a transcriptional regulator of CAT1 in response to iron
Since the C. albicans CCAAT-binding factor has been suggested to function in both transcriptional activation and repression of genes involved in respiratory metabolism and iron homeostasis [34–36], we hypothesized some OSR genes may be coordinately regulated by the same transcription factor to protect cells from reactive oxygen species generated during respiratory metabolism or by iron via the Fenton reaction. To test this hypothesis, we examined the level of CAT1 mRNA in the wild-type versus hap5Δ/Δ mutant after growth in iron replete (YPD) and iron-limiting (YPD+BPS) medium. Total mRNA was isolated from the strains and Northern blots performed. As shown in Fig 1A, the hap5Δ/Δ strain showed a 4- to 5-fold decrease in CAT1 mRNA levels compared to the wild-type strain after growth in iron replete medium. In contrast, when cells were grown in iron-limiting medium, the hap5Δ/Δ strain showed significantly higher expression of CAT1 than the wild-type strain. These data suggest that the CCAAT-binding factor acts as a transcriptional activator in iron replete medium and as a transcriptional repressor during iron limitation. Alternatively, we cannot exclude the indirect possibility of the CCAAT-binding factor activating an as yet unidentified repressor during iron-limited growth. To determine whether the difference in mRNA levels had observable consequences at the phenotypic level, we examined the sensitivity of wild-type and hap5Δ/Δ strains to oxidative stress induced by hydrogen peroxide after the initial growth in iron replete (YPD) versus iron-limiting (YPD + BPS) medium. As shown in Fig 1B, the hap5Δ/Δ strain is more sensitive to oxidative stress after growth in iron replete medium; whereas, the hap5Δ/Δ mutant displayed more resistance to oxidative stress following growth under iron limitation. The phenotype was consistent with the level CAT1 mRNA observed in the hap5Δ/Δ mutant; however, it was plausible that the mRNA levels may not reflect the catalase enzymatic activity in the cells. This is particularly relevant because catalase contains porphyrin heme groups; therefore, the enzymatic activity may be absent due to iron depletion. To examine this possibility, we measured catalase activity from cell extracts prepared from wild-type and hap5Δ/Δ mutant after growth in iron replete and iron-limiting medium. As shown in Fig 1C, catalase activity was qualitatively similar to the CAT1 mRNA levels, with higher activity in the wild type versus the hap5Δ/Δ strain when cells were grown in iron replete medium. Under iron limitation, catalase activity was substantially reduced; however, the activity was reproducibly higher in the hap5Δ/Δ strain, suggesting the increased resistance to hydrogen peroxide stress (Fig 1B) was due to the increase in catalase activity. To demonstrate that the CCAAT-binding factor is regulating CAT1 expression at the transcriptional level, a CAT1-Renilla luciferase reporter plasmid was introduced into a wild-type and hap5Δ/Δ mutant and luciferase activity was determine after growth in iron-replete and iron-limiting conditions. As shown in Fig 1D, the Renilla luciferase activity observed from the CAT1-Rluc reporter supported the Northern blot data (Fig 1A), indicating that mRNA stability is not involved in the levels of CAT1 expression we observed. It is important to note that we have not demonstrated directly CCAAT-binding factor binding to the putative CCAAT sites within the CAT1 promoter; therefore, it remains plausible that the CCAAT-binding factor indirectly influences CAT1 mRNA expression.
Fig 1. The CCAAT-binding factor regulates CAT1 in response to iron availability.
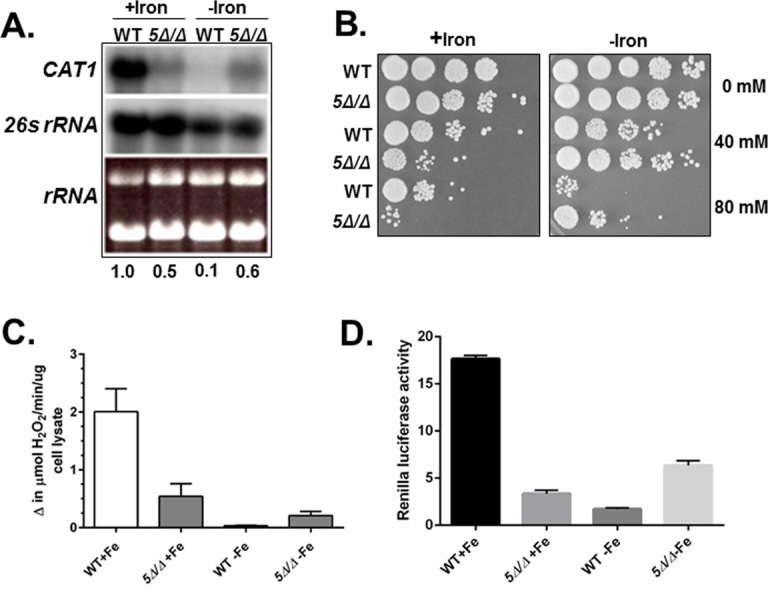
(A) Northern blot analysis of CAT1 mRNA expression in the wild-type (DMC146) and hap5Δ/Δ mutant (DMC117) following growth in iron-replete (+iron) and iron-limiting (-iron) medium. The 26s rRNA was the loading control. mRNA levels were normalized to the 26s rRNA control using the WT as the reference value. (B) C. albicans wild-type (DMC146) and hap5Δ/Δ mutant (DMC117) were grown in iron-replete (+iron) or iron-limited (-iron) medium and subsequently exposed to hydrogen peroxide at the indicated concentrations for 2 h at 30°C. Ten-fold serial dilutions were spotted to YPD medium and incubated at 30°C for 3 days. (C) Catalase enzymatic activity in cell extracts derived from the wild type (DMC146) and hap5Δ/Δ mutant (DMC117) following growth in iron-replete (+iron) and iron-limiting (-iron) medium. The enzymatic assays are the average of three independent experiments with the error bars indicating the standard error. (D) C. albicans wild-type (DMC356) and hap5Δ/Δ mutant (DMC357) were grown in iron-replete (+iron) or iron-limited (-iron) medium and subsequently assayed for expression of Renilla luciferase driven by the CAT1 promoter. The luciferase assays are the average of three independent experiments with the error bars indicating the standard error.
Regulation of other OSR genes by the CCAAT-binding factor
Since the CCAAT-binding factor was involved in the regulation of CAT1, it was reasonable to predict that other OSR genes, such as SOD1-6 (superoxide dismutases), GRX1-4 (glutaredoxins) and/or TRX1 (thioredoxin), could be coordinately regulated. We examined the mRNA levels of SOD1 through SOD6, GRX1, GRX2, GRX3, GRX5 and TRX1 in the wild type and hap5Δ/Δ mutant strains grown in iron replete and iron-limiting YPD medium. We observed that SOD1, SOD2, and SOD3 were repressed by the CCAAT-binding factor; however, the repression was not dependent on iron (Fig 2). In contrast, the expression of SOD4 was only observed under iron-limitation in the hap5Δ/Δ mutant, suggesting the CCAAT-binding factor may be involved in the regulation of SOD4 in an iron-dependent manner. It should also be noted that the mRNA levels of SOD5 and SOD6 were examined, but expression of neither was altered by iron availability or the CCAAT-binding factor (not shown).
Fig 2. SOD1, SOD2, SOD3, and SOD4 are regulated by the CCAAT-binding factor.
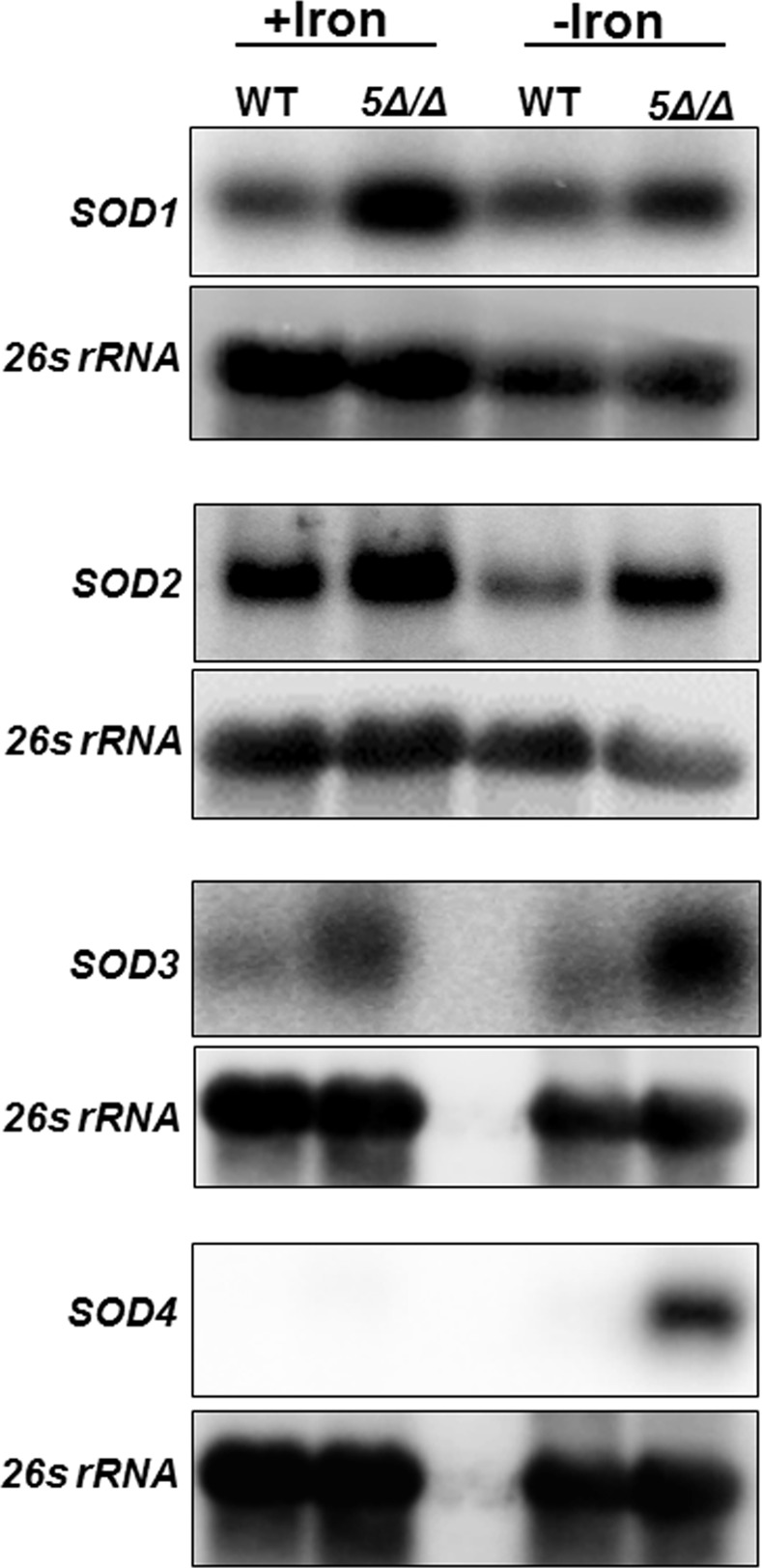
Northern blot analysis was performed to examine the expression of SOD1, SOD2, SOD3, and SOD4 mRNA as indicated in the wild-type (DMC146) and hap5Δ/Δ mutant (DMC117) following growth in iron-replete (+iron) and iron-limiting (-iron) medium. The 26s rRNA was the loading control.
With respect to the expression of the genes encoding the glutaredoxins and thioredoxin, it was found that GRX2 and TRX1 appear to be repressed by the CCAAT-binding factor in iron-replete medium; however, no clear iron-dependent regulation was observed (Fig 3). GRX3 mRNA expression was unchanged in the hap5Δ/Δ mutant or by the iron status of the growth medium. In contrast, GRX5 demonstrated a pattern of mRNA expression reminiscent of CAT1, indicating the CCAAT-binding factor is involved in the activation of GRX5 during iron-replete growth and repression when iron was limiting (Fig 3), indicating GRX5 transcription is coordinately regulated with CAT1. Although we find both genes have a similar CCAAT-binding factor-dependent response to iron availability, it remains to be shown that the regulation involves the direct binding of the transcription factor to its cognate promoter sequence. The GRX1 expression was also examined, but no mRNA was detected under the growth conditions used in this study (not shown).
Fig 3. GRX2, GRX5, and TRX1 expression is regulated by the CCAAT-binding factor.
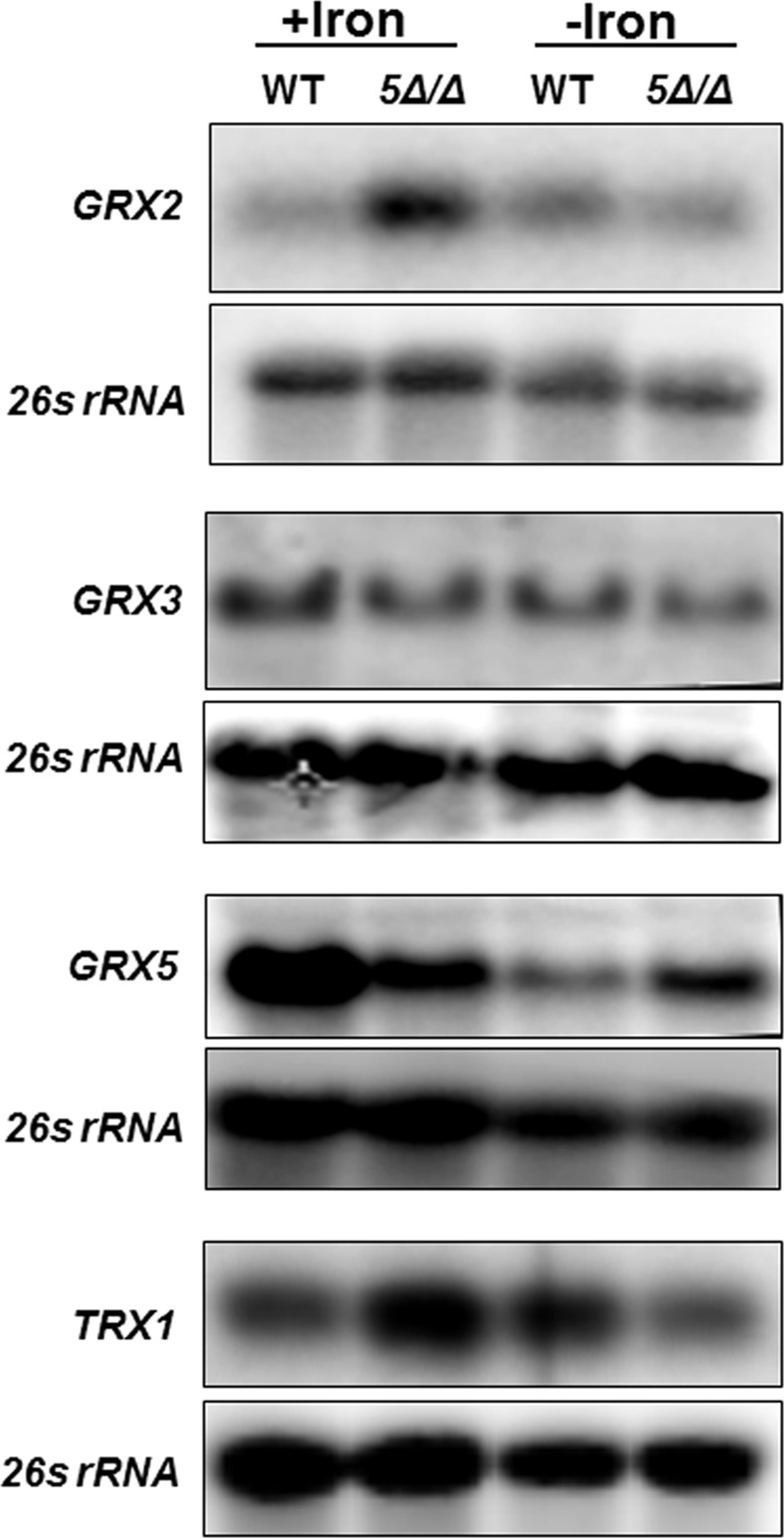
Northern blot analysis was performed to examine the expression of GRX2, GRX3, GRX5, and TRX1 mRNA as indicated in the wild-type (DMC146) and hap5Δ/Δ mutant (DMC117) following growth in iron-replete (+iron) and iron-limiting (-iron) medium. The 26s rRNA was the loading control.
Role of Hap31 and Hap32 in the iron-dependent regulation of CAT1
The CCAAT-binding factor appears to function in the regulation of several OSR genes; however, the expression of CAT1 and GRX5 mRNA in response to iron availability suggested the CCAAT-binding factor may function in transcriptional activation and repression in response to environmental conditions. We wanted to further explore the mechanism behind this dual function. C. albicans encodes two different Hap3 subunits, termed Hap31 and Hap32, and three putative Hap4 subunits, designated Hap41, Hap42, and Hap43. One putative model to explain the dual role of this multi-subunit transcription factor involves the different Hap3 and/or Hap4 subunits interacting with Hap2 and Hap5 in a combinatorial manner to form distinct CCAAT-binding complexes that either promote or repress transcription in response to environmental signals. To explore this possibility, we examined the oxidative stress sensitivity of various null mutants of the CCAAT-binding factor subunits and compared them to the hap5Δ/Δ mutant. Thus, the hap2Δ/Δ, hap31Δ/Δ, hap32Δ/Δ, and hap31Δ/Δ hap32Δ/Δ double mutant were grown in iron replete or iron-limiting medium (YPD or YPD + BPS, respectively) and subsequently exposed to various concentrations of hydrogen peroxide and serial dilutions plated on YPD medium (Fig 4). As expected, the hap2Δ/Δ strain mimicked the phenotype of the hap5Δ/Δ mutant (Fig 4A), with higher sensitivity to oxidative stress than the wild-type after iron-replete growth, yet more resistant to oxidative killing following iron-limited growth. When the hap31Δ/Δ and hap32Δ/Δ mutants were examined, neither demonstrated increased sensitivity to oxidative stress after iron replete growth (Fig 4B); however, the hap31Δ/Δ hap32Δ/Δ double mutant showed sensitivity equivalent to the hap2Δ/Δ and hap5Δ/Δ mutants. After iron-limited growth, the hap31Δ/Δ and the hap31Δ/Δ hap32Δ/Δ double mutant demonstrated increased resistance to oxidative stress, analogous to the hap2Δ/Δ and hap5Δ/Δ mutants, suggesting the loss of Hap31 is key to the oxidative stress resistance observed during iron-limiting growth. Moreover, it was observed that the hap32Δ/Δ mutant was more sensitive than the wild-type strain to oxidative stress after growth under iron limitation. These data suggest a relationship between HAP31 and HAP32 in which hap31Δ/Δ is epistatic to hap32Δ/Δ in iron-limiting conditions since the double mutant shows the same survival phenotype to the hydrogen peroxide treatment as the hap31Δ/Δ mutant. The epistatic relationship among these genes would indicate a cross-regulatory mechanism between the subunits depending of which Hap3 subunit is associated with the CCAAT-binding factor.
Fig 4. Hap31 and Hap32 display a differential response to hydrogen peroxide stress.
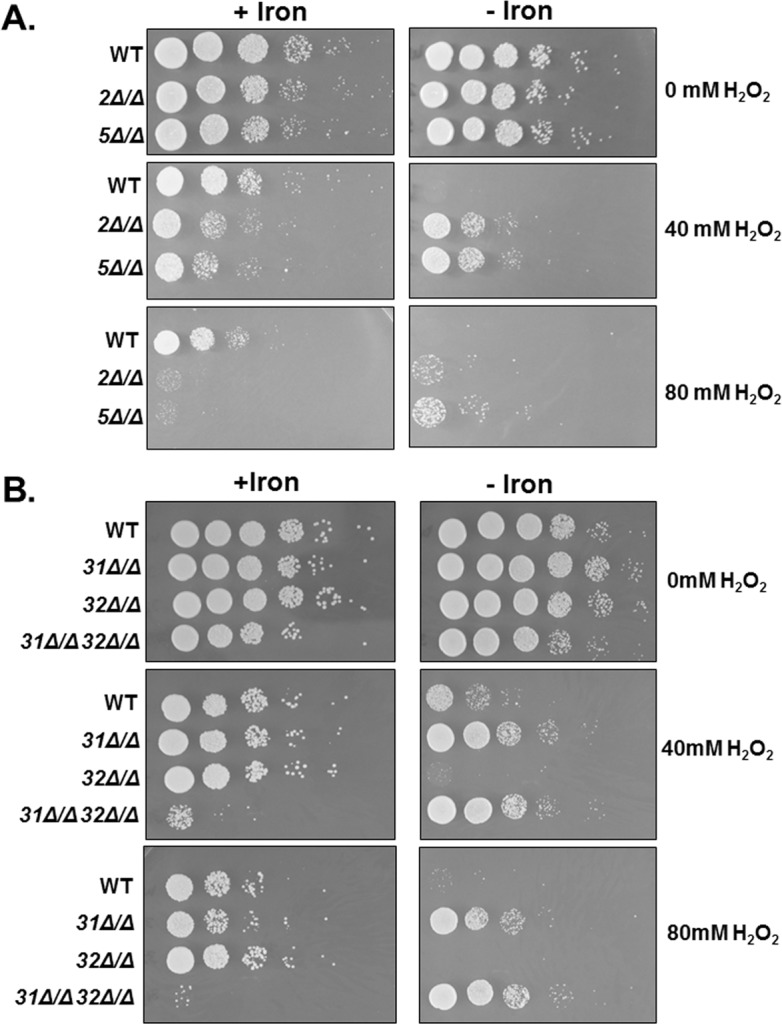
(A) C. albicans wild-type (DMC146), hap2Δ/Δ (DMC249), and hap5Δ/Δ mutant (DMC117), were grown in iron-replete (+iron) or iron-limited (-iron) medium and subsequently exposed to hydrogen peroxide at the indicated concentrations for 2 h at 30°C. Ten-fold serial dilutions were spotted to YPD medium and incubated at 30°C for 2 days. (B) C. albicans wild-type (DMC146), hap31Δ/Δ (DMC280), hap32Δ/Δ (DMC285), and the hap31Δ/Δ hap32Δ/Δ (DMC290) mutants were grown in iron-replete (+iron) or iron-limited (-iron) medium and subsequently exposed to hydrogen peroxide at the indicated concentrations for 2 h at 30°C for 3 days.
When the CAT1 mRNA levels were examined by Northern blotting in the various hap mutants, it was found that the hap2Δ/Δ and hap31Δ/Δ hap32Δ/Δ mutants showed a decrease in expression analogous to the hap5Δ/Δ after iron replete growth, confirming the loss of CCAAT-binding activity reduced the activation of CAT1 (Fig 5A). Moreover, the hap2Δ/Δ and hap31Δ/Δ hap32Δ/Δ mutants demonstrated a similar loss of CAT1 repression after growth under iron limitation, demonstrating the loss of repression is due to the lack of CCAAT-binding activity. Interestingly, neither the hap31Δ/Δ or hap32Δ/Δ mutants showed a major decrease in CAT1 mRNA after iron-replete growth. In fact, the hap32Δ/Δ mutant displayed a three to four-fold higher level of expression than the wild-type strain. When the same strains were grown in iron-limiting medium it was found that the hap31Δ/Δ and hap32Δ/Δ mutants displayed expression similar to the wild-type strain (Fig 5A). To correlate the phenotype of the hap mutants with the catalase activity, we examined the catalase activity of the hap31Δ/Δ, hap32Δ/Δ, and hap31Δ/Δ hap32Δ/Δ mutants after growth in iron-replete or iron-limiting conditions. As shown in Fig 5B, the hap31Δ/Δ hap32Δ/Δ mutant demonstrated a 6-fold reduction in catalase activity that mimicked the hap5Δ/Δ mutant, while the hap31Δ/Δ and hap32Δ/Δ mutants showed only a 2.5- to 3.5-fold decrease in catalase activity. It seems plausible that the catalase activity, although reduced in the hap3 mutants, may remain sufficient for the oxidative stress phenotype comparable to wild-type strain (see Fig 4B). After growth in iron-limiting medium, the lysate from the hap31Δ/Δ hap32Δ/Δ mutant showed catalase activity that was reproducibly higher than the wild-type strain and similar to the hap5Δ/Δ mutant. The residual activity may explain the relative oxidative stress resistance of these strains during iron limitation. The catalase activity of the hap31Δ/Δ and hap32Δ/Δ single mutants observed after growth in iron-limiting conditions was consistently similar to the wild-type strain, yet the hap31Δ/Δ mutant was phenotypically more resistant to oxidative stress under iron limitation and the hap32Δ/Δ mutant was more sensitive. We did observe that the catalase activity of the hap31Δ/Δ mutant was reproducibly higher than the wild-type strain after iron-limiting growth, conversely the hap32Δ/Δ mutant was consistently less than the wild-type; however, these differences were not statistically significant. Nevertheless, there may be a minimum threshold of catalase activity necessary for oxidative stress resistance during iron-limiting growth and the hap31Δ/Δ mutant surpasses that minimum while the hap32Δ/Δ is below the activity necessary for survival of the oxidative stress conditions used in this study. Alternatively, other OSR genes induced by iron starvation may contribute to the enhanced hydrogen peroxide resistance in a hap31Δ/Δ mutant.
Fig 5. Hap31 and Hap32 are interchangeable in the function of the CCAAT-binding factor.
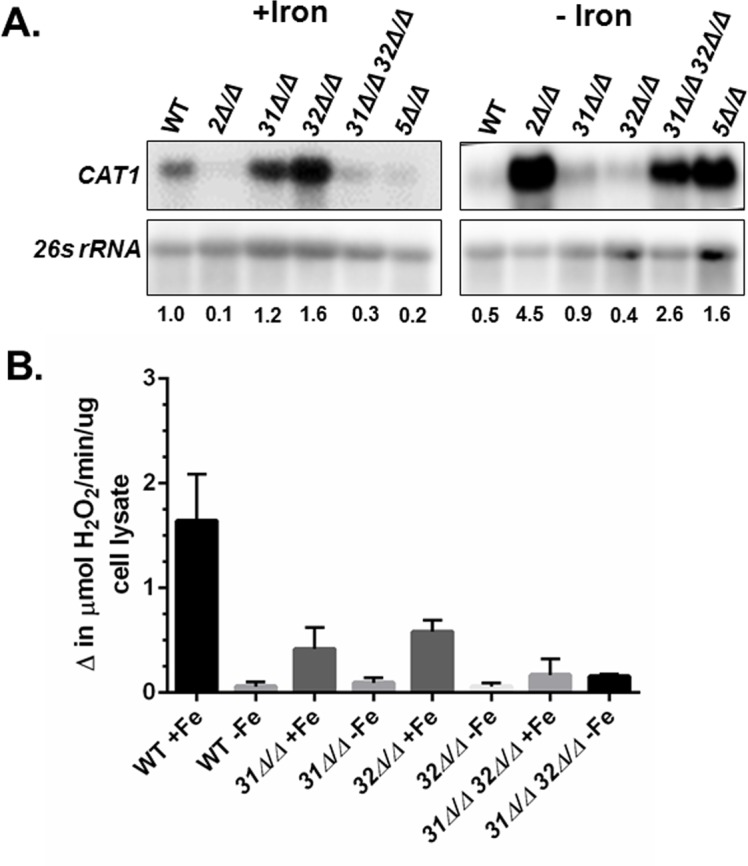
(A) Northern blot analysis of CAT1 mRNA expression in the wild-type (DMC146) and the indicated hap mutants following growth in iron-replete (+iron) and iron-limiting (-iron) medium. The 26s rRNA was the loading control. mRNA levels were normalized to the 26s rRNA control using the WT as the reference value. (B) Catalase activity in cell extracts derived from the wild type (DMC146), hap31Δ/Δ (DMC280), hap32Δ/Δ (DMC285), and hap31Δ/Δ hap32Δ/Δ (DMC290) mutants following growth in iron-replete (+iron) and iron-limiting (-iron) medium. The enzymatic assays are the average of three independent experiments with the error bars indicating the standard error.
Hap43 is solely responsible for the iron-dependent regulation of CAT1
C. albicans is predicted to encode three distinct Hap4-like proteins, termed Hap41, Hap42, and Hap43 [35]. Previous studies have demonstrated that Hap43, as a subunit of the CCAAT-binding factor, is involved in the iron-dependent regulation of multiple genes [25,26,34,37]. We examined the role of the three Hap4 subunits in the OSR of C. albicans. We generated homozygous gene deletions of HAP41, HAP42, and HAP43 and subsequently examined their sensitivity to hydrogen peroxide after growth in iron replete and iron-limiting medium. As shown in Fig 6A, none of the hap4 null mutants showed increased sensitivity to hydrogen peroxide stress relative to the wild-type control following iron replete growth, whereas the hap5Δ/Δ control was highly sensitive. By contrast, the mutants evaluated for oxidative stress sensitivity following iron-limited growth, demonstrated that only the hap43Δ/Δ mutant had resistance comparable to the hap5Δ/Δ strain (Fig 6A). This phenotype indicates that the hap43Δ/Δ and hap5Δ/Δ mutants should have higher catalase activity after iron-limited growth compared to the wild-type, hap41Δ/Δ, and hap42Δ/Δ strains. To confirm this prediction, catalase assays were performed on lysates prepared from each strain following iron replete and iron-limiting growth, and the data confirmed that the hap43Δ/Δ mutant had significantly higher catalase activity, comparable to that of the hap5Δ/Δ mutant (Fig 6B). On the basis of these data, the Hap4 subunits do not appear to play a significant role in the regulation of CAT1 during iron replete growth; however, Hap43 was needed for the repression of CAT1 during growth under iron limitation. As a corollary, Hap41 and Hap42 do not appear to function in the regulation of CAT1.
Fig 6. The role of the individual Hap4-like subunits in regulating the OSR.
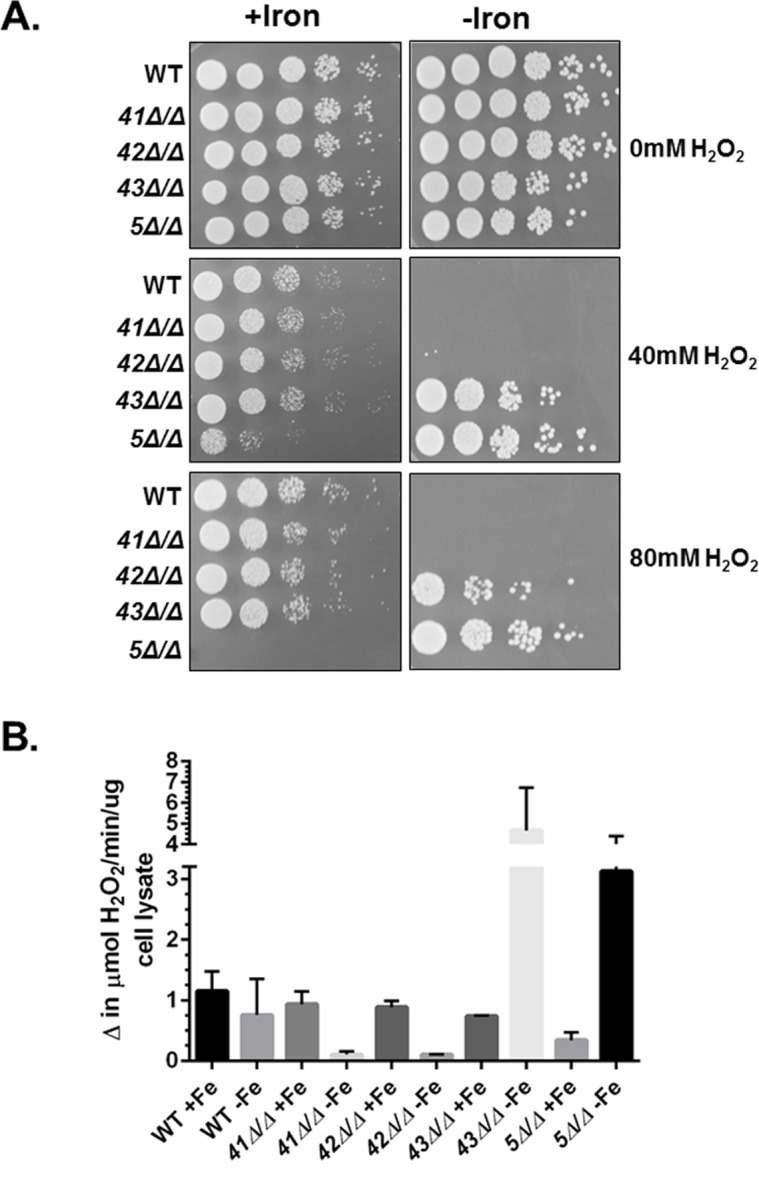
(A) C. albicans wild-type (DMC146), hap41Δ/Δ (DMC190), hap42Δ/Δ (DMC350), hap43Δ/Δ (DMC351) and hap5Δ/Δ mutant (DMC117) were grown in iron-replete (+iron) or iron-limited (-iron) medium and subsequently exposed to hydrogen peroxide at the indicated concentrations for 2 h at 30°C. Ten-fold serial dilutions were spotted to YPD medium and incubated at 30°C for 3 days. (B) Catalase activity assays were performed on extracts from the wild type (DMC146), hap41Δ/Δ (DMC190), hap42Δ/Δ (DMC350), hap43Δ/Δ (DMC351) and hap5Δ/Δ mutant (DMC117) mutants following growth in iron-replete (+iron) and iron-limiting (-iron) medium. The enzymatic assays are the average of three independent experiments with the error bars indicating the standard error.
The experiments above indicate that Hap43 is solely responsible for the iron-dependent oxidative stress phenotype. To further validate this finding, we constructed strains that included all hap4 mutant combinations, including the hap41Δ/Δ hap42Δ/Δ hap43Δ/Δ triple mutant, to determine whether the different Hap4-like proteins could provide compensatory activity and mask potential phenotypes. Previous studies have demonstrated that the hap43Δ/Δ is unable to grow under iron limitation [26,34,36]. We initially examined the growth of the various hap4 mutants for a growth defect on iron-deficient medium. As shown in Fig 7A, only the mutants that included the hap43Δ/Δ knockout were unable to grow on iron-limiting medium, whereas the hap41Δ/Δ hap42Δ/Δ double mutant grew comparable to wild type. These data imply that Hap43 is solely important for growth during iron limitation.
Fig 7. Hap43 is the sole Hap4 subunit involved in the CCAAT-binding factor-mediated oxidative stress response.

(A) C. albicans wild-type (DMC146) and indicated hap4 combination mutants were grown in iron-replete (YPD) or iron-limited (YPD+BPS) liquid medium and ten-fold serial dilutions were spotted to YPD or YPD+BPS, respectively, and incubated at 30°C for 3 days. (B) C. albicans wild-type (DMC146), hap5Δ/Δ (DMC117) and indicated hap4Δ/Δ combination mutants were grown in iron-replete (+iron) or iron-limited (-iron) medium and subsequently exposed to hydrogen peroxide at the indicated concentrations for 2 h at 30°C. Ten-fold serial dilutions were spotted to YPD medium and incubated at 30°C for 3 days.
To examine the oxidative stress phenotype of the various hap4 mutants, the strains were grown in iron replete or iron-limiting medium and subsequently exposed to hydrogen peroxide stress. Serial dilutions of the cells were spotted to rich medium and the survival phenotype examined. When grown in iron replete medium, none of the hap4 null mutants displayed a phenotype that differed significantly from the wild-type strain (Fig 7B), suggesting none of the Hap4 subunits function in the regulation of general OSR in a nutrient-rich environment. In contrast, the iron-limiting growth resulted in significant oxidative stress resistance of strains that contained the hap43Δ/Δ mutation (Fig 7B). Importantly, given that the hap41Δ/Δ hap42Δ/Δ mutant was phenotypically sensitive to oxidative stress comparable to the wild-type strain, these data strongly support the conclusion that Hap43 is the sole subunit critical for the OSR as well as the repression of genes under iron deficient conditions.
Given the phenotypic observations described above for the various hap4 mutants, we hypothesized that the CAT1 mRNA levels would not vary significantly after growth in iron replete medium. As a corollary, we predicted the loss of CAT1 repression during iron-limiting growth in mutants that included the hap43Δ/Δ mutation. To confirm this prediction, we performed Northern blots on RNA isolated from all of the hap4 knockout mutants grown in iron replete and iron-limiting conditions (Fig 8). Although there was some variability in CAT1 expression in the hap4 knockout mutants or combination mutants after growth in iron replete medium (Fig 8A), we did not observe statistically significant differences that were reproducible in three independent Northern blot experiments. This led to the conclusion that none of the Hap4 subunits played a significant role in CAT1 regulation in rich growth medium. In contrast, the expression of CAT1 after iron-limiting growth clearly demonstrated that Hap43 was important for the repression of CAT1 during iron deprivation since the hap43Δ/Δ mutant as well as any combination mutant that included hap43Δ/Δ displayed the loss of CAT1 repression (Fig 8B). These data unambiguously support the role of Hap43 as the effector subunit that interacts with the CCAAT-binding factor to regulate genes involved in the OSR in response to iron.
Fig 8. Hap43 is necessary for the CCAAT-binding factor-mediated regulation of CAT1 in response to iron.
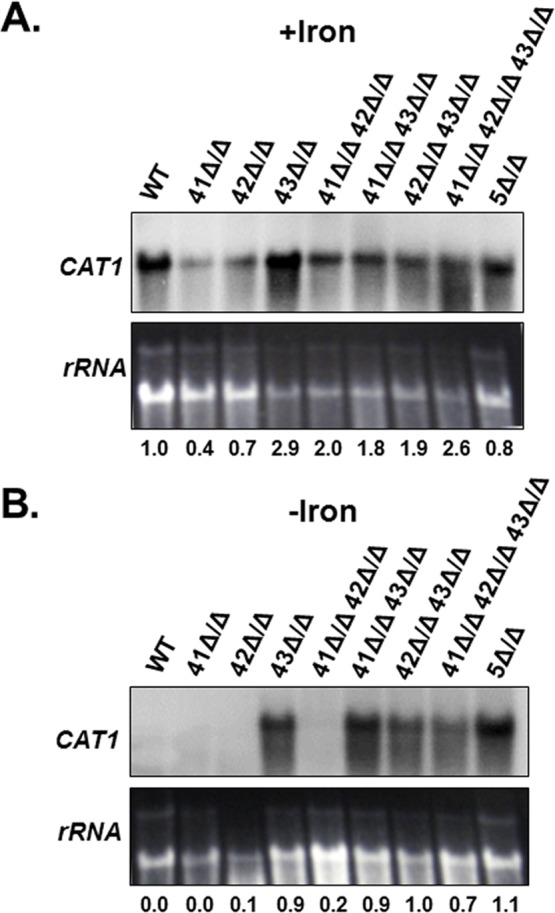
Northern blot analysis of CAT1 mRNA expression in the wild-type (DMC146), hap5Δ/Δ mutant (DMC117) and the indicated hap4Δ/Δ single or combination mutants following growth in (A) iron-replete (+iron) and (B) iron-limiting (-iron) medium. The rRNA was the loading control. mRNA levels were normalized to the rRNA control using the WT as the reference value.
Discussion
The CCAAT-binding factor has been previously shown to be a transcriptional regulator involved in modulating the expression of genes involved in respiratory metabolism [35] and iron acquisition/utilization [25,26,34,36,55]. In addition, prior microarray expression studies have implicated the CCAAT-binding factor in the iron-dependent regulation of genes involved in oxidative stress [34]. In this work we further dissect the requirement of each of the CCAAT-binding factor subunits for the regulated expression of some OSR genes in response to iron. In addition, we connect the mRNA expression of CAT1 to the in vivo activity of catalase, measured by the cell’s ability to overcome oxidative stress. Finally, we present data suggesting the modular nature of the CCAAT-binding complex, with the Hap31 or Hap32 subunits, with or without Hap43, is essential for the assembly of the transcription complexes required for either activation or repression of the indicated OSR genes.
The CCAAT-binding factor has a dual and contrasting role in the regulation of OSR genes in response to iron
During cell metabolism, reactive oxygen species are generated primarily through the mitochondrial electron transport chain via the partial reduction of oxygen through the transfer of one, two, or three electrons, generating superoxide, hydrogen peroxide or hydroxyl radicals. C. albicans, like other eukaryotes, has developed antioxidant mechanisms such as superoxide dismutases, catalase, thioredoxins and glutaredoxins to neutralize the reactive oxygen species and repair cell damage. We hypothesized that the CCAAT-binding factor may play a central role in regulating the oxidative stress response in C. albicans, thereby coordinately regulating iron acquisition/utilization, respiratory metabolism and the oxidative stress response.
In our initial gene expression analysis of CAT1 we found the mRNA levels decreased to a basal level in the hap5Δ/Δ strain as compared to the wild-type, implicating the CCAAT-binding factor in transcriptional activation. However, during iron deficient growth the expression levels reversed; with the wild-type expression nearly null and the hap5Δ/Δ strain showing significant CAT1 mRNA, implicating the CCAAT-binding factor in transcriptional repression. Moreover, the luciferase activity of the CAT1-Rluc promoter fusion shows that the CAT1 promoter drives gene expression in a CCAAT-binding factor-dependent manner. These data demonstrate the role of the CCAAT-binding factor as an activator or repressor of target genes in response to iron. Importantly, these findings complement previous data demonstrating the role of this transcription factor in the regulation of respiratory genes in response to carbon source availability [35]. Moreover, this work is consistent with previous reports that have shown that the CCAAT-binding factor functions as a transcriptional activator/repressor of numerous genes encoding proteins that utilize iron as a cofactor in response to iron availability [25,26,34].
This finding prompted us to ask whether the CCAAT-binding factor also regulates other OSR genes in C. albicans. Among the six genes predicted to encode superoxide dismutase enzymes, three of them, SOD1, SOD2, and SOD3, were repressed by the CCAAT-binding factor, but in an iron-independent manner, as the transcript levels increased in the hap5Δ/Δ strain compared to the wild-type under iron-replete as well as iron-limiting conditions. The CCAAT-binding factor-dependent repression of SOD2 and SOD3 has also been reported by microarray studies used to compare the transcriptional profiling of a wild-type to a hap43 (cap2) mutant under iron limitation [34]. Since Hap43 is an effector subunit of the CCAAT-binding factor [25,26,34,36], one would predict that null mutations in the genes encoding the other Hap subunits would display a similar profile. Both cytoplasmic Sod1 and Sod3 have been shown to be critical for C. albicans virulence although they are induced under different growth conditions, consistent with the low expression levels we observed in exponentially grown wild-type cells. Sod1 is known to be induced under hyphal-promoting conditions and it has been implicated in protection against ROS generated by menadione and macrophages [21,56]. Sod1 expression is repressed during stationary phase as a defense mechanism to evade copper toxicity [21,22]. Sod3 is an unusual cytoplasmic MnSod expressed during stationary phase [57]. The transcript level of the mitochondrial Sod2 was most abundant in the wild-type cells under iron-replete conditions, an observation consistent with the protective role of Sod2 against intracellular superoxide anions. The iron replete growth conditions used in our study would yield normal ROS levels as a byproduct of respiratory metabolism [22]. In agreement with this response, SOD2 mRNA levels are diminished under iron limitation when ROS levels would be reduced. Indeed, exposure to hydrogen peroxide does not seem to increase expression of SOD2, emphasizing its role in intracellular ROS detoxification [15]. No visible transcripts were detected for the cell surface associated Sod4, Sod5 and Sod6 in iron replete conditions. This finding is consistent with the role of these superoxide dismutases in the detoxification of the extracellular ROS threat, especially those generated by the host macrophages and neutrophils [58]. However, under iron deprivation we observed a strong CCAAT-binding factor-dependent repression of SOD4, as evidenced by high mRNA levels present in the hap5Δ/Δ strain. A connection between superoxide stress and intracellular iron levels has been proposed in S. cerevisiae, where mutations in superoxide dismutase genes show altered iron homeostasis [59,60]. Thus, it is possible that a similar mechanism exists in C. albicans, where the repression of the SOD genes by the CCAAT-binding factor during iron limitation permits the mobilization of iron to more essential processes.
We found that two of the glutaredoxin-encoding genes, GRX2 and GRX5, are regulated by the CCAAT-binding factor in a contrasting manner. While the CCAAT-binding complex appeared to repress GRX2, GRX5 was activated by the transcription factor in iron-replete medium. In contrast, during iron limiting growth GRX2 expression appears to be CCAAT-binding factor-independent; while GRX5 was repressed in a manner similar to CAT1. Grx2 has been implicated in the resistance to PMN-mediated killing by the host [23,61]. On the other hand, the Grx5 ortholog in S.cerevisiae is a mitochondrial matrix protein involved in the incorporation of the Fe-S clusters into respiratory chain proteins [61]. Thus, on the basis of cellular function, the CCAAT-binding factor seems to regulate GRX2 and GRX5 expression differentially in the same environmental conditions. GRX5 repression under iron limitation may contribute to increasing intracellular iron, and mobilizing it to essential proteins needed for cell survival. In support of this hypothesis, it has been shown that a S. cerevisiae grx5Δ strain accumulates iron intracellularly [11,61].
We also observed the CCAAT-binding factor-dependent expression of the thioredoxin gene TRX1 in response to iron; however, it was opposite of that seen with CAT1 and GRX5. The repression and activation observed during iron replete and iron limiting growth, respectively, may represent an unexplored function of Trx1 related to iron metabolism. It should be noted that this pattern of TRX1 regulation was also reported in a whole genome transcriptional profile that compared a wild-type to a hap43Δ/Δ mutant under iron limited growth [34]. In S. cerevisiae it has been shown that the thioredoxins and glutaredoxins are relevant for maintaining the cellular thiol-redox system; however, evidence suggests that they operate through different non-redundant pathways [62,63]. For C. albicans, additional work is needed to understand the contribution and regulation of these genes in the OSR and iron homeostasis. What is clear is that the CCAAT-binding factor can specifically activate as well as repress some of these genes in response to the iron available during cell growth.
The CCAAT-binding factor regulates the in vivo OSR
The hydrogen peroxide sensitivity assay allowed us to examine the cellular response of C. albicans when confronted with ROS after prior growth in iron-replete or iron deficient environments. The survival to peroxide treatment correlated with CAT1 mRNA levels and the catalase activity of cells in response to iron. Moreover, the assay demonstrated that the integrity of the CCAAT-binding factor is essential for C. albicans to cope with peroxide stress. This is supported by the fact that any one of the strains carrying a deletion of the subunits essential for the integrity of the CCAAT-binding factor, i.e., hap2Δ/Δ, hap5Δ/Δ, or the double deletion hap31Δ/Δ hap32Δ/Δ, was unable to survive the hydrogen peroxide treatment if the cells were previously grown in iron-replete medium. The opposite was seen when the cells were grown in iron-limited conditions, in perfect agreement with the high CAT1 mRNA levels as well as the catalase activity of the respective strains. This is consistent with prior studies that have shown that the CCAAT-binding factor functions as a transcriptional repressor of numerous genes that utilize iron as a cofactor after exposure to iron-limiting growth conditions [25,26,34]. Moreover, the OSR has been shown to be regulated by the CCAAT-binding factor in the filamentous fungus, Aspergillus nidulans. In A. nidulans, the complex is involved in redox sensing via the oxidative modification of thiol groups in the evolutionarily conserved cysteine residues of the histone fold motif of HapC, which results in the regulation of OSR genes [64]. The HapC orthologs in C. albicans, Hap31 and Hap32, also share the conserved cysteine residues, suggesting a possible mechanism for their participation in the OSR. Since either protein could serve as a putative sensor of the redox state of the cell, it is plausible that their function is dependent on environmental cues.
The modular nature of the CCAAT-binding complex is essential for the differential regulation of CAT1 in response to iron
Since the CCAAT-binding factor serves contrasting roles as an activator or repressor in response to iron, we hypothesized that this may be achieved through the differential recruitment of the Hap31 or Hap32 as well as Hap41, Hap42, or Hap43 to form functional CCAAT-binding complexes. Our data showed that the presence of either Hap31 or Hap32 is absolutely essential for the activation of CAT1 in iron-replete conditions, since the strain lacking both genes had less catalase and recovered poorly from peroxide stress, mimicking the hap5Δ/Δ mutant. Thus, it appears that both subunits are expressed and capable of compensating for each other in the formation and function of the complex. During iron-limited growth the regulation of CAT1 is more complex. The mRNA levels indicate that the presence of either Hap31 or Hap32 represses CAT1 expression to similar levels and comparable to the wild-type levels, again suggesting a compensatory role between these subunits in the formation of an active CCAAT-binding complex. However, in spite of the low levels of catalase the hap31Δ/Δ strain showed resistance to the hydrogen peroxide treatment. This unexpected result may reflect threshold levels of catalase sufficient for the protective function, or alternatively, a catalase-independent mechanism of managing peroxide stress that was only manifested in a hap31Δ/Δ mutant. Such a mechanism could involve other OSR genes that are expressed under iron limitation. The latter possibility has been suggested in C. glabrata where a cta1Δ (cat1Δ) strain remains capable of adaptation to oxidative stress [65].
Our analysis of CAT1 expression and the survival to peroxide stress in the strains containing single, double, and triple combination of hap41Δ/Δ, hap42Δ/Δ and hap43Δ/Δ alleles lead to the conclusion that none of the Hap4-like subunits were necessary for expression during iron-replete growth. In contrast, during iron limitation, Hap43 was the sole Hap4-like subunit responsible for CAT1 repression. This is consistent with the fact that C. albicans Hap43 has been reported to be a global repressor of genes encoding proteins that involve utilization of iron in iron-limiting environments; a mechanism that requires the physical interaction between Hap43 and the Hap5 subunit of the CCAAT-binding complex [34]. Evolutionarily, the Hap43 orthologs from other yeast and fungi, including HapX (A. nidulans, Aspergillus fumigatus and Cryptococcus neoformans) and Php4 (Schizosaccharomyces pombe), have been implicated in the regulation of genes involved in iron transport/utilization [66–69]. Hap43 and HapX share three cysteine-rich protein domains, which have been proposed to mediate iron sensing through the formation of an iron-binding domain [66,70]. Whether this domain coordinates iron or iron-sulfur clusters remains to be established.
One of these genes regulated by the CCAAT-binding factor in C. albicans is CYC1, encoding cytochrome c, which is repressed during iron limitation in a Hap5- and Hap43-dependent manner and activated by the CCAAT-binding factor under iron-replete conditions [51]. This is relevant because previous studies have demonstrated that CYC1 is regulated by the CCAAT-binding factor in a carbon source-dependent manner [35]. Thus, the CCAAT-binding factor serves as a modulator of gene expression in response to both carbon source and iron. Our data indicates that the CCAAT-binding factor regulates the OSR genes via its regulation of iron uptake and utilization; whereas, Hog1 and Cap1 have previously been shown to be direct transcriptional regulators of the OSR genes [16,71–73]. Importantly, fluctuations in the level of intracellular iron have a direct impact on the redox potential within the cell. It makes sense that a multi-subunit transcription factor could sense both redox status and the availability of iron within the cell, and coordinate gene expression accordingly. Therefore, it will be interesting to investigate whether such a coordinated response is regulated in C. albicans by the potential redox sensing via Hap31 and/or Hap32 along with the putative iron-sensing through the cysteine-rich domains of Hap43.
Why is it advantageous to co-regulate genes involved in respiration, iron uptake/utilization, and the OSR? First, the human host is essentially a low iron environment due to sequestration of iron with proteins such as transferrin, ferritin, lactoferrin as well as other iron-binding proteins [28–30]. While C. albicans, like many other pathogens, has evolved multiple sophisticated mechanisms for scavenging iron from the human host [31,32], the organism must adjust its metabolic needs to meet this challenge. Moreover, dependent on the specific micro-environment in the host, the availability of iron can vary dramatically [25,28]. To meet this metabolic challenge, C. albicans uses the CCAAT-binding factor to control the expression of genes involved in iron acquisition/utilization, respiration, and the OSR. Although our in vitro studies do not aim to mimic the host-pathogen interaction, one can envision the iron-limited environment of the human host activating all the fungal iron-scavenging mechanisms, while repressing OSR genes and genes that express iron-requiring proteins. However, once adequate iron levels have been established, C. albicans can quickly express OSR genes and modulate a strong response to the host defense mechanisms, such as the oxidative burst following phagocytosis by neutrophils.
Supporting Information
(DOCX)
(DOCX)
Acknowledgments
The authors wish to thank LaShall Bates for her expertise and guidance during the early stages of this work.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the NIH, R01 A1051470 (http://www.niaid.nih.gov) and the Arkansas Biosciences Institute (http://arbiosciences.org). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Odds FC, editor. Candida and candidosis, 2nd Ed. London, United Kingdom: Bailliere Tindall; 1988. [Google Scholar]
- 2.Calderone RA, editor. Candida and candidiasis. Washington, D.C.: ASM Press; 2002. [Google Scholar]
- 3.Collette JR, Zhou H, Lorenz MC. Candida albicans suppresses nitric oxide generation from macrophages via a secreted molecule. PLoS One. 2014;9: e96203 10.1371/journal.pone.0096203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cuellar-Cruz M, Lopez-Romero E, Ruiz-Baca E, Zazueta-Sandoval R. Differential response of Candida albicans and Candida glabrata to oxidative and nitrosative stresses. Curr Microbiol. 2014;69: 733–739. 10.1007/s00284-014-0651-3 [DOI] [PubMed] [Google Scholar]
- 5.Gazendam RP, van Hamme JL, Tool AT, van Houdt M, Verkuijlen PJ, Herbst M, et al. Two independent killing mechanisms of Candida albicans by human neutrophils: evidence from innate immunity defects. Blood. 2014;124: 590–597. 10.1182/blood-2014-01-551473 [DOI] [PubMed] [Google Scholar]
- 6.Kaloriti D, Jacobsen M, Yin Z, Patterson M, Tillmann A, Smith DA, et al. Mechanisms underlying the exquisite sensitivity of Candida albicans to combinatorial cationic and oxidative stress that enhances the potent fungicidal activity of phagocytes. MBio. 2014;5: e01334–14. 10.1128/mBio.01334-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dantas Ada S, Day A, Ikeh M, Kos I, Achan B, Quinn J. Oxidative stress responses in the human fungal pathogen, Candida albicans. Biomolecules. 2015;5: 142–165. 10.3390/biom5010142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nakamura K, Kanno T, Mokudai T, Iwasawa A, Niwano Y, Kohno M. Microbial resistance in relation to catalase activity to oxidative stress induced by photolysis of hydrogen peroxide. Microbiol Immunol. 2012;56: 48–55. 10.1111/j.1348-0421.2011.00400.x [DOI] [PubMed] [Google Scholar]
- 9.Gleason JE, Galaleldeen A, Peterson RL, Taylor AB, Holloway SP, Waninger-Saroni J, et al. Candida albicans SOD5 represents the prototype of an unprecedented class of Cu-only superoxide dismutases required for pathogen defense. Proc Natl Acad Sci U S A. 2014;111: 5866–5871. 10.1073/pnas.1400137111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gleason JE, Li CX, Odeh HM, Culotta VC. Species-specific activation of Cu/Zn SOD by its CCS copper chaperone in the pathogenic yeast Candida albicans. J Biol Inorg Chem. 2014;19: 595–603. 10.1007/s00775-013-1045-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herrero E, Ros J, Belli G, Cabiscol E. Redox control and oxidative stress in yeast cells. Biochim Biophys Acta. 2008;1780: 1217–1235. 10.1016/j.bbagen.2007.12.004 [DOI] [PubMed] [Google Scholar]
- 12.Ratti BA, Godoy JS, de Souza Bonfim Mendonca P, Bidoia DL, Nakamura TU, Nakamura CV, et al. Microbicidal activity of neutrophils is inhibited by isolates from recurrent vaginal candidiasis (RVVC) caused by Candida albicans through fungal thioredoxin reductase. Cell Immunol. 2015;293: 22–29. 10.1016/j.cellimm.2014.11.004 [DOI] [PubMed] [Google Scholar]
- 13.Toone WM, Jones N. Stress-activated signalling pathways in yeast. Genes Cells. 1998;3: 485–498. [DOI] [PubMed] [Google Scholar]
- 14.Brown AJ, Budge S, Kaloriti D, Tillmann A, Jacobsen MD, Yin Z, et al. Stress adaptation in a pathogenic fungus. J Exp Biol. 2014;217: 144–155. 10.1242/jeb.088930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Enjalbert B, Nantel A, Whiteway M. Stress-induced gene expression in Candida albicans: absence of a general stress response. Mol Biol Cell. 2003;14: 1460–1467. 10.1091/mbc.E02-08-0546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alonso-Monge R, Navarro-Garcia F, Roman E, Negredo AI, Eisman B, Nombela C, et al. The Hog1 mitogen-activated protein kinase is essential in the oxidative stress response and chlamydospore formation in Candida albicans. Eukaryot Cell. 2003;2: 351–361. 10.1128/EC.2.2.351-361.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alonso-Monge R, Carvaihlo S, Nombela C, Rial E, Pla J. The Hog1 MAP kinase controls respiratory metabolism in the fungal pathogen Candida albicans. Microbiology. 2009;155: 413–423. 10.1099/mic.0.023309-0 [DOI] [PubMed] [Google Scholar]
- 18.Adrover MA, Zi Z, Duch A, Schaber J, Gonzalez-Novo A, Jimenez J, et al. Time-dependent quantitative multicomponent control of the G(1)-S network by the stress-activated protein kinase Hog1 upon osmostress. Sci Signal. 2011;4: ra63 10.1126/scisignal.2002204 [DOI] [PubMed] [Google Scholar]
- 19.Babazadeh R, Furukawa T, Hohmann S, Furukawa K. Rewiring yeast osmostress signalling through the MAPK network reveals essential and non-essential roles of Hog1 in osmoadaptation. Sci Rep. 2014;4: 4697 10.1038/srep04697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wysong DR, Christin L, Sugar AM, Robbins PW, Diamond RD. Cloning and sequencing of a Candida albicans catalase gene and effects of disruption of this gene. Infect Immun. 1998;66: 1953–1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hwang CS, Rhie GE, Oh JH, Huh WK, Yim HS, Kang SO. Copper- and zinc-containing superoxide dismutase (Cu/ZnSOD) is required for the protection of Candida albicans against oxidative stresses and the expression of its full virulence. Microbiology. 2002;148: 3705–3713. 10.1099/00221287-148-11-3705 [DOI] [PubMed] [Google Scholar]
- 22.Hwang CS, Baek YU, Yim HS, Kang SO. Protective roles of mitochondrial manganese-containing superoxide dismutase against various stresses in Candida albicans. Yeast. 2003;20: 929–941. 10.1002/yea.1004 [DOI] [PubMed] [Google Scholar]
- 23.Chaves GM, Bates S, Maccallum DM, Odds FC. Candida albicans GRX2, encoding a putative glutaredoxin, is required for virulence in a murine model. Genet Mol Res. 2007;6: 1051–1063. [PubMed] [Google Scholar]
- 24.da Silva Dantas A, Patterson MJ, Smith DA, Maccallum DM, Erwig LP, Morgan BA, et al. Thioredoxin regulates multiple hydrogen peroxide-induced signaling pathways in Candida albicans. Mol Cell Biol. 2010;30: 4550–4563. 10.1128/MCB.00313-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen C, Pande K, French SD, Tuch BB, Noble SM. An iron homeostasis regulatory circuit with reciprocal roles in Candida albicans commensalism and pathogenesis. Cell Host Microbe. 2011;10: 118–135. 10.1016/j.chom.2011.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hsu PC, Yang CY, Lan CY. Candida albicans Hap43 is a repressor induced under low-iron conditions and is essential for iron-responsive transcriptional regulation and virulence. Eukaryot Cell. 2011;10: 207–225. 10.1128/EC.00158-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pierre JL, Fontecave M, Crichton RR. Chemistry for an essential biological process: the reduction of ferric iron. Biometals. 2002;15: 341–346. [DOI] [PubMed] [Google Scholar]
- 28.Weinberg ED. Iron availability and infection. Biochim Biophys Acta. 2009;1790: 600–605. 10.1016/j.bbagen.2008.07.002 [DOI] [PubMed] [Google Scholar]
- 29.Ganz T. Iron in innate immunity: starve the invaders. Curr Opin Immunol. 2009;21: 63–67. 10.1016/j.coi.2009.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sutak R, Lesuisse E, Tachezy J, Richardson DR. Crusade for iron: iron uptake in unicellular eukaryotes and its significance for virulence. Trends Microbiol. 2008;16: 261–268. 10.1016/j.tim.2008.03.005 [DOI] [PubMed] [Google Scholar]
- 31.Almeida RS, Wilson D, Hube B. Candida albicans iron acquisition within the host. FEMS Yeast Res. 2009;9: 1000–1012. 10.1111/j.1567-1364.2009.00570.x [DOI] [PubMed] [Google Scholar]
- 32.Kornitzer D. Fungal mechanisms for host iron acquisition. Curr Opin Microbiol. 2009;12: 377–383. 10.1016/j.mib.2009.05.005 [DOI] [PubMed] [Google Scholar]
- 33.Kuznets G, Vigonsky E, Weissman Z, Lalli D, Gildor T, Kauffman SJ, et al. A relay network of extracellular heme-binding proteins drives C. albicans iron acquisition from hemoglobin. PLoS Pathog. 2014;10: e1004407 10.1371/journal.ppat.1004407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Singh RP, Prasad HK, Sinha I, Agarwal N, Natarajan K. Cap2-HAP complex is a critical transcriptional regulator that has dual but contrasting roles in regulation of iron homeostasis in Candida albicans. J Biol Chem. 2011;286: 25154–25170. 10.1074/jbc.M111.233569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnson DC, Cano KE, Kroger EC, McNabb DS. Novel regulatory function for the CCAAT-binding factor in Candida albicans. Eukaryot Cell. 2005;4: 1662–1676. 10.1128/EC.4.10.1662-1676.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baek YU, Li M, Davis DA. Candida albicans ferric reductases are differentially regulated in response to distinct forms of iron limitation by the Rim101 and CBF transcription factors. Eukaryot Cell. 2008;7: 1168–1179. 10.1128/EC.00108-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hsu PC, Chao CC, Yang CY, Ye YL, Liu FC, Chuang YJ, et al. Diverse Hap43-independent functions of the Candida albicans CCAAT-binding complex. Eukaryot Cell. 2013;12: 804–815. 10.1128/EC.00014-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mantovani R. The molecular biology of the CCAAT-binding factor NF-Y. Gene. 1999;239: 15–27. [DOI] [PubMed] [Google Scholar]
- 39.Forsburg SL, Guarente L. Identification and characterization of HAP4: a third component of the CCAAT-bound HAP2/HAP3 heteromer. Genes Dev. 1989;3: 1166–1178. [DOI] [PubMed] [Google Scholar]
- 40.McNabb DS, Xing Y, Guarente L. Cloning of yeast HAP5: a novel subunit of a heterotrimeric complex required for CCAAT binding. Genes Dev. 1995;9: 47–58. [DOI] [PubMed] [Google Scholar]
- 41.McNabb DS, Pinto I. Assembly of the Hap2p/Hap3p/Hap4p/Hap5p-DNA complex in Saccharomyces cerevisiae. Eukaryot Cell. 2005;4: 1829–1839. 10.1128/EC.4.11.1829-1839.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Forsburg SL, Guarente L. Communication between mitochondria and the nucleus in regulation of cytochrome genes in the yeast Saccharomyces cerevisiae. Annu Rev Cell Biol. 1989;5: 153–180. 10.1146/annurev.cb.05.110189.001101 [DOI] [PubMed] [Google Scholar]
- 43.Zitomer RS, Lowry CV. Regulation of gene expression by oxygen in Saccharomyces cerevisiae. Microbiol Rev. 1992;56: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dang VD, Bohn C, Bolotin-Fukuhara M, Daignan-Fornier B. The CCAAT box-binding factor stimulates ammonium assimilation in Saccharomyces cerevisiae, defining a new cross-pathway regulation between nitrogen and carbon metabolisms. J Bacteriol. 1996;178: 1842–1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.DeRisi JL, Iyer VR, Brown PO. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278: 680–686. [DOI] [PubMed] [Google Scholar]
- 46.Guthrie C, Fink GR. Guide to yeast genetics and molecular biology. San Diego, Calif.: Academic Press; 1991. [Google Scholar]
- 47.Wilson RB, Davis D, Mitchell AP. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J Bacteriol. 1999;181: 1868–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fonzi WA, Irwin MY. Isogenic strain construction and gene mapping in Candida albicans. Genetics. 1993;134: 717–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Garcia MG, O'Connor JE, Garcia LL, Martinez SI, Herrero E, del Castillo Agudo L. Isolation of a Candida albicans gene, tightly linked to URA3, coding for a putative transcription factor that suppresses a Saccharomyces cerevisiae aft1 mutation. Yeast. 2001;18: 301–311. [DOI] [PubMed] [Google Scholar]
- 50.Chibana H, Uno J, Cho T, Mikami Y. Mutation in IRO1 tightly linked with URA3 gene reduces virulence of Candida albicans. Microbiol Immunol. 2005;49: 937–939. [DOI] [PubMed] [Google Scholar]
- 51.Bates LL. The role of multiple CCAAT-binding factors in Candida albicans gene expression. Ph.D., University of Arkansas. 2009. Available: http://search.proquest.com/docview/304845825?accountid=8361.
- 52.Hoffman CS, Winston F. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene. 1987;57: 267–272. [DOI] [PubMed] [Google Scholar]
- 53.Asubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, et al. Current protocols in molecular biology. New York, N.Y.: Greene Publishing Associates and Wiley-Interscience; 1994. [Google Scholar]
- 54.Ueda M, Mozaffar S, Tanaka A. Catalase from Candida boidinii 2201. Methods Enzymol. 1990;188: 463–467. [DOI] [PubMed] [Google Scholar]
- 55.Homann OR, Dea J, Noble SM, Johnson AD. A phenotypic profile of the Candida albicans regulatory network. PLoS Genet. 2009;5: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chaves GM, da Silva WP. Superoxide dismutases and glutaredoxins have a distinct role in the response of Candida albicans to oxicative stress generated by the chemical compounds menadione and diamide. Mem Inst Oswaldo Cruz. 2012;107: 998–1005. [DOI] [PubMed] [Google Scholar]
- 57.Lamarre C, LeMay JD, Deslauriers N, Bourbonnais Y. Candida albicans expresses an unusual cytoplasmic manganese-containing superoxide dismutase (SOD3 gene product) upon the entry and during the stationary phase. J Biol Chem. 2001;276: 43784–43791. 10.1074/jbc.M108095200 [DOI] [PubMed] [Google Scholar]
- 58.Frohner IE, Bourgeois C, Yatsyk K, Majer O, Kuchler K. Candida albicans cell surface superoxide dismutases degrade host-derived reactive oxygen species to escape innate immune surveillance. Mol Microbiol. 2009;71: 240–252. 10.1111/j.1365-2958.2008.06528.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.De Freitas JM, Liba A, Meneghini R, Valentine JS, Gralla EB. Yeast lacking Cu-Zn superoxide dismutase show altered iron homeostasis. Role of oxidative stress in iron metabolism. J Biol Chem. 2000;275: 11645–11649. [DOI] [PubMed] [Google Scholar]
- 60.Srinivasan C, Liba A, Imlay JA, Valentine JS, Gralla EB. Yeast lacking superoxide dismutase(s) show elevated levels of "free iron" as measured by whole cell electron paramagnetic resonance. J Biol Chem. 2000;275: 29187–29192. 10.1074/jbc.M004239200 [DOI] [PubMed] [Google Scholar]
- 61.Rodriguez-Manzaneque MT, Tamarit J, Belli G, Ros J, Herrero E. Grx5 is a mitochondrial glutaredoxin required for the activity of iron/sulfur enzymes. Mol Biol Cell. 2002;13: 1109–1121. 10.1091/mbc.01-10-0517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Toledano MB, Kumar C, Le Moan N, Spector D, Tacnet F. The system biology of thiol redox system in Escherichia coli and yeast: differential functions in oxidative stress, iron metabolism and DNA synthesis. FEBS Lett. 2007;581: 3598–3607. 10.1016/j.febslet.2007.07.002 [DOI] [PubMed] [Google Scholar]
- 63.Toledano MB, Delaunay-Moisan A, Outten CE, Igbaria A. Functions and cellular compartmentation of the thioredoxin and glutathione pathways in yeast. Antioxid Redox Signal. 2013;18: 1699–1711. 10.1089/ars.2012.5033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thon M, Al Abdallah Q, Hortschansky P, Scharf DH, Eisendle M, Haas H, et al. The CCAAT-binding complex coordinates the oxidative stress response in eukaryotes. Nucleic Acids Res. 2010;38: 1098–1113. 10.1093/nar/gkp1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cuellar-Cruz M, Briones-Martin-del-Campo M, Canas-Villamar I, Montalvo-Arredondo J, Riego-Ruiz L, Castano I, et al. High resistance to oxidative stress in the fungal pathogen Candida glabrata is mediated by a single catalase, Cta1p, and is controlled by the transcription factors Yap1p, Skn7p, Msn2p, and Msn4p. Eukaryot Cell. 2008;7: 814–825. 10.1128/EC.00011-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hortschansky P, Eisendle M, Al-Abdallah Q, Schmidt AD, Bergmann S, Thon M, et al. Interaction of HapX with the CCAAT-binding complex—a novel mechanism of gene regulation by iron. EMBO J. 2007;26: 3157–3168. 10.1038/sj.emboj.7601752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schrettl M, Beckmann N, Varga J, Heinekamp T, Jacobsen ID, Jochl C, et al. HapX-mediated adaption to iron starvation is crucial for virulence of Aspergillus fumigatus. PLoS Pathog. 2010;6: e1001124 10.1371/journal.ppat.1001124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jung WH, Saikia S, Hu G, Wang J, Fung CK, D'Souza C, et al. HapX positively and negatively regulates the transcriptional response to iron deprivation in Cryptococcus neoformans. PLoS Pathog. 2010;6: e1001209 10.1371/journal.ppat.1001209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Labbe S, Khan MG, Jacques JF. Iron uptake and regulation in Schizosaccharomyces pombe. Curr Opin Microbiol. 2013;16: 669–676. 10.1016/j.mib.2013.07.007 [DOI] [PubMed] [Google Scholar]
- 70.Gsaller F, Hortschansky P, Beattie SR, Klammer V, Tuppatsch K, Lechner BE, et al. The Janus transcription factor HapX controls fungal adaptation to both iron starvation and iron excess. EMBO J. 2014;33: 2261–2276. 10.15252/embj.201489468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Alarco AM, Raymond M. The bZip transcription factor Cap1p is involved in multidrug resistance and oxidative stress response in Candida albicans. J Bacteriol. 1999;181: 700–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Enjalbert B, Smith DA, Cornell MJ, Alam I, Nicholls S, Brown AJ, et al. Role of the Hog1 stress-activated protein kinase in the global transcriptional response to stress in the fungal pathogen Candida albicans. Mol Biol Cell. 2006;17: 1018–1032. 10.1091/mbc.E05-06-0501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang Y, Cao YY, Jia XM, Cao YB, Gao PH, Fu XP, et al. Cap1p is involved in multiple pathways of oxidative stress response in Candida albicans. Free Radic Biol Med. 2006;40: 1201–1209. 10.1016/j.freeradbiomed.2005.11.019 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.


