Abstract
Nuclear factor E2-related factor 2 (Nrf2), a transcription factor, controls the expression of genes encoding cytoprotective proteins, including antioxidant enzymes that combat oxidative and electrophilic stress to maintain redox homeostasis. However, recent studies demonstrated that, in cancer, aberrant activation of Nrf2 by epigenetic alterations promotes high expression of cytoprotective proteins, which can decrease the efficacy of anticancer drugs used for chemotherapy. In this review, we summarize recent findings regarding the relationship between oxidative stress, Nrf2, epigenetic modification, and anticancer drug resistance, which should aid in development of new strategies to improve chemotherapeutic efficacy.
Keywords: Anticancer drug resistance, Oxidative stress, Nrf2 transcription factor, Epigenetic modification
INTRODUCTION
Nuclear factor E2-related factor 2 (Nrf2), a transcription factor that regulates multiple antioxidant and detoxifying enzymes, is primarily involved in adaption to various cellular stresses. High Nrf2 activity makes a critical contribution to cancer cell resistance to various anticancer therapies (1,2). Epigenetic modifications, which are heritable alterations in gene expression not accompanied by changes in the primary DNA sequence, are intimately involved in oxidative stress and Nrf2 expression (3). In this review, we discuss the latest findings regarding epigenetic regulation of Nrf2 expression by DNA demethylation and histone methylation in 5-fluorouracil-resistant cells. Studies of the epigenetic modification in Nrf2 expression will provide new candidate therapeutic targets for anticancer drug resistance.
ANTICANCER DRUG RESISTANCE AND OXIDATIVE STRESS
Anticancer drugs rely primarily on induction of cell death via apoptosis, mitotic catastrophe, or premature senescence. However, anticancer drug resistance and dose-limiting toxicities are major obstacles to achieving successful cancer chemotherapy (4). The mechanisms underlying the development of anticancer drug resistance are complex and often involve activation of antioxidant or detoxifying enzymes (5,6). Studies of anticancer drug resistance have yielded valuable information about how to improve cancer chemotherapy by circumventing such resistance.
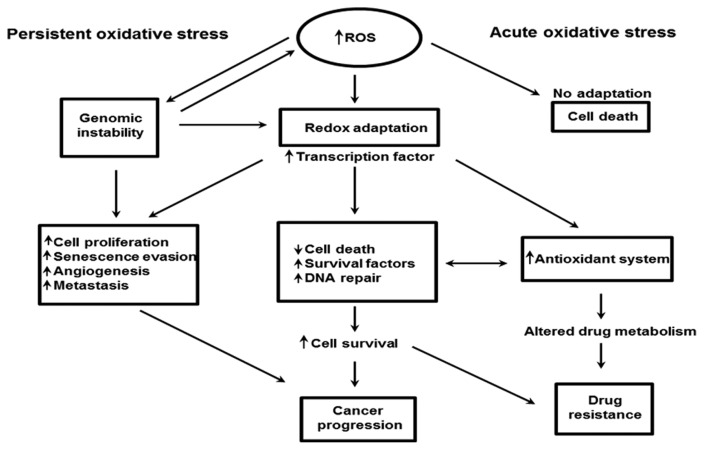
Reactive oxygen species (ROS) comprise hydrogen peroxide (H2O2), hydroxyl radical (•OH), superoxide (O2−), and singlet oxygen (1O2). Under physiological conditions, cells can maintain a balance between cellular oxidants and antioxidants, a state referred to as redox homeostasis. Many types of tumor cells contain higher levels of ROS than normal cells, promoting cancer progression and development (7). On the other hand, when ROS concentrations become extremely high, they cause tumor cell death (8). Thus, a variety of drugs with direct or indirect effects on ROS levels have been employed as effective cancer therapies. In certain cancer cells, persistent oxidative stress may induce adaptive stress responses, including activation of redox-sensitive transcription factors such as Nrf2, leading to an increase in the expression of ROS-scavenging enzymes, elevated levels of survival factors, and suppression of cell death factors. ROS-mediated DNA mutations or deletions promote genomic instability, providing an additional mechanism for stress adaptation. Together, these events enable tumor cells to maintain cellular viability with high levels of ROS. Because these transcription factors also regulate the expression of genes that are responsible for proliferation, evasion of senescence, angiogenesis, and metastasis, redox adaptation may promote cancer development. The resultant alterations in drug metabolism, together with elevated cell survival, may render cancer cells more resistant to chemotherapeutic agents (8–10) (Fig. 1). Accordingly, manipulating ROS levels via redox modulation represents a promising strategy for selectively killing cancer cells without causing significant toxicity in normal cells.
Fig. 1.
Oxidative stress status in anticancer drug resistance (8). Drug-induced persistent oxidative stress can be overcome by increasing genomic instability and by redox adaption. Both processes are implicated in cancer progression and drug resistance.
NUCLEAR FACTOR E2-RELATED FACTOR 2 (Nrf2)
The transcription factor Nrf2 binds to small Maf proteins at the antioxidant response element (ARE) in the promoter regions of target genes, and Kelch-like ECH-associated protein 1 (Keap1), a cytoplasmic anchor of Nrf2, promotes Nrf2 degradation by the ubiquitin proteasome pathway (11, 12). The target genes for Nrf2 are multiple antioxidant enzymes, including heme oxygenase 1 (HO-1), catalase, superoxide dismutase, NAD(P)H quinone oxidoreductase 1, glutamate-cysteine ligase, glutathione S-transferase, and glutathione synthetase.
Several studies reported that suppression of carcinogenesis by chemopreventive drugs is mediated through activation of Nrf2 (13,14). Paradoxically, however, recent studies showed that oncogene-induced Nrf2 promotes both ROS detoxification and tumorigenesis (12,15). Moreover, genetic analyses of human cancers revealed that Nrf2 may be oncogenic and cause resistance to chemotherapy (16). Na and Surh reviewed the evidence that the cellular stress response or cytoprotective signaling mediated via the Nrf2-HO-1 axis is exploited by cancer cells to promote their proliferation and increase their ability to survive chemotherapy (17). Therefore, Nrf2 and its downstream target HO-1 represent candidate therapeutic targets for the management of cancer.
EPIGENETIC MODIFICATIONS
Epigenetics is the molecular phenomenon by which phenotypic changes are transmitted from one generation to another with no accompanying alterations in the DNA sequence. Epigenetic mechanisms include DNA methylation/demethylation, histone modifications, and microRNA (miRNA)-mediated regulation. Although many types of histone modifications have been reported, here we focus on methylation/demethylation of DNA and histones.
DNA methylation/demethylation
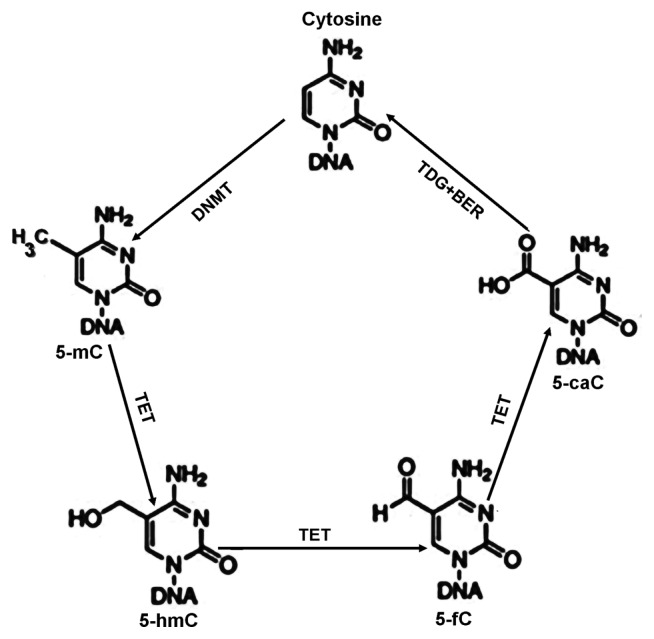
DNA methylation occurs when methyl groups are incorporated into cytosine molecules by DNA methyltransferases (DNMTs), forming 5-methylcytosine (5-mC). In particular, two de novo DNMTs (DNMT3A, DNMT3B) and a maintenance DNMT (DNMT1) transfer methyl groups from S-adenosyl-methionine to cytosine in CpG dinucleotides. This process contributes to the suppression of transcription. Methylation can be reversed by DNA demethylases, the ten-eleven translocation enzymes (TETs): TET1, TET2, and TET3. These enzymes convert 5-mC to 5-hydroxymethylcytosine (5-hmC), 5-formylcytosine (5-fC), and 5-carboxylcytosine (5-caC), which are subsequently excised from the DNA by glycosylases (18,19) (Fig. 2). Generally, DNA demethylation contributes to the activation of transcription.
Fig. 2.
Cytosine methylation and demethylation (18,19). DNMTs catalyze conversion of cytosine into 5-mC. TET proteins can iteratively oxidize 5-mC to 5-hmC, 5-fC, and 5-caC. Thymine DNA glycosylase (TDG) can decarboxylate either 5-fC or 5-caC and replace it with an unmodified cytosine through base excision repair (BER), completing the cycle of dynamic cytosine removal.
Histone methylation/demethylation
Histone methylation can either repress or activate transcription, depending on several factors: arginine methylation of histone H3 and H4 promotes transcriptional activation, whereas lysine methylation of histone H3 and H4 is implicated in both transcriptional activation and repression, depending on the methylation site. In addition, lysine residues can be mono-, di-, or tri-methylated, providing further functional diversity to each methylation site. Tri-methylation on K4 of histone H3 (H3K4Me3) by the mixed lineage leukemia (MLL) protein is generally associated with transcriptional activation (20), whereas H3K9Me2, H3K9Me3, and H3K27Me3 generated by G9a or enhancer of zeste homolog 2 (EZH2) are generally associated with transcriptional repression (21). Histone demethylases, e.g., the lysine-specific demethylase 1 and Jumonji domain-containing histone demethylases, are involved in demethylation of mono-, di-, and tri-methylated lysines (22).
RELATIONSHIP BETWEEN DRUG RESISTANCE, OXIDATIVE STRESS, Nrf2, AND EPIGENETIC MODIFICATIONS
The epigenetic regulation of Keap1 has been investigated in various cancers. Wang and colleagues first reported that Keap1 is highly expressed in BEAS-2B human normal bronchial epithelial cells but is down-regulated in a series of lung cancer cell lines and human lung cancer tissues; this down-regulation was accompanied by the hypermethylation of CpG sites in the Keap1 promoter region and was restored by 5-aza treatment (23). Similar results have been obtained in various cancers, including malignant gliomas and breast, colorectal, prostate, thyroid, and head and neck cancer cells. Activation of Nrf2 signaling by hypermethylation of Keap1 promoter is associated with tumor progression and resistance to therapies (24). Li and colleagues showed that decreased EZH2 expression significantly correlated with elevated expression of Nrf2, NQO1, and HO-1 in lung cancer tissues and cell lines, which was mainly attributed to a decrease in H3K27Me3 in the Nrf2 promoter (25). Recently, Kang and colleagues focused on the causative relationship between Nrf2 expression and epigenetic alterations, especially DNA methylation at cytosines during 5-fluorouracil (FU)-induced oxidative stress (26). They found that 5-FU-induced ROS activate DNA demethylases (TETs), resulting in hypomethylation of the Nrf2 promoter and activation of Nrf2 expression. This, in turn, upregulates expression of the antioxidant enzyme HO-1, a Nrf2 target, resulting in resistance to 5-FU. In addition, in mice implanted with shNrf2-transfected 5-FU-resistant SNUC5 (SNUC5/5-FUR) colon cancer cells, 5-FU treatment decreased tumor volume, size, and weight, and increased the proportion of apoptotic cells to a greater extent than in mice implanted with shControl-transfected SNUC5/5-FUR cells. Thus, high Nrf2 expression resulting from oxidative stress-induced DNA demethylation may promote resistance to 5-FU (26). Kang and colleagues demonstrated that, in addition to DNA methylation, histone methylation status under 5-FU-induced oxidative stress also influences expression of Nrf2 and its target antioxidant enzymes, resulting in resistance to 5-FU. Levels of MLL and the modification it catalyzes (H3K4Me3) were higher in SNUC5/5-FUR cells than in SNUC5 cells, and siRNA knockdown of MLL in these cells significantly decreased expression of Nrf2 and HO-1 (27). Furthermore, Kang and colleagues demonstrated that the TETs and MLL interact with O-GlcNAc transferase (OGT) and host cell factor 1 (HCF1) to regulate Nrf2 expression (27). Several reports showed that the TETs-OGT interaction increases TET DNA demethylation activity and recruit histone methyltransferase complex required to a high H3K4Me3 chromatin environment, resulting in transcriptional activation (28–30).
MLL is a component of the complex of proteins associated with Set1 (COMPASS) like complex. HCF1, a component of the MLL complex, is critical for the integrity of the MLL/COMPASS-like complex. Capotosti and colleagues suggested that HCF1 is GlcNAcylated and undergoes proteolytic maturation in the active site of OGT. This OGT link is associated with activation of the regulatory functions of HCF1 (31). In addition, a previous study suggested that HCF1 functions as a transcriptional switch for the action of MLL (32). siRNA knockdown of HCF1 decreases the level of MLL mRNA and protein in SNUC5/5-FUR cells, resulting in reduced expression of Nrf2 protein (27). These results suggest that the MLL and HCF1 components of the MLL/COMPASS-like complex are necessary for Nrf2 expression.
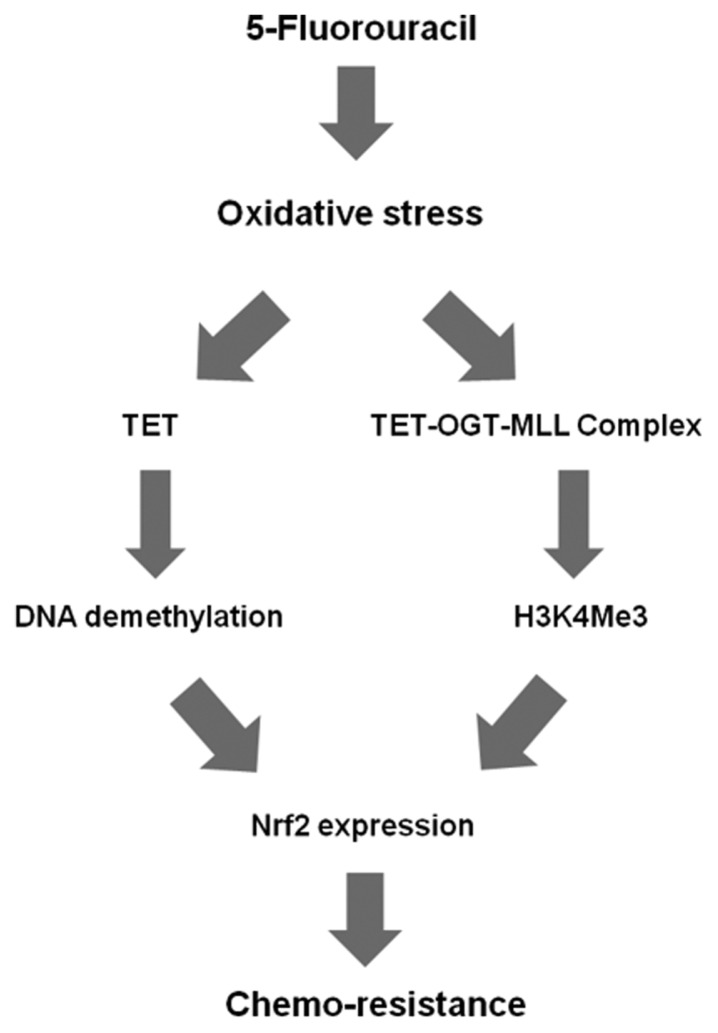
Recent work showed that OGT stabilizes the MLL protein: depletion of OGT in cells decreased the level of MLL via ubiquitin/proteasome-dependent proteolytic degradation, whereas ectopic expression of OGT protein suppressed MLL ubiquitylation (33). siRNA-mediated knockdown of OGT decreased mRNA and protein levels of MLL in SNUC5/5-FUR cells, resulting in reduced expression of HCF1 and Nrf2 proteins (27). In addition, upregulation of Nrf2 by the TET1-OGT-MLL complex was confirmed by ChIP assay: the level of H3K4Me3 on the Nrf2 promoter was significantly elevated in SNUC5/5-FUR cells, whereas the H3K4Me3 level on the β-actin promoter, which is unrelated to 5-FU resistance, was unchanged (27). Thus, oxidative stress-activated TET1 demethylates DNA in the promoter of Nrf2. In addition, TET1 recruits OGT and the MLL/COMPASS-like complex to the promoter. The complex methylates H3K4, resulting in transcriptional activation of Nrf2, ultimately leading to chemo-resistance (Fig. 3).
Fig. 3.
Model: the roles of DNA demethylation and histone methylation in the mechanism of resistance to 5-FU (26,27). 5-FU induces oxidative stress via generation of ROS, which increase TET1 activity. Induction of TET1 leads to activation of Nrf2, ultimately causing drug resistance. Moreover, oxidative stress-activated TET1 recruits OGT to the Nrf2 promoter. OGT GlcNAcylates HCF1, a component of the MLL/COMPASS-like complex. This complex methylates H3K4, resulting in transcriptional activation of Nrf2, leading to chemo-resistance.
CONCLUSION
A recent study elucidated the epigenetic regulation of Nrf2 expression in 5-FU-resistant cancer cells (26,27). As depicted schematically in Figure 3, Nrf2 expression can be activated by DNA demethylation in CpGs of promoter regions, as well as histone methylation. However, although oxidative stress-induced anticancer drug resistance is involved in these epigenetic modifications, it is not clear whether epigenetic processes other than DNA demethylation and histone methylation also contribute. The exact mechanisms of epigenetic regulation of Nrf2 expression in anticancer drug resistance, as well as its physiological significance, remain open for further investigation.
ACKNOWLEDGMENTS
This study was supported by a grant from the National R&D Program for Cancer Control, Ministry for Health and Welfare, Republic of Korea (1120340) and by the National Research Foundation of Korea Grant funded by the Korean Government (MEST) (NRF-2016R1A2B4007934).
REFERENCE
- 1.Bai X, Chen Y, Hou X, Huang M, Jin J. Emerging role of NRF2 in chemoresistance by regulating drug-metabolizing enzymes and efflux transporters. Drug Metab Rev. 2016;48:541–567. doi: 10.1080/03602532.2016.1197239. [DOI] [PubMed] [Google Scholar]
- 2.Furfaro AL, Piras S, Domenicotti C, Fenoglio D, De Luigi A, Salmona M, Moretta L, Marinari UM, Pronzato MA, Traverso N, Nitti M. Role of Nrf2, HO-1 and GSH in neuroblastoma cell resistance to bortezomib. PLoS ONE. 2016;11:e0152465. doi: 10.1371/journal.pone.0152465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guo Y, Yu S, Zhang C, Kong AN. Epigenetic regulation of Keap1-Nrf2 signaling. Free Radic Biol Med. 2015;88:337–349. doi: 10.1016/j.freeradbiomed.2015.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seton-Rogers S. Chemotherapy: preventing competitive release. Nat Rev Cancer. 2016;16:199. doi: 10.1038/nrc.2016.28. [DOI] [PubMed] [Google Scholar]
- 5.Holohan C, Van Schaeybroeck S, Longley DB, Johnston PG. Cancer drug resistance: an evolving paradigm. Nat Rev Cancer. 2013;13:714–726. doi: 10.1038/nrc3599. [DOI] [PubMed] [Google Scholar]
- 6.Ramos P, Bentires-Alj M. Mechanism-based cancer therapy: resistanceto therapy, therapy for resistance. Oncogene. 2015;34:3617–3626. doi: 10.1038/onc.2014.314. [DOI] [PubMed] [Google Scholar]
- 7.Gorrini C, Harris IS, Mak TW. Modulation of oxidative stress as an anticancer strategy. Nat Rev Drug Discov. 2013;12:931–947. doi: 10.1038/nrd4002. [DOI] [PubMed] [Google Scholar]
- 8.Trachootham D, Alexandre J, Huang P. Targeting cancercells by ROS-mediated mechanisms: a radical therapeutic approach? Nat Rev Drug Discov. 2009;8:579–591. doi: 10.1038/nrd2803. [DOI] [PubMed] [Google Scholar]
- 9.Liu Y, Li Q, Zhou L, Xie N, Nice EC, Zhang H, Huang C, Lei Y. Cancer drug resistance: redox resetting renders a way. Oncotarget. 2016;7:42740–42761. doi: 10.18632/oncotarget.8600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Debatin KM, Krammer PH. Death receptors in chemotherapy and cancer. Oncogene. 2004;23:2950–2966. doi: 10.1038/sj.onc.1207558. [DOI] [PubMed] [Google Scholar]
- 11.Ren D, Villeneuve NF, Jiang T, Wu T, Lau A, Toppin HA, Zhang DD. Brusatol enhances the efficacy of chemotherapy by inhibiting the Nrf2-mediated defense mechanism. Proc Natl Acad Sci USA. 2011;108:1433–1438. doi: 10.1073/pnas.1014275108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeNicola GM, Karreth FA, Humpton TJ, Gopinathan A, Wei C, Frese K, Mangal D, Yu KH, Yeo CJ, Calhoun ES, Scrimieri F, Winter JM, Hruban RH, Iacobuzio-Donahue C, Kern SE, Blair IA, Tuveson DA. Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature. 2011;475:106–109. doi: 10.1038/nature10189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Y, Gordon GB. A strategy for cancer prevention: stimulation of the Nrf2-ARE signaling pathway. Mol Cancer Ther. 2004;3:885–893. [PubMed] [Google Scholar]
- 14.Giudice A, Arra C, Turco MC. Review of molecular mechanisms involved in the activation of the Nrf2-ARE signaling pathway by chemopreventive agents. Methods Mol Biol. 2010;647:37–74. doi: 10.1007/978-1-60761-738-9_3. [DOI] [PubMed] [Google Scholar]
- 15.Chio II, Jafarnejad SM, Ponz-Sarvise M, Park Y, Rivera K, Palm W, Wilson J, Sangar V, Hao Y, Öhlund D, Wright K, Filippini D, Lee EJ, Da Silva B, Schoepfer C, Wilkinson JE, Buscaglia JM, DeNicola GM, Tiriac H, Hammell M, Crawford HC, Schmidt EE, Thompson CB, Pappin DJ, Sonenberg N, Tuveson DA. NRF2 promotes tumor maintenance by modulating mRNA translation in pancreatic cancer. Cell. 2016;166:963–976. doi: 10.1016/j.cell.2016.06.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sporn MB, Liby KT. NRF2 and cancer: the good, the bad and the importance of context. Nat Rev Cancer. 2012;12:564–571. doi: 10.1038/nrc3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Na HK, Surh YJ. Oncogenic potential of Nrf2 and its principal target protein heme oxygenase-1. Free Radic Biol Med. 2014;67:353–365. doi: 10.1016/j.freeradbiomed.2013.10.819. [DOI] [PubMed] [Google Scholar]
- 18.Shen L, Song CX, He C, Zhang Y. Mechanism and function of oxidative reversal of DNA and RNA methylation. Annu Rev Biochem. 2014;83:585–614. doi: 10.1146/annurev-biochem-060713-035513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shilatifard A. Molecular implementation and physiological roles for histone H3 lysine 4 (H3K4) methylation. Curr Opin Cell Biol. 2008;20:341–348. doi: 10.1016/j.ceb.2008.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coward WR, Feghali-Bostwick CA, Jenkins G, Knox AJ, Pang L. A central role for G9a and EZH2 in the epigenetic silencing of cyclooxygenase-2 in idiopathic pulmonary fibrosis. FASEB J. 2014;28:3183–3196. doi: 10.1096/fj.13-241760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang D, Okamura H, Teramachi J, Haneji T. Histone demethylase Jmjd3 regulates osteoblast apoptosis through targeting anti-apoptotic protein Bcl-2 and pro-apoptotic protein Bim. Biochim Biophys Acta. 2016;1863:650–659. doi: 10.1016/j.bbamcr.2016.01.006. [DOI] [PubMed] [Google Scholar]
- 23.Wang R, An J, Ji F, Jiao H, Sun H, Zhou D. Hypermethylation of the Keap1 gene in human lung cancer cell lines and lung cancer tissues. Biochem Biophys Res Commun. 2008;373:151–154. doi: 10.1016/j.bbrc.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 24.Muscarella LA, Barbano R, D’Angelo V, Copetti M, Coco M, Balsamo T, la Torre A, Notarangelo A, Troiano M, Parisi S, Icolaro N, Catapano D, Valori VM, Pellegrini F, Merla G, Carella M, Fazio VM, Parrella P. Regulation of Keap1 expression by promoter methylation in malignant gliomas and association with patient’s outcome. Epigenetics. 2011;6:317–325. doi: 10.4161/epi.6.3.14408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Z, Xu L, Tang N, Xu Y, Ye X, Shen S, Niu X, Lu S, Chen Z. The polycomb group protein EZH2 inhibits lung cancer cell growth by repressing the transcription factor Nrf2. FEBS Lett. 2014;588:3000–3007. doi: 10.1016/j.febslet.2014.05.057. [DOI] [PubMed] [Google Scholar]
- 26.Kang KA, Piao MJ, Kim KC, Kang HK, Chang WY, Park IC, Keum YS, Surh YJ, Hyun JW. Epigenetic modification of Nrf2 in 5-fluorouracil-resistant colon cancer cells: involvement of TET-dependent DNA demethylation. Cell Death Dis. 2014;5:e1183. doi: 10.1038/cddis.2014.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kang KA, Piao MJ, Ryu YS, Kang HK, Chang WY, Keum YS, Hyun JW. Interaction of DNA demethylase and histone methyltransferase upregulates Nrf2 in 5-fluorouracil-resistant colon cancer cells. Oncotarget. 2016;7:40594–40620. doi: 10.18632/oncotarget.9745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mohan M, Herz HM, Smith ER, Zhang Y, Jackson J, Washburn MP, Florens L, Eissenberg JC, Shilatifard A. The COMPASS family of H3K4 methylases in Drosophila. Mol Cell Biol. 2011;31:4310–4318. doi: 10.1128/MCB.06092-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deplus R, Delatte B, Schwinn MK, Defrance M, Méndez J, Murphy N, Dawson MA, Volkmar M, Putmans P, Calonne E, Shih AH, Levine RL, Bernard O, Mercher T, Solary E, Urh M, Daniels DL, Fuks F. TET2 and TET3 regulate GlcNAcylation and H3K4 methylation through OGT and SET1/COMPASS. EMBO J. 2013;32:645–655. doi: 10.1038/emboj.2012.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ardehali MB, Mei A, Zobeck KL, Caron M, Lis JT, Kusch T. Drosophila Set1 is the major histone H3 lysine 4 trimethyltransferase with role in transcription. EMBO J. 2011;30:2817–2828. doi: 10.1038/emboj.2011.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Capotosti F, Guernier S, Lammers F, Waridel P, Cai Y, Jin J, Conaway JW, Conaway RC, Herr W. O-GlcNAc transferase catalyzes site-specific proteolysis of HCF-1. Cell. 2011;144:376–388. doi: 10.1016/j.cell.2010.12.030. [DOI] [PubMed] [Google Scholar]
- 32.Yokoyama A, Wang Z, Wysocka J, Sanyal M, Aufiero DJ, Kitabayashi I, Herr W, Cleary ML. Leukemia proto-oncoprotein MLL forms a SET1-like histone methyltransferase complex with menin to regulate Hox gene expression. Mol Cell Biol. 2004;24:5639–5649. doi: 10.1128/MCB.24.13.5639-5649.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ding X, Jiang W, Zhou P, Liu L, Wan X, Yuan X, Wang X, Chen M, Chen J, Yang J, Kong C, Li B, Peng C, Wong CC, Hou F, Zhang Y. Mixed lineage leukemia 5 (MLL5) protein stability is cooperatively regulated by O-GlcNac transferase (OGT) and ubiquitin specific protease 7 (USP7) PLoS ONE. 2015;10:e0145023. doi: 10.1371/journal.pone.0145023. [DOI] [PMC free article] [PubMed] [Google Scholar]