Abstract
Low-molecular-weight metabolites produced by the intestinal microbiome play a direct role in health and disease. However, little is known about the ability of the colon to absorb these metabolites. It is also unclear whether these metabolites are bioavailable. Here, metabolomics techniques (capillary electrophoresis with time-of-flight mass spectrometry, CE-TOFMS), germ-free (GF) mice, and colonized (Ex-GF) mice were used to identify the colonic luminal metabolites transported to colonic tissue and/or blood. We focused on the differences in each metabolite between GF and Ex-GF mice to determine the identities of metabolites that are transported to the colon and/or blood. CE-TOFMS identified 170, 246, 166, and 193 metabolites in the colonic feces, colonic tissue, portal plasma, and cardiac plasma, respectively. We classified the metabolites according to the following influencing factors: (i) the membrane transport system of the colonocytes, (ii) metabolism during transcellular transport, and (iii) hepatic metabolism based on the similarity in the ratio of each metabolite between GF and Ex-GF mice and found 62 and 22 metabolites that appeared to be absorbed from the colonic lumen to colonocytes and blood, respectively. For example, 11 basic amino acids were transported to the systemic circulation from the colonic lumen. Furthermore, many low-molecular-weight metabolites influenced by the intestinal microbiome are bioavailable. The present study is the first to report the transportation of metabolites from the colonic lumen to colonocytes and somatic blood in vivo, and the present findings are critical for clarifying host-intestinal bacterial interactions.
Introduction
The intestinal microbiome plays an important role in health and disease [1] because it influences pathological and normal homeostatic functions involved in obesity [2, 3], immune disease [4, 5], colon cancer [6], brain function [7], behavior [8], and life-span [9, 10]. We propose that these interactions depend upon direct stimulation of bacterial cell components and the effects of bacterial metabolites. Bacterial cell components influence the physiological and pathological functions of the immune system through the direct stimulation of Toll-like receptors expressed by colonocytes and dendritic cells that reside in the colonic mucosa [11, 12]. In contrast, the relationships between bacterial metabolites and health/disease are not well known, except for specific metabolites such as short-chain fatty acids (SCFA) [13]. It is also unclear what metabolites are transported to the body from the colonic lumen. Although many blood metabolites have been reported to be different between GF and Ex-GF mice [14], no comprehensive data are available to indicate that blood and colonocytes contain bacterial metabolites derived from the colonic lumen because appropriate analytical methods are not available. There is a general concept that the role of the colon is mainly to absorb water, electrolytes, and some vitamins; thus, in the field of gastrointestinal physiology and nutrition, there have been few reports indicating that low-molecular-weight chemicals are absorbed from the large intestine, and physiologists and nutritionists have typically not considered metabolites produced by intestinal bacteria as research targets. In this study, we tried to identify the bioavailable low-molecular-weight metabolites transported to middle colonic tissue and/or blood from the colonic lumen.
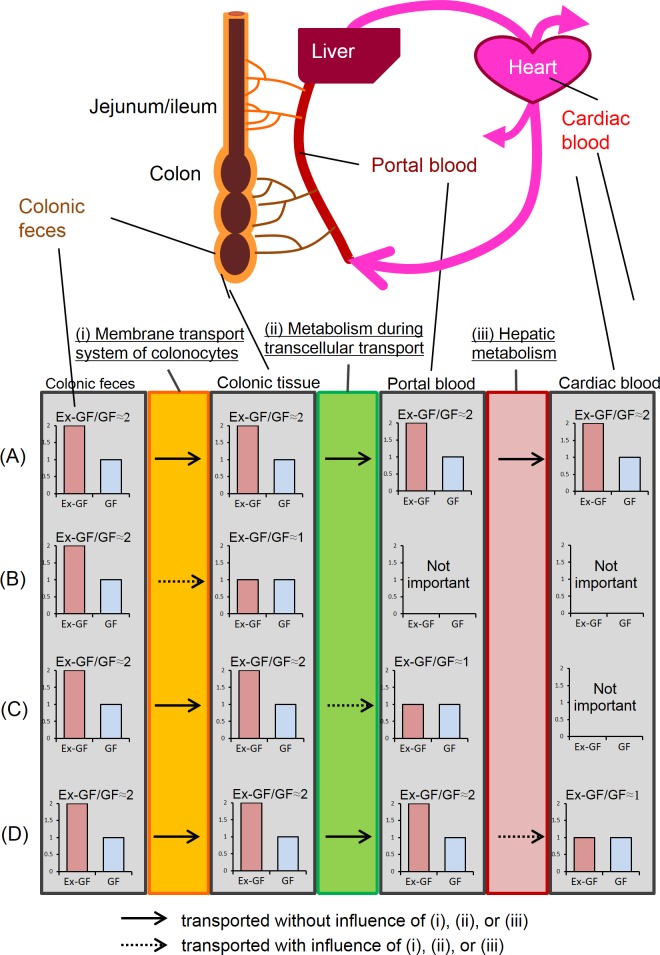
It is theoretically possible for transportation of specific substances to be detected using stable or radioactive isotopes [15, 16]; however, it is impossible to comprehensively detect a large number of metabolites using these methods. Using capillary electrophoresis combined with time-of-flight mass spectrometry (CE-TOFMS), which analyzes and differentially displays metabolic profiles [17], we previously demonstrated that 125 low-molecular-weight metabolites that showed significant differences in the colonic feces of germ-free (GF) and ex-germ-free (Ex-GF) mice (i.e., mice that were previously GF-free but now harbored specific pathogen-free mouse intestinal microbiota) were influenced by the intestinal microbiome [18]. Here, we focused on these differences between GF and Ex-GF mice to determine the identities of low-molecular-weight metabolites transported to body from colonic lumen. In brief, the blood Ex-GF/GF ratio of metabolites that are transported to the blood from the colonic lumen is similar to the Ex-GF/GF ratio in colonic feces (Fig 1). We determined the Ex-GF/GF ratio for each metabolite derived from the colonic feces, colonic tissue (colonocytes), portal plasma, and cardiac plasma of GF and Ex-GF mice and searched for metabolites with similar Ex-GF/GF ratios in the colonic feces and colonic tissue, portal plasma, and cardiac plasma.
Fig 1. Estimation of metabolites to transport to body from colonic lumen using the difference of concentration between GF and Ex-GF mice (Ex-GF/GF ratio).
(A) Metabolites with similar Ex-GF/GF ratios in colonic feces, colonic tissue, portal plasma, and cardiac plasma were considered to be transported from the colonic lumen to cardiac blood. (B) Metabolites with different Ex-GF/GF ratios between colonic feces and colonic tissue are controlled by (i) membrane transport system (C) Metabolites with different Ex-GF/GF ratios between colonic tissue and portal plasma are controlled by (ii) metabolism during transcellular transport. (D) Metabolites with different Ex-GF/GF ratios between portal plasma and portal.
Materials and Methods
Mice and diets
Germ-free (GF) BALB/c mice were purchased from Japan Clea Inc. (Tokyo, Japan) and bred at the Department of Infectious Diseases, Tokai University School of Medicine, Kanagawa, Japan. Six male litters were divided into GF and Ex-GF (see below) groups to remove effects caused by inherited individual distinctions. Mice were housed in Trexler-type flexible film plastic isolators with sterilized tips (CLEA Japan, Inc., Tokyo, Japan) as bedding. The mice were provided with water and commercial CL-2 pellets (CLEA Japan, Inc.) that were sterilized using an autoclave (121°C, 30 min). Bacteriological contamination of feces was analyzed throughout the cultivation procedure using Gifu Anaerobe Medium (GAM) agar (Nissui, Tokyo, Japan). Using a gastric gavage tube, we inoculated the stomachs of Ex-GF mice (4 weeks of age) with 0.5 mL of a 1:10 dilution of feces obtained from specific-pathogen-free BALB/c mice and housed them mice of various for 3 weeks until collecting specimens at 7 weeks of age. This study was carried out in strict accordance with the recommendations in the guidelines of the Animal Care Committee of Tokai University. The protocol was approved by the Kyodo Milk Animal Use Committee (Permit Number: 2009–03).
Specimen preparation
Cardiac and portal blood samples were collected from mice anesthetized with inhalation of isoflurane using small animal anesthetizer MK-A110 (Muromachi Kikai Co. Ltd., Tokyo, Japan) (7 weeks of age) into tubes containing sodium ethylenediamine tetraacetate (final concentration, 0.13%). Portal blood (50–100 μl) and cardiac blood (50–100 μl) were collected using a needle. The blood samples were centrifuged for 20 min at 2,300 × g at 4°C. The samples were stored at −80°C. The mice were sacrificed by cervical dislocation, and the middle colonic tissues and content (colonic feces) were collected. Middle colonic tissues were removed from a region containing feces and were frozen immediately in liquid nitrogen and stored at –80°C. Colonic feces and blood were prepared according to a published procedure [19]. Colons were suspended in methanol (500 μl) with 50 μM internal standard and vortexed vigorously five times for 60 s using a MicroSmash MS-100R (Tomy Digital Biology Co., Ltd., Tokyo, Japan) at 4,000 rpm at 4°C. The resulting colon sample served as the crude metabolome.
Capillary electrophoresis–time-of-flight mass spectrometry (CE-TOFMS)
Metabolomic measurements were performed using an Agilent Capillary Electrophoresis System, and data were processed by Huma Metabolome Technologies Inc. (Tsuruoka, Japan) according to a published method [20]. All pretreated metablome samples were centrifugally filtered through a 5-kDa cutoff filter Ultrafree-MC (Millipore). The values of the peaks were normalized to those of the internal standards methionine sulfone (cationic) and D-camphor-10-sulfonic acid (anionic), respectively.
Ratio of values in GF mice to those in Ex-GF mice (Ex-GF/GF ratio)
In this study, we calculated the ratio of values in GF mice to those in Ex-GF mice (Ex-GF/GF ratio) and classified the metabolites. We defined the threshold Ex-GF/GF ratio to determine whether metabolites were present at higher or lower concentrations in Ex-GF mice than in GF mice. The metabolites were measured in the metabolome of the cardiac plasma, which was influenced by all gateways: (i) the membrane transport system of colonocytes, (ii) metabolism during transcellular transport, and (iii) hepatic metabolism (Fig 1). The highest Ex-GF/GF ratio for metabolites present at higher concentrations in Ex-GF than in GF mice (where the difference was still significantly different) was 1.259 (S1 Table). The lowest Ex-GF/GF ratio for metabolites present at lower concentrations in Ex-GF than in GF mice was 0.866 (S1 Table). Therefore, in this study, the Ex-GF/GF ratio of metabolites present at higher concentrations in Ex-GF mice than in GF mice was defined as ≥1.25, and the Ex-GF/GF ratio of metabolites present at lower concentrations in Ex-GF mice than in GF mice was defined as < 0.8.
Statistical analysis
IBM SPSS Statistics (Japan IBM, Tokyo) software was used to conduct statistical analyses. Metabolome data were analyzed using PCA and clustering analysis. Differences in relative quantities between GF and Ex-GF mice were evaluated for each metabolite using Welch’s t test. The levels of individual metabolites in GF and Ex-GF mice were compared using Fisher’s exact test.
Results
Analysis of the metabolomes of the colonic feces, colon, portal plasma, and cardiac plasma of GF and Ex-GF mice
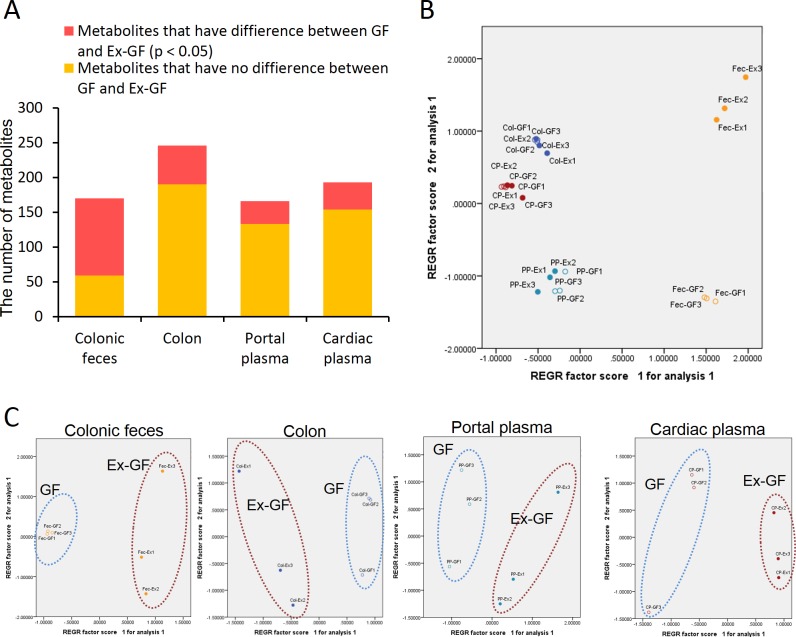
CE-TOFMS identified 170, 246, 166, and 193 metabolites from the colonic feces, colonic tissue, portal plasma, and cardiac plasma, respectively. Significant differences (p < 0.05) were observed between GF and Ex-GF mice in each specimen; the number of metabolites with differences in the Ex-GF/GF ratio was as follows: colonic feces, 111 (65.7%); colonic tissue, 56 (22.8%); portal plasma, 33 (19.9%); and cardiac plasma, 39 (20.2%) (Fig 2A). Principal component analysis (PCA) showed patterns for metabolites in all samples (Fig 2B). The metabolomic profiles of colonic feces showed the greatest difference between the two groups of mice. However, the metabolome difference for colonic tissue, portal and cardiac plasma was not significant. Based on the PCA results of each specimen, it was possibles to assign all metabolomic profiles into two clusters corresponding to GF mice and Ex-GF mice (Fig 2C).
Fig 2. Differences in the metabolomes of GF and Ex-GF mice.
(A) Number of metabolites that were significantly (p < 0.05) or not significantly different between GF and Ex-GF mice.(B) PCA of metabolome profiles. Fec-Ex, colonic feces of Ex-GF mice; Fec-GF, colonic feces of GF mice; Col-Ex, colon of Ex-GF mice; Col-GF, colon of GF mice; PP-Ex, portal plasma of Ex-GF mice; PP-GF, portal plasma of GF mice; CP-Ex, cardiac plasma of Ex-GF mice; CP-GF, cardiac plasma of GF mice.(C) PCA of metabolome profiles in each specimen.
Classification of metabolites according to the Ex-GF/GF ratio
In the present study, 146 colonic fecal metabolites detected in all mice were selected for analysis (122 metabolites that were detected in colonic feces and colonic tissue are shown in Table 1, and 24 metabolites that were detected in colonic feces but not in colonic tissue are shown in Table 2). The table shows that the levels of most of the selected metabolites were influenced by the intestinal microbiome in the colonic lumen because they showed significant differences between GF and Ex-GF mice. Transport of low-molecular-weight metabolites to the cardiac blood from the colonic lumen appeared to be influenced by 3 gateways: (i) the membrane transport system of colonocytes, (ii) metabolism during transcellular transport, and (iii) hepatic metabolism (Fig 1). Therefore, we estimated the effect of these gateways for each metabolite using the Ex-GF/GF ratio (Table 1). In total, 62 metabolites (No. 1 to No. 62) had Ex-GF/GF ratios that were similar between the colonic feces and colonic tissue, indicating that their levels are not influenced by the membrane transport system of colonocytes. Twenty-two metabolites (No. 1 to No. 22) had similar Ex-GF/GF ratios for colonic feces, colonic tissue, portal plasma, and cardiac plasma, and were thus probably transported from the colonic lumen to the systemic blood supply independently of factors (i), (ii), and (iii) defined above. The other 40 metabolites (No. 23 to No. 62), which had Ex-GF/GF ratios that were similar between the colonic feces and colonic tissue but not portal blood, were transported to colonic tissue, although their levels were controlled by (ii) metabolism during transcellular transport and/or (iii) hepatic metabolism. In contrast, 60 metabolites (No. 63 to No. 122) with different Ex-GF/GF ratios between the colonic feces and colonic tissue were also detected (Table 1). Because these metabolites are influenced by (i) the membrane transport system of colonocytes, their concentrations in the colonic tissue were independent of their concentrations in the colonic lumen. Twenty-four metabolites were not detected in the colonic tissue (Table 2). Therefore, these metabolites are not transported to the body from the colonic lumen. Eight of 11 peptides detected were in this group.
Table 1. The ExGF/GF ratio and the influence of 3 gateways on each metabolite detected in the colonic feces, colon, portal plasma, and cardiac plasma.
| Metbolites | Category | ExGF/GF ratio | Membrane transport system of colonocytes | ExGF/GF ratio | Metabolism during transcellular transport | ExGF/GF ratio | Hepatic metabolism | ExGF/GF ratio | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| KEGG ID | Colonic feces | Colon | Portal plasma | Cardiac plasma | |||||||
| (HMDB ID) | |||||||||||
| 1 | Cytosine | Base/nucleotide | C00380 | → | 1.43 | → | unclear | NA | |||
| 2 | Cholic acid | C00695 | → | → | → | 1.71 | |||||
| 3 | Pipecolic acid | C00408 | → | 2.45 | → | 1.61 | → | 1.66 | |||
| 4 | Stachydrine | C10172 | 1.14 | → | 0.94 | → | 0.84 | → | 0.90 | ||
| 5 | Phe* | Amino acid | C00079 | 0.76 | → | 0.64 | → | 0.76 | → | 0.83 | |
| 6 | Val | Amino acid | C00183 | 0.66 | → | 0.66 | → | 0.61 | → | 0.64 | |
| 7 | Lys | Amino acid | C00047 | 0.66 | → | 0.41 | → | 0.70 | → | 0.74 | |
| 8 | Ile | Amino acid | C00407 | 0.64 | → | 0.56 | → | 0.57 | → | 0.60 | |
| 9 | 5-Hydroxylysine | C16741 | 0.60 | → | 0.50 | → | 0.59 | → | 0.72 | ||
| 10 | Leu | Amino acid | C00123 | 0.58 | → | 0.60 | → | 0.63 | → | 0.65 | Ex-GF/GF |
| 11 | N8-Acetylspermidine* | C01029 | 0.51 | → | 0.37 | → | 0.67 | →/→ | 0.82 | 3 | |
| 12 | Ser | Amino acid | C00065 | 0.50 | → | 0.36 | → | 0.56 | → | 0.59 | 2.5 |
| 13 | N-Acetylhistidine | C02997 | 0.44 | → | 0.78 | → | 0.87 | → | 0.69 | 2 | |
| 14 | Gly | Amino acid | C00037 | 0.44 | → | 0.75 | → | 0.56 | → | 0.55 | 1.5 |
| 15 | His | Amino acid | C00135 | 0.39 | → | 0.43 | → | 0.76 | → | 0.87 | 1 |
| 16 | Thr | Amino acid | C00188 | 0.18 | → | 0.30 | → | 0.50 | → | 0.54 | 0.67 |
| 17 | Arg | Amino acid | C00062 | 0.15 | → | 0.28 | → | 0.37 | →/→ | 0.89 | 0.5 |
| 18 | Pro | Amino acid | C00148 | 0.14 | → | 0.38 | → | 0.57 | → | 0.63 | 0.4 |
| 19 | Hydroxyproline | Amino acid | C01015 | 0.05 | → | 0.22 | → | 0.47 | → | 0.54 | 0 |
| 20 | Creatinine | C00791 | 0.01 | → | 0.34 | → | 0.66 | → | 0.77 | ||
| 21 | N-Acetyl-β-alanine | Alkylamino acid | C01073 | → | 0.40 | → | 0.73 | →/→ | 1.52 | ||
| 22 | 1-Methylnicotinamide | Alkylamino acid | C02918 | → | 0.51 | → | 0.50 | → | 0.45 | ||
| 23 | Glutaric acid | C00489 | → | unclear | NA | X | ND | ||||
| 24 | Valeric acid | Fatty acid | C00803 | → | 39.53 | →/→ | 1.22 | X | ND | ||
| 25 | 2-Aminobutyric acid | Amino acid | C02261 | → | 1.89 | →/→ | 0.73 | → | 0.78 | ||
| 26 | Sarcosine | Choline | C00213 | → | 2.43 | unclear | NA | unclear | 0.42 | ||
| 27 | 1-Methyl-4-imidazoleacetic acid | Alkylamino acid | C05828 | 19.20 | → | 7.02 | →/→ | 1.27 | → | 1.19 | |
| 28 | Lactic acid | Energy | C00186 | 1.04 | → | 0.93 | →/→ | 1.29 | → | 1.19 | |
| 29 | Thiamine | Co-enzyme/its derivatives | C00378 | 0.98 | → | 1.18 | →/→ | 1.73 | → | 1.75 | |
| 30 | 5-Oxoproline | C01879 | 0.88 | → | 0.75 | →/→ | 1.43 | → | 2.11 | ||
| 31 | Trp | Amino acid | C00078 | 0.83 | → | 0.89 | →/→ | 0.75 | →/→ | 0.88 | |
| 32 | Glu | Amino acid | C00025 | 0.78 | → | 0.89 | →/→ | 1.94 | → | 1.29 | |
| 33 | Carnitine | C00318 | 0.26 | → | 0.69 | →/→ | 0.99 | → | 1.02 | ||
| 34 | Glucosamine | C00329 | 0.05 | → | 0.09 | unclear | NA | unclear | NA | ||
| 35 | Gluconic acid | C00257 | 0.01 | → | 0.20 | →/→ | 1.52 | →/→ | 1.10 | ||
| 36 | 4-Guanidinobutyric acid | C01035 | → | 0.19 | →/→ | 0.86 | → | 0.86 | |||
| 37 | Urea | C00086 | → | 0.71 | →/→ | 0.96 | → | 0.99 | |||
| 38 | Ophthalmic acid | Peptide | (05765) | → | 0.21 | unclear | NA | unclear | 0.47 | ||
| 39 | Asp | Amino acid | C00049 | 1.24 | → | 1.10 | →/→ | 2.04 | → | 1.35 | |
| 40 | 2'-Deoxycytidine | C00881 | 0.18 | → | 0.56 | →/→ | 1.23 | → | 1.16 | ||
| 41 | 5-Methoxyindoleacetic acid | Alkylamino acid | C05660 | → | 0.67 | →/→ | 1.31 | X | ND | ||
| 42 | S-Adenosylmethionine | Co-enzyme/its derivatives | C00019 | → | 0.74 | unclear | NA | unclear | 1.18 | ||
| 43 | Nicotinic acid | Co-enzyme/its derivatives | C00253 | 33.97 | → | X | ND | unclear | ND | ||
| 44 | 5-Aminovaleric acid | Amino acid | C00431 | → | 32.55 | X | ND | unclear | ND | ||
| 45 | 2,6-Diaminopimelic acid | Amino acid | C00666 | → | X | ND | unclear | ND | |||
| 46 | p-Hydroxyphenylacetic acid | C00642 | → | X | ND | unclear | ND | ||||
| 47 | 1H-Imidazole-4-propionic acid | - | → | X | ND | unclear | ND | ||||
| 48 | Piperidine | C01746 | → | X | ND | unclear | ND | ||||
| 49 | Adenine | C00147 | → | 1.72 | X | ND | unclear | ND | |||
| 50 | Prostaglandin E2 | Neurotransmitter | C00584 | → | 1.66 | X | ND | unclear | ND | ||
| 51 | N-Acetylglutamic acid | Alkylamino acid | C00624 | → | 1.62 | X | ND | unclear | 0.99 | ||
| 52 | 4-Pyridoxic acid | C00847 | 4.46 | → | X | ND | unclear | ND | |||
| 53 | N-Acetylornithine | Alkylamino acid | C00437 | 2.17 | → | 1.62 | X | ND | unclear | ND | |
| 54 | N-Acetylputrescine | C02714 | 1.51 | → | 1.28 | X | ND | unclear | ND | ||
| 55 | Methionine sulfoxide | C02989 | 1.01 | → | 1.18 | X | ND | unclear | 0.49 | ||
| 56 | Thr-Asp | Peptide | - | 0.76 | → | 0.48 | X | ND | unclear | ND | |
| 57 | Ser-Glu | Peptide | - | 0.73 | → | 0.47 | X | ND | unclear | ND | |
| 58 | SDMA | (03334) | 0.57 | → | 0.59 | X | ND | unclear | 1.07 | ||
| 59 | N6,N6,N6-Trimethyllysine | Alkylamino acid | C03793 | 0.24 | → | 0.11 | X | ND | unclear | ND | |
| 60 | Guanosine | C00387 | 0.12 | → | 0.69 | X | ND | unclear | NA | ||
| 61 | N-Acetylmethionine | Alkylamino acid | C02712 | → | 0.57 | X | ND | unclear | ND | ||
| 62 | Gly-Asp | Peptide | - | → | 0.47 | X | ND | unclear | ND | ||
| 63 | β-Ala | Amino acid | C00099 | →/→ | 1.11 | → | 1.18 | → | 1.24 | ||
| 64 | Pantothenic acid | Co-enzyme/its derivatives | C00864 | →/→ | 0.85 | → | 1.07 | →/→ | 1.45 | ||
| 65 | Taurine | Amino acid | C00245 | 16.86 | →/→ | 1.19 | → | 1.08 | → | 1.09 | |
| 66 | Citrulline | Amino acid | C00327 | 9.98 | →/→ | 0.68 | → | 0.68 | → | 0.73 | |
| 67 | Ornithine | Amino acid | C00077 | 7.38 | →/→ | 0.94 | → | 1.18 | →/→ | 0.69 | |
| 68 | 3-Methylhistidine | Alkylamino acid | C01152 | 2.52 | →/→ | 1.04 | → | 0.90 | → | 0.94 | |
| 69 | Pyridoxal | C00250 | 1.82 | →/→ | 1.05 | → | 0.92 | → | 0.97 | ||
| 70 | N6-Acetyllysine | Alkylamino acid | C02727 | 1.39 | →/→ | 0.46 | → | 0.51 | → | 0.68 | |
| 71 | Met | Amino acid | C00073 | 1.02 | →/→ | 0.41 | → | 0.52 | → | 0.57 | |
| 72 | Isethionic acid | C05123 | 1.00 | →/→ | 0.69 | → | 0.65 | → | 0.45 | ||
| 73 | Tyr | Amino acid | C00082 | 0.98 | →/→ | 0.66 | → | 0.70 | → | 0.75 | |
| 74 | Ala | Amino acid | C00041 | 0.90 | →/→ | 0.79 | → | 0.66 | → | 0.57 | |
| 75 | Gly-Leu | Peptide | C02155 | 0.86 | →/→ | 0.25 | → | 0.66 | → | 0.47 | |
| 76 | Choline | Choline | C00114 | 0.26 | →/→ | 1.24 | → | 0.94 | →/→ | 1.46 | |
| 77 | Taurocholic acid | C05122 | 0.26 | →/→ | 1.01 | → | 1.19 | →/→ | 0.18 | ||
| 78 | Betaine | Choline | C00719 | 0.49 | →/→ | 0.99 | → | 0.82 | → | 0.84 | |
| 79 | Trimethylamine N-oxide | C01104 | →/→ | 2.54 | → | 2.23 | → | 2.99 | |||
| 80 | O-Acetylcarnitine | C02571 | 0.14 | →/→ | 0.88 | → | 1.20 | → | 1.19 | ||
| 81 | 1-Methyladenosine | C02494 | →/→ | 0.82 | → | 0.89 | → | 1.06 | |||
| 82 | Nicotinamide | Co-enzyme/its derivatives | C00153 | →/→ | 0.89 | → | 0.98 | →/→ | 0.62 | ||
| 83 | N-Acetylaspartic acid | Neurotransmitter | C01042 | →/→ | 1.21 | unclear | NA | X | ND | ||
| 84 | N,N-Dimethylglycine | Choline | C01026 | →/→ | 0.93 | →/→ | 0.65 | → | 0.70 | ||
| 85 | Succinic acid | Energy | C00042 | →/→ | 1.00 | →/→ | 2.18 | →/→ | 0.88 | ||
| 86 | 3-Phenylpropionic acid | C05629 | →/→ | 0.67 | →/→ | 1.13 | X | ND | |||
| 87 | Cytidine | C00475 | →/→ | 0.72 | →/→ | 1.65 | → | 1.32 | |||
| 88 | Ribulose 5-phosphate | Energy | C00199 | →/→ | 0.81 | →/→ | 1.29 | →/→ | 1.17 | ||
| 89 | Homoserine | Amino acid | C00263 | →/→ | 0.91 | unclear | NA | unclear | 0.82 | ||
| 90 | Putrescine | C00134 | 17.15 | →/→ | 1.06 | unclear | NA | unclear | 1.02 | ||
| 91 | γ-Butyrobetaine | C01181 | 8.97 | →/→ | 1.08 | →/→ | 0.53 | → | 0.57 | ||
| 92 | Glyceric acid | C00258 | 7.33 | →/→ | 0.49 | →/→ | 1.10 | X | ND | ||
| 93 | Hypoxanthine | C00262 | 5.01 | →/→ | 0.94 | →/→ | → | ||||
| 94 | GABA | Neurotransmitter | C00334 | 3.66 | →/→ | 1.11 | →/→ | 1.26 | → | 4.20 | |
| 95 | Urocanic acid | C00785 | 3.52 | →/→ | 1.02 | →/→ | 1.30 | →/→ | 0.57 | ||
| 96 | Spermidine | C00315 | 2.85 | →/→ | 0.96 | unclear | NA | unclear | 0.57 | ||
| 97 | Glycerol 3-phosphate | C00093 | 0.53 | →/→ | 1.08 | →/→ | 1.92 | →/→ | 0.98 | ||
| 98 | Uric acid | C00366 | 0.44 | →/→ | 1.00 | →/→ | 1.54 | →/→ | 0.80 | ||
| 99 | Gln | Amino acid | C00064 | 0.27 | →/→ | 1.01 | →/→ | 2.09 | → | 1.80 | |
| 100 | Creatine | C00300 | 0.22 | →/→ | 1.07 | →/→ | 1.42 | →/→ | 1.14 | ||
| 101 | Inosine | C00294 | 0.16 | →/→ | 0.82 | →/→ | → | ||||
| 102 | Uridine | C00299 | →/→ | 1.03 | unclear | NA | unclear | 1.55 | |||
| 103 | Cystine | Peptide | C00491 | →/→ | NA | →/→ | 0.47 | → | 0.59 | ||
| 104 | N-Methylproline | Alkylamino acid | - | →/→ | NA | X | ND | unclear | ND | ||
| 105 | Guanine | C00242 | →/→ | 0.85 | X | ND | unclear | ND | |||
| 106 | Arg-Glu | Peptide | - | →/→ | 0.48 | X | ND | unclear | ND | ||
| 107 | β-Ala-Lys | Peptide | C05341 | →/→ | 0.37 | X | ND | unclear | ND | ||
| 108 | Tyramine | Co-enzyme/its derivatives | C00483 | →/→ | 1.19 | X | ND | unclear | NA | ||
| 109 | Uracil | C00106 | →/→ | 1.04 | X | ND | unclear | NA | |||
| 110 | CMP | C00055 | →/→ | 1.09 | X | ND | unclear | 1.07 | |||
| 111 | Propionic acid | Fatty acid | C00163 | →/→ | NA | X | ND | unclear | 1.03 | ||
| 112 | Adenosine | C00212 | →/→ | 0.53 | X | ND | unclear | NA | |||
| 113 | N-Acetylglucosamine | Alkylamino acid | C00140 | 7.41 | →/→ | 1.01 | X | ND | unclear | 1.14 | |
| 114 | N-Acetylneuraminic acid | Alkylamino acid | C00270 | 2.95 | →/→ | 1.25 | X | ND | unclear | 0.57 | |
| 115 | 3-Aminoisobutyric acid | Amino acid | C05145 | 2.26 | →/→ | 0.81 | X | ND | unclear | ||
| 116 | Glu-Glu | Peptide | C01425 | 0.83 | →/→ | NA | X | ND | unclear | ND | |
| 117 | His-Glu | Peptide | - | 0.55 | →/→ | NA | X | ND | unclear | ND | |
| 118 | Betaine aldehyde_+H2O | Choline | C00576 | 0.41 | →/→ | 1.74 | X | ND | unclear | ND | |
| 119 | Spermine | C00750 | 0.28 | →/→ | 0.95 | X | ND | unclear | ND | ||
| 120 | S-Lactoylglutathione | Peptide | C03451 | →/→ | 3.11 | X | ND | unclear | ND | ||
| 121 | Thymidine | C00214 | →/→ | 1.04 | X | ND | unclear | NA | |||
| 122 | Glucuronic acid | C00191 | →/→ | NA | X | ND | unclear | 0.70 |
→: metabolites are transported without selection by the gateway. →/→: metabolites are influenced by selection by the gateway. X: metabolites are blocked by the gateway. Metabolites detected in only Ex-GF or GF mice are indicated in red or green without the number, respectively.
*Although the Ex-GF/GF ratio was >0.8 in cardiac plasma, the ratio was judged as similar, because there was no significant difference between portal and cardiac plasma.
Table 2. Metabolites detected in colonic feces but not in colonic tissue.
| Metbolites | ExGF/GF ratio | ||||||
|---|---|---|---|---|---|---|---|
| Category | KEGG ID | Colonic feces | Colon | Portal plasma | Cardiac plasma | ||
| 4-Methylbenzoic acid | - | 20.00 | ND | ND | ND | ||
| Hydroxyindole | C03766 | 20.00 | ND | ND | ND | Ex-GF/GF | |
| 3'-CMP; Cytidine 2'-monophosphate | Base/nucleotide | C03104 | 20.00 | ND | ND | ND | 3 |
| Pyridoxamine | C13710 | 20.00 | ND | ND | ND | 2.5 | |
| 1,3-Diaminopropane | - | 20.00 | ND | ND | ND | 2 | |
| Cadaverine | C01672 | 20.00 | ND | ND | ND | 1.5 | |
| 3-Methylbenzoic acid | C01454 | 20.00 | ND | ND | ND | 1 | |
| 3-(4-Hydroxyphenyl)propionic acid | C01456 | 20.00 | ND | ND | ND | 0.67 | |
| Saccharopine | C00449 | 1.70 | ND | ND | ND | 0.5 | |
| Indole-3-acetamide | - | 1.25 | ND | ND | ND | 0.4 | |
| Oxypurinol | C07599 | 1.21 | ND | ND | ND | 0 | |
| Allantoin | - | 0.12 | ND | ND | ND | ||
| Galactosamine | C16675 | 0.09 | ND | ND | ND | ||
| 3-Nitrotyrosine | - | 0.00 | ND | ND | ND | ||
| O-Succinylhomoserine | C03776 | 0.00 | ND | ND | ND | ||
| γ-Glu-2-aminobutanoic acid | 0.00 | ND | ND | ND | |||
| 3-Hydroxy-3-methylglutaric acid | C03761 | 0.00 | ND | ND | ND | ||
| Glucaric acid | C00767 | 0.00 | ND | ND | ND | ||
| Quinic acid | C00296 | 0.00 | ND | ND | ND | ||
| 2'-Deoxyguanosine | Base/nucleotide | C08507 | 0.41 | ND | ND | ND | |
| 2-Oxoglutaric acid | C00026 | 20.00 | ND | 1.03 | ND | ||
| 4-Methyl-2-oxovaleric acid/ 3-Methyl-2-oxovaleric acid | Alkylamino acid | C00233 | 20.00 | ND | 0.70 | ND | |
| Xanthine | Base/nucleotide | C00385 | 1.42 | ND | ND | 1.18 | |
| Butyric acid | Fatty acid | C00246 | 20.00 | ND | 1.26 | ND | |
Metabolites detected in only Ex-GF or GF mice are indicated red or green without number, respectively.
Discussion
This is the first study to clarify the effect of both the absorption system and intestinal microbiome on the bioavailability of colonic luminal low-molecular-weight metabolites. First, we explain why we believe this method was able to accurately detect the transport of metabolites from colonic lumen. If the colon does not contribute to the absorption of colonic luminal metabolites, as required by the notion that the colon absorbs only water, minerals, and some vitamins, the difference in the metabolite concentration in colonic tissue, portal plasma, and cardiac plasma between GF mice and Ex-GF mice would not be apparent. However, in the present study, we detected differences in the concentration of each metabolite between GF and Ex-GF mice in all specimens, and these Ex-GF/GF ratios were similar among the colonic feces, colonic tissue, portal plasma, and cardiac plasma. Thus, it is evident that this method is able to accurately detect low-molecular-weight metabolites transported to the body from the colonic lumen. In fact, the similarity of the Ex-GF/GF ratio of Arg, which is known to be absorbed by colonic epithelial cells [21], between colonic feces and colonic tissue supports this method.
The Ex-GF/GF ratio of 62 metabolites (No. 1 to No. 62) was similar between the colonic feces and colonic tissue, indicating that many metabolites are bioavailable and revising the general concept that the role of the colon is mainly to absorb water [22], electrolytes [22], and some vitamins [23]. In fact, some metabolites belonging to this group have been known as substances specifically absorbed from the colon, for example, amino acids [21, 24]. Furthermore, 56 metabolites (all of the metabolites except for stachydrine, lactic acid, thiamine, 5-oxoproline, Trp, and methionine sulfoxide) showed differences between GF and Ex-GF in colonic feces, indicating that many low-molecular-weight metabolites influenced by the intestinal microbiome are bioavailable.
From the classification of metabolites based on the Ex-GF/GF ratio of each specimen, we estimated the influence of these metabolites in health and disease. Twenty-two metabolites (No. 1 to No. 22) (Table 1), including 11 basic amino acids, that had similar Ex-GF/GF ratios for all specimens were transported to the systemic circulation from the colonic lumen. Therefore, these metabolites are probably directly involved in health and disease. Forty metabolites (No. 23 to No. 62) with similar Ex-GF/GF ratios between the colonic feces and colonic tissue but not portal blood are proposed to be key factors that influence colonic barrier function and the intestinal mucosal immune system. In contrast, 60 metabolites (No. 63 to No. 122) with different Ex-GF/GF ratios between the colonic feces and colonic tissue are stringently regulated by cellular homeostasis. For example, significant differences were observed in the Ex-GF/GF ratio of GABA (No. 94) between colonic feces and colonic tissue, and between portal plasma and cardiac plasma. Therefore, GABA must be regulated by the membrane transport system of colonocytes and by the hepatic metabolism because it functions as a neurotransmitter. Significant differences were also observed in the Ex-GF/GF ratios of Glu (No. 32), Asp (No. 39), and Tyr (No. 73), which function as neurotransmitters or precursors of neurotransmitters, in colonic feces and other specimens, although many basic amino acids are transported from the colonic lumen to the systemic blood with the same Ex-GF/GF ratio. Twenty-four metabolites that were not detected in the colonic tissue (Table 2) are not transported to the body from the colonic lumen. Eight of 11 peptides detected were in this group, indicating that nearly all peptides are not absorbed by colonocytes, although previous studies suggested that peptides are absorbed through peptide transporter 1 (PEPT1) [25, 26]. However, it was reported that PEPT1 is present in the distal part of the colon but not in the proximal colon [26]. Unfortunately, since the middle colon was analyzed in this study, the presence of PEPT1 is unclear. Surprisingly, butyrate, which is involved in the metabolism and normal development of colonic epithelial cells [27, 28], also belongs to this group, indicating that butyrate absorbed from the lumen is immediately metabolized in colonocytes because in vivo studies demonstrated that butyrate in the colonic lumen is absorbed and is metabolized to CO2, 3-hydroxybutyrate, and lactate [29, 30]. Colonic gene expression of butyrate transporter SLC5A8 is markedly lower in GF mice than in conventional mice [31], indicating that the stimulation of intestinal bacteria is an important factor in the expression of transporter. On the other hand, Jakobsdottir et al. [32] reported that the concentrations of acetate, propionate, and butyrate in the cecal content correlated significantly with those in portal serum after the oral administration of dietary fiber. Based on these reports, parts of the colon (distal, middle, and proximal) and effects of bacterial stimulation are also important factors to understand the colonic transport system of low-molecular-weight metabolites. Further study is required to clarify the bioavailability of peptides and SCFA in colonic tissue considering these points and using analytical instruments such as liquid chromatography for SCFA. Regarding polyamines, Ex-GF/GF ratios of fecal putrescine, spermidine, and spermine were 17.15, 2.85, and 0.28, respectively (Table 1), demonstrating that the colonic luminal putrescine and spermidine are produced by the colonic microbiome. In contrast, colonic luminal spermine is probably derived from live or exfoliated epithelial cells and is absorbed by the colonic microbiome. On the other hand, since polyamines have to be strictly controlled because of physiological function and toxicity in the body, Ex-GF/GF ratios of all polyamines in colon tissue were around 1 (putrescine: 1.06, spermidine: 0.96, and spermine: 0.95).
To the best of our knowledge, little is known about the colonic transporter that absorbs metabolites. Although it is known that lipophiles are delivered by passive diffusion across the enterocyte membrane [33],water-soluble substances detected using CE-TOFMS are probably absorbed through transporters using energy produced by Na+/K+ATPase. For example, amino acids are absorbed through the Na+ pump transporter [34]. Here, we show that 14 basic amino acids were absorbed into colonocytes (colonic tissue) with an Ex-GF/GF ratio similar to that of colonic feces, indicating that the transporters mediate the uptake of amino acids by colonocytes. The present study indicates that known/unknown transporters are used to absorb water-soluble metabolites.
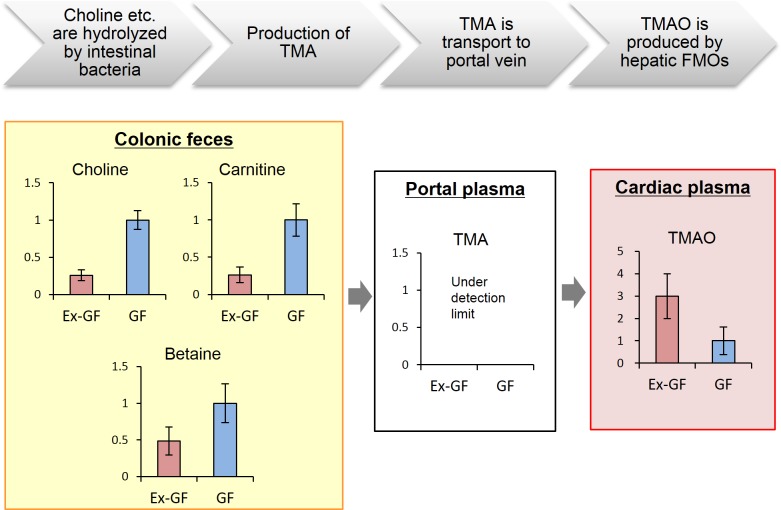
We confirmed the validity of this study using snapshots of the metabolome corresponding to the known relationship among the intestinal microbiome, intestinal bacterial metabolites, and health/disease using bacterial metabolism of trimethylamine N-oxide (TMAO) from choline, which is associated with cardiovascular disease risk [35]. Here, we showed that the colonic fecal concentrations of choline (No. 76) and carnitine (No. 33) in Ex-GF mice were lower than those in GF mice because they are metabolized by the intestinal microbiota (Fig 3). Furthermore, the TMAO (No. 79) concentrations in the cardiac plasma were higher in Ex-GF mice because TMAO was produced through liver metabolism of trimethylamine (TMA) produced by the intestinal microbiota in Ex-GF mice (the TMA concentration was below the limit of detection). Therefore, we consider that the metabolome data obtained from the present study are useful for understanding the mechanism of diseases caused by intestinal microbiome.
Fig 3. Metabolomic snapshots of the known phenomenon of intestinal microbiome-dependent production of pro-atherogenic trimethylamine N-oxide by degradation of carnitine/choline in the intestinal tract.
FMO: flavin-containing monooxygenase.
In the present study, we succeeded in the estimation of low-molecular-weight metabolites transported to the rest of the body from the colonic lumen by focusing on differences in the Ex-GF/GF ratio between GF and Ex-GF mice and have thus revised the general concept of the role of the colon. We also found that many low-molecular-weight metabolites influenced by the intestinal microbiome appeared to be absorbed from the colonic lumen. In the field of gastrointestinal physiology, it is very important to note that many low-molecular-weight chemicals influenced by intestinal microbiome in the large intestine appeared to be absorbed from the colonic lumen. Although this study does not provide direct evidence that low-molecular-weight metabolites influenced by intestinal microbiome are absorbed into the body from the large intestine, it contributes to the field of nutrition by providing a list of low-molecular-weight metabolites that are transported to the body from the colonic lumen. In other fields such as medicine, immunology, physiology, pharmacology, bacteriology, and nutrition, these findings may also contribute new insight into the function of the colon and its interactions with the intestinal microbiome and provide candidate chemicals whose roles in the body can be elucidated in future studies. In addition, further studies are required to investigate the colonic absorption of low-molecular-metabolites in mice of various strains and ages and in female mice.
Supporting Information
(DOCX)
Acknowledgments
This work in part was supported by the Programme for Promotion of Basic and Applied Researches for Innovations in Bio-oriented Industry by the Bio-oriented Technology Research Advancement Institution (BRAIN), JAPAN. We also thank Ms. Ayako Fujita for the PCA analyses.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was in part supported by the Programme for Promotion of Basic and Applied Researches for Innovations in Bio-oriented Industry by the Bio-oriented Technology Research Advancement Institution (BRAIN), Japan, and by Kyodo Milk Industry Co. Ltd. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. Kyodo Milk Industry Co. Ltd. and Human Metabolome Technologies, Inc. provided support in the form of salaries for authors MM and TO, but did not have any additional role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. The specific roles of these authors are articulated in the ‘author contributions’ section.
References
- 1.Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science 2005;307: 1915–1920. 10.1126/science.1104816 [DOI] [PubMed] [Google Scholar]
- 2.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 2006;444: 1027–1031. 10.1038/nature05414 [DOI] [PubMed] [Google Scholar]
- 3.Ley RE, Backhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci U S A 2005;102: 11070–11075. 10.1073/pnas.0504978102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A 2007;104: 13780–13785. 10.1073/pnas.0706625104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kirjavainen PV, Arvola T, Salminen SJ, Isolauri E. Aberrant composition of gut microbiota of allergic infants: a target of bifidobacterial therapy at weaning? Gut 2002;51: 51–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sobhani I, Tap J, Roudot-Thoraval F, Roperch JP, Letulle S, Langella P, et al. Microbial Dysbiosis in Colorectal Cancer (CRC) Patients. PLoS ONE 2011;6: e16393 10.1371/journal.pone.0016393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sudo N, Chida Y, Aiba Y, Sonoda J, Oyama N, Yu XN, et al. Postnatal microbial colonization programs the hypothalamic-pituitary-adrenal system for stress response in mice. J Physiol 2004;558: 263–275. 10.1113/jphysiol.2004.063388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diaz Heijtz R, Wang S, Anuar F, Qian Y, Bjorkholm B, Samuelsson A, et al. Normal gut microbiota modulates brain development and behavior. Proc Natl Acad Sci U S A 2011;108: 3047–3052. 10.1073/pnas.1010529108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matsumoto M, Kurihara S, Kibe R, Ashida H, Benno Y. Longevity in mice is promoted by probiotic-induced suppression of colonic senescence dependent on upregulation of gut bacterial polyamine production. PloS one 2011;6: e23652 10.1371/journal.pone.0023652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kibe R, Kurihara S, Sakai Y, Suzuki H, Ooga T, Sawaki E, et al. Upregulation of colonic luminal polyamines produced by intestinal microbiota delays senescence in mice. Scientific reports 2014;4: 4548 10.1038/srep04548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol 2009;9: 313–323. 10.1038/nri2515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tannock GW. Molecular analysis of the intestinal microflora in IBD. Mucosal Immunol 2008;1 Suppl 1: S15–18. [DOI] [PubMed] [Google Scholar]
- 13.Kim CH, Park J, Kim M. Gut microbiota-derived short-chain Fatty acids, T cells, and inflammation. Immune network 2014;14: 277–288. 10.4110/in.2014.14.6.277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, Peters EC, et al. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci U S A 2009;106: 3698–3703. 10.1073/pnas.0812874106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jandacek RJ, Rider T, Yang Q, Woollett LA, Tso P. Lymphatic and portal vein absorption of organochlorine compounds in rats. Am J Physiol Gastrointest Liver Physiol 2009;296: G226–234. 10.1152/ajpgi.90517.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cabre E, Hernandez-Perez JM, Fluvia L, Pastor C, Corominas A, Gassull MA. Absorption and transport of dietary long-chain fatty acids in cirrhosis: a stable-isotope-tracing study. Am J Clin Nutr 2005;81: 692–701. [DOI] [PubMed] [Google Scholar]
- 17.Monton MR, Soga T. Metabolome analysis by capillary electrophoresis-mass spectrometry. J Chromatogr A 2007;1168: 237–246; discussion 236. 10.1016/j.chroma.2007.02.065 [DOI] [PubMed] [Google Scholar]
- 18.Matsumoto M, Kibe R, Ooga T, Aiba Y, Kurihara S, Sawaki E, et al. Impact of intestinal microbiota on intestinal luminal metabolome. Scientific reports 2012;2: 233 10.1038/srep00233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matsumoto M, Kibe R, Ooga T, Aiba Y, Sawaki E, Koga Y, et al. Cerebral low-molecular metabolites influenced by intestinal microbiota: a pilot study. Front Syst Neurosci 2013;7: 9 10.3389/fnsys.2013.00009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ooga T, Sato H, Nagashima A, Sasaki K, Tomita M, Soga T, et al. Metabolomic anatomy of an animal model revealing homeostatic imbalances in dyslipidaemia. Molecular bioSystems 2011;7: 1217–1223. 10.1039/c0mb00141d [DOI] [PubMed] [Google Scholar]
- 21.Singh K, Coburn LA, Barry DP, Boucher JL, Chaturvedi R, Wilson KT. L-arginine uptake by cationic amino acid transporter 2 is essential for colonic epithelial cell restitution. Am J Physiol Gastrointest Liver Physiol 2012;302: G1061–1073. 10.1152/ajpgi.00544.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cummings JH. Absorption and secretion by the colon. Gut 1975;16: 323–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Said HM, Mohammed ZM. Intestinal absorption of water-soluble vitamins: an update. Curr Opin Gastroenterol 2006;22: 140–146. 10.1097/01.mog.0000203870.22706.52 [DOI] [PubMed] [Google Scholar]
- 24.Ugawa S, Sunouchi Y, Ueda T, Takahashi E, Saishin Y, Shimada S. Characterization of a mouse colonic system B(0+) amino acid transporter related to amino acid absorption in colon. Am J Physiol Gastrointest Liver Physiol 2001;281: G365–370. [DOI] [PubMed] [Google Scholar]
- 25.Daniel H. Molecular and integrative physiology of intestinal peptide transport. Annu Rev Physiol 2004;66: 361–384. 10.1146/annurev.physiol.66.032102.144149 [DOI] [PubMed] [Google Scholar]
- 26.Wuensch T, Schulz S, Ullrich S, Lill N, Stelzl T, Rubio-Aliaga I, et al. The peptide transporter PEPT1 is expressed in distal colon in rodents and humans and contributes to water absorption. American journal of physiology Gastrointestinal and liver physiology 2013;305: G66–73. 10.1152/ajpgi.00491.2012 [DOI] [PubMed] [Google Scholar]
- 27.Mortensen PB, Clausen MR. Short-chain fatty acids in the human colon: relation to gastrointestinal health and disease. Scand J Gastroenterol Suppl 1996;216: 132–148. [DOI] [PubMed] [Google Scholar]
- 28.Hague A, Singh B, Paraskeva C. Butyrate acts as a survival factor for colonic epithelial cells: further fuel for the in vivo versus in vitro debate. Gastroenterology 1997;112: 1036–1040. [DOI] [PubMed] [Google Scholar]
- 29.Fitch MD, Fleming SE. Metabolism of short-chain fatty acids by rat colonic mucosa in vivo. Am J Physiol 1999;277: G31–40. [DOI] [PubMed] [Google Scholar]
- 30.Jorgensen JR, Fitch MD, Mortensen PB, Fleming SE. In vivo absorption of medium-chain fatty acids by the rat colon exceeds that of short-chain fatty acids. Gastroenterology 2001;120: 1152–1161. 10.1053/gast.2001.23259 [DOI] [PubMed] [Google Scholar]
- 31.Cresci GA, Thangaraju M, Mellinger JD, Liu K, Ganapathy V. Colonic gene expression in conventional and germ-free mice with a focus on the butyrate receptor GPR109A and the butyrate transporter SLC5A8. J Gastrointest Surg 2010;14: 449–461. 10.1007/s11605-009-1045-x [DOI] [PubMed] [Google Scholar]
- 32.Jakobsdottir G, Jadert C, Holm L, Nyman ME. Propionic and butyric acids, formed in the caecum of rats fed highly fermentable dietary fibre, are reflected in portal and aortic serum. Br J Nutr 2013;110: 1565–1572. 10.1017/S0007114513000809 [DOI] [PubMed] [Google Scholar]
- 33.Hofmann AF, Borgstrom B. Physico-chemical state of lipids in intestinal content during their digestion and absorption. Fed Proc 1962;21: 43–50. [PubMed] [Google Scholar]
- 34.MLT style. Press Release: The 1997 Nobel Prize in Chemistry".: Nobelprize.org. Nobel Media AB 2014; [19 May 2015]. Available from: <http://www.nobelprize.org/nobel_prizes/chemistry/laureates/1997/press.html>
- 35.Koeth RA, Wang Z, Levison BS, Buffa JA, Org E, Sheehy BT, et al. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat Med 2013;19: 576–585. 10.1038/nm.3145 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.