Abstract
Integration of functional and genomic screening strategies reveals clinically actionable genetic events that impact the effectiveness of cancer treatment regimens and the outcomes of cancer patients.
MOLECULAR STRATIFICATION AND TARGETING IN CANCER
Molecular characterization is changing our conceptual understanding of cancer, the process of cancer diagnosis, and the therapeutic interventions that are prescribed for individual patients. Hematologic malignancies were first described in the mid-1800s as an improper expansion of white blood cells. By the early 1900s, hematologic malignancies were classified into four distinct categories. Over the past 100 years, the stratification of this disease has increased from those four categories to more than 150 types of hematologic malignancy that are now recognized by the World Health Organization. A few of these current diagnostic subsets are already defined according to genetic parameters; however, the vast majority of diagnoses are still classified according to patterns of neoplastic cell lineage, morphology, histology, and immunohistochemistry. Over the coming years, we are poised for a further dramatic expansion of cancer subtypes based on genetic and epigenetic features of these diseases, and this innovation is being spurred by the technological revolution of deep sequencing.
Deep sequencing has enabled the rapid sequencing of entire human genomes and has also resulted in a plummeting of DNA sequencing costs. This acceleration of speed and efficiency has yielded a wealth of new information about the genetic abnormalities that occur in the setting of cancer. The advent of deep sequencing has already profoundly changed our understanding of cancer genetics and will continue to do so for a number of years. In the near future, we will have a deep and comprehensive understanding of the mutations that arise and cause cancer as well as the combinatorial diversity of these mutational events. Although this knowledge is exciting from a biological perspective, full delivery of the promise of genetically driven medicine will require that we make the leap from knowledge of cancer genetic events and translate this information into new and personalized therapeutic regimens for patients.
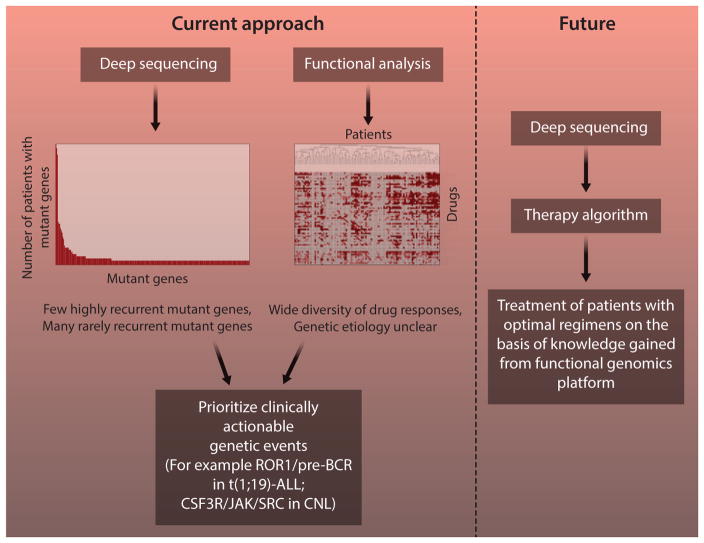
Therapies that are targeted to cancer-causing genetic abnormalities have improved patient outcomes for cancers with well-defined genetic drivers. Extrapolation of this paradigm of targeted therapy to all malignancies will require the subdivision of cancer into distinct entities with operationally important mutant genes, knowledge of the signaling pathways that become dysregulated as a consequence of these mutant genes, and the ability to match the dysregulated pathways with cocktails of effective, targeted therapeutic agents. This task of matching genetic information from deep sequencing with clinically meaningful therapies will require the integration of genomic data with functional information regarding drug-sensitivity patterns of cancer cells. Integration of data in this manner has been the primary emphasis of my research program (Fig. 1), and this article will highlight some of the findings that have been made with such an integrated approach.
Fig. 1. Functional genomics approach: current and future.
To comprehensively define the functional genomic landscape of cancer, we are currently performing parallel analyses of primary specimens from patients with hematologic malignancies. Genomic analysis reveals a large number of genetic events, most of which are seen in very small fractions of patients. Functional screening shows heterogeneous patterns of drug sensitivity with unclear genetic etiology. By integrating these data streams, we have identified pathogenetically important and clinically actionable genetic events, such as dependence on ROR1 and the pre–B cell receptor in t(1;19)-ALL and mutant CSF3R signaling in CNL. As we learn more from this platform, we build toward a future in which personalized, targeted therapies are prescribed solely on the basis of rapid genomic analysis of individual patient tumor specimens.
FUNCTIONAL GENOMICS FOR PERSONALIZED MEDICINE
With the goal of quickly identifying actionable gene targets in individual hematologic malignancy patients, I sought to develop a RNA interference (RNAi)–based screen that could be applied directly to primary cells from patients. One of the early specimens I analyzed in this manner showed a dramatic sensitivity to silencing of the nonreceptor tyrosine kinase JAK2. Sequence analysis of JAK2 and other genes that can regulate JAK2 revealed an abnormal two-base-pair insertion in the thrombopoietin receptor MPL, which signals upstream of JAK2. I confirmed the transformative capacity of this mutant gene as well as its capacity to dysregulate JAK2 signaling. The patient from whom this specimen was drawn was treated on a clinical trial with a kinase inhibitor, midostaurin, that exhibits potent activity against JAK2, and the patient exhibited a dramatic response to this therapeutic agent (1). A further search for patient specimens harboring this two-base-pair insertion in MPL revealed no other patients out of thousands screened, suggesting that functional testing can identify important gene targets irrespective of their disease frequency, but that effective use of this technology to unveil all relevant gene targets will require screening of much larger cohorts of patients. As such, I developed a high-throughput platform with which my research team has collectively analyzed nearly 1000 cases with RNAi and/or a parallel small-molecule inhibitor screening approach (2). We have additionally analyzed many of these same specimens with deep-sequencing technology. This platform has led to the discovery of numerous actionable genetic lesions, with two of the most notable described here.
T(1;19)-POSITIVE ACUTE LYMPHOBLASTIC LEUKEMIA
Functional screening of specimens from patients with acute lymphoblastic leukemia (ALL) harboring a 1;19 chromosomal translocation showed consistent sensitivity to silencing of the cell surface receptor ROR1 and to the U.S. Food and Drug Administration–approved kinase inhibitor dasatinib. Genomic analysis revealed that ROR1 expression is elevated in all t(1;19)-ALL cases compared with other ALL subsets. Follow-up validation work revealed that dasatinib sensitivity in this setting is mediated through dependence of t(1;19)-ALL cells on signaling from the pre-B cell receptor, which signals through SRC family kinases that are potent targets of dasatinib. We also discovered that ROR1 and the pre-BCR signal cooperatively to promote the viability and growth of t(1;19)-positive ALL cells. Collectively, these findings revealed new therapeutic targets and regimens that can be used for t(1;19)-ALL patients, who comprise ~5% of pediatric ALL cases and ~1 to 2% of adult ALL cases (3).
In addition to t(1;19)-ALL cases, we also showed that a related subset, t(17;19)-ALL, exhibits high levels of ROR1 as well as sensitivity to pre-BCR antagonists such as dasatinib. The 17;19-translocation is extremely rare and carries a dismal prognosis with no reported cases of survivorship. One patient with t(17;19)-ALL has been treated with dasatinib with clinical benefit (4), but this patient’s clinical response was transient, highlighting the need for combination therapy approaches. These approaches are also being advanced in the setting of chronic lymphocytic leukemia, in which similar observations of elevated ROR1 expression and dependence on B cell receptor signaling have been made, and major efforts are underway to use antibodies and chimeric antigen T cell receptors that can target ROR1 (5, 6). All together, these strategies of simultaneous antagonism of ROR1 and the B cell receptor are poised to greatly improve clinical treatment options for patients with t(1;19)-ALL, t(17;19)-ALL, and mature B cell neoplasms.
PHILADELPHIA-NEGATIVE CHRONIC MYELOID LEUKEMIA
Chronic neutrophilic leukemia (CNL) and atypical (BCR-ABL1–negative) chronic myeloid leukemia (aCML) are both hematologic malignancies that have historically been diagnosed on the basis of neoplastic expansion of granulocytic cells and exclusion of genetic drivers known to occur in other myeloproliferative neoplasms (MPNs). The primary genetic basis of both CNL and aCML has remained unknown, and this absence of defining genetic lesions has made diagnosis of these diseases challenging, resulting in a dearth of effective therapeutic options for patients. To identify driver kinase oncogenes in CNL/aCML, we applied our functional genomics approach to interrogate primary cells from a cohort of patients with CNL/aCML. In this disease setting, our functional genomics approach led to the identification of gain-of-function CSF3R mutations in ~60% of CNL and aCML patients (7).
CSF3R is the receptor for colony-stimulating factor 3 (CSF3; GCSF) and is known to play a prominent role in the growth and differentiation of granulocytes. CSF3R mutations have been described in patients with severe congenital neutropenia (SCN), which can evolve into acute myeloid leukemia (AML) (8), but they had not previously been described in de novo leukemia. CSF3R has been shown to signal through both the Janus kinase/signal transducers and activators of transcription (JAK/STAT) pathway and SRC family kinases. CSF3R mutations cluster into two distinct regions of the receptor, with the majority of mutations falling in the extracellular domain (membrane proximal mutations) and the remainder being nonsense/missense mutations that truncate the cytoplasmic tail (truncation mutations). Our parallel functional analysis of specimens harboring these two distinct mutation types revealed differential mechanisms of activation, resulting in divergent downstream signaling and sensitivity to small-molecule inhibitors. We found that membrane proximal mutations conferred ligand independence, which resulted in sensitivity to inhibitors of JAK kinases. We further showed that the ligand independence observed in the setting of these membrane proximal mutations is due to constitutive receptor dimerization and that the mutated residues are sites of O-linked glycosylation, which is abrogated in the context of these mutations (9). In contrast, truncation mutations conferred receptor overexpression and ligand hypersensitivity, resulting in sensitivity to inhibitors of SRC family kinases as well as a new CSF3R signaling mediator, TNK2.
Multiple CSF3R-mutant CNL patients have now been treated with single-agent kinase inhibitors, resulting in dramatic and durable clinical responses, although CSF3R mutations can occur in the context of additional genetic events such as SETBP1 (10), suggesting an ultimate need for combination therapies. Cumulatively, these findings have defined a new subtype of hematologic malignancy and have enabled an efficient genetic test that can be used to identify patients who can now be treated with targeted therapeutic regimens.
Taken together, functional genomic screening has already enabled the definition of multiple new subtypes of cancer, and these subtypes can be matched with immediately available targeted therapies. Continuation of this strategy holds tremendous promise for the further refinement of our understanding of specific cancer subsets based on genetic events, and matching these fine-tuned patient subsets with drug sensitivity patterns marks a major step toward optimal use of comprehensive genomic information for cancer therapy.
Acknowledgments
I thank Martin Wachtel, Rose Wachtel, and AAAS for awarding me the 2014 AAAS Martin and Rose Wachtel Cancer Research Award. Funding: My research is supported by grants from the V Foundation for Cancer Research, The Leukemia & Lymphoma Society, the Gabrielle’s Angel Foundation for Cancer Research, and the National Cancer Institute (5R00CA151457-04; 1R01CA183974-01).
Footnotes
Competing interests: I have received research funding from Incyte Pharmaceuticals. A patent is pending on the use of CSF3R mutation as a diagnostic tool in hematologic malignancies.
REFERENCES AND NOTES
- 1.Tyner JW, Deininger MW, Loriaux MM, Chang BH, Gotlib JR, Willis SG, Erickson H, Kovacsovics T, O’Hare T, Heinrich MC, Druker BJ. RNAi screen for rapid therapeutic target identification in leukemia patients. Proc Natl Acad Sci USA. 2009;106:8695–8700. doi: 10.1073/pnas.0903233106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tyner JW, Yang WF, Bankhead A, 3rd, Fan G, Fletcher LB, Bryant J, Glover JM, Chang BH, Spurgeon SE, Fleming WH, Kovacsovics T, Gotlib JR, Oh ST, Deininger MW, Zwaan CM, Den Boer ML, van den Heuvel-Eibrink MM, O’Hare T, Druker BJ, Loriaux MM. Kinase pathway dependence in primary human leukemias determined by rapid inhibitor screening. Cancer Res. 2013;73:285–296. doi: 10.1158/0008-5472.CAN-12-1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bicocca VT, Chang BH, Masouleh BK, Muschen M, Loriaux MM, Druker BJ, Tyner JW. Crosstalk between ROR1 and the pre-B cell receptor promotes survival of t(1;19) acute lymphoblastic leukemia. Cancer Cell. 2012;22:656–667. doi: 10.1016/j.ccr.2012.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Glover JM, Loriaux M, Tyner JW, Druker BJ, Chang BH. In vitro sensitivity to dasatinib in lymphoblasts from a patient with t(17;19)(q22;p13) gene rearrangement pre-B acute lymphoblastic leukemia. Pediatr Blood Cancer. 2012;59:576–579. doi: 10.1002/pbc.23383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hudecek M, Schmitt TM, Baskar S, Lupo-Stanghellini MT, Nishida T, Yamamoto TN, Bleakley M, Turtle CJ, Chang WC, Greisman HA, Wood B, Maloney DG, Jensen MC, Rader C, Riddell SR. The B-cell tumor-associated antigen ROR1 can be targeted with T cells modified to express a ROR1-specific chimeric antigen receptor. Blood. 2010;116:4532–4541. doi: 10.1182/blood-2010-05-283309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Widhopf GF, 2nd, Cui B, Ghia EM, Chen L, Messer K, Shen Z, Briggs SP, Croce CM, Kipps TJ. ROR1 can interact with TCL1 and enhance leukemogenesis in Eμ-TCL1 transgenic mice. Proc Natl Acad Sci USA. 2014;111:793–798. doi: 10.1073/pnas.1308374111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maxson JE, Gotlib J, Pollyea DA, Fleischman AG, Agarwal A, Eide CA, Bottomly D, Wilmot B, Mc-Weeney SK, Tognon CE, Pond JB, Collins RH, Goueli B, Oh ST, Deininger MW, Chang BH, Loriaux MM, Druker BJ, Tyner JW. Oncogenic CSF3R mutations in chronic neutrophilic leukemia and atypical CML. N Engl J Med. 2013;368:1781–1790. doi: 10.1056/NEJMoa1214514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dong F, Brynes RK, Tidow N, Welte K, Löwenberg B, Touw IP. Mutations in the gene for the granulocyte colony-stimulating-factor receptor in patients with acute myeloid leukemia preceded by severe congenital neutropenia. N Engl J Med. 1995;333:487–493. doi: 10.1056/NEJM199508243330804. [DOI] [PubMed] [Google Scholar]
- 9.Maxson JE, Luty SB, Macmaniman J, Abel ML, Druker BJ, Tyner JW. Ligand-independence of the colony stimulating factor 3 receptor (CSF3R) T618I mutation results from loss of O-linked glycosylation and increased receptor dimerization. J Biol Chem. 2014;289:5820–5827. doi: 10.1074/jbc.M113.508440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Piazza R, Valletta S, Winkelmann N, Redaelli S, Spinelli R, Pirola A, Antolini L, Mologni L, Donadoni C, Papaemmanuil E, Schnittger S, Kim DW, Boultwood J, Rossi F, Gaipa G, De Martini GP, di Celle PF, Jang HG, Fantin V, Bignell GR, Magistroni V, Haferlach T, Pogliani EM, Campbell PJ, Chase AJ, Tapper WJ, Cross NC, Gambacorti-Passerini C. Recurrent SETBP1 mutations in atypical chronic myeloid leukemia. Nat Genet. 2013;45:18–24. doi: 10.1038/ng.2495. [DOI] [PMC free article] [PubMed] [Google Scholar]