Light polymer nanotransducers control cell functions in vivo.
Keywords: polymer nanoparticles, bio interfaces, bio photonics, Photophysics
Abstract
Current implant technology uses electrical signals at the electrode-neural interface. This rather invasive approach presents important issues in terms of performance, tolerability, and overall safety of the implants. Inducing light sensitivity in living organisms is an alternative method that provides groundbreaking opportunities in neuroscience. Optogenetics is a spectacular demonstration of this, yet is limited by the viral transfection of exogenous genetic material. We propose a nongenetic approach toward light control of biological functions in living animals. We show that nanoparticles based on poly(3-hexylthiophene) can be internalized in eyeless freshwater polyps and are fully biocompatible. Under light, the nanoparticles modify the light response of the animals, at two different levels: (i) they enhance the contraction events of the animal body, and (ii) they change the transcriptional activation of the opsin3-like gene. This suggests the establishment of a seamless and biomimetic interface between the living organism and the polymer nanoparticles that behave as light nanotransducers, coping with or amplifying the function of primitive photoreceptors.
INTRODUCTION
Inducing light sensitivity in living organisms provides groundbreaking opportunities in neuroscience. Optogenetics is a spectacular demonstration of this, yet it is limited by the viral transfection of exogenous genetic material (1). The use of photosensitizers (2) represents an alternative tool for photostimulation of living tissues (3). This led to the visionary proposal that the injection of dyes into the muscles of paraplegics could restore their functioning (4). However, molecular dyes are difficult to localize in the hosting body and can be toxic or phototoxic. Conversely, nanoparticles (NPs) with size comparable to cell organelles can be easily delivered also in vivo, through water suspensions, and functionalized on the surface to somewhat control their localization and function. Metal NPs for photothermal stimulation, inorganic semiconducting rods for electrical stimulation, and inorganic NPs for magnetothermal stimulation (5, 6) have been recently reported. Here, we report on the use of NPs based on semiconducting polymers. These materials have shown excellent biocompatibility, and their possible use in combination with living systems (both in vitro and in vivo conditions) has been intensively investigated, mainly for drug delivery and imaging applications (7–11). However, the exploitation of their light absorption in the visible range and charge photogeneration capability was reported for opto-neuromodulation in very few cases (12), mainly focusing on photothermal effects (9, 13–16). Moreover, to the best of our knowledge, conjugated polymer NPs were never reported to play a direct role in vivo in the control of the animal physiology.
Here, we show that NP based on poly(3-hexylthiophene) (P3HT) can be internalized in eyeless freshwater polyps (17) and are fully biocompatible. Under light, the NPs modify the light response of the animals and enhance opsin expression. This suggests a seamless and biomimetic interface between the polymer NP and the living organism. P3HT is a prototype conjugated polymer used in photodetection, photonics, and photovoltaics (18, 19). Recently, it has been shown that P3HT is highly biocompatible and can transduce a light signal into a cell stimulus (20, 21). Its application in a retinal prosthesis implanted in blind animals is under investigation (22, 23). Geometrically, isotropic nanospheres of P3HT (P3HT-NPs) with a hydrodynamic diameter between 150 and 300 nm, and a relatively low polydispersity index (0.07), have been realized in the absence of surfactants (Fig. 1, A and C). The optical absorption and emission spectra of the aqueous dispersion, peaking at 520 and 640 nm, respectively, are red-shifted and slightly broader compared to the spectra from polymer chains in solution (fig. S1). The zeta potential has been calculated by the phase analysis light scattering technique on the basis of the Smoluchowski equation and amounts to −41.8 ± 0.4 mV, indicating excellent colloidal stability in an aqueous environment. Here, P3HT-NPs are tested as photoactuators in the freshwater polyp Hydra vulgaris, a primitive animal whose simple anatomy can be compared to a tissue. The direct exposure of the outer cell layer, the ectoderm, to the bathing medium enables testing of direct interaction with medium-suspended NPs using a variety of methods (24, 25), including tools for testing the toxicity (26–28) and the bioactivity of nanomaterials (29–31). This allows low-cost, high-throughput investigations, decreases vertebrate experimentation, and avoids ethical issues. Despite the nonvisual photic behavior of this organism, the sequenced genome revealed the presence of all complex repertoire genes involved in vertebrate vision (32–35). We find that soaking Hydra polyps with P3HT-NPs leads to efficient internalization. Using different approaches, we assess the nontoxicity of the P3HT-NPs and investigate their photo-bioactivity. Our results show that P3HT-NP photostimulation elicits two phenomena: the modulation of the photo-induced Hydra contraction behavior and the enhancement of an opsin-like gene transcription.
Fig. 1. Characterization of P3HT-NPs and in vivo toxicological analysis.
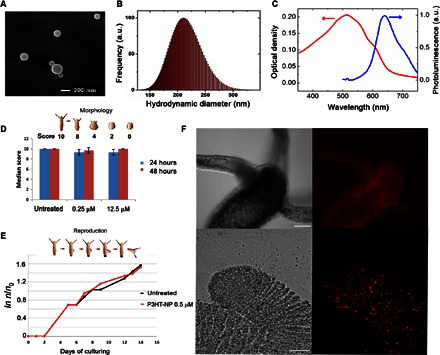
(A and B) Characterization of P3HT-NPs by scanning electron microscopy and dynamic light scattering (DLS) analysis. (C) Optical absorption (left axis) and photoluminescence spectra (right axis) of P3HT-NPs in an aqueous dispersion. Fluorescence emission excitation wavelength, 470 nm. (D and E) Determination of P3HT-NP toxicity endpoints in H. vulgaris. Numerical scores ranging from 10 (healthy polyp) to 0 (dead polyp) were assigned to progressive morphological changes possibly induced by P3HT-NP treatments, at the indicated doses, and recorded every 24 hours. No significant difference is evidenced between treated and untreated polyps (D). P3HT-NP treatment does not affect the reproduction rate of Hydra. The logarithmic growth curve of a treated Hydra population (red) is fully comparable to the one obtained from a control population (E) (n0 is the number of polyps at time 0, and n is the number of animals recorded at time t). (F) In vivo bright-field (left) and fluorescence (right) imaging of a living polyp treated with P3HT-NPs. Soaking the polyps with 0.25 μM P3HT-NPs causes a fluorescent staining of all tissues. NPs after a few hours appear as fluorescent spots located inside the ectodermal cells. The top images show a head with a crown of tentacles around the mouth. Bottom images show details of the tentacle tip. Scale bars, 200 and 50 μm (top and bottom images, respectively). a.u., arbitrary units.
RESULTS
Hydra can react to chemical stressors displaying a broad range of morphological changes, grouped according to numerical values ranging from 10 (normal) to 0 (disintegrated) (36). We assessed P3HT-NP toxicity in the dark against tissue integrity/polyp morphology by treating groups of 10 living polyps with increasing concentrations of P3HT-NPs (0.25 to 12.5 μM). Median morphological scores recorded at 24-hour intervals did not show the induction of morphological aberrations (such as contracted body or tentacles, or tissue damages), indicating the absence of macroscopic toxicity signs (Fig. 1D). We next evaluated the impact of P3HT-NPs on Hydra reproduction by budding. Because the proliferation rate of ectodermal and endodermal cells depends on feeding regime and environmental condition (37), any compound affecting this process will in turn affect the growth rate of the population founded by the exposed polyp. Figure 1E shows no significant differences between population growth rates of untreated (black line) and treated polyps (red line), indicating that P3HT-NP exposure does not undermine Hydra growth. Estimation of population growth rate performed at higher concentration did not show any marked effect at up to 50 μM concentration, confirming the good biocompatibility of polymer beads, at least in physiological processes linked to cell proliferation (fig. S2).
Biodistribution of P3HT-NPs in living polyps was inspected by fluorescence microscopy. Images from Fig. 1F show an efficient uptake of P3HT-NPs in Hydra tissue throughout the body column, especially into tentacles. According to previous experiments carried out using fluorescent inorganic nanocrystals (38), gold NPs of different shapes (30, 39, 40), silica oxide NPs (28), and iron oxide NPs (41), we speculate that the same internalization route might be active, that is, macropinocytosis of medium-suspended NPs and accumulation into cytoplasmic vacuoles.
The natural Hydra behavior at ambient light is characterized by a continuous periodic alternation of body contractions and elongations (42, 43) at an observed average of two contractions in 10 min (44, 45). Here, we compare observations in ambient light with observations under white light-emitting diode (LED). Groups of 20 polyps were incubated with P3HT-NPs (0.25 μM) for 2 and 24 hours, and animal behavior was monitored and recorded every 30 s, starting with exposure for 1 min to ambient light, 3 min to white light (at maximum power density of 530 nm, 0.43 mW/mm2), and 4 min back to ambient light condition (Fig. 2A). To quantify the contraction behavior, we introduced a scoring system, assigning progressive numerical scores to behavioral traits, ranging from 6 (maximal body contraction) to 11 (maximal body elongation). Figure 2B shows the average contraction score (n = 60) obtained for the two sets of animals (red and blue curves) and the comparison with untreated polyps (black curve). During the first minute of recording, all polyps are in the relaxed, physiological state. Under white light illumination, untreated Hydra tends to elongate, whereas P3HT-NP–treated animals tend to contract or at least suppress the elongation. A slightly stronger response is induced in animals treated for 2 hours with P3HT-NPs, compared to those treated for 24 hours, perhaps because at 2 hours, most P3HT-NPs are still located in the external cell layer, whereas afterward, they migrate into the internal layer, as reported for other fluorescent nanocrystals (38). When the light is switched off, the untreated polyps clearly recover the original periodic elongation/contraction behavior (42), whereas the treated ones continue the light-induced contraction. Figure 2C shows that upon illumination, the average number of contractions significantly increases in treated polyps. Analog experiments carried out on light-insensitive, electrically inert NPs made of polystyrene (with comparable dimensions and zeta potential values) did not show any sizable effect (fig. S3).
Fig. 2. P3HT-NPs induce a behavioral response in H. vulgaris.
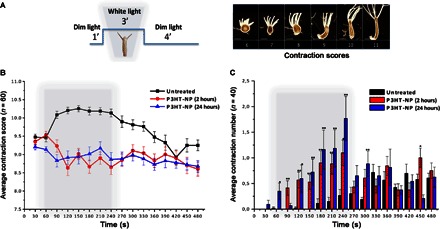
(A) The behavioral response of Hydra to P3HT-NPs was evaluated using a contraction scoring system ranging from 6 (highly contracted) to 11 (elongated polyp). Animal behavior was monitored by continuous video recording for 8 min, according to the illumination protocol shown in the scheme, that is, 1-min dim light, 3-min white light illumination, 4-min dim light. Scale bars, 500 μm. (B) Average contraction scores and SD resulting from behavioral analysis (n = 60 polyps). Gray boxes indicate the 3-min light illumination period; black curves show contraction behavior of untreated polyps; red and blue curves show the contraction behavior of polyps treated with P3HT-NPs for 2 or 24 hours, respectively. SD values are reported as bars for each data set. (C) Average number of contraction events and relative SD estimated on treated (2 and 24 hours, red and blue lines, respectively) and untreated (black) polyps (n = 40, each condition). In most cases, statistically significant differences are observed (Student’s t test, *P < 0.05, **P < 0.01).
We next carried out expression profiling of genes possibly involved in the P3HT-NP–mediated phototransduction. We used quantitative reverse transcription polymerase chain reaction (qRT-PCR) to reveal expression changes of genes involved (in mammals), in response to light, heat, and oxidative stress. Gene selection was performed, whenever possible, among genes previously characterized in Hydra: hsp70 (heat shock protein 70) is well characterized in Hydra for heat responsiveness (46), and its expression has been profiled in response to temperatures ranging from 26° to 37°C (41); Cu-Zn SOD (copper-zinc superoxide dismutase), encoding an antioxidant enzyme and converting superoxide radical into hydrogen peroxide, showed increased expression in Hydra in response to both heat (34°C) (47) and reactive oxygen species (ROS) (48). Eventually, for opsin3-like and trpa1-like (transient receptor potential A-subfamily) selected genes, we characterized the transcriptional modulation mediated by both light and heat stresses.
Animals were treated for 24 hours with P3HT-NPs, equilibrated for 30 min at room temperature under the cold light of a stereomicroscope, and then exposed for 30 min to white light illumination of the same power density as that used in behavioral assays. After recovering for 30 and 120 min, animals were processed for qRT-PCR. Figure 3 shows a significant increase of opsin3-like expression in polyps treated with P3HT-NPs and exposed to light, at both time points, compared to animals not illuminated. Opsin3-like gene expression levels increased, albeit in a lesser amount, also in untreated polyps, under LED illumination at a 2-hour time point, and this light responsiveness was further confirmed in polyps under normal dark/light cycling conditions (fig. S4). We found that hsp70 expression levels were only slightly up-regulated and did not show a significant correlation with P3HT-NP treatment or light exposure. Because hsp70 is involved in regulating protein stability and maturation, the slight overexpression observed in all experimental conditions may indicate the possible involvement of this chaperonin in P3HT-NP general “sensing” and involvement in maintaining cell homeostasis, rather than its participation in a specific light-activated process. trpa1-like and SOD gene transcription levels were not significantly changed in response to P3HT-NP treatment or LED irradiation. Experiments carried out with control polystyrene NPs, in the same experimental conditions and under the same experimental protocol, did not show marked effects on opsin expression, thus confirming the active role played by the chemical/physical properties of P3HT-NPs in the intracellular light transduction pathways (fig. S3). Molecular analysis performed under the illumination condition used for behavioral analysis (3 min) revealed significant expression changes for opsin3-like gene at a 2-hour time point (fig. S5), confirming modulation of opsin transcription by light and indicating the capability of a short illumination period to trigger the molecular response.
Fig. 3. P3HT-NP photoactivation enhances opsin3-like gene transcript levels.
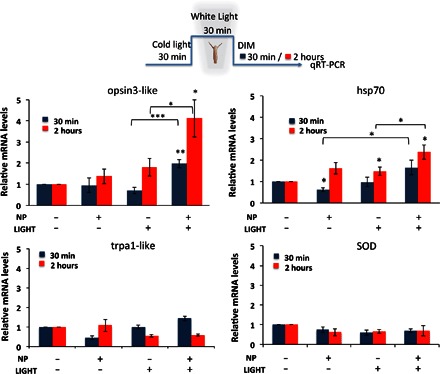
Hydra exposure to P3HT-NPs and/or white light illumination elicits a number of molecular reactions, which may provide useful clues on the transduction pathways activated by each stimulus. Genes selected in this study belong to light transductions pathway (opsin3-like), heat response (hsp70, trpa1-like), and oxidative stress (Cu-Zn SOD). Gene accession numbers for the Hydra homologous genes are reported in table S1. The expression profile of selected genes was investigated by qRT-PCR analysis using elongation factor 1α (HyEf-1α) as reference gene. Polyps treated with 0.25 μM P3HT-NPs for 24 hours underwent the illumination protocol shown on the upper part of the panel, then they were allowed to recover for either 30 min or 2 hours and were processed for RNA extraction and qRT-PCR analysis using specific primers (see table S1). opsin3-like gene expression shows great activation in response to both NPs and white light illumination, as compared to other conditions (untreated, not illuminated; treated, not illuminated). Data are presented as means ± SE of three technical repeats from two biological replicates. Statistical comparisons are performed using unpaired t test, *P < 0.05, **P < 0.01.
To disentangle direct effects of light excitation from light-mediated thermal processes, we investigated gene responsiveness to heat. Results of gene profiling in fig. S6 confirmed heat responsiveness in transcription of hsp70, trpa1-like, and SOD genes, whereas opsin3-like transcript levels were not affected by heat stress. This strongly argues against a thermal effect consequent to photostimulation, although it does not necessarily rule out the involvement of ROS or other molecular transducers.
DISCUSSION
Here, we investigate whether nanostructured conjugated polymers could enhance light photosensitivity in vivo, providing a handle for controlling living organisms with light. We use H. vulgaris as a model system. This is a simple water invertebrate, which evolved before vision was established, yet it is able to react to light illumination, for instance, by heliotropism. First, we demonstrate the successful uptake of polymer NPs in the living animal. Then, we explore the functional interaction of the light-activated NPs with the cells in living Hydra by a behavioral analysis and a quantitative estimation of expression fold changes of genes involved in light, heat, or ROS responses. Overall, both the behavioral and molecular outcomes indicate that P3HT-NPs enhance light-mediated processes boosting the animal’s photosensitivity.
In more detail, we find two effects. First, we observe that NPs photoexcitation modulates the elongation/contraction behavior of P3HT-NP–treated animals. The contraction of the body column in Hydra is regulated at two levels: (i) epitheliomuscular cells are connected by gap junctions and can propagate an electrical signal mediating contraction in response to a stimulus and (ii) nerve cells located in the lower body control the spontaneous initiation of epitheliomuscular cell contraction (49, 50). We suggest that the photoexcitation both activates the surface of P3HT-NPs inducing electrochemical reactions and creates a localized electrical dipole, which causes the synchronous firing of nerve cells and the subsequent coordinated activation of effector epitheliomuscular cells.
The second effect of photostimulation we report is the enhancement of opsin expression levels in treated Hydra, a downstream effect of single or multiple molecular cascades. The reason for such transcriptional activation, although not straightforward, unequivocally links the P3HT-NP photoexcitation to the opsin, the best investigated player underlying phototransduction.
The opsin3-like gene belongs to a seven-transmembrane G protein–coupled receptor superfamily, which plays a key role in vision (51). All known visual pigments in Neuralia (Cnidaria, Ctenophora, and Bilateria) are composed of an opsin and a light-sensitive chromophore, generally a retinal, linked through a Schiff base involving a lysine found at position 296 (K296) of the reference bovine rhodopsin sequence. Considering the conserved position of this amino acid residue in the Hydra opsin3-like predicted protein sequence (figs. S7 and S8), this protein is likely to be involved in light reception in Hydra as well. Furthermore, the increased opsin3-like gene expression under white light illumination in untreated polyps suggests the “natural” responsiveness of the opsin3 gene transcription to white light stimulation. This is also confirmed in polyps under normal light cycle (12-hour light/12-hour dark ) (fig. S3). Other evidence supporting light dependence of Hydra opsin3-like protein comes from functional studies on the opsin subgroup 3 (opn3), including both vertebrate and invertebrate members. Originally named encephalopsin or panopsin, as expressed in the brain, liver, and other nonphotoreceptive tissues, they can form functional photosensitive pigments and activate G proteins in a light-dependent manner (52). Similarly, the Hydra opsin3 homolog investigated here may act as a light sensor, mediating the intracellular transduction of the light signal. Together, these observations support the conjecture that P3HT-NPs behave as light nanotransducers, somewhat mimicking the role of natural photoreceptors and boosting Hydra’s photosensitivity.
Although a definitive assignment of the coupling mechanism between the photoexcited NPs and the Hydra is beyond the scope of this work, we would like to briefly discuss three plausible effects: (i) thermal release, (ii) photochemical reactions, and (iii) electrical polarization (Fig. 4).
Fig. 4. Mechanisms underlying photoexcitation of P3HT-NPs.
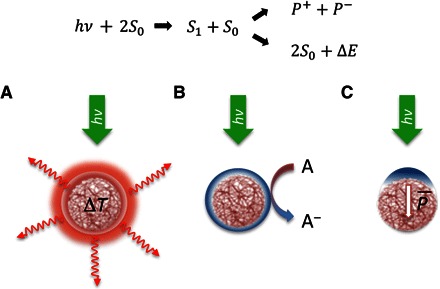
Top: Upon visible light illumination, primary photoexcitation states S1 are created within the polymer NPs, which can give rise to the creation of polaronic charged states P± or to nonradiative release of the excess energy ΔE. Bottom: different decay paths of the NP photoexcited states are shown: (A) photothermal excitation and heat release, (B) photoelectrochemical oxidation processes, and (C) electrical polarization.
(i) Assuming a maximum light intensity of I = 1 mW/mm2, the expected rise in temperature ΔT of a NP at thermodynamic equilibrium in a medium is given by , where R (~100 nm) is the NP radius and κ (~0.6 W/mK) is the water thermal conductivity. We obtain ΔT = 0.04 mK, which is at least four orders of magnitude below the threshold of interest. NP aggregation might increase the amount of energy deposited in a limited volume, thus enhancing the local temperature. However, given the linear dependence of ΔT with R, even an aggregate of 100 NPs would not be sufficient to increase the temperature by 1 K. These considerations rule out photothermal effects as a possible cause of interaction. Note that considering the estimated number of NPs up taken by a Hydra on average and the animal volume, even the total heating of the animal turns out to be negligible.
(ii) A P3HT chain is composed of conjugated segments of variable length. By and large, each segment behaves like an organic molecule supporting singlet states as primary photoexcitation. In the NPs, the polymer chains are closely packed, allowing interchain interactions. In addition, in the presence of oxygen, the particles are p-doped, due to electron trapping at oxygen complexes. Light, oxygen, and moisture boost the doping process, and the radiative decay path of the singlet state can be totally quenched in favor of charge separation. Charged states, known as polarons, become the dominant photoexcitation. They can live up to the millisecond time domain, contributing a steady-state population under continuous wave light, that can feed electrochemical reactions occurring at the NP surface. If we consider an average time constant τ =1 μs for the polaron states and an intensity of I = 0.5 mW/mm2 at an average photon energy of 2 eV, the steady-state population density N on a single NP is N ≈ 1.5× 1011 m−2, corresponding to about 50 charged pairs per particle, which can easily undergo surface photoelectrochemical phenomena.
(iii) The steady-state population of charges can also contribute to build up an electrical polarization of the NP. After photoexcitation, we expect that negative charges appear at the surface of the particle, due to water polarization. Because of the spherical shape of the particle, the surface charge is distributed according to the cosine law with respect to light direction. A trivial integration on the light-exposed hemisphere thus provides a formula for the effective dipole , where d is the charge separation, building up the dipole layer. For σ = 2.5 × 10−5 cm−2, according to I = 0.5 mW/mm2, the maximum electric field at a 10-nm distance from the NP surface is in the order of 5 × 104 V/m.
From the rough evaluation of the order of magnitude involved in the different phenomena, the thermal effect can be ruled out, in agreement with the lack of response of the heat markers (trpa1-like and SOD).
Regarding electrical polarization, this is expected to lead to very localized effects, strongly depending on the specific position of P3HT-NPs with respect to target membranes or organelles. Epitheliomuscular cells have contracting fibers extending parallel to the main body axis. Ultrastructural evidence (49) suggests that the contractile activity is controlled by neuronal cells, establishing chemical and electrical synaptic connection with epitheliomuscular cells. These cells are also connected to each other by gap junctions and can respond synchronously to electrical signaling, mediating contraction (50). When polymeric P3HT-NPs are added to the medium, they are possibly internalized into epitheliomuscular cells and elicit contraction events (Fig. 5A). Both the creation of a localized electric dipole at the NPs surface and the activation of photoelectrochemical reactions may possibly account for this phenomenon.
Fig. 5. Biological pathways activated by P3HT-NP photostimulation.
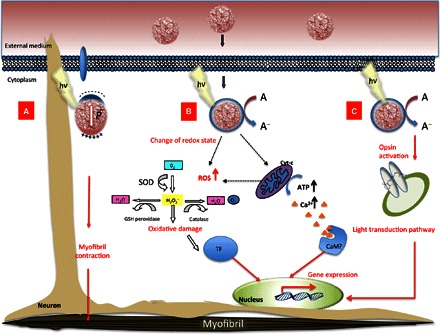
The sketch depicts a general epitheliomuscular ectodermal cell, presenting nucleus, mitochondria, and organelles. When light irradiates P3HT-NPs, both free and/or aggregated forms, electrons are generated in the photosensitive polymer, causing multiple biological responses. (A) NPs can localize within epitheliomuscular cells, and upon photoexcitation, a localized electrical dipole is established. In addition, photoelectrochemical reactions are promoted at the NP surface. These effects may locally act on neurons, leading to myofibril contraction and modulating the animal contracting behavior, as observed in behavioral studies. (B) Charged states sustain photoelectrochemical reactions, which increase the cytoplasmic concentration of ROS. This induces enzymatic reactions and acts on transcription factors (TF), or alternatively activates redox reactions of the respiratory chain and calcium binding transcription factors (CaM). In any case, targeted gene transcription is enhanced. (C) The retinal moiety of opsin3-like molecules or the P3HT-NPs themselves may initiate a light-dependent molecular cascade. GSH, glutathione.
On the other side, the enhancement of the transcription of genes involved in the light transduction pathway would not necessarily require a spatially localized activation mechanism and a close contact between the NPs and the targeted organelle. Thus, although we cannot completely exclude a possible contribution of local polarization in this case as well, we believe that the most probable photoexcitation mechanism is sustained here by photoelectrochemical reactions occurring at the P3HT-NP surface. These can trigger specific bioelectrical signaling processes (Fig. 5, B and/or C). Photogenerated charged states feed electrochemical reactions occurring at the NP surface, increasing the concentration of cytoplasmic ROS, as recently reported also for polythiophene-tamoxifen drug conjugates (53). Changes in cellular redox may induce detoxifying enzymatic reactions to eliminate dangerous species or act on transcription factors, driving gene transcription. In the proximity of mitochondria, electrochemical reactions may excite flavins and cytochrome, activating redox reactions of the respiratory chain. The increased adenosine triphosphate (ATP) levels boost all the ATP-driven ion carriers such as Na+/K+-ATPase (sodium-potassium adenosine triphosphatase) and Ca2+ pumps. The increase in the cytoplasmic Ca2+ levels activates calcium binding transcription factors, such as a hypothetical calmodulin (CaM)–like protein and, in turn, enhances expression of target genes (Fig. 5B). Alternatively and/or in a complementary way, through either a capacitive or an inductive coupling, the retinal moiety of opsin-like molecules or the P3HT-NPs themselves may initiate a light-dependent molecular cascade (Fig. 5C). We observe that the lack of SOD transcriptional activation (Fig. 3) could stem from the limited number of ROS involved or be a consequence of the specific transducing path triggered by the P3HT-NPs, which fits the natural one.
The complexity of the scenario reflects the variety of possible biological responses, whose elucidation will be an object of future investigations. Overall, we believe that P3HT-NPs behave as light nanotransducers, coping with or amplifying the function of primitive photoreceptors and leading to the enhancement of the light sensitivity in the whole animal.
Our results put forward the potential of this approach to control physiological functions in living organisms with light. In perspective, the demonstration that light-sensitive polymer NPs are well tolerated within animal models, and do play a functional role in enhancing the perception of light, may open interesting opportunities in biomedical applications. In particular, we recently demonstrated that P3HT thin films are able to partially restore light sensitivity in explanted blind retinas (22) and are very well tolerated once implanted in the rat retina, in place of the photoreceptors (23). Despite the excellent mechanical flexibility and reduced thickness of the implant, a very complex and rather invasive procedure is required for subretinal implantation. The availability of fully biocompatible nanomaterials with comparable optoelectronic properties would allow circumvention of this major issue. To this goal, specific targeting capability of NPs should be properly implemented, and detailed long-term localization analysis of the NPs should be carried out. The results here presented demonstrate the possibility of exploiting P3HT-NP visible light absorption for transduction of the light stimuli into the complexity level of a living animal. This first proof of concept is key to proceed on the route of “injectable electronics,” as it was recently defined (54).
MATERIALS AND METHODS
P3HT-NPs were prepared from freshly synthesized P3HT by the reprecipitation method in the absence of surfactants (55). The polymer was dissolved in tetrahydrofuran, and the solution was added to distilled water (solvent/nonsolvent volume in a 1:20 ratio). By means of differential centrifugation, the NPs were separated into fractions of different sizes and characterized by DLS with a NanoBrook Omni particle size analyzer, with a wavelength of 659 nm in back scattering mode (see also the Supplementary Materials). Ultraviolet (UV)–visible (vis) spectroscopy measurements were carried out using a PerkinElmer Lambda 1050 UV/Vis/NIR spectrophotometer and a Horiba Jobin Yvon NanoLog fluorometer. Scanning electron microscopy images were acquired with a Tescan MIRA3.
Animal culture
H. vulgaris (strain Zurich) were asexually cultured in Hydra medium [1 mM CaCl2 and 0.1 mM NaHCO3 (pH 7)] according to the method of Loomis and Lenhoff (56) with minor modifications. The animals were kept at 18° ± 1°C and fed three times per week with freshly hatched Artemia salina nauplii.
For morphological evaluation tests, groups of 20 animals were collected in plastic multiwells and allowed to equilibrate at room temperature in 300 μl of Hydra medium. The test was initiated by adding P3HT-NPs (0.5 μM) to each well and incubating as necessary. NP uptake was monitored by a stereomicroscope (Olympus SZX-RFL2). After extensive washes, in vivo imaging was accomplished using a standard inverted fluorescence microscope (Nikon Eclipse Ti-S, equipped with 4×/20× objective) and filtering the light with a bandpass filter (cut-on wavelength, 550 nm). Possible morphological changes induced by P3HT-NPs were monitored by using the scoring system introduced by Wilby (36) and assigning a numerical score to progressive morphological alterations [from 10 (healthy polyp) to 0 (disintegrated polyp)].
A healthy and well-fed Hydra population expands exponentially following the formula ln(n/n0) = kt, where n is the number of animals at time t, n0 is the number of animals at t0, and k is the growth constant (37). To assess the effect of P3HT-NPs on Hydra growth rate, five Hydra (population founders) were treated with P3HT-NPs for 24 hours, washed, and, in the following day, placed in 3.5-cm petri dishes (one Hydra per dish). Control polyps at the same developmental stage were not treated. Both treated and untreated Hydra were fed once daily for 14 days. The number of individuals was recorded daily and reported as the normalized logarithmic growth curves of treated Hydra population compared to a control population.
Behavioral activity
Groups of two Hydra were placed in a plastic petri dish containing Hydra medium to carry out the experiments. Hydra behavior upon white light illumination (Thorlabs LED MCWHL2-C4; power density, 0.43 mW/mm2) was investigated in the presence and absence of P3HT-NPs of 200-nm approximate diameter. P3HT-NPs were added to every single polyp at a concentration of 0.25 μM and incubated for 2 and 24 hours. After incubation, the illumination protocol illustrated in Fig. 2 was adopted. Experiments were performed before addition of P3HT-NPs, after 2 and 24 hours of P3HT-NP incubation on 24-hour–starved animals at room temperature. Both the contraction per elongation average degree of the Hydra body and the total number of contractions were evaluated before (1 min), during (3 min), and after (4 min) visible light excitation.
RNA extraction and qRT-PCR
Total RNA from treated and untreated animals was purified using TRI Reagent (Molecular Research Center), and its concentration was determined on the NanoDrop ND-1000 spectrophotometer (Thermo Scientific). The first-strand complementary DNA (cDNA synthesis) was carried out with the SuperScript II retrotranscriptase (Invitrogen) and oligo(dT)s, using 0.5 μg of DNA-free RNA in a final volume of 25 μl, according to the manufacturer’s instructions. qRT-PCR was performed in 25 μl of reaction mixture consisting of 1x Express SYBR GreenER qPCR SuperMix with premixed ROX (Invitrogen), serial cDNA dilutions, and 0.3 μM of each primer. The reactions were processed using the StepOne Real-Time PCR System (Applied Biosystem) under the following fast cycling steps: initial denaturation for 2 min at 94°C, followed by 40 cycles at 94°C for 2 s, 59°C for 30 s. In addition, melting curves (20 min, from 59° to 90°C) were generated to check any spurious amplification products. To normalize RNA levels, Hydra elongation factor 1α gene (HyEf-1α) was used as an internal calibrator. Nucleotide sequences and alignments were obtained from the Hydra genome database (http://www.compagen.org/). Specific primers of Hydra homolog genes of opsin (opsin3-like), SOD, hsp70, transient receptor potential-A family (trpa1-like) were designed using the Primer3 software (http://frodo.wi.mit.edu/primer3/) and are listed in table S1, together with the corresponding GenBank accession numbers. Further information is provided in the Supplementary Materials section (table S2). At least three technical repeats from three biological replicates were carried out. Here, the 2−ΔΔCT method, for comparing relative expression results between treatments, was applied (57).
The Supplementary Materials include details about NP synthesis and characterization, Hydra growth rates at 50 μM P3HT-NP concentration, control measurements with polystyrene NPs, transcriptional profiling of opsin3-like gene under physiological light cycle, transcriptional analysis of opsin3-like and hsp70 genes under the illumination condition used for behavioral analysis, transcriptional profiling of hsp70, opsin3-like, trpa1-like, and SOD genes under thermal stress, list of forward and reverse primers used in the qRT-PCR analysis, structural features of Hydra predicted opsin3-like protein, list of the opsin-like gene sequences present in the H. vulgaris genome database, alignment of opsin chromophore-binding transmembrane domain proteins from different animal species, and alignment of the predicted protein sequences representative of different classes of Hydra opsins.
Supplementary Material
Acknowledgments
This work was partially supported by the European Commission (project FP7-PEOPLE-212-ITN 316832 “OLIMPIA”) and by a national grant from the Fondazione Cariplo (project ON-IRIS 2013–0738). Author contributions: C.T., M.R.A., and G.L. planned and supervised the work. A.T., V.M., and A.B. performed toxicological studies, molecular analysis, and relative statistical and data analyses. C.B. and S.V.M. carried out UV-vis spectroscopy experiments and behavioral studies. M.Z. provided P3HT-NPs and carried out DLS measurements. All authors contributed to data analysis and manuscript writing. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/3/1/e1601699/DC1
fig. S1. Characterization of P3HT-NPs.
fig. S2. Impact of P3HT-NPs on Hydra population growth.
fig. S3. Evaluation of polystyrene NP impact on Hydra behavior and gene expression.
fig. S4. Light responsiveness of Hydra opsin3-like gene.
fig. S5. Gene profiling under short illumination condition.
fig. S6. Hydra opsin3-like gene transcription does not respond to heat stress.
fig. S7. Alignment of opsin chromophore-binding transmembrane domain proteins from different animal species.
fig. S8. Alignment of the predicted protein sequences representative of different classes of Hydra opsins.
table S1. List of forward and reverse primers used in the qRT-PCR analysis.
table S2. List of the opsin-like gene sequences present in the H. vulgaris genome database.
REFERENCES AND NOTES
- 1.Deisseroth K., Optogenetics: 10 years of microbial opsins in neuroscience. Nat. Neurosci. 18, 1213–1225 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Spikes J. D., Applications of dye-sensitized photoreactions in neurobiology. Photochem. Photobiol. 54, 1079–1092 (1991). [DOI] [PubMed] [Google Scholar]
- 3.Chalazonitis N., Light energy conversion in neuronal membranes. Photochem. Photobiol. 3, 539–559 (1964). [Google Scholar]
- 4.Kerkut G. A., Can fibre optic systems drive lower motoneurones? Prog. Neurobiol. 14, 1–23 (1980). [DOI] [PubMed] [Google Scholar]
- 5.Chen R., Romero G., Christiansen M. G., Mohr A., Anikeeva P., Wireless magnetothermal deep brain stimulation. Science 347, 1477–1480 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Huang H., Delikanli S., Zeng H., Ferkey D. M., Pralle A., Remote control of ion channels and neurons through magnetic-field heating of nanoparticles. Nat. Nanotechnol. 5, 602–606 (2010). [DOI] [PubMed] [Google Scholar]
- 7.Feng L., Zhu C., Yuan H., Liu L., Lv F., Wang S., Conjugated polymer nanoparticles: Preparation, properties, functionalization and biological applications. Chem. Soc. Rev. 42, 6620–6633 (2013). [DOI] [PubMed] [Google Scholar]
- 8.Feng L., Liu L., Lv F., Bazan G. C., Wang S., Preparation and biofunctionalization of multicolor conjugated polymer nanoparticles for imaging and detection of tumor cells. Adv. Mat. 26, 3926–3930 (2014). [DOI] [PubMed] [Google Scholar]
- 9.Miyako E., Russier J., Mauro M., Cebrian C., Yawo H., Ménard-Moyon C., Hutchison J. A., Yudasaka M., Iijima S., De Cola L., Bianco A., Photofunctional nanomodulators for bioexcitation. Angew Chem. 53, 13121–13125 (2014). [DOI] [PubMed] [Google Scholar]
- 10.Peters M., Zaquen N., D’Olieslaeger L., Bové H., Vanderzande D., Hellings N., Junkers T., Ethirajan A., PPV-based conjugated polymer nanoparticles as a versatile bioimaging probe: A closer look at the inherent optical properties and nanoparticle–cell interactions. Biomacromolecules 17, 2562–2571 (2016). [DOI] [PubMed] [Google Scholar]
- 11.Pennakalathil J., Özgün A., Durmaz I., Cetin-Atalay R., Tuncel D., pH-responsive near-infrared emitting conjugated polymer nanoparticles for cellular imaging and controlled-drug delivery. J. Polym. Sci. A Polym. Chem. 53, 114–122 (2015). [Google Scholar]
- 12.Colombo E., Feyen P., Antognazza M. R., Lanzani G., Benfenati F., Nanoparticles: A challenging vehicle for neural stimulation. Front. Neurosci. 10, 105 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takano Y., Numata T., Fujishima K., Miyake K., Nakao K., Grove W. D., Inoue R., Kengaku M., Sakaki S., Mori Y., Murakamia T., Imahori H., Optical control of neuronal firing via photoinduced electron transfer in donor–acceptor conjugates. Chem. Sci. 7, 3331–3337 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lyu Y., Xie C., Chechetka S. A., Miyako E., Pu K., Semiconducting polymer nanobioconjugates for targeted photothermal activation of neurons. J. Am. Chem. Soc. 138, 9049–9052 (2016). [DOI] [PubMed] [Google Scholar]
- 15.Li W., Luo R., Lin X., Jadhav A. D., Zhang Z., Yan L., Chan C. Y., Chen X., He J., Chen C. H., Shi P., Remote modulation of neural activities via near-infrared triggered release of biomolecules. Biomaterials 65, 76–85 (2015). [DOI] [PubMed] [Google Scholar]
- 16.Ye F., Wu C., Jin Y., Chan Y.-H., Zhang X., Chiu D. T., Ratiometric temperature sensing with semiconducting polymer dots. J. Am. Chem. Soc. 133, 8146–8149 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Technau U., Steele R. E., Evolutionary crossroads in developmental biology: Cnidaria. Development 138, 1447–1458 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baeg K.-J., Binda M., Natali D., Caironi M., Noh Y.-Y., Organic light detectors: Photodiodes and phototransistors. Adv. Mater. 25, 4267–4295 (2013). [DOI] [PubMed] [Google Scholar]
- 19.Brabec C. J., Gowrisanker S., Halls J. J., Laird D., Jia S., Williams S. P., Polymer-fullerene bulk-heterojunction solar cells. Adv. Mater. 22, 3839–3856 (2010). [DOI] [PubMed] [Google Scholar]
- 20.Ghezzi D., Antognazza M. R., Dal Maschio M., Lanzarini E., Benfenati F., Lanzani G., Hybrid bioorganic interface for neuronal photoactivation. Nat. Commun. 2, 166 (2011). [DOI] [PubMed] [Google Scholar]
- 21.Benfenati V., Martino N., Antognazza M. R., Pistone A., Toffanin S., Ferroni S., Lanzani G., Muccini M., Photostimulation of whole-cell conductance in primary rat neocortical astrocytes mediated by organic semiconducting thin films. Adv. Healthc. Mater. 3, 392–399 (2014). [DOI] [PubMed] [Google Scholar]
- 22.Ghezzi D., Antognazza M.R., Maccarone R., Bellani S., Lanzarini E., Martino N., Mete M., Pertile G., Bisti S., Lanzani G., Benfenati F., A polymer optoelectronic interface restores light sensitivity in blind rat retinas. Nat. Photonics 7, 400–406 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Antognazza M. R., Di Paolo M., Ghezzi D., Mete M., Di Marco S., Maya-Vetencourt J. F., Maccarone R., Desii A., Di Fonzo F., Bramini M., Russo A., Laudato L., Donelli I., Cilli M., Freddi G., Pertile G., Lanzani G., Bisti S., Benfenati F., Characterization of a polymer-based, fully organic prosthesis for implantation into the subretinal space of the rat. Adv. Healthc. Mater. 5, 2271–2282 (2016). [DOI] [PubMed] [Google Scholar]
- 24.C. Tortiglione, in Biomedical Engineering: From Theory to Application, R. Fazel-Rezai, Ed. (InTech, 2011), pp. 225–252. [Google Scholar]
- 25.Ambrosone A., Tortiglione C., Methodological approaches for nanotoxicology using cnidarian models. Toxicol. Mech. Methods 23, 207–216 (2013). [DOI] [PubMed] [Google Scholar]
- 26.Ambrosone A., Mattera L., Marchesano V., Quarta A., Susha A. S., Tino A., Rogach A. L., Tortiglione C., Mechanisms underlying toxicity induced by CdTe quantum dots determined in an invertebrate model organism. Biomaterials 33, 1991–2000 (2012). [DOI] [PubMed] [Google Scholar]
- 27.Ambrosone A., Marchesano V., Bartelmess J., Strisciante F., Tino A., Echegoyen L., Tortiglione C., Giordani S., Impact of carbon nano-onions on Hydra vulgaris as a model organism for nanoecotoxicology. Nanomaterials 5, 1331–1350 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ambrosone A., Scotto di Vettimo M. R., Malvindi M. A., Roopin M., Levy O., Marchesano V., Pompa P. P., Tortiglione C., Tino A., Impact of amorphous SiO2 nanoparticles on a living organism: Morphological, behavioral, and molecular biology implications. Front. Bioeng. Biotechnol. 2, 37 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Malvindi M. A., Carbone L., Quarta A., Tino A., Manna L., Pellegrino T., Tortiglione C., Rod-shaped nanocrystals elicit neuronal activity in vivo. Small 4, 1747–1755 (2008). [DOI] [PubMed] [Google Scholar]
- 30.Conde J., Ambrosone A., Sanz V., Hernandez Y., Marchesano V., Tian F., Child H., Berry C. C., Ibarra M. R., Baptista P. V., Tortiglione C., de la Fuente J. M., Design of multifunctional gold nanoparticles for in vitro and in vivo gene silencing. ACS Nano 6, 8316–8324 (2012). [DOI] [PubMed] [Google Scholar]
- 31.Ambrosone A., Marchesano V., Carregal-Romero S., Intartaglia D., Parak W. J., Tortiglione C., Control of Wnt/β-catenin signalling pathway in vivo via light responsive capsules. ACS Nano 10, 4828–4834 (2016). [DOI] [PubMed] [Google Scholar]
- 32.Plachetzki D. C., Degnan B. M., Oakley T. H., The origins of novel protein interactions during animal opsin evolution. PLOS ONE 2, e1054 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Plachetzki D. C., Fong C. R., Oakley T. H., The evolution of phototransduction from an ancestral cyclic nucleotide gated pathway. Proc. Biol. Sci. 277, 1963–1969 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Terakita A., Kawano-Yamashita E., Koyanagi M., Evolution and diversity of opsins. Wiley Interdiscip. Rev. Membr. Transp. Signal. 1, 104–111 (2012). [Google Scholar]
- 35.Porter M. L., Blasic J. R., Bok M. J., Cameron E. G., Pringle T., Cronin T. W., Robinson P. R., Shedding new light on opsin evolution. Proc. Biol. Sci. 279, 3–14 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilby O., Tesh J. M., The Hydra assay as an early screen for teratogenic potential. Toxicol. In Vitro 4, 582–583 (1990). [DOI] [PubMed] [Google Scholar]
- 37.Bosch T. C. G., David C. N., Growth regulation in Hydra: Relationship between epithelial cell cycle length and growth rate. Dev. Biol. 104, 161–171 (1984). [DOI] [PubMed] [Google Scholar]
- 38.Tortiglione C., Quarta A., Malvindi M. A., Tino A., Pellegrino T., Fluorescent nanocrystals reveal regulated portals of entry into and between the cells of Hydra. PLOS ONE 4, e7698 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marchesano V., Hernandez Y., Salvenmoser W., Ambrosone A., Tino A., Hobmayer B., de la Fuente J. M., Tortiglione C., Imaging inward and outward trafficking of gold nanoparticles in whole animals. ACS Nano 7, 2431–2442 (2013). [DOI] [PubMed] [Google Scholar]
- 40.Ambrosone A., del Pino P., Marchesano V., Parak W. J., de la Fuente J. M., Tortiglione C., Gold nanoprisms for photothermal cell ablation in vivo. Nanomedicine 9, 1913–1922 (2014). [DOI] [PubMed] [Google Scholar]
- 41.Moros M., Ambrosone A., Stepien G., Fabozzi F., Marchesano V., Castaldi A., Tino A., de la Fuente J. M., Tortiglione C., Deciphering intracellular events triggered by mild magnetic hyperthermia in vitro and in vivo. Nanomedicine 10, 2167–2183 (2015). [DOI] [PubMed] [Google Scholar]
- 42.Passano L. M., McCullough C. B., The light response and the rhythmic potentials of Hydra. Proc. Natl. Acad. Sci. U.S.A. 48, 1376–1382 (1962). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Passano L. M., McCullough C. B., Coordinating systems and behavior in Hydra. I. Pacemaker system of the periodic contractions. J. Exp. Biol. 41, 643–644 (1964). [Google Scholar]
- 44.Taddei-Ferretti C., Musio C., Photobehaviour of Hydra (Cnidaria, Hydrozoa) and correlated mechanisms: A case of extraocular photosensitivity. J. Photochem. Photobiol. B 55, 88–101 (2000). [DOI] [PubMed] [Google Scholar]
- 45.Guertin S., Kass-Simon G., Extraocular spectral photosensitivity in the tentacles of Hydra vulgaris. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 184, 163–170 (2015). [DOI] [PubMed] [Google Scholar]
- 46.Gellner K., Praetzel G., Bosch T. C. G., Cloning and expression of a heat-inducible hsp70 gene in two species of Hydra which differ in their stress response. Eur. J. Biochem. 210, 683–691 (1992). [DOI] [PubMed] [Google Scholar]
- 47.Malafoglia V., Traversetti L., Del Grosso F., Scalici M., Lauro F., Russo V., Persichini T., Salvemini D., Mollace V., Fini M., Raffaeli W., Muscoli C., Colasanti M., Transient receptor potential melastatin-3 (TRPM3) mediates nociceptive-like responses in Hydra vulgaris. PLOS ONE 11, e0151386 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Woo S., Lee A., Won H., Ryu J. C., Yum S., Toxaphene affects the levels of mRNA transcripts that encode antioxidant enzymes in Hydra. Comp. Biochem. Physiol. 156, 37–41 (2012). [DOI] [PubMed] [Google Scholar]
- 49.Westfall J. A., Kinnamon J. C., Sims D. E., Neuro-epitheliomuscular cell and neuro-neuronal gap junctions in Hydra. J. Neurocytol. 9, 725–732 (1980). [DOI] [PubMed] [Google Scholar]
- 50.Takaku Y., Hwang J. S., Wolf A., Böttger A., Shimizu H., David C. N., Gojobori T., Innexin gap junctions in nerve cells coordinate spontaneous contractile behavior in Hydra polyps. Sci. Rep. 4, 3573 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Terakita A., The opsin. Genome Biol. 6, 213 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Koyanagi M., Takada E., Nagata T., Tsukamoto H., Terakita A., Homologs of vertebrate Opn3 potentially serve as a light sensor in nonphotoreceptive tissue. Proc. Natl. Acad. Sci. U.S.A. 110, 4998–5003 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang B., Yuan H., Zhu C., Yang Q., Lv F., Liu L., Wang S., Polymer-drug conjugates for intracellar molecule-targeted photoinduced inactivation of protein and growth inhibition of cancer cells. Sci. Rep. 2, 766–773 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu J., Fu T.-M., Cheng Z., Hong G., Zhou T., Jin L., Duvvuri M., Jiang Z., Kruskal P., Xie C., Suo Z., Fang Y., Lieber C. M., Syringe-injectable electronics. Nat. Nanotechnol. 10, 629–636 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pecher J., Mecking S., Nanoparticles of conjugated polymers. Chem. Rev. 110, 6260–6279 (2010). [DOI] [PubMed] [Google Scholar]
- 56.Loomis W. F., Lenhoff H. M., Growth and sexual differentiation of Hydra in mass culture. J. Exp. Zool. 132, 555–573 (1956). [Google Scholar]
- 57.Livak K. J., Schmittgen T. D., Analysis of relative gene expression data using real-time quantitative PCR and the 2–CT method. Methods 25, 402–408 (2001). [DOI] [PubMed] [Google Scholar]
- 58.Senthilan P. R., Piepenbrock D., Ovezmyradov G., Nadrowski B., Bechstedt S., Pauls S., Winkler M., Mobius W., Howard J., Göpfert M. C., Drosophila auditory organ genes and genetic hearing defects. Cell 150, 1042–1054 (2012). [DOI] [PubMed] [Google Scholar]
- 59.Shen W. L., Kwon Y., Adegbola A. A., Luo J., Chess A., Montell C., Function of rhodopsin in temperature discrimination in Drosophila. Science 331, 1333–1336 (2011). [DOI] [PubMed] [Google Scholar]
- 60.Pennisi E., Physiology. Opsins: Not just for eyes. Science 339, 754–755 (2013). [DOI] [PubMed] [Google Scholar]
- 61.Holstein T. W., A view to kill. BMC Biol. 10, 18 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Plachetzki D. C., Fong C. R., Oakley T. H., Cnidocyte discharge is regulated by light and opsin-mediated phototransduction. BMC Biol. 10, 17 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Papadopoulos J. S., Agarwala R., COBALT: Constraint-based alignment tool for multiple protein sequences. Bioinformatics 23, 1073–1079 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/3/1/e1601699/DC1
fig. S1. Characterization of P3HT-NPs.
fig. S2. Impact of P3HT-NPs on Hydra population growth.
fig. S3. Evaluation of polystyrene NP impact on Hydra behavior and gene expression.
fig. S4. Light responsiveness of Hydra opsin3-like gene.
fig. S5. Gene profiling under short illumination condition.
fig. S6. Hydra opsin3-like gene transcription does not respond to heat stress.
fig. S7. Alignment of opsin chromophore-binding transmembrane domain proteins from different animal species.
fig. S8. Alignment of the predicted protein sequences representative of different classes of Hydra opsins.
table S1. List of forward and reverse primers used in the qRT-PCR analysis.
table S2. List of the opsin-like gene sequences present in the H. vulgaris genome database.


