Abstract
We evaluated the antioxidant activity and anti-melanogenic effects of Oenothera laciniata methanol extract (OLME) in vitro by using melan-a cells. The total polyphenol and flavonoid content of OLME was 66.3 and 19.0 mg/g, respectively. The electron-donating ability, 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) (ABTS) radical-scavenging activity, and superoxide dismutase (SOD)-like activity of OLME (500 μg/mL) were 94.5%, 95.6%, and 63.6%, respectively. OLME and arbutin treatment at 50 μg/mL significantly decreased melanin content by 35.5% and 14.2%, respectively, compared to control (p < 0.05). OLME and arbutin treatment at 50 μg/mL significantly inhibited intra-cellular tyrosinase activity by 22.6% and 12.6%, respectively, compared to control (p < 0.05). OLME (50 μg/mL) significantly decreased tyrosinase, tyrosinase-related protein-1 (TRP-1), TRP-2, and microphthalmia-associated transcription factor-M (MITF-M) mRNA expression by 57.1%, 67.3%, 99.0%, and 77.0%, respectively, compared to control (p < 0.05). Arbutin (50 μg/mL) significantly decreased tyrosinase, TRP-1, and TRP-2 mRNA expression by 24.2%, 42.9%, and 48.5%, respectively, compared to control (p < 0.05). However, arbutin (50 μg/mL) did not affect MITF-M mRNA expression. Taken together, OLME showed a good antioxidant activity and anti-melanogenic effect in melan-a cells that was superior to that of arbutin, a well-known skin-whitening agent. The potential mechanism underlying the anti-melanogenic effect of OLME was inhibition of tyrosinase activity and down-regulation of tyrosinase, TRP-1, TRP-2, and MITF-M mRNA expression.
Keywords: Anti-melanogenic effect, Antioxidant activity, Melan-a cells, Tyrosinase activity, Tyrosinase-related protein
INTRODUCTION
Melanin is one of the mostly widely distributed pigments, and is found in bacteria, fungi, plants, and animals. Increased production and accumulation of melanin is a characteristic of many skin diseases, including acquired hyperpigmentation, melasma, postinflammatory melanoderma, and solar lentigo (1). Melanin production occurs predominantly in a lysosome-like structure known as the melanosome in melanocytes (2). Melanin synthesis occurs with the participation of the tyrosinase family of proteins, including tyrosinase, tyrosinase-related protein-1 (TRP-1), and tyrosinase-related protein-2 (TRP-2)/dopachrome tautomerase (DCT) (3).
Tyrosinase is a key enzyme that catalyzes the synthesis of melanin in melanocytes (4). The biochemical pathway begins with two early reactions catalyzed by tyrosinase: the hydroxylation of l-tyrosine to l-3,4-dihydroxyphenylalanine (l-DOPA) and the oxidation of l-DOPA to dopaquinone. Dopaquinone spontaneously converts to dopachrome. TRP-2 catalyzes the oxidation of dopachrome to 5,6-dihydroxyindole-2-carboxylic acid (DHICA) and TRP-1 catalyzes the oxidation of DHICA to indole-5,6-quinone-2-carboxylic acid (5). Microphthalmia-associated transcription factor (MITF) consists of at least five isomers with distinct amino-termini, including MITF-A, MITF-B, MITF-C, MITF-H, and MITF-M (6). In particular, M promoter has the melanocyte-specific promoter function (7).
Tyrosinase inhibitors, including hydroquinone, kojic acid, and arbutin, have been used for treatment of hyperpigmentation disorders, but have recently been found to be unsafe for use in humans (8,9). Therefore, the use of natural products for depigmentation has recently been the focus of much attention.
Evening primrose (Oenothera odorata Jacquin) is a biennial herb that belongs to the Onagraceae family. This plant is native to South America and four species, including O. biennis L., O. erythrosepala Borbas, O. laciniata Hill, and O. stricta Ledeb, are wild-growing plants in Korea. The root or whole evening primrose plant has been used in Chinese medicine as a medicinal plant and by American Indians for inflammation and skin rash or dry cough. Evening primrose has also been used to control hypertension and blood cholesterol levels and to prevent or relieve myocardial infarction, rheumatoid arthritis, obesity, multiple sclerosis, eczema, acne, hyperactivity in children, premenstrual discomfort, alcoholism, and many other conditions (10). In addition, antitumor and antiviral components, including the oenotheins B and C, which are tannins, have been isolated from O. erythrosepala Borbas (11). O. laciniata (O. laciniata Hill) is a species endemic to Jeju Island, Korea with a seaside habitat. O. laciniata extract has been reported to contain bioactive substances with antioxidative activity, cytotoxic activity against a leukemia cell line, and antibacterial activity (12,13).
In our recent study (14), we found that O. laciniata methanol extract (OLME) showed anti-aging activity in human dermal fibroblasts. In the present study, the whitening effect of OLME was evaluated regarding melanin synthetic ability, tyrosinase activity, and gene expression of melanogenic enzymes and related factors in melan-a cells.
MATERIALS AND METHODS
Reagents and apparatus
Dimethyl sulfoxide (DMSO), 2,6-di-tert-butylate hydroxytoluene (BHT), l-DOPA, 1,1-diphenyl-2-picryl hydrazyl (DPPH), tannic acid, L-tyrosine, Folin-Ciocalteu’s phenol reagent, 3-(4,5-dimethyl-thiazol-2-yl)-2,5-diphenyl-tetrazolium bromide (MTT), MITF-M, TRP-1 and TRP-2 were obtained from Amersham Company (Bucks, UK). 2,2′-Azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) (ABTS) and 12-o-tetradecanoyl-phorbol-13-acetate (TPA) were obtained from Sigma-Aldrich (St. Louis, MO, USA). Dulbecco’s modified Eagle’s medium (DMEM), Roswell Park Memorial Institute (RPMI)-1640 medium, fetal bovine serum (FBS), and penicillin/streptomycin mixture (P/S) were obtained from Lonza (Cascade, MD, USA). An inverted microscope (CKX41, Olympus, Tokyo, Japan) was used to observe cell growth and a CO2 incubator (MCO-17AC, Sanyo Electric, Osaka, Japan) was used for cell culture.
Preparation of OLME
Methanol extract of the O. laciniata plant was obtained from the Korea Plant Extract Bank (Daejeon, Korea). Whole O. laciniata plants were collected from Sanyang-ri, Hangyeong-myeon, Buk Jeju-gun, Jeju-city, Korea. Pulverized samples (70 g) were added to a flask and subjected to extraction thrice for 24 hr each time with 1 L of 100% methanol at 45°C. The extract was filtered through filter paper, concentrated using a rotary vacuum evaporator, and dried. The yield of the O. laciniata methanol extract was 12.26%. This specimen was dissolved in DMSO before use.
Total polyphenol content
The total polyphenol content of OLME was measured using the Folin and Denis assay (15). OLME samples were dissolved in 1 mL of distilled water and placed in test tubes. Folin-reagent (1 mL) was added and the tubes were left to stand for 3 min. Adding 1 mL of 10% Na2CO3, the mixture was shaken vigorously and allowed to stand for 60 min. The absorbance was measured at 725 nm and the polyphenol content was quantified using a standard curve prepared using tannic acid.
Total flavonoid content
The total flavonoid content of OLME was measured using the method described by Davies et al. (16) with modification. OLME samples (100 μL) were mixed with 1 mL of di-ethylene glycol reagent and 100 μL of 1 N NaOH in test tubes. The mixture was shaken vigorously and allowed to react at 37°C for 60 min before the absorbance was measured at 420 nm. The standard curve used to quantify flavonoid content was prepared using rutin.
Electron-donating ability
The DPPH radical-scavenging effect was evaluated using the method described by Blois (17). OLME was dissolved in distilled water to a range of final concentrations (100, 500, and 1000 μg/mL). Test agent solution (1 mL) was placed in each test tube, followed by the addition of 4 mL of 4 × 10−4 M DPPH. The mixture was shaken vigorously, then left for 10 sec at 60°C before the absorbance was measured at 525 nm. BHT was used as the positive control.
ABTS radical-scavenging ability
Antioxidant activity was measured with the ABTS radical by using the ABTS+ cation decolorization assay method (18). ABTS+ was produced by mixing 7 mM ABTS with 2.4 mM of potassium peroxodisulfate (final concentration) and allowing the mixture to stand in the dark at room temperature for 12 hr. Thereafter, the mixture was diluted with ethanol to an absorbance of 0.7 (± 0.02) at 734 nm. After preparing 100 μg/mL samples at the targeted concentration in a 96-well plate, the diluted 100 μg/mL ABTS was added to the 96-well plate. The plate was then allowed to stand in the dark at room temperature for 7 min and the percentage of inhibition of absorbance at 734 nm was calculated.
Superoxide dismutase (SOD)-like activity
SOD-like activity was measured using the method described by Marklund-Marklund (19). A solution of 0.2 mL of sample and 2.6 mL of tris-HCl buffer (50 mM tris + 10 mM EDTA, pH 8.5) was allowed to react at 25°C for 10 min. Thereafter, 0.1 mL of 1 M HCl was added to stop the reaction. The amount of oxidized pyrogallol in the solution was measured at an absorbance of 420 nm.
Cell culture
The melan-a cells (passages 30–37) used in this study were obtained from Dr. Dorothy Bennett (St. George’s Hospital, London, UK). These highly pigmented and immortalized cells were derived from C57BL/6 mice. Cells were grown in RPMI-1640 medium, supplemented with 10% FBS, 1% P/S, and 200 nM TPA at 37°C and 10% CO2 in an incubator for 72 hr.
MTT assay
Cell viability was assessed using an MTT assay. Melan-a cells were seeded in a 96-well plate (0.5 × 104 cells/well) and grown in the incubator at 37°C and 10% CO2 for 24 hr. Aliquots (200 μL) of OLME, diluted with RPMI-1640 medium to a range of concentrations (25, 50, 100, and 200 μg/mL), were added to the wells and the cells were grown in the incubator at 37°C and 10% CO2 for 48 hr. Cells were then placed in a medium containing 0.5 μg/mL MTT and grown in the incubator at 37°C and 10% CO2 for 3 hr. Following centrifugation of the plate at 200 ×g for 10 min, the cells settled and the medium was removed. DMSO (200 μL) was added to each well and the cells were resuspended for 15 min using a plate-shaker. Absorbance was measured at 540 nm by using a plate reader (680, Bio-Rad, Tokyo, Japan).
Melanin assay
Melan-a cells were distributed in a 48-well plate (2 × 104 cells/well) and grown in the incubator at 37°C and 10% CO2 for 24 hr. Following incubation, 500 μL of OLME diluted with RPMI-1640 medium to a range of concentrations (12.5, 25, 50, and 100 μg/mL) were added to the wells and the cells were grown in the incubator at 37°C and 10% CO2 for 72 hr. The cells were washed and the same treatment was repeated once. Thereafter, melanin was dissolved in 1 N NaOH and the absorbance was measured at 490 nm by using a plate reader.
Intra-cellular tyrosinase activity assay
Melan-a cells were distributed in a 60-mm cell culture dish (4 × 105 cells/well) and grown in the incubator at 37°C and 10% CO2 for 24 hr. OLME (5 mL) was diluted with RPMI-1640 medium to a range of concentrations (12.5, 25, 50, and 100 μg/mL) and added to the dish. Cells were grown in the incubator at 37°C and 10% CO2 for 48 hr. The cells were washed with phosphate-buffered saline (PBS), detached with 200 μL of 1% Triton X-100, transferred to Eppendorf tubes, subjected to extraction on ice with agitation six times every 10 min, and then centrifuged at 4°C and 18,000 ×g for 20 min. Following centrifugation, 100 μL of 0.2% L-DOPA was added and the mixture was placed in the incubator at 37°C and 10% CO2 for 1 hr. Absorbance was measured at 490 nm by using a plate reader.
Reverse transcription-polymerase chain reaction (RT-PCR)
Total RNA was isolated using Trizol-Reagent (Invitrogen, New York, NY, USA) according to the manufacturer’s instructions. Total RNA (5 μg) was reverse transcribed using 8 μL of Molony murine leukemia virus RT (M-MLV RT) 5 × buffer, 3 μL of 10 mM dNTPs, 0.45 μL of 40 U/μL RNasein inhibitor, 0.3 μL of 200 U/μL M-MLV RT (Promega, Madison, WI, USA), and 1.5 μL of 50 μM oligo dT (Bioneer, Daejeon, Korea) in a 40 μL volume. Single-stranded cDNA was then amplified by PCR using 4 μL of 5 × green Go Taq flexi buffer, 0.4 μL of 10 mM dNTPs, 0.1 μL of 5 U/μL Taq polymerase, 1.2 μL of 25 mM MgCl2 (Promega), and 0.4 μL of 20 μM of specific sense and anti-sense primers of tyrosinase, TRP-1, TRP-2, MITF-M, or β-actin. The primer sequences used for PCR were as follows: tyrosinase forward 5′-CAT TTT TGA TTT GAG TGT CT-3′, reverse 5′-TGT GGT AGT CGT CTT TGT CC-3′; TRP-1 forward 5′-GCT GCA GGA GCC TTC TTT CTC-3′, reverse 5′-AAG ACG CTG CAC TGC TGG TCT-3′; TRP-2 forward 5′-GGA TGA CCG TGA GCA ATG GCC-3′, reverse 5′-CGG TTG TGA CCA ATG GGT GCC-3′; MITF-M forward 5′-TAC AGA AAG TAG AGG GAG GAG GAC TAA G-3′, reverse 5′-CAC AGT TGG AGT TAA GAG TGA GCA TAG CC-3′; β-actin forward 5′-ACC GTG AAA AGA TGA CCC AG-3′, reverse 5′-TAC GGA TGT CAA CGT CAC AC-3′. The expected sizes of the PCR products of tyrosinase, TRP-1, TRP-2, MITF-M, and β-actin, respectively, were 1192, 268, 1044, 326, and 528 base pairs. The PCR conditions were as follows: tyrosinase and TRP-1, 28 cycles of denaturation at 94°C for 60 sec, annealing at 56°C for 60 sec, and extension at 72°C for 60 sec; TRP-2, 28 cycles of denaturation at 94°C for 60 sec, annealing at 64°C for 60 sec, and extension at 72°C for 60 sec; MITF-M, 30 cycles of denaturation at 94°C for 30 sec, annealing at 54°C for 30 sec, and extension at 72°C for 30 sec; β-actin, 30 cycles of denaturation at 94°C for 30 sec, annealing at 51°C for 30 sec, and extension at 72°C for 60 sec. The PCR products were analyzed on 1.2% agarose gel. β-Actin was used as an internal control to evaluate the relative expression of tyrosinase, TRP-1, TRP-2, and MITF-M.
Statistical analysis
Statistical analysis was performed using SPSS 19.0 for Windows (IBM, Armonk, NY, USA). Differences between the groups were evaluated using one-way analysis of variance (ANOVA) followed by Duncan’s multiple range test for a post hoc comparison. Statistical significance was set at p < 0.05.
RESULTS
Antioxidant activity of OLME
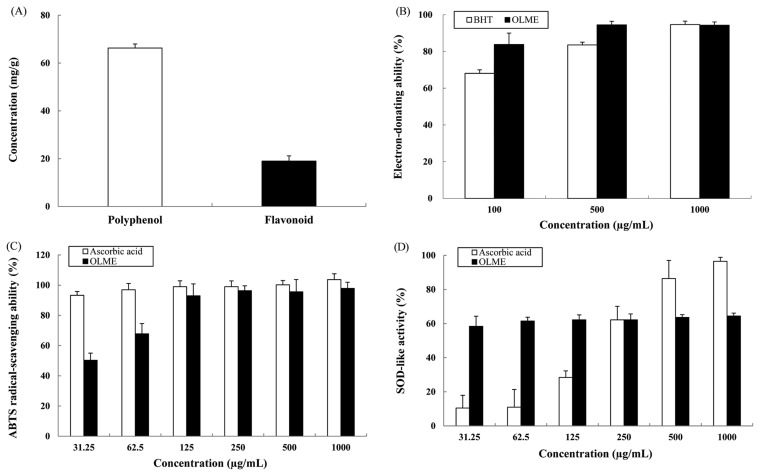
The total polyphenol content of OLME was 66.3 ± 1.9 mg/g according to the standard curve obtained using tannic acid (Fig. 1A). The total flavonoid content of OLME was 19.0 ± 2.3 mg/g according to the standard linear curve constructed using rutin (Fig. 1A). The electron-donating ability of BHT (positive control, PC) at 500 and 1000 μg/mL was 83.6% and 94.7%, respectively, whereas the electron-donating ability of OLME at 500 and 1000 μg/mL was 94.5% and 94.3%, respectively. The electron-donating ability of OLME was similar to or better than that of BHT (Fig. 1B). The ABTS radical-scavenging ability of ascorbic acid (PC) at 500 and 1000 μg/mL was 100.2% and 103.7%, respectively, whereas the ABTS radical-scavenging ability of OLME at 500 and 1000 μg/mL was 95.6% and 97.9%, respectively. The ABTS radical-scavenging ability of OLME was relatively lower than that of ascorbic acid (Fig. 1C). The SOD-like activity of ascorbic acid (PC) at 500 and 1000 μg/mL was 86.4% and 96.5%, respectively, whereas the SOD-like activity of OLME at 500 and 1000 μg/mL was 63.6% and 64.4%, respectively. The SOD-like activity of OLME was lower than that of ascorbic acid at the higher concentration (500 and 1000 μg/mL). However, the SOD-like activity of OLME was higher than that of ascorbic acid at the lower concentration (31.25~250 μg/mL), showing a concentration-independent reaction (Fig. 1D).
Fig. 1.
Antioxidant activity of Oenothera laciniata methanol extract. (A) Total polyphenol and flavonoid contents. (B) Electron-donating ability. (C) ABTS radical-scavenging ability. (D) SOD-like activity. BHT: 2,6-di-tert-butylate hydroxytoluene, OLME: Oenothera laciniata methanol extract. Values are the mean ± SD of three independent experiments.
Effect of OLME on melan-a cell viability
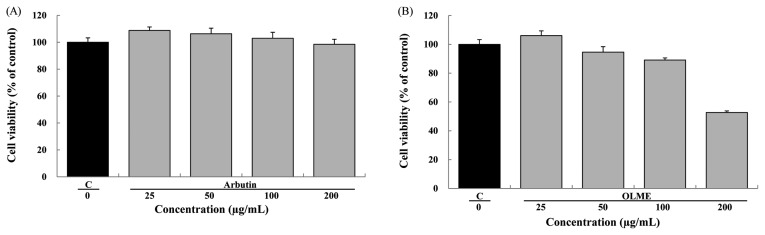
An MTT assay was used to determine the maximum permissible level (MPL) of OLME in melan-a cells. Arbutin (PC) did not reduce cell viability at concentrations between 25 and 200 μg/mL and cell viability remained above 98%. The MPL for arbutin application to melan-a cells was greater than 200 μg/mL (Fig. 2A). Similarly, OLME did not reduce cell viability at concentrations between 25 and 100 μg/mL and cell viability remained above 89%. However, cell viability was reduced to 53% at 200 μg/mL of OLME. The MPL for OLME application to melan-a cells was 100 μg/mL (Fig. 2B). Arbutin treatment of melan-a cells at 200 μg/mL resulted in no diminution of dendrites or melanin deposition compared to control (Figure not shown). OLME treatment of melan-a cells at 200 μg/mL resulted in more diminution of dendrites and melanin deposition compared to control (Figure not shown).
Fig. 2.
Effect of test materials on the viability of melan-a cells. Viability of melan-a cells treated with arbutin (A) and Oenothera laciniata methanol extract (B) (0~200 μg/mL) for 48 hr was analyzed using MTT assay. C: control, OLME: Oenothera laciniata methanol extract. Values are the mean ± SD of three independent experiments.
Effect of OLME on melanin synthesis
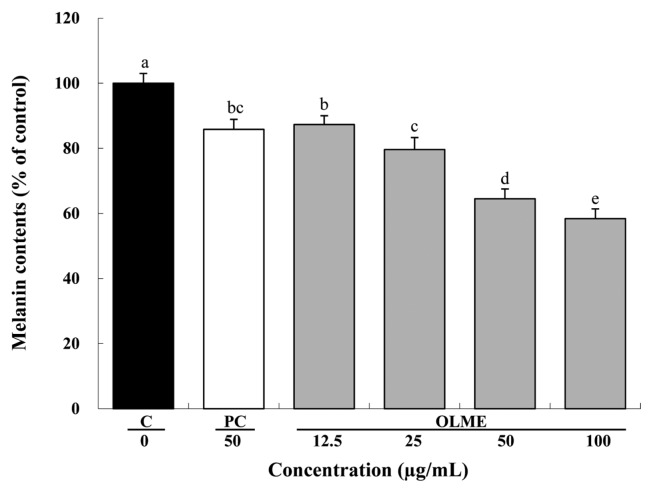
Compared to control, OLME treatment at 12.5, 25, 50, and 100 μg/mL significantly decreased melanin content by 12.7%, 20.4%, 35.5%, and 41.6%, respectively, in a dose-dependent manner (p < 0.05). Arbutin treatment (PC) at 50 μg/mL also significantly decreased melanin content by 14.2% compared to control (p < 0.05). Compared to arbutin, OLME treatment at 50 μg/mL resulted in significantly lower melanin content (p < 0.05) (Fig. 3).
Fig. 3.
Anti-melanogenic effect of Oenothera laciniata methanol extract. Melanin production in melan-a cells treated with the test materials for 72 hr was measured with an ELISA. C: control, PC: arbutin, OLME: Oenothera laciniata methanol extract. Values are the mean ± SD of three independent experiments. Statistically significant differences (p < 0.05) identified using an ANOVA and Duncan’s multiple range test are indicated by different letters (a>b>c>d>e).
Effect of OLME on tyrosinase activity
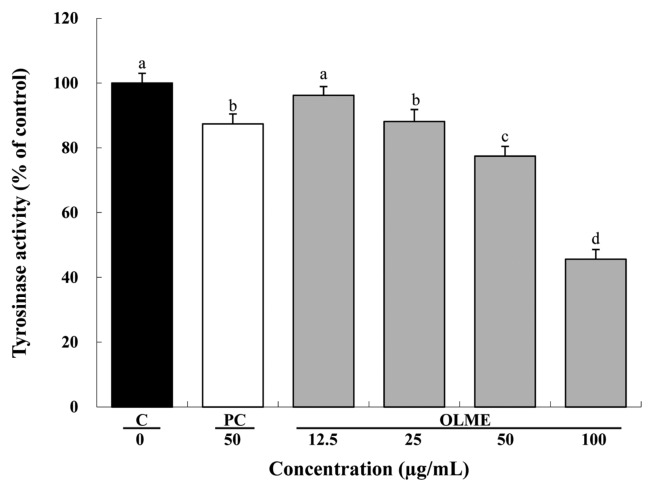
Compared to control, OLME treatment at 12.5, 25, 50, and 100 μg/mL decreased tyrosinase activity in a dose-dependent manner by 3.8%, 11.9% (p < 0.05), 22.6% (p < 0.05), and 54.4% (p < 0.05), respectively. Arbutin treatment (PC) at 50 μg/mL also significantly decreased tyrosinase activity by 12.6% compared to control (p < 0.05). Compared to arbutin, OLME treatment at 50 μg/mL significantly decreased tyrosinase activity (p < 0.05) (Fig. 4).
Fig. 4.
Effect of Oenothera laciniata methanol extract on intracellular tyrosinase activity in melan-a cells. Tyrosinase activity in the cells treated with the test materials for 48 hr was measured with an ELISA. C: control, PC: arbutin, OLME: Oenothera laciniata methanol extract. Values are the mean ± SD of three independent experiments. Statistically significant differences (p<0.05) identified using an ANOVA and Duncan’s multiple range test are indicated by different letters (a>b>c>d).
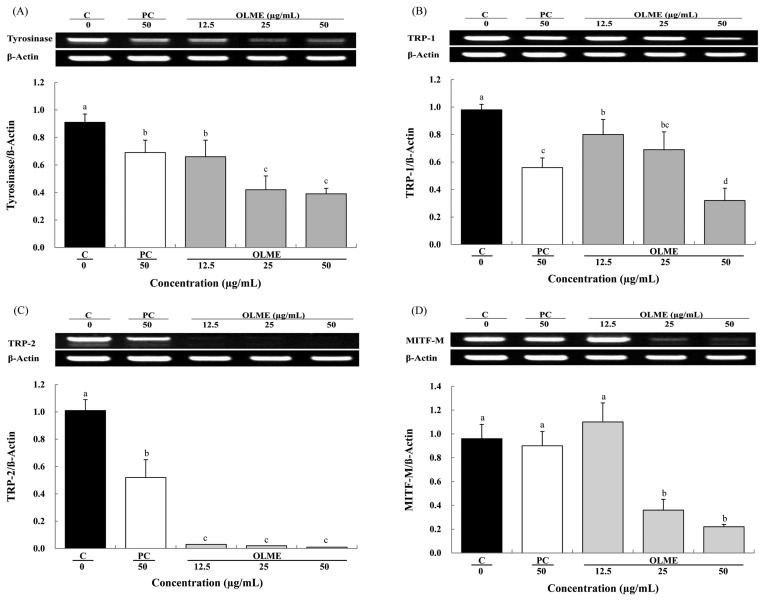
Effect of OLME on tyrosinase, TRP-1, TRP-2, and MITF-M mRNA expression
We evaluated the mRNA expression of tyrosinase, TRP-1, TRP-2, and MITF-M after treatment with OLME at 12.5, 25, and 50 μg/mL and arbutin at 50 μg/mL. Compared to control, OLME significantly decreased tyrosinase mRNA expression in a dose-dependent manner by 27.5%, 53.8%, and 57.1%, respectively (p < 0.05). Arbutin (PC) also significantly decreased tyrosinase mRNA expression by 24.2% compared to control (p < 0.05). Compared to arbutin, OLME treatment at 50 μg/mL resulted in significantly lower tyrosinase mRNA expression (p < 0.05) (Fig. 5A). OLME significantly decreased TRP-1 mRNA expression in a dose-dependent manner by 18.4%, 29.6%, and 67.3%, respectively (p < 0.05). Arbutin (PC) also significantly decreased TRP-1 mRNA expression by 42.9% (p < 0.05). Compared to arbutin, OLME treatment at 50 μg/mL resulted in significantly lower TRP-1 mRNA expression (p < 0.05) (Fig. 5B). OLME significantly decreased TRP-2 mRNA expression by 97.0%, 98.0%, and 99.0%, respectively (p < 0.05). Arbutin (PC) also significantly decreased TRP-2 mRNA expression by 48.5% (p < 0.05). Compared to arbutin, OLME treatment at 50 μg/mL resulted in significantly lower TRP-2 mRNA expression (p < 0.05) (Fig. 5C). OLME treatment at 12.5 μg/mL increased MITF-M mRNA expression by 14.5%, whereas OLME treatment at 25 and 50 μg/mL significantly decreased MITF-M mRNA expression by 62.5% and 77.0%, respectively (p < 0.05). Arbutin treatment (PC) at 50 μg/mL decreased MITF-M mRNA expression by 6%. Compared to arbutin, OLME treatment at 50 μg/mL resulted in significantly lower MITF-M mRNA expression (p < 0.05) (Fig. 5D).
Fig. 5.
Effect of Oenothera laciniata methanol extract on mRNA expression of tyrosinase, TRP-1, TRP-2, and MITF-M in melan-a cells. mRNA expression in the cells treated with the test materials for 48 hr was analyzed using RT-PCR. (A) A representative gel (upper) and semi-quantification of tyrosinase mRNA expression by normalization of β-actin (lower). (B) A representative gel (upper) and semi-quantification of TRP-1 mRNA expression by normalization of β-actin (lower). (C) A representative gel (upper) and semi-quantification of TRP-2 mRNA expression by normalization of β-actin (lower). (D) A representative gel (upper) and semi-quantification of MITF-M mRNA expression by normalization of β-actin (lower). C: control, PC: arbutin, OLME: Oenothera laciniata methanol extract. Values are the mean ± SD of three independent experiments. Statistically significant differences (p < 0.05) identified using an ANOVA and Duncan’s multiple range test are indicated by different letters (a > b > c > d).
DISCUSSION
Antioxidants protect against potentially damaging oxidative stress, which is the result of an imbalance between the formation of reactive oxygen species (ROS) and the body’s antioxidant defense. Plant polyphenols (PPs), or flavonoids, have been shown to act as antioxidants because of their capacity to scavenge free radicals, chelate metal ions, and prevent free radical formation (20). PPs are plant metabolites characterized by the presence of several phenol groups (i.e., aromatic rings with hydroxyls) that are derived from L-phenylalanine (21). As a polyphenol compound, a flavonoid has antioxidant, anti-inflammatory, and antiviral effects (22). In this study, the total amount of polyphenol compounds and flavonoid compounds in OLME was 66.3 mg/g and 19.0 mg/g, respectively.
To determine antioxidant activity, many tests have been used to accelerate oxidative conditions that provoke lipid oxidation by means of both a high temperature and a high oxygen supply. DPPH used as an index of active oxygen activity is one of the experimental methods used to generally evaluate the erasure reaction of the free radical that receives electrons from lipid peroxidation and implies a color according to the redox reaction (23). In this study, the electron-donating ability of OLME at 500 μg/mL was 94.5%, which was superior to that of BHT (83.6%), the positive control. The extent of decolorization as a percentage of the inhibition of the ABTS radical cation was determined as a function of concentration and time and calculated relative to the reactivity of Trolox as a standard, under the same conditions (18). The ABTS radical-scavenging ability of OLME at 500 μg/mL was 95.6%, slightly lower than that of ascorbic acid (100.2%), the positive control. SOD converts the superoxide radical into hydrogen peroxide in the organism, acting to prevent oxidation and aging. SOD, catalase, and glutathione peroxidase are antioxidative enzymes that are associated with important defensive mechanisms that protect from the insults of solar damage (24). The SOD-like activity of OLME at 500 μg/mL was 63.6%, lower than that of ascorbic acid (86.4%), the positive control. These results confirmed that OLME has good antioxidative ability.
Hyperpigmentation disorders, including melasma, freckles, and senile lentigines, are associated with abnormal accumulation of melanin, and these disorders have a psychosocial and cosmetic impact (25). Because OLME distinguishably inhibited melanin synthesis and dendrite development upon gross observation, quantitative analysis of the melanin synthetic ability of OLME in melan-a cells was conducted. To investigate the cytotoxicity of OLME in melan-a cells, an MTT assay was carried out, which indicated that the MPL of OLME was 100 μg/mL. Compared to control group, the melanin content in the OLME treatment group (12.5~100 μg/mL) was significantly reduced (p < 0.05). This agreed with the results of gross observation. Compared to control group, arbutin at 50 μg/mL decreased melanin content by 14.2% (p < 0.05), whereas OLME at 50 μg/mL decreased melanin content by 35.5% (p < 0.05). Therefore, OLME more effectively inhibited melanin synthesis in melan-a cells than did arbutin, the positive control. These results clearly indicated that OLME may be effective as a whitening agent.
Tyrosinase is the core enzyme in the melanin generation process from l-tyrosine. Therefore, the inhibition of tyrosinase is closely related to the whitening effect of certain compounds. Compared to control group, arbutin at 50 μg/mL inhibited intra-cellular tyrosinase activity by 12.6% (p < 0.05), whereas OLME at 50 μg/mL inhibited intra-cellular tyrosinase activity by 22.6% (p < 0.05). We also obtained similar values of cell-extracted tyrosinase activity for arbutin and OLME (data not shown). These results indicated that OLME regulated tyrosinase activity and subsequently inhibited melanin synthesis in melanocytes. OLME more effectively inhibited tyrosinase activity than arbutin, the positive control. Because antioxidants could be good inhibitors of tyrosinase (26), the anti-melanogenic effect of OLME might be contributed to its potential antioxidative ability. Tyrosinase is a glycoprotein located in the melanosomal membrane (27). It has an inner melanosomal domain that contains the catalytic region (approximately 90% of the protein), followed by a short trans-membrane domain and a cytoplasmic domain composed of approximately 30 amino acids (28). In addition, the enzyme phenylalanine hydroxlyase contributes to tyrosinase activation because it catalyzes the conversion of l-phenylalanine to l-tyrosine, providing a substrate for tyrosinase (29).
TRPs are also considered important melanogenic enzymes and at least two isozymes of TRP, TRP-1 (DHICA oxidase) and TRP-2 (DOPAchrome tautomerase), have been identified. TRP-1 and TRP-2 reside within melanosomes and, like tyrosinase, span the melanosomal membrane (30). Mutations of TRP-1, such as those present in a specific type of oculocutaneous albinism, result in pale skin and hair color and demonstrate that TRP-1 is required for optimal melanin synthesis (31). TRP-2 converts DOPAchrome to the carboxylated derivative, DHICA, during one of the later stages of melanin biosynthesis (5). MITF, a master transcriptional regulator for melanogenic enzymes, is involved in the differentiation, growth, and survival of pigment cells (32). MITF-M transactivates tyrosinase, TRP-1, and TRP-2 melanogenic genes in vitro via binding to an M-box motif present in the promoters of those genes (33). Therefore, the effects of OLME on the expression of melanogenic enzymes and factors were determined at mRNA levels. Compared to control group, tyrosinase mRNA expression in the arbutin and OLME treatment groups at 50 μg/mL concentration significantly decreased by 24.2% and 57.1%, respectively (p < 0.05). The expression of TRP-1, TRP-2, and MITF-M mRNA in the arbutin and OLME treatment groups at 50 μg/mL concentration significantly decreased compared to those in the control group (p < 0.05) and the values for the OLME treatment group were lower than those for the arbutin treatment group. Therefore, it was confirmed that OLME more effectively down-regulated the gene expression of tyrosinase, TRP-1, TRP-2, and MITF-M than did arbutin in melan-a cells. The increase in MITF-M expression induces the up-regulation of the tyrosinase gene family, which leads to increased melanin synthesis (32). In the current study, we also found that OLME decreased MITF-M expression, which induced the down-regulation of tyrosinase, TRP-1, and TRP-2 transcription.
In conclusion, OLME suppressed melanin synthesis in melan-a cells by inhibiting tyrosinase activity and the mRNA expression of tyrosinase, TRP-1, TRP-2, and MITF-M. The potential anti-melanogenic ingredients in OLME need to be further investigated.
REFERENCES
- 1.Kang HY, Yoon TJ, Lee GJ. Whitening effects of marine Pesudomonas extract. Ann Dermatol. 2011;23:144–149. doi: 10.5021/ad.2011.23.2.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hearing VJ. Biogenesis of pigment granules: a sensitive way to regulatemelanocyte function. J Dermatol Sci. 2005;37:3–14. doi: 10.1016/j.jdermsci.2004.08.014. [DOI] [PubMed] [Google Scholar]
- 3.Huang YH, Lee TH, Chan KJ, Hsu FL, Wu YC, Lee MH. Anemonin is a natural bioactive compound that can regulate tyrosinase-related proteins and mRNA in human melanocytes. J Dermatol Sci. 2008;49:115–123. doi: 10.1016/j.jdermsci.2007.07.008. [DOI] [PubMed] [Google Scholar]
- 4.Sturm RA, Teasdale RD, Box NF. Human pigmentation genes: identification, structure and consequences of polymorphic variation. Gene. 2001;277:49–62. doi: 10.1016/S0378-1119(01)00694-1. [DOI] [PubMed] [Google Scholar]
- 5.Mallick S, Singh SK, Sarkar C, Saha B, Bhadra R. Human placental lipid induces melanogenesis by increasing the expression of tyrosinase and itsrelated proteins in vitro. Pigment Cell Res. 2005;18:25–33. doi: 10.1111/j.1600-0749.2004.00193.x. [DOI] [PubMed] [Google Scholar]
- 6.Udono T, Yasumoto K, Takeda K, Amae S, Watanabe K, Saito H, Fuse N, Tachibana M, Takahashi K, Tamai M, Shibahara S. Structural organization of the human microphthalmia-associated transcription factor gene containingfour alternative promoters. Biochim Biophys Acta. 2000;1491:205–219. doi: 10.1016/S0167-4781(00)00051-8. [DOI] [PubMed] [Google Scholar]
- 7.Fuse N, Yasumoto K, Suzuki H, Takahashi K, Shibahara S. Identificationof a melanocyte-type promoter of the microphthalmia-associated transcription factor gene. Biochem Biophys Res Commun. 1996;219:702–707. doi: 10.1006/bbrc.1996.0298. [DOI] [PubMed] [Google Scholar]
- 8.Draelos ZD. Skin lightening preparations and the hydroquinone controversy. Dermatol Ther. 2007;20:308–313. doi: 10.1111/j.1529-8019.2007.00144.x. [DOI] [PubMed] [Google Scholar]
- 9.Lim JT. Treatment of melasma using kojic acid in a gel containing hydroquinone and glycolic acid. Dermatol Surg. 1999;25:282–284. doi: 10.1046/j.1524-4725.1999.08236.x. [DOI] [PubMed] [Google Scholar]
- 10.Varro ET, Lynn RB, James ER. Pharmacognosy. 9th edition. Lea and Febriger; Philadelphia: 1988. p. 471. [Google Scholar]
- 11.Hatano T, Yasuhara T, Fukuda T, Noro T, Okuda T. Phenolic constituents of licorice. II: Structures of licopyranocoumarin, licoarylcoumarin and glisoflavone, and inhibitory effects of licorice phenolics on xanthine oxidase. Chem Pharm Bull. 1989;37:3005–3009. doi: 10.1248/cpb.37.3005. [DOI] [PubMed] [Google Scholar]
- 12.Kim JY, Lee JA, Park SY. Antibacterial activities of Oenothera laciniata extracts. J Korean Soc Food Sci Nutr. 2007;36:255–259. doi: 10.3746/jkfn.2007.36.3.255. [DOI] [Google Scholar]
- 13.Lee JA, Kim JY, Yoon WJ, Oh DJ, Jung YH, Lee WJ, Park SY. Biological activities of Oenothera laciniata extracts (Onagraceae, Myrtales) Korean J Food Sci Technol. 2006;38:810–815. [Google Scholar]
- 14.Kim SE, Lee CM, Kim YC. Anti-wrinkle efficacy of Oenothera laciniata methanol extract in human dermal fibroblasts. J Invest Cosmetol. 2016;12:197–203. doi: 10.15810/jic.2016.12.3.001. [DOI] [Google Scholar]
- 15.Folin O, Denis W. On phosphotungstic-phosphomolybdic compounds as color reagents. J Biol Chem. 1912;12:239–243. [Google Scholar]
- 16.Davies R, Massey RC, Mcweeny DJ. The catalisis of the N-nitrosation of secondary amines by nitrosophenols. Food Chem. 1980;6:115–122. doi: 10.1016/0308-8146(80)90027-8. [DOI] [Google Scholar]
- 17.Blois MS. Antioxidant determinations by the use of a stable freeradical. Nature. 1958;181:1199–1200. doi: 10.1038/1811199a0. [DOI] [Google Scholar]
- 18.Re R, Pellegrini N, Proteggente A, Pannala A, Yang M, Rice-Evans C. Antioxidant activity applying an improved ABTS radical cation decolorization assay. Free Radic Biol Med. 1999;26:1231–1237. doi: 10.1016/S0891-5849(98)00315-3. [DOI] [PubMed] [Google Scholar]
- 19.Marklund S, Marklund G. Involvement of the superoxide anion radical in the autoxidation of pyrogallol and a convenient assay for superoxide dismutase. Eur J Biochem. 1974;47:469–474. doi: 10.1111/j.1432-1033.1974.tb03714.x. [DOI] [PubMed] [Google Scholar]
- 20.Rice-Evans CA, Miller NJ, Bolwell PG, Bramley PM, Pridham JB. The relative antioxidant activities of plant-derived polyphenolic flavonoids. Free Radic Res. 1995;22:375–383. doi: 10.3109/10715769509145649. [DOI] [PubMed] [Google Scholar]
- 21.Boudet AM. Evolution and current status of research in phenolic compounds. Phytochemistry. 2007;68:2722–2735. doi: 10.1016/j.phytochem.2007.06.012. [DOI] [PubMed] [Google Scholar]
- 22.Seyoum A, Asres K, El-Fiky FK. Structure-radical scavenging activity relationships of flavonoids. Phytochemistry. 2006;67:2058–2070. doi: 10.1016/j.phytochem.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 23.Bondet V, Brand-Williams W, Berset C. Kinetics and mechanisms of antioxidant activity using the DPPH· free radical method. Food Sci Technol. 1997;30:609–615. [Google Scholar]
- 24.Kondo S. The roles of cytokines in photoaging. J Dermatol Sci. 2000;23:S30–S36. doi: 10.1016/S0923-1811(99)00076-6. [DOI] [PubMed] [Google Scholar]
- 25.Kang HY, Yoon TJ, Lee GJ. Whitening effects of marine Pesudomonas extract. Ann Dermatol. 2011;23:144–149. doi: 10.5021/ad.2011.23.2.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chae GY, Ha BJ. The comparative evaluation of fermented and non-fermented soybean extract on antioxidation and whitening. Toxicol Res. 2011;27:205–209. doi: 10.5487/TR.2011.27.4.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park HY, Kosmadaki M, Yaar M, Gilchrest BA. Cellularmechanisms regulating human melanogenesis. Cell Mol Life Sci. 2009;66:1493–1506. doi: 10.1007/s00018-009-8703-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kwon BS, Haq AK, Pomerantz SH, Halaban R. Isolation and sequence of a cDNA clone for human tyrosinase that maps at the mouse c-albino locus. Proc Natl Acad Sci USA. 1987;84:7473–7477. doi: 10.1073/pnas.84.21.7473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schallreuter KU, Kothari S, Chavan B, Spencer JD. Regulation of melanogenesis-controversies and new concepts. Exp Dermatol. 2008;17:395–404. doi: 10.1111/j.1600-0625.2007.00675.x. [DOI] [PubMed] [Google Scholar]
- 30.del Marmol V, Beermann F. Tyrosinase and related proteins in mammalian pigmentation. FEBS Lett. 1996;381:165–168. doi: 10.1016/0014-5793(96)00109-3. [DOI] [PubMed] [Google Scholar]
- 31.Boissy RE, Zhao H, Oetting WS, Austin LM, Wildenberg SC, Boissy YL, Zhao Y, Sturm RA, Hearing VJ, King RA, Nordlund JJ. Mutation in and lack of expression of tyrosinase-related protein-1(TRP-1) in melanocytes from an individual with brown oculocutaneous albinism: a new subtype of albinism classified as “OCA3”. Am J Hum Genet. 1996;58:1145–1156. [PMC free article] [PubMed] [Google Scholar]
- 32.Levy C, Khaled M, Fisher DE. MITF: master-regulator of melanocyte development and melanoma oncogene. Trends Mol Med. 2006;12:406–414. doi: 10.1016/j.molmed.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 33.Park HY, Wu C, Yonemoto L, Murphy-Smith M, Wu H, Stachur CM, Gilchrest BA. MITF mediates cAMP-induced protein kinase C-β expression in human melanocytes. Biochem J. 2006;395:571–578. doi: 10.1042/BJ20051388. [DOI] [PMC free article] [PubMed] [Google Scholar]