Abstract
Purpose
The recent discovery and characterization of an oncogenic ROS1 gene rearrangement has raised significant interest because small molecule inhibitors are effective in these tumors. The aim of this study was to determine frequency and clinicopathological features associated with ROS1 rearrangement in patients with cholangiocarcinoma (CCA).
Materials and Methods
A total of 261 patients who underwent surgery for CCA between October 1997 and August 2013 were identified from an international, multi-institutional database. ROS1 rearrangement was evaluated by break-apart fluorescence in situ hybridization using tissue microarrays of these patients.
Results
Of 261 CCA evaluated, three cases (1.1%) showed ROS1 rearrangement by fluorescence in situ hybridization (FISH), all of which were derived from intrahepatic origin. ROS1 protein expression was observed in 38 samples (19.1%). Significantly larger tumor size was observed in ROS1 immunohistochemistry (IHC)–negative patients compared with ROS1 IHC–positive patients. ROS1 FISH–positive patients had a single tumor with a median size of 4 cm and well-to-moderate differentiation. Overall, there was no difference in terms of baseline characteristics, overall survival, and recurrence-free survival between ROS1-positive and -negative patients.
Conclusion
ROS1 rearrangement was detected in 1.1% of CCA patients. Although rare, conduct of clinical trials using ROS1 inhibitors in these genetically unique patients is warranted.
Keywords: Cholangiocarcinoma, ROS1, Fluorescent in situ hybridization
Introduction
Cholangiocarcinoma (CCA) is a lethal biliary tract cancer [1]. The age-adjusted incidence rate is 3.3 per 100,000 among Asians, and despite the relatively low incidence, the overall incidence of CCA is increasing worldwide [2,3]. CCA is generally regarded as a surgical disease, and surgical resection offers the only chance for long-term survival [4]. However, most patients are diagnosed with advanced-stage disease, which limits the treatment options. In addition, even after curative-intent surgery, the survival outcomes are disappointing with a 5-year survival rate of 20%-30% [5]. Combination chemotherapy with gemcitabine and a platinum agent is regarded as a standard first-line treatment [6]; however, the prognosis is still poor and overall survival (OS) remains less than 12 months. Therefore, efforts to improve outcomes for CCA by incorporating novel targeted therapeutics are urgently needed.
Contemporary research techniques have enabled identification of several markers involved in molecular pathogenesis of CCA [7]. In a recent study from Singapore, 206 somatic mutations were identified through exome sequencing of liver-fluke associated CCA [8]. TP53 accounted for the most common cancer-related gene (44%), followed by KRAS (17%) and SMAD4 (17%). Genetic alterations associated with deactivation of histone modifiers including ARID1A (9%) were also identified. However, none of these genetic alterations is amenable to targeted therapy. Recently, in the first phase 3 trial to assess a targeted therapy plus chemotherapy combination, improvement in OS was not observed by treatment with gemcitabine and oxaliplatin with or without erlotinib. Only a small improvement in median progression-free survival was observed in a subset of patients with CCA (5.9 months vs. 3.0 months) [9], and there was no distinct predictive biomarker for erlotinib response in this study.
ROS1 rearrangements were recently detected in CCA patient tissues, suggesting that this could play a role as a driver oncogene. The reported incidence of ROS1 rearrangement was 8.7% of CCA patients [10]. Interestingly, fusedin-glioblastoma-c-ros-oncogene 1 (FIG-ROS) fusion accelerated cholangiocarcinogenesis in mouse models [11], supporting ROS1 as a novel therapeutic target in CCA. ROS1 rearrangement was previously detected in other solid tumors including non-small-cell lung cancer (NSCLC) and glioblastoma [12-14]. To date, nine different ROS1 fusion partners have been identified, all of which are potentially targetable due to the intact cytoplasmic portion of the ROS1 tyrosine kinase domain [15]. Due to the biological similarity of ROS1 and anaplastic lymphoma kinase (ALK), inhibition of ROS1 by several ALK inhibitors has been demonstrated [16]. Recent data from a phase 1 trial of crizotinib in the ROS1-positive NSCLC expansion cohort showed an overall response rate of 72% [17]. Therefore, identification of ROS1 rearrangement in CCA could offer a new therapeutic option in treatment of this fatal disease.
The current study uses the largest data set yet generated of CCA. The aim of this study was to determine the frequency of ROS1 rearrangement and to evaluate clinicopathological features associated with ROS1 rearrangement in CCA patients.
Materials and Methods
1. Patients
A total of 261 patients with CCA who underwent surgical resection with curative intent between October 1997 and August 2013 were identified at two institutions (Yonsei University College of Medicine, Seoul, Korea; National University of Singapore, Singapore). The database was reviewed retrospectively and only patients with histologically confirmed CCA and available tissue for ROS1 analysis were included. Patient records/information was anonymized and de-identified prior to analysis. This study was approved by the Institutional Review Boards of Severance Hospital and Singapore General Hospital and the requirement for consent was waived.
Out of 261 patients, 242 patients (93%) were available for ROS1 analysis. Standard demographic and clinicopathologic data were collected, including sex, age, and primary tumor characteristics. Specifically, data were collected on primary tumor location, size, and number as well as morphologic subtype and presence of vascular invasion, biliary invasion, lymph node involvement, and stage. Staging was done according to the seventh edition of the American Joint Committee on Cancer/Union for International Cancer Control staging manual. Dates related to treatment including surgery, recurrence, and last follow-up were collected on all patients. OS was measured from the date of diagnosis until the date of death and recurrence-free survival (RFS) was measured from the date of surgery until the date of recurrence.
2. Tissue microarrays
The tumor samples were fixed in 10% buffered formalin, processed, and embedded in paraffin using the standard protocol [18]. All hematoxylin and eosin–stained slides were reviewed, and representative areas were carefully selected and marked on individual paraffin blocks. Three 3.0-mm tissues cores were taken from each tumor specimen.
3. ROS1 fluorescence in situ hybridization
For identification of ROS1 rearrangement, fluorescence in situ hybridization (FISH) assays were performed using a break-apart probe to ROS1 (Break-Apart Rearrangement Probe, Abbott Molecular, Des Plaines, IL) according to the manufacturer’s instructions using tissue microarrays. At least 100 nuclei of tumor cells per case were evaluated. FISH positivity for ROS1 rearrangement was defined as > 15% of tumor cells with a split signal or > 15% of single green signals. FISH studies were interpreted by two experienced evaluators (J.E.Y. and Y.N.P.) who were blinded to the clinical data.
4. Immunohistochemical staining
Protein expression of ROS1 was detected by immunohistochemical staining in the Korean cohort, and it was not performed in the Singapore cohort due to lack of unstained slides. Tissue microarray sections were stained using the Ventana automated immunostainer BenchMark XT (Ventana Medical Systems, Tucson, AZ). The slides were dried at 60°C for 1 hour and deparaffinized using EZ Prep (Ventana Medical Systems) at 75°C for 4 minutes. Cell conditioning was performed using CC1 solution (Ventana Medical Systems) at 100°C for 8 minutes. ROS1 antibody (rabbit monoclonal, clone D4D6, Cell Signaling Technology, Danvers, MA) was diluted to 1:10, followed by treatment, and incubation at 37°C for 2 hours. Signals were detected using the OptiView DAB IHC Detection Kit (Ventana Medical Systems). Counterstaining was performed using Hematoxylin I (Ventana Medical Systems) for 4 minutes at room temperature.
5. Statistical analysis
Statistics were obtained using established methods and presented as percentages, mean, or median values. Survival was estimated using the Kaplan-Meier method for the median and 95% confidence interval (CI); comparison between groups was performed using a two-sided log-rank test. Fisher exact test, t test, or Mann-Whitney U test were used for comparison of differences between groups. p-values of ≤ 0.05 were considered statistically significant. Statistical analyses were performed using SPSS ver. 20.0 (IBM Co., Armonk, NY).
Results
1. Patient characteristics
The clinicopathological features of 261 patients including 208 intrahepatic CCAs and 53 extrahepatic CCAs are shown in Table 1. The median age of all patients was 65.6 years and males (56%) were predominant compared with females (44%). The majority of patients presented with intrahepatic CCAs (80%), a single tumor (81%) with a median tumor size of 4.6 cm. The tumor was well-to-moderately differentiated in 80% of patients. Lymphadenectomy was performed in 209 patients (80%), and 67 patients (32%) presented with lymph node positive disease. The median preoperative carbohydrate antigen 19-9 level was 71.9 U/mL (range, 0 to 20,000 U/mL; reference value, ≤ 37 U/mL). In comparison of samples from the Korea and Singapore cohorts (Supplementary Table 1), no significant difference in clinicopathological features was observed between the two cohorts.
Table 1.
Demographic and clinical characteristics of patients
| Variable | No. (%) (n=261) |
|---|---|
| Age, median (range, yr) | 65.6 (33-91) |
| Sex | |
| Male | 148 (56) |
| Female | 113 (44) |
| Tumor location | |
| Intrahepatic | 208 (80) |
| Extrahepatic | 53 (20) |
| Tumor size, median (range, cm) | 4.6 (0.8-15) |
| Multiple tumors | 51 (19) |
| Tumor differentiation | |
| Well-moderately differentiated | 211 (80) |
| Poorly differentiated | 50 (20) |
| Lymph node disease | 67/209 (32) |
| CA19-9, median (range, U/mL) | 71.9 (0-20,000) |
| Stage | |
| I | 88 (34) |
| II | 55 (21) |
| III | 23 (9) |
| IV | 95 (36) |
CA19-9, carbohydrate antigen 19-9.
2. Break-apart FISH and immunohistochemical analysis of ROS1
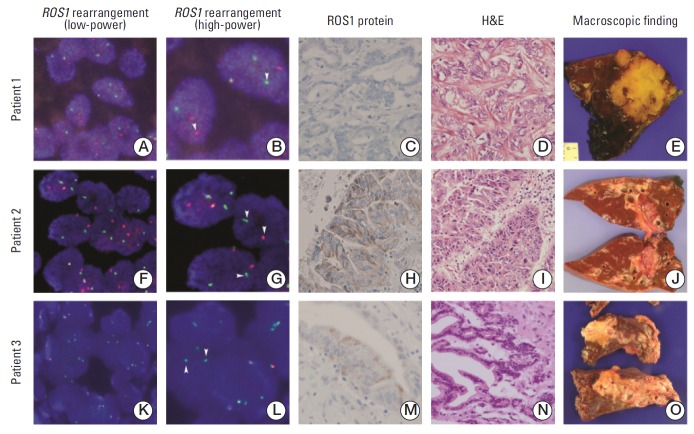
Among 261 patients screened for ROS1 rearrangement, 245 cases were analyzed by FISH (detection rate, 93.9%), and FISH signal was not detected in 16 cases due to sample quality. Three cases showed ROS1-positivity (1.2%) by FISH; patients 1 and 2 showed break-apart signals, while patient 3 showed a single signal as shown in Fig. 1. Comparison of ROS1-negative and -positive patients showed no significant differences in baseline demographic and clinical characteristics. For immunohistochemistry (IHC) analysis, 198 cases were available for analysis of ROS1 protein expression. ROS1 protein expression was observed in 38 samples (19.1%). Comparison of ROS1-negative and -positive patients showed a significant difference in tumor size, while there was no difference in other parameters. ROS1-negative patients had a significantly larger tumor size than ROS1-positive patients (p=0.025) (Supplementary Table 2). Of note, among three ROS1 FISH–positive cases, two patients (66.7%) showed ROS1 immunoreactivity (Fig. 1). The sensitivity for the detection of ROS1 rearranged CCA was 66.7% with 81.5% specificity.
Fig. 1.
Representative ROS1 rearrangement features in cholangiocarcimomas. Fluorescence in situ hybridization (FISH) showing ROS1 rearrangement with break-apart signals in patients 1 and 2 and single green signals in patient 3. FISH showing ROS1 rearrangement with break-apart signals in low-power field (A, F, K) and high-power field (B, G, L). FISH showing ROS1 rearrangement with single green signals (K, L). The arrows indicate ROS1 break-apart or single green signals (A, F, K, ×400; B, G, L, ×630). Immunohistochemical stain for ROS1 in ROS1 rearranged cholangiocarcinoma (C, H, M, ×200). Microscopic features (D, I, N, H&E staining, ×200) and photographs of surgical specimens (E, J, O) are also shown.
3. Clinical outcome of patients
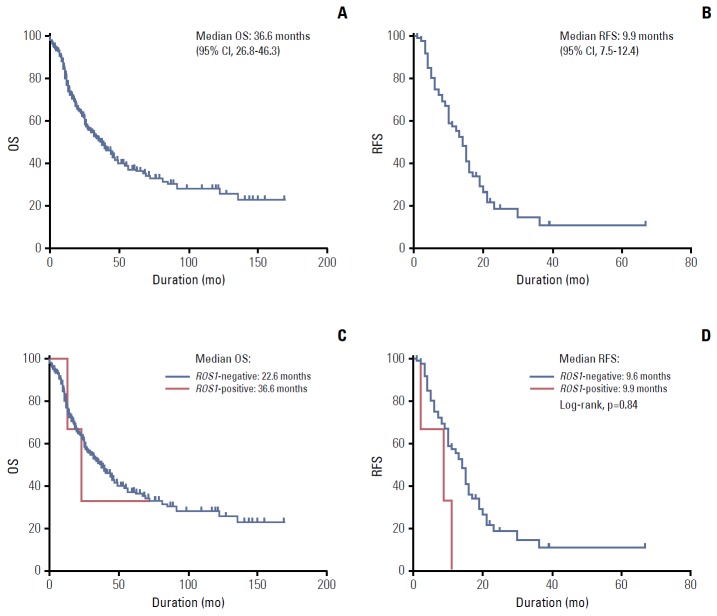
The clinical outcomes of all patients and ROS1-positive versus ROS1-negative patients were compared. The median OS was 36.6 months (95% CI, 26.8 to 46.3 months), and the median RFS was 9.9 months (95% CI, 7.5 to 12.4 months) for all patients (Fig. 2A and B). There were no statistical differences in terms of OS and RFS (Fig. 2C and D) between ROS1 FISH–positive and ROS1 FISH–negative patients, and there was no statistical difference in terms of OS and RFS between ROS1 IHC–positive and ROS1 IHC–negative patients.
Fig. 2.
Survival analysis. (A) Kaplan-Meier curve of median overall survival (OS) of all patients. (B) Kaplan-Meier curve of median recurrence-free survival (RFS) of all patients. (C) Comparison of OS between ROS1-positive and -negative patients. (D) Comparison of RFS between ROS1-positive and -negative patients. CI, confidence interval.
4. Analysis of ROS1 FISH–positive patients
A detailed summary of the clinicopathological data of three patients with ROS1 rearrangement is shown in Table 2. These patients were all intrahepatic CCA, and no signs of hepatolithiasis and parasitic infection were detected.
Table 2.
Clinicopathological features of ROS1-rearranged cholangiocarcinoma patients
| No. | Age (yr) | Sex | ROS1 FISH pattern | ROS1 protein expression | Tumor size (cm) | Location | Gross type | Histologic type | Differentiation | Non-tumor liver | Stage |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 66 | F | Break-apart | Negative | 6.5 | Intrahepatic | Mass forming | AC | MD | Chronic hepatitis, B-viral | pT1N1M0 |
| 2 | 73 | M | Break-apart | Positive | 3.0 | Intrahepatic | Intraductal growth | AC | WD | Macrovesicular steatosis, mild | pT1N0M0 |
| 3 | 60 | M | Single-green | Positive | 3.1 | Intrahepatic | Periductal infiltrative growth | AC | MD | Non-specific reactive hepatitis, mild | pT4N1M0 |
FISH, fluorescence in situ hybridization; F, female; AC, adenocarcinoma; MD, moderately differentiated; M, male; WD, well-differentiated.
Patient 1 had a 6.5-cm-sized mass forming tumor with moderate differentiation. Patient 2 had a 3-cm-sized CCA with intraductal growth and well differentiation. Patient 3 had a 3-cm-sized CCA with periductal infiltrative growth and moderate differentiation. Patients 1 and 3 showed lymph node metastasis of CCA.
Discussion
In this study, we screened CCA tissue archives and found that approximately 1.1% of CCA patients harbored ROS1 rearrangement. Of note, all ROS1-positive patients had intrahepatic CCA. To date, this is the first large-scale screening effort to examine the frequency of ROS1 rearrangement in CCA patient tissue.
CCA is a fatal disease with a huge unmet need for novel therapeutics and efforts have been made to define genetic sub-classification of CCA. The potentially targetable signaling pathways in CCA include EGFR, VEGF, HER2/neu, MET, AKT, PI3K/mTOR, MEK, PARP1/2, IDH1, and IDH2 [1]. However, clinical trials targeting these genetic alterations have not been successful in proving survival benefit. Combinations of doublet chemotherapy with anti-EGFR agents have shown modest results, and responses with small molecule inhibitors are limited. Use of erlotinib, lapatinib, sorafenib, cetuximab, and bevacizumab has been attempted in biliary tract cancer patients, but the response rates are between 0%-12%, median progression-free survival of 1.8 to 3.7 months, and median OS of 4.4 to 9.8 months. Therefore, greater effort is required for identification of genomically homogenous patient subsets who would benefit from specific targeted agents.
The discovery and characterization of ROS1 rearrangement in CCA has attracted significant clinical interest because small molecule inhibitors have effective antitumor activity. Because ALK and ROS1 share an approximately 49% amino acid sequence in the kinase domain, ALK inhibitors have been proved to be effective in inhibiting ROS1 activity [16]. In addition, an orthotopic allograft mouse model of intrahepatic CCA validated FIG-ROS1 fusion as a potent oncogene where it cooperates with KRAS and mutant p53 to accelerate tumor onset [11]. These results suggest that ROS1 rearrangement in CCA may be a promising druggable target requiring further investigation.
While nine fusion partners to ROS1 have been identified (FIG, CCDC6, CD74, EZR, KDELR2, LRIG3, SLC34A2, SDC4, and TPM3), all of which retain the ROS1 cytoplasmic kinase domain [15,16], only FIG-ROS1 fusion transcript has been identified in CCA so far. The frequency of ROS1 rearrangement in CCA has not yet been well-defined. Gu et al. [10], who screened for ROS1 rearrangement by reverse transcriptase-polymerase chain reaction (RT-PCR) in CCA patients, reported the frequency of FIG-ROS1 rearrangement as 8.7% (2 out of 23). However, Arai et al. [18] recently reported that the FIG-ROS1 fusion in CCA was 0% when screened by RT-PCR in 102 patients. We report the frequency of ROS1 rearranged tumors as 1.1% in the largest cohort ever. Despite its low incidence, we agree that ROS1 rearrangements warrant a new molecular subtype of CCA. Indeed, the results of an ongoing clinical trial of a drug targeting ROS1 rearrangement in CCA patients are eagerly awaited (NCT02374489).
Regarding the screening method of ROS1 rearrangement, FISH and RT-PCR have been commonly used, although time consuming, costly, and not suitable for massive screening. Accurate identification of ROS1-rearranged cancers by IHC analyses using an anti-ROS1 rabbit monoclonal antibody (D4D6) showing 100% (8/8) sensitivity and 100% (138/138) specificity when compared with break-apart FISH in lung cancer was recently reported [19]. However, there are no concrete data validating the screening utility of IHC in CCA. In a recent study IHC screening of ROS1 expression in intrahepatic CCA showed 37.1% (72/194) positivity, but all were FISH-negative [20]. Similarly, in our study incidence of ROS1 protein expression was significantly higher than gene rearrangement (19.1% vs. 1.2%). As protein expression of ROS1 may result from amplification and epigenetic changes, we should be cautious in interpreting results of ROS1 immunohistochemistry in CCA. For now, IHC cannot be an efficient screening method, and further validation with respect to ROS1 diagnostics should be established in the future.
The limitation of this study is the lack of information on the fusion variants in ROS1-positive samples, which was due to insufficient tissue samples and poor quality of extracted RNA in formalin-fixed, paraffin-embedded tumors.
Conclusion
In conclusion, we screened for ROS1 rearrangement in the largest-ever international of cohort of 261 CCAs including 208 intrahepatic CCAs and 53 extrahepatic CCAs. ROS1 rearrangement was detected in 1.2% of total CCAs and 1.4% of intrahepatic CCAs. Our results indicate that future screening efforts may offer a new therapeutic option for advanced CCA patients. Further investigation on the impact of ROS1 inhibition on the survival of CCA patients harboring ROS1 rearrangements is necessary.
Acknowledgments
This research was supported by grants from the National Research Foundation of Korea (NRF) funded by the Korean government (MSIP) (grant number: NRF-2013R1A2A2A 05005990, NRF-2012M3A9B6055350, NRF-2011-0030086) and the Korea Institute of Science and Technology Institutional Program (2Z04690) to Park YN.
We would also like to thank Wei Keat Wan and Wei Qiang Leow for their assistance in the work described in this publication.
Footnotes
We deeply appreciate Abbott for providing the ROS1 FISH probes.
Electronic Supplementary Material
Supplementary materials are available at Cancer Research and Treatment website (http://www.e-crt.org).
References
- 1.Razumilava N, Gores GJ. Cholangiocarcinoma. Lancet. 2014;383:2168–79. doi: 10.1016/S0140-6736(13)61903-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Everhart JE, Ruhl CE. Burden of digestive diseases in the United States Part III: liver, biliary tract, and pancreas. Gastroenterology. 2009;136:1134–44. doi: 10.1053/j.gastro.2009.02.038. [DOI] [PubMed] [Google Scholar]
- 3.Khan SA, Davidson BR, Goldin RD, Heaton N, Karani J, Pereira SP, et al. Guidelines for the diagnosis and treatment of cholangiocarcinoma: an update. Gut. 2012;61:1657–69. doi: 10.1136/gutjnl-2011-301748. [DOI] [PubMed] [Google Scholar]
- 4.Summersgill BM, Shipley JM. Fluorescence in situ hybridization analysis of formalin fixed paraffin embedded tissues, including tissue microarrays. Methods Mol Biol. 2010;659:51–70. doi: 10.1007/978-1-60761-789-1_4. [DOI] [PubMed] [Google Scholar]
- 5.Rosen CB, Heimbach JK, Gores GJ. Liver transplantation for cholangiocarcinoma. Transpl Int. 2010;23:692–7. doi: 10.1111/j.1432-2277.2010.01108.x. [DOI] [PubMed] [Google Scholar]
- 6.Valle J, Wasan H, Palmer DH, Cunningham D, Anthoney A, Maraveyas A, et al. Cisplatin plus gemcitabine versus gemcitabine for biliary tract cancer. N Engl J Med. 2010;362:1273–81. doi: 10.1056/NEJMoa0908721. [DOI] [PubMed] [Google Scholar]
- 7.Zabron A, Edwards RJ, Khan SA. The challenge of cholangiocarcinoma: dissecting the molecular mechanisms of an insidious cancer. Dis Model Mech. 2013;6:281–92. doi: 10.1242/dmm.010561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ong CK, Subimerb C, Pairojkul C, Wongkham S, Cutcutache I, Yu W, et al. Exome sequencing of liver fluke-associated cholangiocarcinoma. Nat Genet. 2012;44:690–3. doi: 10.1038/ng.2273. [DOI] [PubMed] [Google Scholar]
- 9.Lee J, Park SH, Chang HM, Kim JS, Choi HJ, Lee MA, et al. Gemcitabine and oxaliplatin with or without erlotinib in advanced biliary-tract cancer: a multicentre, open-label, randomised, phase 3 study. Lancet Oncol. 2012;13:181–8. doi: 10.1016/S1470-2045(11)70301-1. [DOI] [PubMed] [Google Scholar]
- 10.Gu TL, Deng X, Huang F, Tucker M, Crosby K, Rimkunas V, et al. Survey of tyrosine kinase signaling reveals ROS kinase fusions in human cholangiocarcinoma. PLoS One. 2011;6:e15640. doi: 10.1371/journal.pone.0015640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saborowski A, Saborowski M, Davare MA, Druker BJ, Klimstra DS, Lowe SW. Mouse model of intrahepatic cholangiocarcinoma validates FIG-ROS as a potent fusion oncogene and therapeutic target. Proc Natl Acad Sci U S A. 2013;110:19513–8. doi: 10.1073/pnas.1311707110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bergethon K, Shaw AT, Ou SH, Katayama R, Lovly CM, McDonald NT, et al. ROS1 rearrangements define a unique molecular class of lung cancers. J Clin Oncol. 2012;30:863–70. doi: 10.1200/JCO.2011.35.6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sharma S, Birchmeier C, Nikawa J, O'Neill K, Rodgers L, Wigler M. Characterization of the ros1-gene products expressed in human glioblastoma cell lines. Oncogene Res. 1989;5:91–100. [PubMed] [Google Scholar]
- 14.Kim HR, Lim SM, Kim HJ, Hwang SK, Park JK, Shin E, et al. The frequency and impact of ROS1 rearrangement on clinical outcomes in never smokers with lung adenocarcinoma. Ann Oncol. 2013;24:2364–70. doi: 10.1093/annonc/mdt220. [DOI] [PubMed] [Google Scholar]
- 15.Chin LP, Soo RA, Soong R, Ou SH. Targeting ROS1 with anaplastic lymphoma kinase inhibitors: a promising therapeutic strategy for a newly defined molecular subset of non-small-cell lung cancer. J Thorac Oncol. 2012;7:1625–30. doi: 10.1097/JTO.0b013e31826baf83. [DOI] [PubMed] [Google Scholar]
- 16.Ou SH, Tan J, Yen Y, Soo RA. ROS1 as a 'druggable' receptor tyrosine kinase: lessons learned from inhibiting the ALK pathway. Expert Rev Anticancer Ther. 2012;12:447–56. doi: 10.1586/era.12.17. [DOI] [PubMed] [Google Scholar]
- 17.Shaw AT, Ou SH, Bang YJ, Camidge DR, Solomon BJ, Salgia R, et al. Crizotinib in ROS1-rearranged non-small-cell lung cancer. N Engl J Med. 2014;371:1963–71. doi: 10.1056/NEJMoa1406766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arai Y, Totoki Y, Hosoda F, Shirota T, Hama N, Nakamura H, et al. Fibroblast growth factor receptor 2 tyrosine kinase fusions define a unique molecular subtype of cholangiocarcinoma. Hepatology. 2014;59:1427–34. doi: 10.1002/hep.26890. [DOI] [PubMed] [Google Scholar]
- 19.Davies KD, Le AT, Theodoro MF, Skokan MC, Aisner DL, Berge EM, et al. Identifying and targeting ROS1 gene fusions in non-small cell lung cancer. Clin Cancer Res. 2012;18:4570–9. doi: 10.1158/1078-0432.CCR-12-0550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee KH, Lee KB, Kim TY, Han SW, Oh DY, Im SA, et al. Clinical and pathological significance of ROS1 expression in intrahepatic cholangiocarcinoma. BMC Cancer. 2015;15:721. doi: 10.1186/s12885-015-1737-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.