Abstract
Transmembrane protein 39A (TMEM39A) belongs to the TMEM39 family. TMEM39A gene is a susceptibility locus for multiple sclerosis. In addition, TMEM39A seems to be implicated in systemic lupus erythematosus. However, any possible involvement of TMEM39A in cancer remains largely unknown. In the present report, we provide evidence that TMEM39A may play a role in brain tumors. Western blotting using an anti-TMEM39A antibody indicated that TMEM39A was overexpressed in glioblastoma cell lines, including U87-MG and U251-MG. Deep-sequencing transcriptomic profiling of U87-MG and U251-MG cells revealed that TMEM39A transcripts were upregulated in such cells compared with those of the cerebral cortex. Confocal microscopic analysis of U251-MG cells stained with anti-TMEM39A antibody showed that TMEM39A was located in dot-like structures lying close to the nucleus. TMEM39A probably located to mitochondria or to endosomes. Immunohistochemical analysis of glioma tissue specimens indicated that TMEM39A was markedly upregulated in such samples. Bioinformatic analysis of the Rembrandt knowledge base also supported upregulation of TMEM39A mRNA levels in glioma patients. Together, the results afford strong evidence that TMEM39A is upregulated in glioma cell lines and glioma tissue specimens. Therefore, TMEM39A may serve as a novel diagnostic marker of, and a therapeutic target for, gliomas and other cancers.
Keywords: TMEM39a, Multiple sclerosis, Systemic lupus erythematosus, Glioma
INTRODUCTION
Glioblastoma multiform (GBM) is the most aggressive form of glioma that arises from astrocytes (1). Approximately 15% of all primary brain tumors are GBMs, which may arise de novo or from low-grade astrocytomas. Genetic abnormalities are common in GBM patients (2). GBM is malignant, and patients typically die within 1 year of diagnosis (3). GBMs are characterized by the presence of a solid mass of extremely circuitous large-diameter vessels with abnormally thickened basement membranes (4). Functionally, the tumor vasculature is unusual. Vessel permeability is increased, triggering vasogenic edema and hemorrhage (5). As the vessels of the vasculature are highly disorganized, the efficacies of radio- and chemotherapy may be compromised by the abnormal blood flow (6). Recent preclinical and clinical studies have discovered new molecular GBM targets, which may afford opportunities to improve anti-angiogenic strategies (7).
Presently, the drug most often used to target vascular endothelial growth factor (VEGF) is bevacizumab (BEV), a recombinant humanized monoclonal antibody against VEGF-A (8). However, the initially reported beneficial effects of BEV may be (at least in part) attributed to imaging limitations caused by reduced neoangiogenesis and vascular permeability. These lead to an apparent (but controversial) decrease in the contrast-enhanced tumor volume (9). Recent prospective phase III trials (using avastin and radiation therapy) sought to validate the efficacy of temozolomide (TMZ)-based radio-chemotherapy, combined with BEV, in GBM patients (10). The “Avastin in GBM” (AVAglio) study showed that progression-free survival (PFS) was significantly prolonged (by 4.4 months) upon BEV co-treatment. In the “Radiation Therapy Oncology Group (RTOG) 0825” trial, no significant benefits were evident in terms of either PFS or overall survival. However, PFS benefits did not translate into improvements in overall survival (11). Therefore, it is necessary to find novel therapeutic targets in GBM patients, since TMZ and anti-VEGF drugs are not adequately efficacious.
Transmembrane protein 39A (TMEM39A) belongs to the TMEM39 family, consisting of TMEM39A and TMEM39B. The two TMEM39 isoforms are produced via alternative splicing (12). Transmembrane proteins extend from one side of the plasma membrane to the other. Many transmembrane proteins control the transport of materials across biological membranes (13). The TMEM39A-encoding gene may be a susceptibility locus for multiple sclerosis (14,15). Furthermore, TMEM39A is associated with systemic lupus erythematosus (16–18). However, no study has yet investigated any possible role for TMEM39A in cancer. Therefore, we sought a putative role for TMEM39A in GBM. Novel therapeutic markers for this brain cancer are needed urgently. Here, we provide clear evidence that TMEM39A is upregulated in GBM cell lines and GBM tissues from patients. This affords novel insight into the role played by TMEM39A in GBM. TMEM39A may be a useful new therapeutic target for brain cancer.
MATERIALS AND METHODS
Antibodies and reagents
Anti-TMEM39A antibody and Anti-Actin antibodies were purchased from Sigma-Aldrich (St. Louis, MO, USA). Horseradish peroxidase-conjugated anti-mouse IgG or anti-rabbit IgG secondary antibodies were purchased from Komabiotech (Seoul, Korea).
Patient samples
The study was approved by the Hospital Institutional Review Board (approval number CNUH 2013-11-006) according to the Declaration of Helsinki at Chungnam National University Hospital (Daejeon, Korea), and written informed consent was obtained from each patient by research team before surgery. Normal brain tissue samples were obtained from cadavers alternatively, from autopsy of surrounding normal brain of glioblastoma patient who underwent surgery.
Cell culture
The glioblastoma cells (U87-MG, U251-MG, U343-MG and U373-MG) and non-glioblastoma cell (HEK-293A) were maintained in medium (RPMI) supplemented with 10% FBS, 25 mM HEPES (Thermo Scientific), 1% Antibiotics-Antimycotics (Life Technologies, CA, USA).
Immunoblot analysis
The western blot analysis was performed as the described previously (19–21). Briefly, cells were placed on ice and extracted with lysis buffer containing 50 mM Tris-HCl, pH 7.5, 1% v/v Nonidet P-40, 120 mM NaCl, 25 mM sodium fluoride, 40 mM β-glycerol phosphate, 0.1 mM sodium orthovanadate, 1 mM phenylmethylsulfonyl fluoride, 1 mM benzamidine, and 2 mM microcystin-LR. Lysates were centrifuged for 15 min at 12,000 g. The cell extracts were resolved by 10~15% SDS-PAGE, and transferred to Immobilon-P membranes (Millipore, MA, USA). The filters were blocked for 1 hr in 1 X tri-buffered saline buffer (TBS-140 mM NaCl, 2.7 mM KCl, 250 mM Tris-HCl, pH 7.4), containing 5% skimmed milk and 0.2% Tween-20, followed by an overnight incubation with the anti-TMEM39A and anti-Actin antibodies diluted 1000-fold at 4°C. The secondary antibody was horseradish peroxidase-conjugated anti-mouse IgG or antirabbit IgG (Komabiotech, Seoul, Korea), diluted 5000-fold in the blocking buffer. The detection of protein expression was visualized by enhanced chemiluminescence, according to the manufacturer’s instructions (Thermo Fisher Scientific, CA, USA).
Real-time quantitative reverse transcription-polymerase chain reaction (qRT-PCR)
Total RNA was extracted from frozen tissue samples or from cells using the PureHelix RNA Extraction Solution (Nanohelix, Seoul, Korea). The cDNA was synthesized from total RNA with the Super-Script III First-Strand Synthesis System for qRT-PCR (Invitrogen, Grand Island, NY, USA). The qRT-PCR measurement of individual cDNAs was performed using SYBR green dye to measure duplex DNA formation with the StepOne Plus real-time PCR system (Invitrogen) and normalized to the expression of glyceraldehyde 3-phosphate dehydrogenase (GAPDH) RNA. The following primers were used in the qRT-PCR (F: Forward, R: Reverse); Human TMEM39A: F-5′-CCCACCTATCACAGCCTTAATC/R-5′-AAAGAGCAACCAACAGGT AGAT; human GAPDH : F-5′-TCGACAGTCAGCCGCATCTTCTTT/R-5′-TACGACCA AATCCGTTGACTCCGA.
RNA sequencing and RNA-Seq data analysis
Total RNA of U87-MG, U251-MG and normal brain was extracted using Trizol reagent (Invitrogen) following the manufacturer’s procedures. The total RNA quantity and purity were analysis of Bioanalyzer 2100 and RNA 6000 Nano Lab-Chip Kit (Agilent, Santa Clara, CA, USA). Roughly 10 μg of total RNA was subjected to isolate Poly (A) mRNA with poly-T oligo attached magnetic beads (Invitrogen). Following purification, the mRNA is fragmented into small pieces using divalent cations under raised temperature. Then the cleaved RNA fragments were reverse-transcribed to create the final cDNA library in accordance with the protocol for the mRNA-Seq sample preparation kit (Illumina, CA, USA). The average insert size for the paired-end libraries was 300 bp (± 50 bp). Next we performed the paired-end sequencing on an Illumina Hiseq 2000 system at Macrogen (Seoul, Korea) following the vendor’s recommended protocol. For each sample, sequenced reads were aligned to the UCSC human reference genome (22) using the Tophat package (23), which initially removes a portion of the reads based on quality information accompanying each read and then maps the reads to the reference genome. FPKM (fragments per kilobase of exon per million fragments mapped) were calculated to compare the expression level of TMEM39A mRNA variants in each sample.
Confocal imaging analysis and indirect immunofluorescence
U251-MG cells were grown on glass coverslips until they were 50~70% confluent. After 24 hrs, the cells were fixed in 4% paraformaldehyde at room temperature for 10 min and permeabilized in 0.2% Triton X100 for 5 min at room temperature. Then cells were incubated in blocking buffer containing 5% bovine serum albumin (Sigma-Aldrich) in 1 X TBS for 1 hr at 37°C. The rabbit polyclonal anti-TMEM39A was diluted 200-fold for primary antibody and incubated for overnight. The secondary antibody, FITC-conjugated anti-rabbit antibody (BD Biosciences, NJ, USA) was used. After appropriate rinsing, cover slips were mounted with Vectashield (Vector Laboratories, CA, USA) and visualized using a Zeiss confocal microscope.
Immunohistochemistry
The analysis of Immunohistochemistry was performed as the described previously (24,25). A human cancer tissue array slide with paraffin sections was purchased from Bio Max (US Biomax Inc., MD, USA). Histostain-Plus kits (Zymed Laboratories Inc., CA, USA) were used in accordance with the manufacturer’s instructions for the immunohistochemistry of tissue array. Briefly, paraffin sections were deparaffinized with xylene and rehydrated in a graded series of ethanol. The slide was submerged in peroxidase quenching solution for 10 min. After it was washed twice with PBS for 5 min, it was added with 2 drops of Reagent A for blocking and incubated for 30 min. Following two washes with PBS, the primary antibody, anti-TMEM39A antibody, was applied at 4°C for overnight. Then biotinylated secondary antibody, Reagent B, was added after rinsing with PBS. It was incubated at room temperature for 1 hr. It was rinsed with PBS and dropped with enzyme conjugated Reagent C. After it was washed with PBS, DAB chromogen, and a mixture of Reagent D1, D2, and D3, it was dropped, and signals were observed with a florescence microscope (Zeiss, Oberkochen, Germany). Then the reaction was stopped with distilled water, and pictures were taken with a microscope.
Bioinformatics data set
Glioma data sets and corresponding clinical data were downloaded from the publicly available databases (446 cases from the Repository of Molecular Brain Neoplasia Data (REMBRANDT; http://www.betastasis.com/glioma/rembrandt/). Normal; n = 21, GBM; n = 214, oligodendroglioma; n = 66, Astrocytomas; n = 145.
Statistical analysis
Data are expressed as the mean ± S.D. from at least three separate experiments performed triplicate. The differences between groups were analyzed using a Student’s t test and p < 0.05 (*) was considered significant, and p < 0.01 (**) was highly significant compared with corresponding control values. Comparison of TMEM39A expression in various gliomas was carried out by one-way ANOVA with Dunn’s post-test (one variable). Statistical analyses were carried out using SPSS software ver. 13.0 (SPSS Inc., NY, USA). For the analysis of Kaplan-Meier survival curve, p values were obtained from log-rank test, while hazard ratio (HR) and 95% confidence interval (CI) were determined by univariate Cox regression model.
RESULTS
Upregulation of TMEM39A expression in glioblastoma cell lines
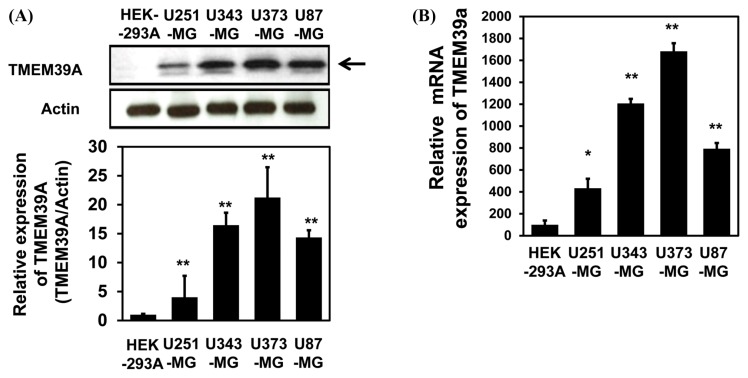
To explore a putative role for TMEM39A in brain cancer, we performed Western blotting using an anti-TMEM39A antibody. As shown in Fig. 1A, TMEM39A expression was markedly enhanced in U343-MG and U373-MG GBM cells compared with other cell type non-GBM cells, HEK-293A cells. Quantitative real-time PCR (qRT-PCR) of glioblastoma cell lines also showed that the levels of mRNA encoding TMEM39A were elevated in U343-MG and U373-MG cells (Fig. 1B).
Fig. 1.
TMEM39A expression in glioblastoma (GBM) cell lines. (A) Lysates were prepared from four established GBM cell lines (U87-MG, U251-MG, U373-MG, and U343-MG) and one established non-GBM cell lines (HEK-293A). These samples were subjected to Western blotting using anti-TMEM39A and anti-actin antibodies. The results are representative of those of three independent experiments (top panel). Relative densities were obtained by densitometry. Relative differences in TMEM39A expression levels (and the associated statistics) were calculated by normalizing all densitometric values to that of actin (in each lane) and setting the values from HEK-293A cells to 1 (bottom panel). Results are presented as the means ± SDs of data from three independent experiments. (B) Total RNA extracted from each GBM cell line was analyzed by real-time quantitative reverse transcription-polymerase chain reaction (qRT-PCR) using human TMEM39A-specific primers, as described in Materials and Methods. The results are presented as means ± SDs of data from three independent experiments. *p< 0.05, **p< 0.01.
TMEM39A transcription is enhanced in U87-MG cells and U251-MG cells
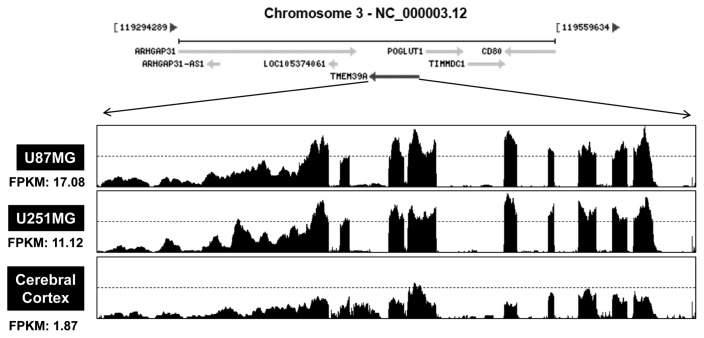
Based on the above observations, TMEM39A mRNA levels were measured by RNA sequencing of glioblastoma cell lines. Total RNA were isolated from two cell lines (U87-MG and U251-MG), which showed the low expression of TMEM39A in Fig. 1A and 1B. Also, we isolated total RNA from normal brain cells. The numbers of “fragments per kilobase of exon per million fragments mapped” (FPKMs) were calculated to compare the expression levels of TMEM39A mRNA among the various samples. As shown in Fig. 2, the FPKMs were markedly higher in U87-MG cells (17.08) and U251-MG cells (11.12) than in cerebral cortex cells (1.87), indicating that TMEM39A is transcriptionally upregulated in GBM cells.
Fig. 2.
Relative differences in TMEM39A transcript levels in GBM cells. Total RNAs were isolated from two GBM cell lines (U87-MG and U251-MG) and normal brain tissue. These samples were analyzed by standard RNA deep-sequencing (RNA-seq), as described in Materials and Methods. RNA-seq read densities of TMEM39A transcripts were plotted against relative RNA-seq read coverages (counts). “Fragments per kilobase of exon per million fragments mapped” (FPKMs) were calculated to compare the expression levels of TMEM39A mRNA variants among various sample.
Subcellular localization of TMEM39A in U251-MG cells
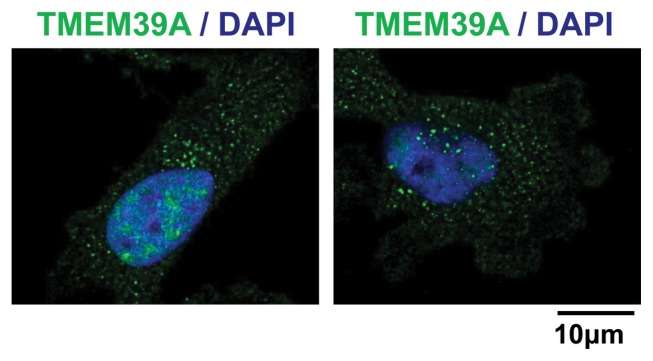
We used immunocytochemistry to determine the subcellular location of TMEM39A in U251-MG cells. Interestingly TMEM39A was found located in dot-like structures lying close to the nucleus, likely mitochondria and endosomes (Fig. 3). This suggested that the membrane-bound form of TMEM39A was functional in GBM cells.
Fig. 3.
Subcellular localization of TMEM39A in U251-MG cells. (A) U251-MG cells were grown on glass coverslips, fixed, and permeabilized with 0.2% (v/v) Triton X-100. After immunostaining with anti-TMEM39A antibody, the cover slips were mounted on Vectashield and examined using a Zeiss confocal microscope. Scale bars: 10 μm.
TMEM39A is expressed in GBM tissue
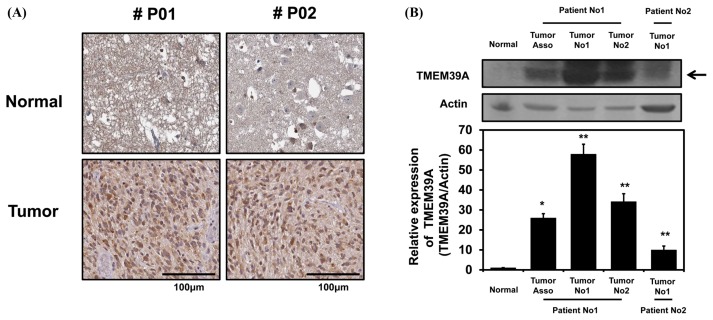
To determine if the above observations (Fig. 1 and 2) were clinically relevant, we subjected a human cancer tissue array to immunohistochemical analysis. As shown in Fig. 4A, the tumor tissues were stained with an anti-TMEM39A antibody to a greater extent than were the surrounding normal tissues. In addition, total cell lysates from normal and cancerous tissues derived from two GBM patients during surgery were subjected to Western blotting using an anti-TMEM39A antibody. As expected, GBM tissues expressed TMEM39A more prominently than did the surrounding normal tissues, indicating that TMEM39A expression is upregulated in GBM patients.
Fig. 4.
TMEM39A expression levels in human brain tumors. (A) Human glioma tissue arrays were immunohistochemically analyzed in terms of TMEM39A staining. Representative images from samples from two patients are shown. Scale bars: 100 μm. (B) Total cell lysates from normal and GBM tissues (tumors no. 1 and 2) from two patients were analyzed in terms of TMEM39A expression (top panel). Tumor-associated normal tissue served as a control (Tumor Asso.). Relative densities were calculated by densitometry. Relative differences in TMEM39A expression levels were determined by normalizing all densitometric values to those of actin (in each lane) and setting the control values to 1 (bottom panel). The results are presented as means ± SDs of data from three independent experiments. *p< 0.05, **p< 0.01.
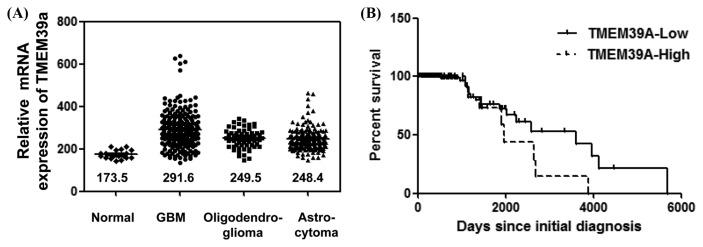
Differential TMEM39A mRNA expression and validation of the prognostic value of this observation in the REMBRANDT cohort
Building upon the above observations (Fig. 1–3), we assessed TMEM39A expression levels, and the prognostic significance of any differences, in gliomas from patients of the REMBRANDT cohort (http://www.betastasis.com/glioma/rembrandt/). Consistent with the above results (derived using a tissue microarray, Western blotting, and RNA sequencing), TMEM39A expression was markedly increased in various gliomas (GBMs, n = 214; oligodendroglioma, n = 66; and astrocytomas, n = 145) compared with normal control tissues (n = 21; one way ANOVA P-value < 0.0001; Fig. 5A). Moreover, high-level TMEM39A mRNA expression (in 167 patients), compared with low-level expression (in 31 patients), was significantly associated with poor survival (log-rank P value = 0.012; HR = 2.17, 95% confidence interval [CI] 0.80–2.89; Fig. 5B), suggesting that the TMEM39A expression level is correlated with the clinical prognosis of glioma patients.
Fig. 5.
Differential expression of TMEM39A mRNA in glioma specimens from the REMBRANDT cohort. (A) TMEM39A mRNA levels were measured in various normal (n = 21) and glioma (GBMs, n = 214; oligodendrogliomas, n = 66; and astrocytomas, n = 145) tissues. The median values for each group are indicated by lines and numbers. (B) The Kaplan-Meier curve compares the level of TMEM39A expression with overall survival. P values were obtained using the log-rank test, and hazard ratios (HRs) with 95% confidence intervals (CIs) were determined using the aid of a univariate Cox’s regression model.
DISCUSSION
GBM is the most aggressive type of brain tumor. Prognosis is poor even when multiple therapies are applied. Molecular targeting may be important when developing efficient GBM treatment strategies. Genetic abnormalities are common in GBM patients (2). Therefore, prognostic biomarkers and potential molecular targets must be identified immediately if the disease is to be overcome.
In the present study, we showed that TMEM39A is a novel GBM prognostic marker. TMEM39A is upregulated in GBM cell lines and patient tissues. Genome-wide association studies (GWASs) recently indicated that TMEM39A is involved in multiple sclerosis (14,15). A GWAS also revealed the involvement of TMEM39A in systemic lupus erythematosus (SLE), a chronic heterogeneous autoimmune disorder characterized by loss of tolerance to self-antigens and dysregulation of interferon synthesis (16–18,26). However, TMEM39A has not yet been implicated in any other disease. Therefore, we investigated whether TMEM39A is involved in the development of brain tumors including GBM. Interestingly, TMEM39A was highly expressed in GBM cell lines, including U343-MG and U373-MG cells (Fig. 1A). qRT-PCR using a TMEM39A-specific primer revealed that these cells express more TMEM39A mRNA (Fig. 1B) than do non-GBM cells, HEK-293A cells. In addition, RNA deep-sequencing of U87-MG and U251-MG cells showed that the FPKMs of TMEM39A mRNA were higher in these cells than in normal brain cells (Fig. 2). Many transmembrane proteins function as gateways or “loading docks”, permitting or forbidding the transport of specific substances across biological membranes. Both entry into the cell and exit from the cell (for example of waste byproducts) are thus regulated. Depending on the shapes of the molecules to be transported, “freight-handling” transmembrane proteins may fold or bend the molecules in specific ways (27). We therefore identified the subcellular location of TMEM39A in U251-MG cells. As expected, the protein was present in dot-like structures (probably mitochondria and endosomes, Fig. 3) lying close to the nucleus, suggesting that membrane-binding of TMEM39A is important when the protein plays a role in GBM.
To further evaluate the role played by TMEM39A in GBM progression, we used a tissue microarray and performed Western blotting of patient tissue samples. As shown in Fig. 4A, TMEM39A was upregulated in GBM tissues. This was confirmed by Western blotting of total cell lysates from normal and cancerous tissues sampled during surgery on two GBM patients (Fig. 4B). We also found that TMEM39A was highly expressed in all gliomas (including GBMs) of patients of the REMBRANDT cohort (Fig. 5A). Furthermore, high-level TMEM39A mRNA expression was correlated with poor overall survival (Fig. 5B), clearly indicating that TMEM39A expression is associated with the clinical prognosis of glioma. Together, the data indicate that TMEM39A is associated with glioma progression, and this molecule may serve as a novel prognostic biomarker of GBM.
ACKNOWLEDGMENTS
This work was financially supported by a research fund from Chungnam National University in 2014 (grant to S.H. Kim) and by National Research Foundation of Korea (NRF) grants from the Korean Government (MEST) (NRF-2012M3A9B6055302, NRF-2014R1A1A3050752, NRF-2015R1A2A2A01003597, NRF-2015R1D1A3A01015694).
REFERENCES
- 1.Friedmann-Morvinski D, Bushong EA, Ke E, Soda Y, Marumoto T, Singer O, Ellisman MH, Verma IM. Dedifferentiation of neurons and astrocytes by oncogenes can induce gliomas in mice. Science. 2012;338:1080–1084. doi: 10.1126/science.1226929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arif SH, Pandith AA, Bhat AR, Ramzan AU, Malik NK, Chibber SS, Wani AA, Tabasum R, Kirmani A. EGFR and PTEN gene mutation status in glioblastoma patients and their prognostic impact on patient’s survival. J Carcinog Mutagen. 2015;6:218. [Google Scholar]
- 3.Virk SM, Gibson RM, Quinones-Mateu ME, Barnholtz-Sloan JS. Identification of variants in primary and recurrent glioblastoma using a cancer-specific gene panel and whole exome sequencing. PLoS ONE. 2015;10:e0124178. doi: 10.1371/journal.pone.0124178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wesseling P, Ruiter DJ, Burger PC. Angiogenesis in brain tumors; pathobiological and clinical aspects. J Neurooncol. 1997;32:253–265. doi: 10.1023/A:1005746320099. [DOI] [PubMed] [Google Scholar]
- 5.Jain RK, di Tomaso E, Duda DG, Loeffler JS, Sorensen AG, Batchelor TT. Angiogenesis in brain tumours. Nat Rev Neurosci. 2007;8:610–622. doi: 10.1038/nrn2175. [DOI] [PubMed] [Google Scholar]
- 6.Das S, Marsden PA. Angiogenesis in glioblastoma. N Engl J Med. 2013;369:1561–1563. doi: 10.1056/NEJMcibr1309402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carmeliet P, Jain RK. Molecular mechanisms and clinical applications of angiogenesis. Nature. 2011;473:298–307. doi: 10.1038/nature10144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Norden AD, Drappatz J, Wen PY. Novel antiangiogenic therapies for malignant gliomas. Lancet Neurol. 2008;7:1152–1160. doi: 10.1016/S1474-4422(08)70260-6. [DOI] [PubMed] [Google Scholar]
- 9.Chinot OL, Macdonald DR, Abrey LE, Zahlmann G, Kerloeguen Y, Cloughesy TF. Response assessment criteria for glioblastoma: practical adaptation and implementation in clinical trials of antiangiogenic therapy. Curr Neurol Neurosci Rep. 2013;13:347. doi: 10.1007/s11910-013-0347-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chinot OL, Wick W, Mason W, Henriksson R, Saran F, Nishikawa R, Carpentier AF, Hoang-Xuan K, Kavan P, Cernea D, Brandes AA, Hilton M, Abrey L, Cloughesy T. Bevacizumab plus radiotherapy-temozolomide for newly diagnosed glioblastoma. N Engl J Med. 2014;370:709–722. doi: 10.1056/NEJMoa1308345. [DOI] [PubMed] [Google Scholar]
- 11.Gilbert MR, Dignam JJ, Armstrong TS, Wefel JS, Blumenthal DT, Vogelbaum MA, Colman H, Chakravarti A, Pugh S, Won M, Jeraj R, Brown PD, Jaeckle KA, Schiff D, Stieber VW, Brachman DG, Werner-Wasik M, Tremont-Lukats IW, Sulman EP, Aldape KD, Curran WJ, Jr, Mehta MP. A randomized trial of bevacizumab for newly diagnosed glioblastoma. N Engl J Med. 2014;370:699–708. doi: 10.1056/NEJMoa1308573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hao Y, Colak R, Teyra J, orbi-Verge C, Ignatchenko A, Hahne H, Wilhelm M, Kuster B, Braun P, Kaida D, Kislinger T, Kim PM. Semi-supervised learning predicts approximately one third of the alternative splicing isoforms as functional proteins. Cell Rep. 2015;12:183–189. doi: 10.1016/j.celrep.2015.06.031. [DOI] [PubMed] [Google Scholar]
- 13.Moon CP, Fleming KG. Side-chain hydrophobicity scale derived from transmembrane protein folding into lipid bilayers. Proc Natl Acad Sci USA. 2011;108:10174–10177. doi: 10.1073/pnas.1103979108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.International Multiple Sclerosis Genetics Consortium (IMSGC) Comprehensive follow-up of the first genome-wide association study of multiple sclerosis identifies KIF21B and TMEM39A as susceptibility loci. Hum Mol Genet. 2010;19:953–962. doi: 10.1093/hmg/ddp542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Varadé J, Comabella M, Ortiz MA, Arroyo R, Fernández O, Pinto-Medel MJ, Fedetz M, Izquierdo G, Lucas M, Gómez CL, Rabasa AC, Alcina A, Matesanz F, Alloza I, Antigüedad A, García-Barcina M, Otaegui D, Olascoaga J, Saiz A, Blanco Y, Montalbán X, Vandenbroeck K, Urcelay E. Replication study of 10 genes showing evidence for association with multiple sclerosis: validation of TMEM39A, IL12B and CBLB [correction of CLBL] genes. Mult Scler. 2012;18:959–965. doi: 10.1177/1352458511432741. [DOI] [PubMed] [Google Scholar]
- 16.You Y, Zhai ZF, Chen FR, Chen W, Hao F. Autoimmune risk loci of IL12RB2, IKZF1, XKR6, TMEM39A and CSK in Chinese patients with systemic lupus erythematosus. Tissue Antigens. 2015;85:200–203. doi: 10.1111/tan.12522. [DOI] [PubMed] [Google Scholar]
- 17.Sheng YJ, Xu JH, Wu YG, Zuo XB, Gao JP, Lin Y, Zhu ZW, Wen LL, Yang C, Liu L, Cheng YY, Chang Y, Yang LL, Zhou FS, Tang XF, Zheng XD, Yin XY, Tang HY, Sun LD, Cui Y, Yang S, Zhang XJ. Association analyses confirm five susceptibility loci for systemic lupus erythematosus in the Han Chinese population. Arthritis Res Ther. 2015;17:85. doi: 10.1186/s13075-015-0602-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lessard CJ, Adrianto I, Ice JA, Wiley GB, Kelly JA, Glenn SB, Adler AJ, Li H, Rasmussen A, Williams AH, Ziegler J, Comeau ME, Marion M, Wakeland BE, Liang C, Ramos PS, Grundahl KM, Gallant CJ, Alarcón-Riquelme ME, Alarcón GS, Anaya JM, Bae SC, Boackle SA, Brown EE, Chang DM, Cho SK, Criswell LA, Edberg JC, Freedman BI, Gilkeson GS, Jacob CO, James JA, Kamen DL, Kimberly RP, Kim JH, Martin J, Merrill JT, Niewold TB, Park SY, Petri MA, Pons-Estel BA, Ramsey-Goldman R, Reveille JD, Scofield RH, Song YW, Stevens AM, Tsao BP, Vila LM, Vyse TJ, Yu CY, Guthridge JM, Kaufman KM, Harley JB, Wakeland EK, Langefeld CD, Gaffney PM, Montgomery CG, Moser KL. Identification of IRF8, TMEM39A, and IKZF3-ZPBP2 as susceptibility loci for systemic lupus erythematosus in a large-scale multiracial replication study. Am J Hum Genet. 2012;90:648–660. doi: 10.1016/j.ajhg.2012.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim AY, Kwak JH, Je NK, Lee YH, Jung YS. Epithelial-mesenchymal transition is associated with acquired resistance to 5-fluorocuracil in HT-29 colon cancer cells. Toxicol Res. 2015;31:151–156. doi: 10.5487/TR.2015.31.2.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Y, Park J, Piao L, Kong G, Kim Y, Park KA, Zhang T, Hong J, Hur GM, Seok JH, Choi SW, Yoo BC, Hemmings BA, Brazil DP, Kim SH, Park J. PKB-mediated PHF20 phosphorylation on Ser291 is required for p53 function in DNA damage. Cell Signal. 2013;25:74–84. doi: 10.1016/j.cellsig.2012.09.009. [DOI] [PubMed] [Google Scholar]
- 21.Kim IS, Yang SY, Han JH, Jung SH, Park HS, Myung CS. Differential gene expression in GPR40-overexpressing pancreatic β-cells treated with linoleic acid. Korean J Physiol Pharmacol. 2015;19:141–149. doi: 10.4196/kjpp.2015.19.2.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bioinformatics, U.G. Human Genome Sequencing Consortium. 2015 Available from: http://genome.ucsc.edu/
- 23.Kim D, Salzberg S. TopHat 210 release. 2015 [cited 2015 Jun 29]. Available from: https://ccb.jhu.edu/software/tophat/index.shtml/
- 24.Na CH, Hong JH, Kim WS, Shanta SR, Bang JY, Park D, Kim HK, Kim KP. Identification of protein markers specific for papillary renal cell carcinoma using imaging mass spectrometry. Mol Cells. 2015;38:624–629. doi: 10.14348/molcells.2015.0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheon JM, Kim DI, Kim KS. Insulin sensitivity improvement of fermented Korean Red Ginseng (Panax ginseng) mediated by insulin resistance hallmarks in old-aged ob/ob mice. J Ginseng Res. 2015;39:331–337. doi: 10.1016/j.jgr.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang J, Ma Y, Qiu M, Yang L, Bu Y, Tang X. Correlation between TMEM39A gene polymorphism and systemic lupus erythematosus in Chinese Han patients. Nan Fang Yi Ke Da Xue Xue Bao. 2014;34:556–559. [PubMed] [Google Scholar]
- 27.Goodman SR. In: Medical cell biology in Medical Cell Biology. Goodman SR, editor. Academic Press; 2008. p. 37. [Google Scholar]