Abstract
Glycosylation of proteins is one of the most prevalent post-translational modifications occurring in nature, with a wide repertoire of biological implications. Pathways for the main types of this modification, the N- and O-glycosylation, can be found in all three domains of life—the Eukarya, Bacteria and Archaea—thereby following common principles, which are valid also for lipopolysaccharides, lipooligosaccharides and glycopolymers. Thus, studies on any glycoconjugate can unravel novel facets of the still incompletely understood fundamentals of protein N- and O-glycosylation. While it is estimated that more than two-thirds of all eukaryotic proteins would be glycosylated, no such estimate is available for prokaryotic glycoproteins, whose understanding is lagging behind, mainly due to the enormous variability of their glycan structures and variations in the underlying glycosylation processes. Combining glycan structural information with bioinformatic, genetic, biochemical and enzymatic data has opened up an avenue for in-depth analyses of glycosylation processes as a basis for glycoengineering endeavours. Here, the common themes of glycosylation are conceptualised for the major classes of prokaryotic (i.e. bacterial and archaeal) glycoconjugates, with a special focus on glycosylated cell-surface proteins. We describe the current knowledge of biosynthesis and importance of these glycoconjugates in selected pathogenic and beneficial microbes.
Keywords: glycan biosynthesis, glycoengineering, glycoproteins, prokaryotes, secondary cell-wall polymers, surface (S-) layer
Introduction
Glycobiology is one of the rapidly growing fields in the natural sciences with implications to many areas of basic research, biomedicine and biotechnology (for review, see Varki et al. 2015). In fact, glycosylation of proteins is not only the most common but probably also the most important post-translational modification process occurring in nature. It is known to affect the expression, localisation and life time of numerous proteins, which, in turn, might be of relevance for protein function as well as downstream biological events such as the immune behaviour of a cell (Corfield and Berry 2015; Lyons, Milner and Rosenzweig 2015; Valguarnera, Kinsella and Feldman 2016). It is estimated that at least 40% of the human proteome undergoes glycosylation, with glycans comprising as much as up to 90% of the overall molecular mass of certain glycoproteins. It is important to note that glycans are secondary gene products, meaning that instead of being synthesised in a template-driven manner as it is well known from protein biosynthesis, they are built up along an assembly line in which numerous proteins are involved in a sequential order, with the glycosyltransferases being the best-characterised examples. The protein glycosylation mechanism is further characterised by the phenomenon of microheterogeneity, in which variant glycan structures are found at specific attachment sites of a given glycoprotein (Johannessen, Koomey and Børud 2012). Overall, protein glycosylation is an energetically costly cellular process and approximately 2% of the human genome encodes proteins involved in glycosylation events.
While research on glycoproteins of higher organisms has flourished since Neuberger’s glycopeptide preparations of oval-bumin in the late 1930s (Neuberger 1938), the question of whether carbohydrates are integral components of prokaryotic proteins had not convincingly approached until studies on the surface (S-) layer glycoproteins of the halophile Halobacterium salinarum (Mescher and Strominger 1976) and Gram-positive clostridia (Sleytr and Thorne 1976). However, due the markedly structural differences between the glycans and linkage regions of prokaryotic glycoproteins in comparison to eukaryotic and viral glycoproteins (Gabius 2015; Varki et al. 2015), it needed a few decades after the first description of a prokaryotic glycoprotein until their existence was fully accepted by the scientific community. Despite much effort has been put on the studies of prokaryotic glycoproteins since then, our understanding of prokaryotic glycoprotein glycan structures is still limited. Thus, any detailed analysis of a bacterial or archeal glycoconjugate can significantly contribute to the advancement of the field (Ristl et al. 2011).
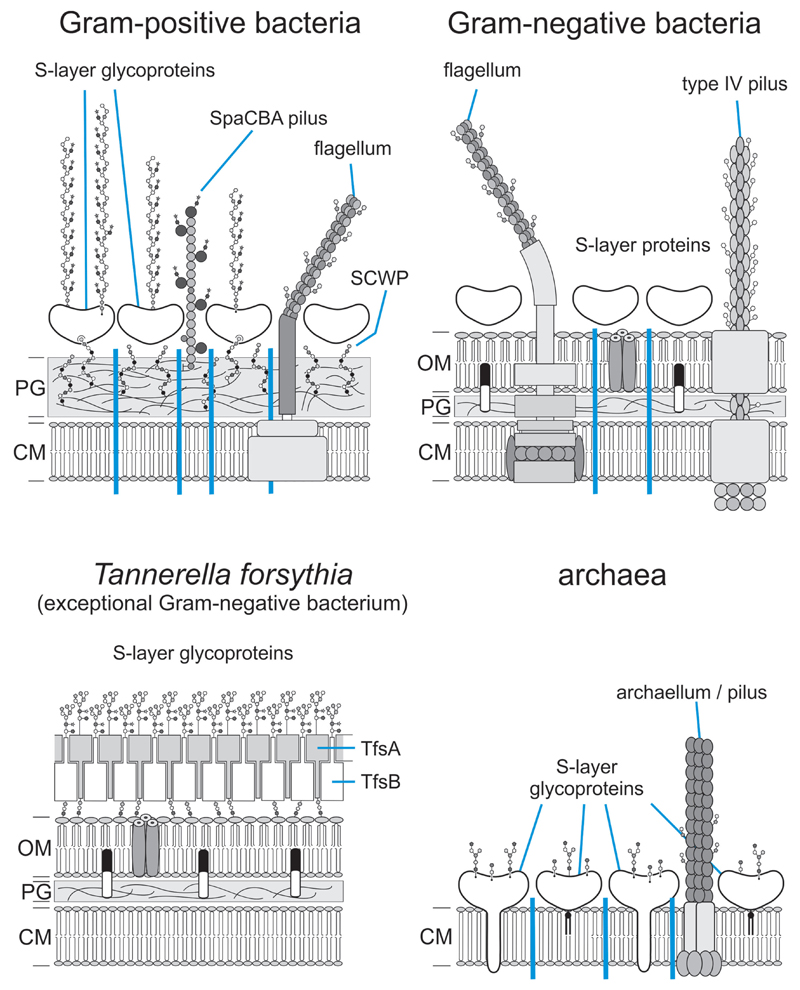
Particularly, the cell surface of prokaryotes (Fig. 1) is rich in glycans where these play fundamental roles in cell physiology (Doyle 2000). As yet relatively unexplored, these modifications have the potential to impact important processes such as microbe–host interactions and immune escape mechanisms. Mostly driven by the discovery of protein glycans in diverse pathogenic microbes (Morrison and Imperiali 2014), there is now an increasing body of evidence of protein glycosylation, both from the domains of Bacteria and Archaea (here is a selection of most relevant reviews from the past five years: (Børud et al. 2011; Giltner, Nguyen and Burrows 2012; Wang et al. 2012; Eichler 2013; Iwashkiw et al. 2013; Messner, Schäffer and Kosma 2013; Meyer and Albers 2013; Nothaft and Szymanski 2013; Jarrell et al. 2014; Tytgat and Lebeer 2014; Kandiba and Eichler 2015; Naegeli and Aebi 2015; Lu, Li and Shao 2015). As more prokaryotic protein glycosylation systems are being identified and characterised, the central question arises as to what governs the biosynthesis and prevalence of particular protein glycans (Table 1). In a recent review Tan, Tang and Exley (2015) discussed different glycoprotein biosynthesis pathways and hypotheses relating to the roles of this post-translational modification in specific host–microbe interactions.
Figure 1.
Scheme of cell envelopes of prokaryotic organisms, showing representative glycoconjugates (different scenarios of cell envelope architecture are separated by blue perpendicular lines), including S-layer glycoproteins (Messner, Schäffer and Kosma 2013), SCWPs (Messner, Schäffer and Kosma 2013), bacterial flagella (Cullen and Trent 2010; Mukherjee and Kearns 2014) and bacterial pili (Korotkov, Sandkvist and Hol 2012; Reunanen et al. 2012) and archaella and archaeal pili (Pohlschroder et al. 2011; Korotkov, Sandkvist and Hol 2012). CM, cytoplasmic membrane; PG, peptidoglycan;  , peptidoglycan strand; OM, outer membrane;
, peptidoglycan strand; OM, outer membrane;  , outer membrane lipid A;
, outer membrane lipid A;  , bacterial and archaeal membrane phospholipid;
, bacterial and archaeal membrane phospholipid;  , archaeal membrane tetraetherlipid;
, archaeal membrane tetraetherlipid;  ,
,  ,
,  , different S-layer glycans; SCWP, secondary cell wall polymer. In different archaeal species, S-layer (glyco)protein anchoring to the cell envelope has been suggested either by a protein transmembrane anchor
, different S-layer glycans; SCWP, secondary cell wall polymer. In different archaeal species, S-layer (glyco)protein anchoring to the cell envelope has been suggested either by a protein transmembrane anchor  (Lechner and Wieland 1989) or in an archaeosortase-dependent process by a lipid anchor
(Lechner and Wieland 1989) or in an archaeosortase-dependent process by a lipid anchor  (Abdul Halim et al. 2016). (Extended and modified from Messner, Schäffer and Kosma 2013. With permission from Elsevier).
(Abdul Halim et al. 2016). (Extended and modified from Messner, Schäffer and Kosma 2013. With permission from Elsevier).
Table 1.
| Organism | Glycosyltransferase | Linkage | Substrates | Residues modified | Glycan | Reference |
|---|---|---|---|---|---|---|
| Campylobacter jejuni | PglB (GT66) | N | ~60 substrates | Asn | Heptasaccharide | Nothaft and Szymanski (2010) |
| Helicobacter pullorum | PglB1 | N | Peptides obtained from a reaction mixture of C. jejuni membranes | Asn | Pentasaccharide | Jervis et al. (2010) |
| Haemophilus influenzae | HMW1C and HMW2C (GT41) | N | HMW1/2 adhesins | Asn | Hex and di-Hex (Hex: Glc or Gal) | Kawai et al. (2011) |
| Actinobacillus pleuropneumoniae | ApNGT | N / O | Autotransporter adhesins (preferred) | Asn, Gln, Ser | Glc and Gal | Naegeli et al. (2014) |
| Campylobacter jejuni 11168 | NC | O | Flagellin FlaA | Ser, Thr | Pse and derivatives | Zampronio et al. (2011) |
| Campylobacter coli VC167 | NC | O | Flagellin | Ser, Thr | Leg and derivatives | Morrison and Imperiali (2014) |
| Burkholderia cepacia K56-2 | bcal0960, PglLBc | O | Relaxed specificity for sugar donor and protein acceptor | Serine | Trisaccharide: HexNAc–HexNAc–Hex | Lithgow et al. (2014) |
| Aeromonas hydrophila AH-3 | OTase-like protein | O | Polar flagella | Ser, Thr | Pse-containing heptasaccharide | Merino et al. (2014) |
| Clostridium difficile 630 | CD0240 | O | Flagellin FliC | Ser, Thr | Modified HexNAc residues | Twine et al. (2009) |
| Paenibacillus alvei CCM 2051T | PAV2c_01630 and PAV2c_01640 | O | Flagellin Hag | Ser, Thr | Trisaccharide: Hex-HexNAc–HexNAc | Janesch et al. (2016) |
| Neisseria meningitidis | PglL | O | Type IV pilin | Ser | Disaccharide: Bac and Gal | Faridmoayer et al. (2007) |
| Neisseria gonorrhoeae | PglO | O | Pilin PilE | Ser | Trisaccharide: Bac and Gal2 | Hartley et al. (2011) |
| Pseudomonas aeruginosa Pa5196 | TfpW (GT-C) | O | Type IVa pilin | Thr, Ser | α1,5-d-Araf residues | Harvey et al. (2011) |
| Mycobacterium tuberculosis | Rv1002c | O | Several cell-surface glycoproteins, including protein Apa | Thr | α1,2-D-mannobioses | Liu et al. (2013) |
| Staphylococcus aureus | GtfA and GtfB | O | Human platelets incl. serine-rich adhesin for platelets (SraP) | Ser | GlcNAc-containing oligosaccharides | Li et al. (2014) |
| Bacillus anthracis | BA3668? | O | BclA spore glycoprotein | Ser, Thr | Serum-specific short O-glycans and large oligosaccharides | Maes et al. (2016) |
| Bacillus subtilis 168 | SunS | S | SunA precursor peptide of sublancin | Cys | Glc | Wang and van der Donk (2011) |
| Haloferax volcanii | AglB | N | Major archaellin FlgA1 | Asn | Pentasaccharide with proximal Glc | Tripepi et al. (2012) |
| Methanococcus maripaludis S2 | AglB | N | Major archaellins FlaB1 and FlaB2, and minor archaellin FlaB3 | Asn | Complex tetra- saccharide with proximal GalNAc | Siu et al. (2015) |
| Methanococcus maripaludis S2 | AglB | N | Major pilin EpdE | Asn | Archaellin tetra- saccharide + Hex, branching off from GalNAc | Siu et al. (2015) |
| Haloferax volcanii | AglB | N | Major adhesion pilins PilA1 and PilA2 | Asn | Like archaellin penta- saccharide | Esquivel et al. (2016) |
| Thermoplasma acidophilum | AglB | N | 6-C-sulfofucose containing cell-surface glycoproteins | Asn | Okta- and heptasac-charide with proximal β-Gal | Vinogradov et al. (2012) |
AglB, archaeal N-oligosaccharyltransferase; Asn, asparagine; ApNGT, A. pleuropneumoniae N-glycosyltransferase; Bac, bacillosamine; BclA, Bacillus collagen-like protein of anthracis; Cys, cysteine; Gal, galactose; Glc, glucose; GalNAc, N-acetylgalactosamine; GlcNAc, N-acetylglucosamine; Gln, glutamine; GT, glycosyltransferase family; Gtf, glycosyltransferase; Hex, hexose; HexNAc, N-acetylhexosamine; HMW, high molecular weight protein; Leg, legionaminic acid; NC, not classified; OTase, oligosaccharyltransferase; Pgl, protein glycosylation; PglB, bacterial N-oligosaccharyltransferase; Pse, pseudaminic acid; SunS, S-glycosyl-transferase; Ser, serine; SraP, serine-rich adhesin for platelets; Thr, threonine.
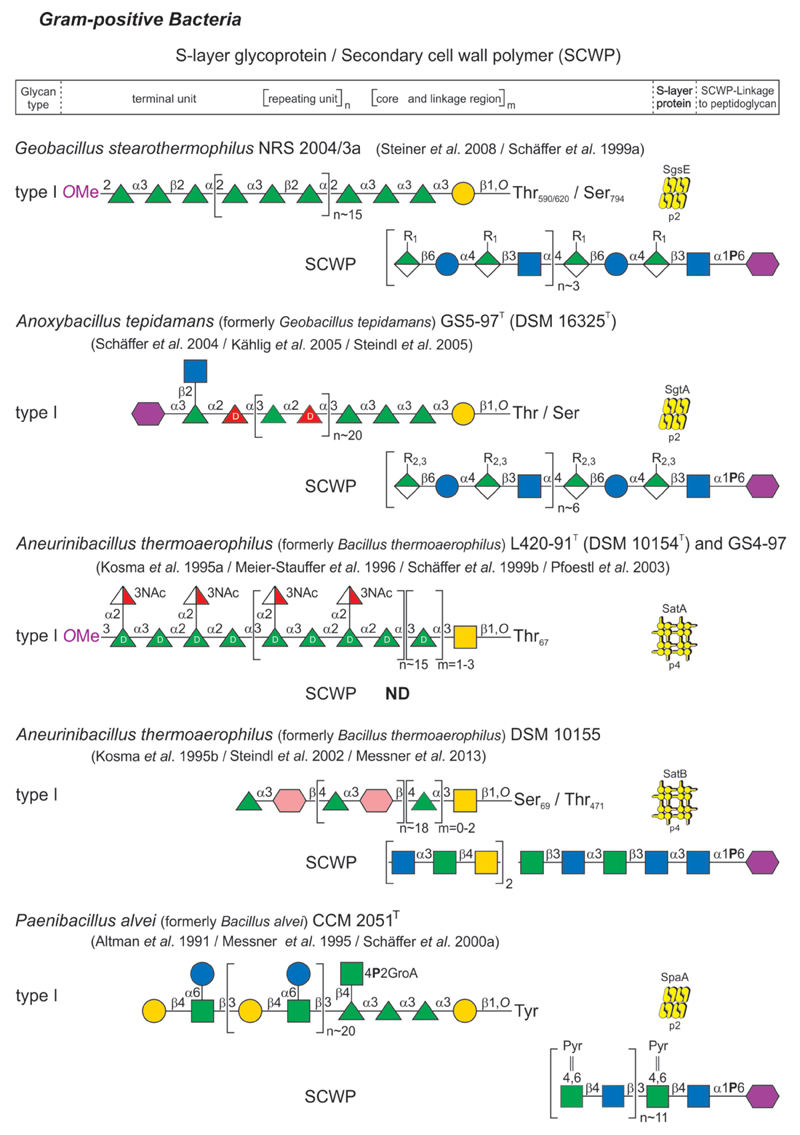
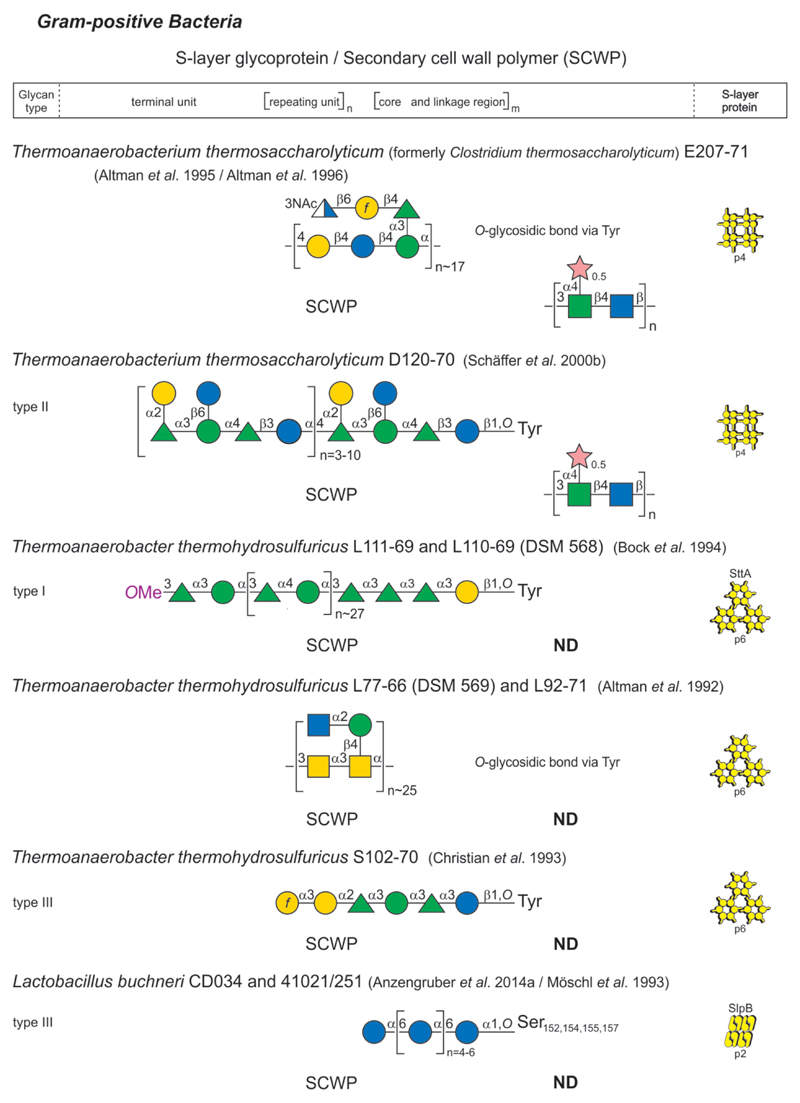
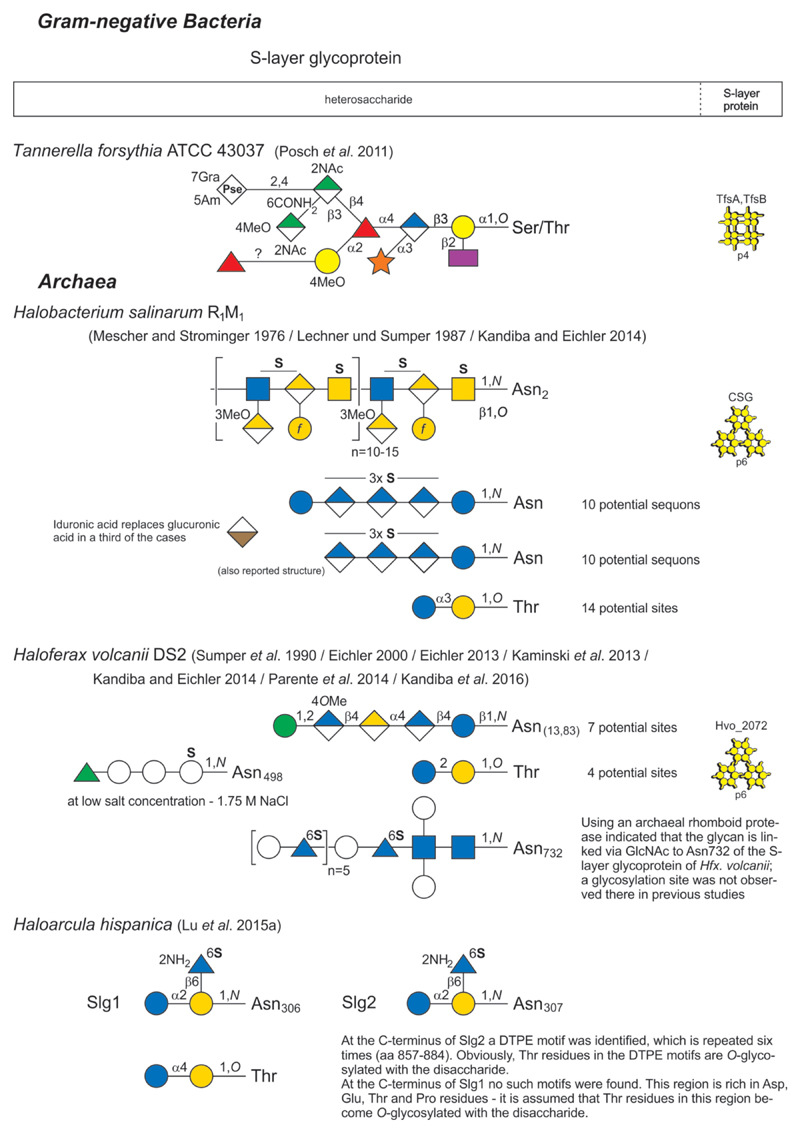
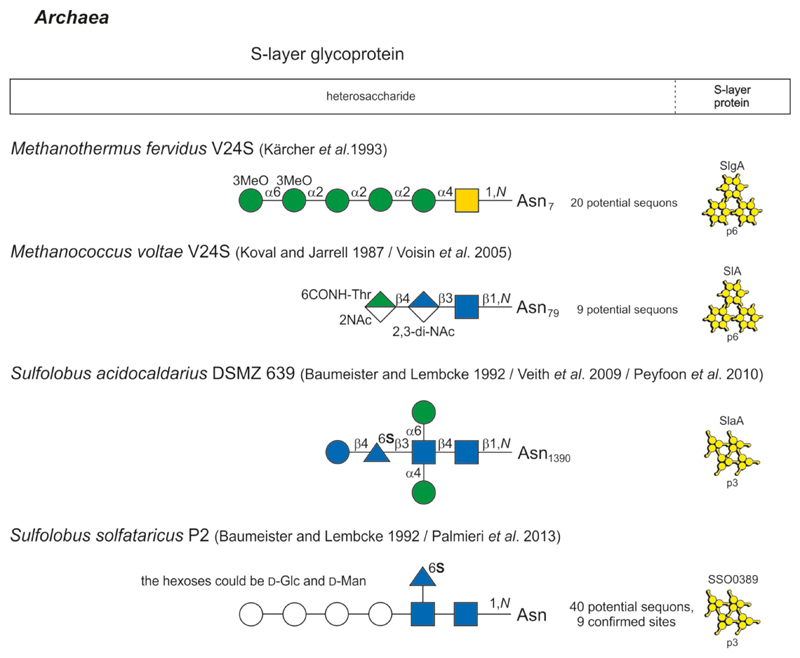
Glycan structure and composition of bacterial and archaeal S-layer glycoproteins and SCWPs are provided in Figs 2-1 to 2-4, and these systems are described in greater detail in specific chapters of the review.
Nowadays it is accepted knowledge that protein N- and O-glycosylation systems are present in both eukaryotes and prokaryotes (for review, see Spiro 2002; Logan 2006; Messner et al. 2010; Jarrell et al. 2014; Varki et al. 2015). While numerous documentations about eukaryotic glycosylation processes predict more than two-thirds of all eukaryotic proteins to be glycosylated (Apweiler, Hermjakob and Sharon 1999), no such estimate is currently available for prokaryotic glycoproteins, whose understanding is lagging behind, mainly due to the enormous variability of glycan structures and underlying glycosylation processes, in several cases accompanied by a lack of tools for genetic manipulation.
The detailed understanding of the impact of glycosylation on structure and function of a protein is often also hampered by the inavailability of well-defined homogeneous glycoproteins/glycopeptides for analysis, because these are mostly difficult to obtain from natural sources in sufficient quantity. To overcome this hurdle, the development of various chemical, enzymatic, chemoenzymatic and bioengineering methods for the synthesis of homogeneous samples is currently a major research goal within the glycobiology research community.
N-Linked Bacterial Glycoproteins
In the following, a description of the currently best investigated N-linked bacterial glycosylation systems is given according to organims, accompanied with glycoeongineering approaches based on distinct modules of these systems.
Protein N-glycosylation in Campylobacter strains
As of yet the best-investigated prokaryotic glycoproteins containing N-linked glycans originate from Campylobacter jejuni, C. lari and other closely related Gram-negative members of the δ-/ε-subdivision of the Proteobacteria family (Szymanski and Wren 2005; Nothaft and Szymanski 2010; Jervis et al. 2012). At the turn of the century, the first general protein glycosylation system was identified in C. jejuni (Szymanski et al. 1999). This bacterium is the etiological agent of human bacterial gastroenteritis worldwide; the consumption of contaminated chicken products is thought to be the principal mode of C. jejuni transmission to the human population. Campylobacter jejuni devotes a large proportion of its small genome (1640 kb) to carbohydrate biosynthesis (Parkhill et al. 2000), yielding an amazing repertoire of glycoconjugates. This includes lipooligosaccharides (LOSs) mimicking human glycolipids, capsular polysaccharides with complex and unusual sugars and about 60 proteins that are post-translationally modified with either N- or O-linked glycans, with the crucial roles of these glycans in the biology and pathogenesis of C. jejuni remaining to be fully deciphered (Alemka et al. 2013; Lu, Li and Shao 2015). It is hypothesised that N-linked glycosylation of surface proteins may enhance C. jejuni fitness by protecting bacterial proteins from cleavage by gut proteases (Alemka et al. 2013). The glycome of this important food-borne pathogen turned out to be an excellent toolbox to unravel the fundamentals of prokaryotic glycosylation pathways and their roles in host–microbe interactions and to exploit these pathways for novel diagnostics and therapeutics (Guerry and Szymanski 2008).
Generally, in N-linked protein glycosylation of eukaryotes and prokaryotes, an oligosaccharide is transferred by a membrane-bound oligosaccharyltransferase (N-OST) in a canonical, highly conserved pathway from a lipid donor to asparagines within the sequon N-X-(S/T) of polypeptides (Burda and Aebi 1999; Weerapana and Imperiali 2006). N-Glycosylation in C. jejuni implies the transfer of the heptasaccharide α-GalNAc-(1→4)-α-GalNAc-(1→4)-[β-Glc-(1→3)]-α-GalNAc-(1→4)-α-GalNAc-(1→4)-α-GalNAc-(1→3)-α-Bac to proteins, where N,N′-diacetylbacillosamine (Bac, diNAcBac) is 2,4-diacetamido-2,4,6-trideoxy-d-Glc (Young et al. 2002). This glycose is also found at the reducing end of O-linked oligosaccharides in C. jejuni as well as in Neisseria gonorrhoeae, another Gram-negative pathogen (Morrison and Imperiali 2014). Biosynthesis of diNAcBac starts from UDP-GlcNAc involving three conserved enzymes in a sequential order—a dehydratase, an aminotransferase and an acetyltransferase—all of which have been extensively studied in C. jejuni (Olivier et al. 2006; Schoenhofen et al. 2006; Vijayakumar et al. 2006). The biosynthesis of the full C. jejuni heptasaccharide following the general N-linked protein glycosylation pathway is encoded in the pgl glycosylation gene cluster; pathway genes are remarkably conserved among the Proteobacteria and do not seem to have the potential for phase variation in C. jejuni (Szymanski and Wren 2005). Using chemically synthesised substrates, B. Imperiali´s group demonstrated the complete enzymatic set-up involved in the C. jejuni N-linked glycosylation process (Weerapana and Imperiali 2006). That study also discovered conceptual differences between prokaryotic and eukaryotic N-glycosylation systems (Larkin and Imperiali 2011). While prokaryotes (i.e.bacteria and archaea) display a certain degree of variability in N-linked glycan structures due to the utilisation of unique monosaccharide building blocks during the assembly process (Larkin and Imperiali 2011; Schwarz and Aebi 2011; Nothaft et al. 2012), nearly all eukaryotes produce the same nascent tetradekasaccharide (Glc3Man9GlcNAc2); heterogeneity is introduced into this glycan structure after it is transferred to the protein through a complex series of glycosyl trimming and addition steps (Burda and Aebi 1999).
Following up fundamental studies of the C. jejuni N-linked glycosylation system, a milestone for prokaryotic glycoengineering had been accomplished with the successful transfer of this N-glycosylation system into Escherichia coli (Wacker et al. 2002). After this ground-breaking observation, an extensive exploitation of the biotechnological application potential of this N-linked glycosylation pathway ensued (e.g. Dürr et al. 2010; Schwarz et al. 2010; Nothaft et al. 2012; Valderrama-Rincon et al. 2012; Naegeli and Aebi 2015; Ollis, Chai and DeLisa 2015), with the N-OST PglB being a key module thereof. In these various glycoengineering approaches, PglB proved to possesses remarkably relaxed substrate specificity.
In 2011, Locher and colleagues published the first X-ray crystal structure of a functional bacterial OST, which was the N-OST from C. lari, in complex with an acceptor peptide (Lizak et al. 2011); partial structures of yeast N-OST subunits have been published earlier (Yan, Wu and Lennarz 2005). The protein N-glycosylation reaction in bacteria and archaea is catalysed by a single protein, namely the N-OST, which is in contrast to most eukaryotes, where a membrane protein complex of nine subunits located in the endoplasmic reticulum performs this task (Yan and Lennarz 2002). This central, catalytic enzyme PglB of Proteobacteria resembles the Stt3 subunit of the eukaryotic N-glycosylation machinery, which has homologues in bacteria and archaea (Lizak et al. 2011; Matsumoto et al. 2013). The X-ray structure defined the fold of Stt3 proteins and provided insight into glycosylation sequon recognition and amide nitrogen activation, both of which are prerequisites for the formation of the N-glycosidic linkage. To better understand the mechanism of PglB, quantification of sequon binding and glycosylation turnover in vitro using purified enzyme and fluorescently labelled, synthetic peptide substrates was performed (Gerber et al. 2013). This work revealed the impact of active site residues and divalent metal ions for sequon binding, the specificity of the Ser/Thr binding pocket and bacteria-specific requirements of sequon recognition. The quantitative assessment of peptide binding and catalysis provided insight into the natural selection of the N-X-S/T sequon in eukaryotes and the extended sequon D/E-Y-N-X-S/T(Y, X ≠ P) in Campylobacter strains (Kowarik et al. 2006) where the eukaryotic primary consensus sequence for N-glycosylation is N-terminally extended for recognition by the bacterial N-OST PglB. Thus, bacterial N-glycosylation site selection is more specific than the eukaryotic counterpart with respect to the polypeptide acceptor sequence. In this context, it is of importance that recently the structure of an ATP-binding cassette (ABC) transporter lipid flippase has been determined at high resolution (Perez et al. 2015). A prominent example of the flipping reaction is the translocation of lipid-linked oligosaccharides (LLOs), which serve as donors in N-linked protein glycosylation. In C. jejuni, this process is catalysed by the ABC transporter PglK. Based on crystal structures in distinct process states in combination with a newly devised in vitro flipping assay and in vivo studies, a mechanism of the PglK-catalysed LLO flipping has been inferred (Perez et al. 2015). The proposed mechanism is distinct from the classical alternating-access model applied to other transporters. Two key differences are the proposed recognition of the polyprenyl tail of the LLOs on the PglK surface by means of the external helix EH in the PglK–LLO interaction, and the requirement of a long, sufficiently wide translocation pathway that contains the pyrophosphate and oligosaccharide moieties during flipping. In contrast, the proposed mechanism for MsbA, the bacterial flippase of the lipid A-core, implicates that the entire lipid A-core may enter the nucleotide-free state of MsbA during the flipping reaction (Ward et al. 2007; Eckford and Sharom 2010). With the recent observations, a more detailed molecular basis for understanding the mechanism of N-linked protein glycosylation in C. jejuni is now provided.
In further glycosylation engineering efforts based on this insight, a convergent chemoenzymatic method was developed that permits site-specific enzymatic ligation between an activated glycan oxazoline and a GlcNAc-peptide/protein to yield homogeneous glycopeptide/proteins (Schwarz et al. 2010; Schwarz and Aebi 2011; Wang 2011). The method involves glycosylation pathway engineering and functional transfer of the C. jejuni glycosylation machinery (Wacker et al. 2002) into E. coli to express a glycoprotein, in which the Asn-linked monosaccharide bacillosamine was changed to the eukaryotic GlcNAc moiety. Fortunately, PglBC. jejuni accepted the resulting GlcNAc(GalNAc)5-containing glycolipid as substrate to glycosylate the target protein. Then the Asn-linked N-glycan was trimmed by α-N-acetylgalactosaminamidase to give a GlcNAc-tagged glycoprotein. Finally, eukaryotic N-glycans of high-mannose or complex type were introduced by an enzymatic transglycosylation reaction to produce homogeneous eukaryotic glycoproteins (Schwarz et al. 2010). Different high-mannose and complex type N-glycans could be readily transferred to the GlcNAc moiety by ENGases to provide full-size glycopeptides. The usefulness of the chemoenzymatic method was exemplified by the efficient synthesis of a complex glycoform of polypeptide C34, a potent HIV inhibitor derived from HIV-1 gp41. Thus, this approach provides an efficient way for introducing complex N-glycans into polypeptides for the gain of novel protein properties that might be valuable for drug discovery (Lomino et al. 2013).
Recently, a mixed approach combining in vivo and in vitro steps for the synthesis of glycoproteins containing the Lewis × antigen was devised by using glycosyltransferases from different bacteria (Hug et al. 2011). Proteins carrying Lewis antigens have been shown to have an application potential in the treatment of diverse autoimmune diseases. The initiating glycosyltransferase WecA from E. coli (Raetz and Whitfield 2002; Ruiz, Kahne and Silhavy 2009) was employed for the addition of the first GlcNAc residue onto the lipid carrier undekaprenyl pyrophosphate (undPP). Glycosyltransferases from the Haemophilus influenzae LOS biosynthesis cluster (lsgc-f) were introduced into E. coli for completion of the precursor glycolipid β-Gal-(1→4)-β-GlcNAc-(1→3)-β-Gal-(1→3)-GlcNAc-undPP. The glycan was then conjugated in the same E. coli cells by the C. jejuni N-OST PglB onto the protein acceptor AcrA. Eventually, the addition of the fucose residue in an α-(1→3) linkage onto the exterior GlcNAc residue by the Helicobacter pylori fucosyltransferase FucT in vitro completed the synthesis. As a result, a Lewis × containing glycoprotein was synthesised using bacterial enzymes from four different species using in vivo and in vitro steps (Hug et al. 2011).
‘Hijacking’ prokaryotic glycosylation systems to exploit them for glycobiotechnological applications including glycoconjugate vaccine and humanised glycoprotein production is a promising perspective for future developments (Cuccui and Wren 2015). The challenges that remain for these approaches to reach full biotechnological maturity will include new strategies for creation of new glycomaterials including glyconanomaterials, evaluation of their targeting potential and their drug delivery properties, as well as characterisation of the internalisation process of these neo-glycoconjugates (Ramström and Yan 2015). Vaccines against both Gram-negative and Gram-positive organisms have already been developed, and efficacy testing has thus far demonstrated that the vaccines are safe and that robust immune responses are being detected. These are likely to complement and reduce the cost of current technologies, thus opening new avenues for glycan-based application strategies. In conclusion, the molecular understanding of bacterial N-glycosylation systems has opened up new avenues for engineering of bacteria to produce glycoproteins that are tailored for specific purposes, displaying either defined eukaryotic-like (such as therapeutic glycoproteins) or completely novel glycan structures (neo-glycoconjugates). Such approaches might also be exploited for screening purposes for improvement and adaptation of the glycosylation machinery to specific applications (Nothaft and Szymanski 2013; Çelik et al. 2015; Cuccui and Wren 2015; Naegeli and Aebi 2015; Vorwerk et al. 2015).
Protein N-glycosylation in Helicobacter, Haemophilus,Pseudomonas and Actinobacillus strains
N-Glycosylation of proteins has also been documented in strains of Helicobacter (Jervis et al. 2010), Haemophilus (St Geme and Yeo 2009) and Pseudomonas (Khemiri et al. 2013), however, without detailed structural characterisation of the synthesised glycans (for review, see Tan, Tang and Exley 2015). In the δ-proteobacterium Actinobacillus pleuropneumoniae, an unusual glycosylation pathway was identified (Naegeli et al. 2014).
Emerging genome sequencing data revealed that pglB orthologues are present in a subset of species from the δ- and ε-Proteobacteria, including three Helicobacter species—H. pullorum, H. canadensis and H. winghamensis. In contrast to C. jejuni, these species contain two unrelated pglB genes (pglB1 and pglB2), neither of which is located within a larger genomic protein glycosylation locus. In complementation experiments, the H. pullorum PglB1 protein, but not PglB2, could transfer the C. jejuni N-linked glycan onto an acceptor protein in E. coli (Jervis et al. 2010). Analysis of N-glycosylation in an in vitro OST assay revealed that a linear pentasaccharide is synthesised in H. pullorum that is processed via PglB1-dependent N-glycosylation. This reaction requires an acidic residue at the –2 position of the N-glycosylation sequon, as for C. jejuni (Kowarik et al. 2006). Attempted insertional knockout mutagenesis of the H. pullorum pglB2 gene was unsuccessful, suggesting that this gene is essential for the organism (Jervis et al. 2010). Up to now, however, no specific function has been assigned to PglB2.
Haemophilus influenzae is a pathogen of the respiratory tract that causes a severe burden of disease in children in both developed and developing countries. Six different capsular serotypes have been identified next to unencapsulated (non-typeable) Ha. Influenzae. Infection by non-typeable Ha. influenzae (NTHi) is the most common cause of exacerbations in chronic obstructive pulmonary disease, a major and growing global health problem in ageing populations (Murphy 2006). Up to 80% of NTHiclinical isolates contain genes encoding the related high-molecular-weight adhesins HMW1 and HMW2 (St Geme and Yeo 2009; Lu, Li and Shao 2015). The Ha. influenzae HMW1 adhesin is glycosylated by HMW1C, a novel glycosyltransferase in the GT41 family. The HMW1C-like proteins share features of glycogen synthases and OSTs, in part accounting for their dual function as glycosyltransferases that catalyse N-linkages of glucose and galactose residues to HMW1 as well as formation of O-glycosidic bonds between glucose residues on HMW1. It was further shown that HMW1 is glycosylated at multiple asparagines within the well-recognised N-X-S/T (X ≠ P) consensus sequence for N-linked glycans and, in one case, at an N-V-E site (Gross et al. 2008; Kawai et al. 2011). Since the modifying carbohydrates at these sites are glucose or galactose residues (for review, see Tan, Tang and Exley 2015) rather than N-acetylated sugars, this revealed an unusual carbohydrate modification suggesting the involvement of a glycosyltransferase with a novel enzymatic activity capable of transferring hexose moieties to asparagine residues (Gawthorne et al. 2014). As mentioned above, in one case of HMWB glycosylation, the glycosylation sequon was found to be N-V-E, yielding a new type of sequon structure. Thus, it will be interesting to see if the common N-glycosylation machinery can modify this site too (Gross et al. 2008).
In A. pleuropneumoniae, protein N-glycosylation was identified to take place in the cytoplasm and to be mediated by a soluble NGT that utilises nucleotide-activated monosaccharides to glycosylate asparagine residues. To characterise this process in detail, the N-glycosylation system of A. pleuropneumoniae was functionally transferred into E. coli using an approach comparable to that described for the transfer of the C. jejuni N-glycosylation machinery into E. coli (Wacker et al. 2002). The A. pleuropneumoniae NGT is an inverting glycosyltransferase that recognises the N-X-(S/T) consensus sequence, thus exhibiting similar acceptor site specificity as the eukaryotic OST despite the unrelated predicted structural architecture and the apparently different catalytic mechanism. The identification of an enzyme that integrates some of the features of an OST in a cytoplasmic pathway defines a novel class of N-linked protein glycosylation found in pathogenic bacteria (Schwarz et al. 2011). This NGT constitutes a general protein glycosylation system with a preference for autotransporter adhesins as protein substrates in vivo, thereby displaying a surprisingly relaxed peptide substrate specificity. Although N-X-(S/T) is the preferred acceptor sequon, glycosylation of alternative sequons was detected, including modification of glutamine and serine residues. The NGT was also able to glycosylate heterologous proteins. Therefore, with A. pleuropneumoniae a novel route for engineering of N-glycoproteins in bacteria has been identified (Naegeli et al. 2014).
Prokaryotic N-linked glycans and key OSTs have been mainly studied in ε-Proteobacteria, including strains from the families Campylobacteraceae and Helicobacteraceae. Information about the N-linked glycoproteome in γ-Proteobacteria such as in members of the order Legionellales or the family Pseudomonadaceae is still scarce. In the literature, there are only two reports on N-glycosylated proteins from different pseudomonads, however, without structural characterisation (Bartels et al. 2011; Khemiri et al. 2013). In the latter report, the FliC flagellin glycopolymorphism in Pseudomonas aeruginosa was investigated after N-glycosidase treatment with 18O labelling. To detect the possible sites of FliC modification, de novo sequencing was performed to discriminate between spontaneous deamidation and N-glycan loss. This approach led to the proposal of three potential N-glycosylated sites on the primary sequence of FliC - Asn26, Asn69 and Asn439—with two of them localised to an N-X-(S/T) consensus sequence (Asn26, Asn439) (Khemiri et al. 2013). Previous analyses of flagellin glycosylation in Pseudomonas strains reported only the presence of O-linked glycans (Takeuchi et al. 2007; Miller et al. 2008). A 2D electrophoresis pattern of FliC, however, strongly suggested that the glycosylation of this protein is more complex (Khemiri et al. 2013). It is assumed that FliC is a polymorphous glycoprotein that also exhibits N-glycoforms, probably arising from macro- and microheterogeneity (Morelle et al. 2006).
O-Linked bacterial glycoproteins
Bacterial O-oligosaccharyl transferases (O-OSTs) constitute a growing family of enzymes that are responsible for protein O-linked glycosylation involving the attachment of glycans to hydroxyl groups of serine, threonine (Power, Seib and Jennings 2006; Aas et al. 2007; Faridmoayer et al. 2007) and tyrosine residues (Zarschler et al. 2010b). On the basis of a sensitive and specific glycan serotyping system, microheterogeneity has been reported for O-linked protein glycosylation systems in different bacteria. Factors underlying microheterogeneity in reconstituted expression systems have been identified and modelled, but those impacting natural systems largely remain enigmatic.
Many proteins from various Gram-negative species, including pathogens such as C. jejuni (Szymanski and Wren 2005; Mahdavi et al. 2014), Burkholderia cenocepacia (Lithgow et al. 2014), Acinetobacter baumannii (Iwashkiw et al. 2012), H. pylori (Hopf et al. 2011), mycoplasmas (Jordan et al. 2013), E. coli (Benz and Schmidt 2001; Sherlock et al. 2006; Charbonneau et al. 2012), Bacteroides fragilis and Tannerella forsythia (Posch et al. 2013b), as well as Gram-positive species including different clostridia (Twine et al. 2008; Twine et al. 2009) and Listeria monocytogenes (Schirm et al. 2004b) are decorated with O-linked glycans of differing chemical composition (for review, see Tan, Tang and Exley 2015).
In the following, bacterial O-linked glycoproteins are described following the categorisation into flagellins, fimbriae and pili, and ‘other glycoproteins’; for the latter, currently only distinct aspects but not an extended picture as for the first two categories is available. The best investigated bacteria of each category are listed and described exemplarily.
Flagellins
Flagella-mediated motility is a common trait among many bacteria. Bacterial flagella are complex nanostructures in which a 10- to 15-μm helical filament extends from the cell surface and is anchored to a rotating basal body spanning the bacterial envelope. The helical filament is composed of repeating subunits known as flagellins (Logan 2006; Erhardt, Namba and Hughes 2010). Flagellin glycosylation has been observed on an increasing number of Gram-negative and Gram-positive bacteria. Many of them are pathogens and despite the enormous differences of the strain-specific glycan structures glycosylation of the flagellar proteins is considered a major virulence factor. Examples of bacteria with glycosylated flagella presented here include Campylobacter (Thibault et al. 2001; Verma et al. 2006), Pseudomonas (Schirm et al. 2004a) and Helicobacter species (Josenhans et al. 2002; Schirm et al. 2003), B. cepacia (Lithgow et al. 2014), Aeromonas spp. (Parker et al. 2012; Wilhelms et al. 2012), different clostridia (Twine et al. 2008; Twine et al. 2009), L. monocytogenes (Schirm et al. 2004b) and Paenibacillus alvei (Janesch et al. 2016).
Of particular interest are C. jejuni and C. coli strains, which possess an O-glycosylation machinery for flagellin glycosylation in addition to the previously described general protein N-glycosylation system (Szymanski and Wren 2005; Guerry et al. 2006; Logan 2006; Guerry and Szymanski 2008). Genes encoding the proteins for the synthesis of the flagellin glycans and the respective protein transferases map adjacent to the flagellin structural genes in one of the most hypervariable regions of the C. jejuni chromosome. All flagellin modifications of C. jejuni are based on 5,7-diacetamido-3,5,7,9-tetradeoxy-l-glycero-l-manno-nonulosonic acid (pseudaminic acid - Pse, Pse5Ac7Ac) (Thibault et al. 2001), a nine-carbon sugar that is structurally similar to sialic acid (Neu5Ac) (Varki and Varki 2007; Chen and Varki 2010). While the major flagellin modification is Pse, also derivatives thereof occur, including an acetamidino form (Pse5Am7Ac), an acetylated form (Pse5Ac7Ac8OAc), a form substituted with hydroxyproprionyl groups (Pse5Pr7Pr) and Pse5Am7Ac to which an N-acetylglutamine residue is attached (Pse5Am7Ac8GlnAc) (Thibault et al. 2001; Schirm et al. 2005). The analysis of flagellin A glycosylation from C. jejuni strain NCTC 11168 showed the presence of dimethylglyceric acid derivatives of Pse in addition to Pse5Am7Ac Pse (Hitchen et al. 2010) and the glycosylation sites were assigned to Ser181, Ser207 and Thr464 or Thr465 (Zampronio et al. 2011). The identification of a large number of frequently unstable homopolymeric tracts of G/C residues within genes from chromosomal loci involved in the biosynthesis of capsular polysaccharide, LOS and the flagellin glycoprotein was the most significant finding of the C. jejuni NCTC 11168 genome sequencing project (Parkhill et al. 2000). The resultant on/off switching in the translational status of homopolymerictract-containing genes is thought to mediate the structural diversity of the bacterial cell surface. In fact, it was observed that genes from the flagellin glycosylation locus are involved in generating flagellin glycan diversity. An important protein in this context is Cj1295, which is based on the presence of an aminopeptidase-like domain, suggested to cleave the acetamido group on carbon 5 of the basal Pse5Ac7Ac/Pse5Ac7Am sugar, enabling the substitution of the acetamido group with a methylglyceroyl group (Hitchen et al. 2010). Another unusual feature of C. jejuni flagellins is the absence of Toll-like receptor 5 (TLR5)-binding sites, which are otherwise highly conserved among bacterial flagellins because of their involvement in subunit interactions in the filament. Flagella from C. jejuni and other members of the ε-Proteobacteria are unusual in that the glycosylation of flagellin is crucial for filament assembly (Guerry 2007), and there is preliminary evidence that the flagellin glycans could contribute to subunit interactions (Guerry and Szymanski 2008). Similarly, surface-exposed glycans mediate autoagglutination and microcolony formation by interactions with other flagellar glycans or other surface structures on adjacent bacteria (Guerry et al. 2006). The flagellin glycans also contribute to virulence of C. jejuni; loss of Pse5Am7Ac from the C. jejuni 81–176 flagellin resulted in reduced adherence to and invasion of intestinal epithelial cells (Guerry and Szymanski 2008). Pse is also important in gastric pathogens such as H. pylori, where it occurs in O-linked flagellin glycosylation that is important to flagellum assembly and motility of H. pylori (Hartley et al. 2011; Morrison and Imperiali 2014).
Studies on the C. coli VC167 flagellin identified a second nine-carbon sugar, legionaminic acid (5,7-diacetamido-3,5,7,9-tetradeoxy-d-glycero-d-galacto-nonulosonic acid; Leg) and its derivatives (McNally et al. 2007). Leg was first characterised in the lipopolysaccharides (LPS) O antigen of Legionella pneumophila (Knirel et al. 1994), where it derived its name from. The Leg biosynthetic pathway is found also in most strains of C. jejuni, except for C. jejuni 81–176 (Schoenhofen et al. 2009). As an altenative, Leg formation can result from modification of diNAcBac through three subsequent enzymatic reactions (Morrison and Imperiali 2014).
Studies on flagellar glycoproteins from other Gram-negative pathogens such as Pseudomonas species revealed further insight into O-linked flagellin glycosylation. Pseudomonas aeruginosa PAK (serotype O6) produces a single polar flagellum containing an a-type flagellin, which has been shown to be post-translationally modified at two sites with heterogeneous O-linked glycans (Schirm et al. 2004a). To determine the potential role of O antigen biosynthetic genes in the synthesis of these O-glycans, flagellin proteins isolated from PAK wild-type strain and three O antigen biosynthesis mutants were compared. Flagellins from wbpL and wbpP mutants had a similar size as the wild-type protein (45 kDa), and both were glycosylated. In contrast, flagellin from the wbpO mutant had an apparent molecular mass of 42 kDa, indicating an obvious defect in glycosylation. Thus, flagellin glycosylation in P. aeruginosa PAK depends on the O antigen biosynthetic enzyme WbpO but not WbpP or WbpL. Kinetic evidence shows that WbpO and WbpP can act in either order in the biosynthesis of UDP-d-GalNAcA; however, there is a strong preference of WbpO to convert UDP-d-GlcNAc into UDP-d-GlcNAcA first, before epimerisation into UDP-d-GalNAcA by WbpP. Since the exact composition of the flagellin glycans is not known, it is not yet possible to propose a pathway for flagellin glycan biosynthesis in P. aeruginosa PAK (Miller et al. 2008).
The flagellins from the phytopathogenic bacteria P. syringae pv. glycinea race 4 and P. syringae pv. tabaci 6605 are glycosylated at six serine residues (i.e.Ser143, Ser164, Ser176, Ser183, Ser193 and Ser201) (Takeuchi et al. 2007). The structure of the flagellin glycan at Ser201 from each pathovar consists of a common unique trisaccharide comprising two Rha residues and one modified 4-amino-4,6-dideoxyglucosyl (Qui4N) residue—β-d-Quip4N(3-hydroxy-1-oxobutyl)2Me-(1→3)-α-l-Rhap-(1→2)-α-l-Rhap—and can be found on each of the six serine residues. Determination of the enantiomeric ratio of the Rha residues showed that the flagellin from P. syringae pv. tabaci 6605 consisted solely of l-Rha, whereas the P. syringae pv. glycinea race 4 flagellin contained both l-Rha and d-Rha at a molar ratio of about 4:1. Glycosylation of flagellin was shown to be essential for virulence and host specificity of P. syringae strains (Takeuchi et al. 2007).
Bacteria of the B. cepacia complex are pathogens of humans, plants and animals. Burkholderia cenocepacia is one of the most common B. cepacia complex species infecting patients with cystic fibrosis and its carriage is associated with poor prognosis (Lithgow et al. 2014). In strain B. cepacia K56-2, a general O-linked protein glycosylation system was characterised. The PglLBc O-OST, encoded by the cloned gene bcal0960, was shown to be capable of transferring a heptasaccharide from the C. jejuni N-glycosylation system to an N. meningitidis-derived acceptor protein in an E. coli background, indicating that the enzyme has relaxed specificity for both the sugar donor and the protein acceptor. Disruption of bcal0960 abolished glycosylation and resulted in reduced swimming motility and attenuated virulence of the bacterium towards both plant and insect model organisms (Lithgow et al. 2014). In B. cepacia K56-2, PglLBc is responsible for the glycosylation of 23 proteins involved in diverse cellular processes. Mass spectrometric analysis revealed that these proteins are modified with a trisaccharide HexNAc–HexNAc–Hex. This trisaccharide is unrelated to the organism’s O antigen, which is a polymer of trisaccharide-repeating unit containing Rha and two N-acetylgalactosamine (GalNAc) residues - →4)-α-l-Rhap-(1→3)-α-d-GalpNAc-(1→3)-β-d-GalpNAc-(1→ (Ortega et al. 2005). For normal motility of B. cepacia, complete O antigen and flagellin glycosylation are required; the flagella also contribute to biofilm production. The key genes of B. cepacia flagellin glycosylation were identified, including a predicted glycosyltransferase gene that is linked to the flagellin biosynthesis gene cluster and a putative acetyltransferase gene located within the O antigen LPS cluster. Another gene from the O antigen cluster, rmlB, which is required for both flagellin glycan and O antigen biosynthesis, is essential for bacterial viability, uncovering a novel target against B. cepacia infections. Using glycosylated and non-glycosylated purified flagellin and a cell reporter system to assess TLR5-mediated responses, it was shown that the presence of the flagellin glycan significantly impaired the inflammatory response of epithelial cells. Based on these data, it is suggested that flagellin glycosylation could provide to B. cepacia a strategy to reduce its recognition by the innate immune system (Hanuszkiewicz et al. 2014). Burkholderia cepacia K56-2 also causes opportunistic infections in plants and insects, suggesting that virulence depends on the host and its innate susceptibility to infection (Khodai-Kalaki et al. 2015). It was hypothesised that modifications in key bacterial molecules recognised by the innate immune system modulate host responses to B. cepacia. Modification of LPS with 4-amino-4-deoxy-l-arabinose (l-Ara4N) and flagellin glycosylation attenuate B. cepacia infection in Arabidopsis thaliana and Galleria mellonella insect larvae. Opportunistic bacteria are a perfect example of microbes whose ability to cause disease is intimately related to the host’s ability to recognise and respond to the infection. LPS and flagellin were further investigated for their contribution to infection and eliciting of a host response. It was shown that flagellin glycosylation and LPS modification with l-Ara4N play significant roles in bacterial survival during the early stage of infection but do not alter the sensing of these molecules by the plant innate immune receptors, indicating that these modifications are only critical for the establishment of the infection. Thus, the microbe’s sensing by the host and establishment of infection are interrelated but independent events (Khodai-Kalaki et al. 2015; Tavares-Carreón, Patel and Valvano 2015).
Flagellin from the rice-avirulent N1141 strain of the Gram-negative phytopathogenic bacterium Acidovorax (formerly Pseudomonas) avenae induces plant immune responses including H2O2 generation, whereas flagellin from the rice-virulent K1 strain of Av. avenae does not induce such a response (Hirai et al. 2011). Structural analyses indicated that the glycan moieties attached to Ser178 or Ser183 in the D2 domain of the K1 flagellin might be involved in flagellin recognition by the plant; plants generally have sensitive systems that detect pathogen-derived molecules to protect against infection (Che et al. 2000). Thus, the glycan moiety attached by the K1 glycosyltransferase disrupts flagellin recognition by rice and causes the induction of immune responses. Elucidating the glycan structure associated with the flagellin in the Av. avenae K1 strain will be important to further understand how flagellin is recognised by rice (Hirai et al. 2011).
Glycosylated flagella of several Gram-negative bacteria such as Aeromonas caviae (Parker et al. 2012) and Ae. hydrophila (Wilhelms et al. 2012; Merino et al. 2014) have been discussed in conjunction with their LPS O antigens (Raetz and Whitfield 2002). Motility in Ae. caviae in broth culture is mediated by a single polar flagellum encoded by the fla genes (Parker et al. 2012). The two flagellin subunits—FlaA and FlaB—undergo O-glycosylation with six to eight Pse residues linked to serine and threonine residues in the central region of the subunits. The flm genetic locus in Av. caviae is required for both flagellin glycosylation and LPS O antigen biosynthesis as evidenced by flm mutants. However, none of the flm genes appeared to encode a candidate glycosyltransferase that might add the Pse moiety to FlaA/B. The motility-associated factors (Maf proteins) were then considered as candidate transferases. Bioinformatic analysis indicated that the genome of Av. caviae encodes a single maf gene homologue (maf1) and that this strain has acquired the minimum gene cluster required for the biosynthesis of Pse5Ac7Ac (Tabei et al. 2009). Phenotypic analysis of a maf mutant showed that it is both non-motile and lacks polar flagella. In contrast to flm mutants, this mutant revealed no change in the LPS O antigen pattern and retained its ability to swarm. Analysis of flaA transcription showed that its transcription was unaltered in the maf mutant, while a His-tagged version of the FlaA flagellin protein produced from a plasmid was detected in unglycosylated intracellular form in the maf mutant strain. Complementation of the maf strain in trans partially restored motility and increased levels of glycosylated flagellin to above wild-type levels. These data provided evidence that maf1 is a pseudaminyl transferase responsible for glycosylation of flagellin and suggest that this event occurs prior to secretion through the flagellar type III secretion system. Based on these data, a glycosylation pathway was proposed for flagellin glycosylation and LPS modification in Av. caviae Sch 3N. The pathway to Pse5Ac7Ac is based on similar predicted functions of Av. caviae proteins as known for corresponding proteins in C. jejuni and H. pylori (McNally et al. 2006; Schoenhofen et al. 2006). Following the biosynthesis of Pse5Ac7Ac by the FlmABD and NeuB enzymes, Pse5Ac7Ac is CMP activated through NeuA activity. CMP-Pse5Ac7Ac is then either transferred onto the flagellin by Maf1, which was predicted to be a polar flagellin-specific glycosyltransferase, or transferred onto a sugar-antigen carrier lipid by Lst (sialyltransferase-like protein) to create an LPS O antigen unit, which is subsequently transported across the cytoplasmic membrane by Lsg (Wzx-like flippase) (Tabei et al. 2009; Parker et al. 2012).
In the related strain Ae. hydrophila AH-3 (serotype O34), polar and lateral flagellin proteins were found to be glycosylated with different carbohydrate moieties. The lateral flagellin was modified in O-linkage with a single Pse derivative of 376 Da at three sites, with one site of modification at Ser178, while the polar flagellin was modified with a heterogeneous, more complex O-glycan (Wilhelms et al. 2012). This heptasaccharide is comprised of three N-acetylhexosamines (with variable addition of O–2 phosphate groups and O–2 methyl groups on each), two hexoses and two unknown monosaccharides of 376 and 102 Da in the sequence –376 Da–Hex–Hex–HexNAc–HexNAc–HexNAc–102 Da. A maximum of six heptasaccharide chains was observed by mass spectrometry (MS) analysis of the polar flagellins, one of them linked to Thr161 on FlaB. Compositional analysis of the flagellin glycan indicated the presence of a GalNAc-containing heptasaccharid, whereas the LPS O34 antigen is built from tetrasaccharide repeats comprised of d-mannose, d-GalNAc and 6-deoxytalose (Knirel et al. 2002). In all cases, the 376-Da monosaccharide is the first sugar attached to the polar flagellin. In-frame deletion mutants of the Pse biosynthetic pseB and pseF orthologues resulted in abolition of polar and lateral flagella formation, obviously due to post-transcriptional regulation of the flagellins. Flagellation could be restored by complementation with wild-type Ae. hydrophila pseB or pseF as well as C. jejuni pseB and pseF (Wilhelms et al. 2012). Mutants unable to produce the WecP (encoding a predicted UDP-HexNAc:polyprenol-P GalNAc1-P transferase) or Gne (encoding a UDP-N-acetylgalactosamine 4-epimerase) enzymes showed altered motility, their glycosylation pattern differed from that observed for wild-type polar flagellin and both mutants were devoid of the O34-antigen LPS. The effects observed upon WecP deletion suggested the involvement of a lipid carrier in the glycosylation process. With WecX, a gene coding for an enzyme linking a sugar to a lipid carrier was identified in strain AH-3 and, indeed, its deletion completely abolished motility, flagella production (according to TEM evidence) and flagellin glycosylation. This was the first report of a lipid carrier involved in flagella O-glycosylation. Furthermore, by comparing mutants with differing degrees of polar flagellin glycosylation, the importance of glycosylation in A. hydrophila flagella formation and motility could be demonstrated (Merino et al. 2014).
Other glycosylated flagellin subunits have been identified on Gram-positive pathogenic clostridia such as Clostridium botulinum (Twine et al. 2008), Cl. difficile (Twine et al. 2009) and L. monocytogenes (Schirm et al. 2004b). In Cl. botulinum, bioinformatic genome analysis identified a flagellar glycosylation island containing homologues of Leg biosynthesis genes previously identified in C. coli (Doig et al. 1996). Indeed, structural characterisation of the Cl. botulinum glycan revealed that it was a novel Leg derivative, 7-acetamido-5-(N-methylglutam-4-yl)-amino-3,5,7,9-tetradeoxy-d-glycero-α-d-galacto-nonulosonic acid (αLeg5GluNMe7Ac) (Twine et al. 2008). In other strains, di-N-acetylhexuronic acids were identified as glycan constituents.
The composition of the Cl. difficile flagellin glycans is quite distinct from those of the related organism Cl. botulinum. All Cl. difficile flagellins examined were shown to carry a glycan that is attached to serine and threonine residues via a HexNAc residue. Clostridium difficile 630 produces flagellin that is O-glycosylated at up to seven sites with a HexNAc residue to which a methylated aspartic acid is linked via a phosphate bond. In contrast, flagellins from a number of Cl. difficile isolates from other outbreaks are decorated with heterogeneous O-linked glycans containing up to five monosaccharide residues with masses of 204 (HexNAc), 146 (deoxyhexose), 160 (methylated deoxyhexose) and 192 (heptose) (Twine et al. 2009).
Post-translational modification of proteins with O-GlcNAc is well known for a number of eukaryotic nuclear and cytoplasmic proteins (Hart et al. 2011) and nowadays emerging also for bacteria, such as Listeria species. The addition of GlcNAc to the Listeria flagellin would not require a dedicated glycan biosynthetic pathway, since GlcNAc is a common biosynthetic precursor of numerous biochemical pathways (Schirm et al. 2004b). Since in eukaryotic systems an important component of the O-GlcNAc modification system is the O-GlcNAc transferase (Janetzko and Walker 2014), an enzyme of equivalent function would be a prerequisite for O-GlcNAcylation of L. monocytogenes flagellin. However, search of the L. monocytogenes EGD genome revealed no obvious candidate gene based on homology to known eukaryotic O-GlcNAc transferases, leaving open the question of how GlcNAcylation would be accomplished in this organism.
Recently, flagellin glycosylation was also investigated in the Gram-positive bacterium Paenibacillus (formerly Bacillus) alvei CCM 2051T (Altman et al. 1991; Forsgren 2010; Janesch et al. 2016). This organism swarms vigorously on solidified culture medium, with swarming relying on functional flagella as evidenced by abolished biofilm formation of a non-motile Pa. alvei mutant defective in the flagellin protein Hag. Investigation of the glycobiology of the polar Pa. alvei flagella showed that the purified 30 kDa Hag protein (PAV_2c01710) is modified with an O-linked trisaccharide comprised of one hexose and two N-acetylhexosamine residues at three glycosylation sites (Janesch et al. 2016). Downstream of the hag gene on the bacterial chromosome, two open reading frames (PAV_2c01630, PAV_2c01640) encoding putative glycosyltransferases were shown to constitute a flagellin glycosylation island. Mutants defective in these genes exhibited altered migration in SDS-PAGE as well as loss of extracellular flagella production and bacterial motility. Thus, flagellin glycosylation in Pa. alvei CCM 2051T is pivotal to flagella formation and bacterial motility in vivo; flagella glycosylation is a second protein O-glycosylation system in this bacterium, in addition to the well-investigated S-layer tyrosine O-glycosylation pathway (vide infra) (Zarschler et al. 2010b).
Pilins and fimbriae
Examples of glycosylated pili and fimbriae included here come from Neisseria spp., P. aeruginosa, Mycobacterium tuberculosis, Francisella tularensis and Porphyromonas gingivalis.
Pilin subunits (Logan 2006), mostly of Gram-negative bacteria including important human pathoges, are frequently decorated with O-linked glycans of strain-specific composition (Castric 1995; Stimson et al. 1995; Castric, Cassels and Carlson 2001; Thibault et al. 2001; Schirm et al. 2003, 2004b; Aas et al. 2007; Kus et al. 2008; Børud et al. 2011; Harvey et al. 2011; Lithgow et al. 2014). Different names have been coined for the O-OSTs involved in pilin O-glycosyltation, such as PglL in N. meningitidis (Faridmoayer et al. 2007; Schulz et al. 2013; Harding et al. 2015), PglO in N. gonorrhoeae (Hartley et al. 2011) and PilO in P. aeruginosa (Castric 1995; Faridmoayer et al. 2007). Whereas PilO activity is restricted to short oligosaccharides, PglL is able to transfer diverse oligo- and polysaccharides. These O-OSTs are inner membrane proteins transfering various glycans from a lipid carrier to different protein acceptors, thus exhibiting relaxed specificities. The activity of wild-type PglL and its mutant derivatives was analysed in vivo in engineered E. coli cells, and in in vitro assays. Limited proteolysis experiments of PglL revealed a conformational change that is triggered upon interaction of the C-terminal region of the enzyme with the LLO substrate; while Gln178 and Tyr405 are required for optimal function, His349 is essential for the enzymatic activity and plays a critical role in the interaction with LLOs (Musumeci et al. 2013, 2014). O-OSTs are in most cases difficult to identify using solely bioinformatic methods because of their sequence similarity with WaaL ligases catalysing the last step in LPS synthesis (Hug and Feldman 2011; Whitfield and Trent 2014). For example, the Wzy_C signature domain common to O-OSTs is also present in WaaL ligases (Gebhart et al. 2012).
Dependent on the neisserial species, pilin is glycosylated at Ser63 with an oligosaccharide, whose structure can vary upon the availability of glycosyltransferases. In addition, the pilins of N. meningitidis are subject to other post-translational modifications including the addition of phosphorylcholine (PC) and phosphoglycerol the pilus of N. gonorrhoeae was found to be modified by the addition of PC and phosphoethanolamine (PE) (Musumeci et al. 2014). In this context, PG was determined to play a role in the detachment of the pilus from bacterial aggregates, allowing for colonisation of new sites. The addition of PC and PE is mediated through the activity of the enzymes PptA and PptB, respectively (Warren and Jennings 2003). The residues within pilin to which the unusual PG post-translational modifications are added via PptB were identified as Ser69 in N. gonorrhoeae and Ser93 in N. meningitidis. The Ser69 has, thus, been found with all three modifications, as this is the site at which PC and PE modifications have previously been identified (Warren and Jennings 2003; Musumeci et al. 2014).
Expression of type IV pili by N. gonorrhoeae plays a critical role in mediating adherence to human epithelial cells. Gonococcal pilin is modified with an O-linked glycan with the structure Gal-(α1→3)-2,4-diacetimido-2,4,6-trideoxyhexose (Gal-DATDH) disaccharide (Hegge et al. 2004), which may be present as a di- or monosaccharide because of phase variation of select pilin glycosylation genes. Pilin glycosylation involves multiple pilin glycosylation (pgl) genes. For example, PglD contributes to the biosynthesis of the basal DATDH sugar after which PglA adds the (first) hexose to the basal monosaccharide (Jennings et al. 1998). Whereas pglA is subject to phase variation (Jennings et al. 1998; Banerjee et al. 2002), pglD expression does not appear to be phase variable (Power et al. 2000). Using primary, human, cervical epithelial (i.e. pex) cells, evidence was provided that the pilin glycan mediates productive cervical infection, with the pilin glycan required for gonococcal binding to the I-domain region of complement receptor 3, which is naturally expressed by pex cells (Jennings et al. 2011).
Pseudomonas aeruginosa strains Pa7 and Pa5196 glycosylate their type IVa pilins with a rare sugar polymer of α1,5-linked d-arabinofuranose (d-Araf) residues, which is identical to that found in the cell walls of Corynebacterineae. Bioinformatic analyses pinpointed a cluster of seven P. aeruginosa genes, including homologues of the M. tuberculosis genes Rv3806c, Rv3790 and Rv3791, required for the synthesis of a polyprenyl-linked d-ribose precursor and its epimerisation to d-Araf. The emergence of multidrug-resistant and extensively drug-resistant strains of M. tuberculosis, one of the world’s most prevalent human pathogens, underlines an urgent need for new antimycobacterials (Manina et al. 2010); this might also translate to P. aeruginosa. Pa5196 mutants lacking the respective orthologues had non-arabinosylated pilins, poor twitching motility, and significantly fewer surface pili than the wild type, even in a retraction-deficient (pilT) background. The Pa5196 mutant pilus system did not assemble heterologous non-glycosylated pilins efficiently, demonstrating that this requires post-translationally modified subunits. Thus, type IV pilins provide a rare case of the requirement of PilA O-glycosylation for efficient pilus assembly. A recombinant P. aeruginosa PAO1 strain coexpressing the genes for d-Araf biosynthesis, the pilin modification enzyme TfpW and the acceptor PilA(IV) produced arabinosylated pili, confirming that the identified Pa5196 genes are both necessary and sufficient for pilin glycosylation. A P. aeruginosa epimerase deletion mutant could be complemented with the corresponding M. smegmatis gene, demonstrating conservation between the systems of the Corynebacterineae and Pseudomonadaceae. This work simultaneously described a novel Gram-negative pathway for the biosynthesis of d-Araf, which in the future may be of therapeutic interest in the context of the emergence of multidrug-resistant strains in Corynebacterineae (Harvey et al. 2011).
While in several bacteria glycosylation has been shown to impact protein function with regard to adhesiveness and invasiveness of host cells, questions about glycosylation of proteins in F. tularensis and its potential connection to bacterial virulence have not been answered yet. Recently, several putative F. tularensis glycoproteins were characterised through the combination of carbohydrate-specific detection and lectin affinity with highly sensitive MS utilising a bottom-up proteomic approach. The pilin protein PilA that was recently found to be glycosylated as well as other novel protein virulence factors were among the identified proteins. Up to 20 putative glycoproteins were detected using fluorescently labelled hydrazide and lectin blotting, while the use of lectin affinity chromatography resulted in the identification of 104 putative F. tularensis subsp. holarctica glycoproteins (Balonova et al. 2010).
The oral mucosal pathogen Po. gingivalis targets the C-type lectin receptor DC-SIGN through its 67-kDa Mfa1 (minor) fimbriae for invasion and persistence within human monocyte-derived dendritic cells (DCs). DCs respond to by inducing an immunosuppressive and Th2-biased CD4+ T-cell response. Biochemical and molecular analyses of native MfA1 fimbriae revealed numerous putative O-glycosylation sites and two putative N-glycosylation sites (Zeituni et al. 2010). It was further demonstrated by ProQ Emerald glycan staining that the minor fimbria is glycosylated and that glycosylation is partially removable by treatment with β-(1→4)-galactosidase, but not by classic N- and O-linked deglycosidases. Monosaccharide analysis by gas chromatography-MS confirmed that the minor fimbria contains the DC-SIGN-targeting carbohydrates Fuc, Man, GlcNAc and GalNAc in a molar ratio of ~2:4:3:1. TEM analysis revealed that the minor fimbria forms fibres of approximately 200 nm in length that could be involved in targeting or cross-linking of DC-SIGN. These findings shed light on molecular mechanisms of invasion and immunosuppression by this unique mucosal pathogen (Zeituni et al. 2010).
Other bacterial glycoproteins
The number of bacterial species known to perform O-linked protein glycosylation is rapidly increasing. While it is evident that many bacterial pathogens are able to O-glycosylate proteins, the involvement of O-glycosylation in bacterial pathogenesis remains unknown; it is conceivable that one such effect could be mediated via the involvement of glycoproteins in biofilm formation. Examples from selected Gram-negative and Gram-positive bacteria presented here include B. thailandensis, Vibrio cholerae, N. gonorrhoeae, Corynebacterium glutamicum, Mycobacterium spp., Streptococcus ssp., Staphylococcus aureus, Bacillus anthracis, Ba. cereus, C. difficile, Bac. fragilis, Po. gingivalis, Aggregatibacter actinomycetemcomitans and Lactobacillus ssp.
The hypothetical proteins BTH I0650 from B. thailandensis E264 and VC0393 from V. cholerae N16961 contain the Wzy_C domain, thus representing putative O-OSTs. Infact, it was demonstrated that both proteins have O-OST activity and therefore they were renamed PglLBt and PglLVc, respectively, in analogy to the N. meningitidis counterpart (PglLNm). In E. coli, PglLBt and PglLVc display relaxed glycan and protein specificity. Glycosylation efficiency depends on the specific combination of the protein acceptor, glycan and O-OSTs involved. The identification of enzymatically active O-OST in members of the Vibrio and Burkholderia genera suggests the presence of still unknown O-glycoproteins in these organisms; these might play roles in bacterial physiology or pathogenesis (Gebhart et al. 2012). The discovery of PglLVc and PglLBt increases the repertoire of enzymes available for glycoengineering (vide supra, PglB, N-glycosylation), which irrespective of the role played by glycosylation in bacterial physiology and/or pathogenesis, may contribute to the design of novel and improved diagnostics tools and conjugate vaccines against bacterial infections (Gebhart et al. 2012).
By employing state-of-the-art MS techniques, it was demonstrated that N. gonorrhoeae expresses a general O-linked protein glycosylation (Pgl) system known to target at least 19 membrane-associated proteins (Anonsen et al. 2012, 2016). No specific target sequon as known from N-glycosylation of both eukaryotic and prokaryotic glycoproteins is apparent (Jarrell et al. 2014; Varki et al. 2015), although glycan attachment sites are most often localised within regions of low-sequence complexity (Anonsen et al. 2012). Moreover, glycan structure variations have been observed in species within the genus Neisseria. To elucidate the underlying biosynthesis mechanism, a hypomorphic allele of pglA (encoding the PglA galactosyltransferase) was identified in a genetic analysis as a significant contributor to the simultaneous expression of multiple glycoforms. These findings showed that the differences in microheterogeneity could be primarily attributed to the pglA allele status and, accordingly, that the corresponding phenotypes resulted from altered pglA-associated activity. Moreover, the high-glycoform phenotype was mapped to a single amino acid polymorphism in PglA; the A350G exchange was both necessary and sufficient for this phenotype. Molecular analyses revealed that many pglA phase-off variants were associated with disproportionally high levels of the N,N′-diacetylbacillosamine-Gal disaccharide glycoform generated by PglA (Johannessen, Koomey and Børud 2012). Thus, the variability in glycan structure and antigenicity is attributable to differences in the content and expression status of glycan synthesis genes; ultimately, this effect could result in a degree of diversity that may exceed the one currently defined (Børud et al. 2014). Unique protein-associated disaccharide glycoforms were identified that carry N-acetylglucosamine at their non-reducing end. This altered structure was correlated with allelic variants of pglH whose product was previously demonstrated to be responsible for the expression of glucose-containing disaccharides. Analogous minimal structural alterations in glycosyltransferases have also been documented in association with LPS and capsular polysaccharide variability (Børud et al. 2011).
Corynebacterium glutamicum and M. tuberculosis are affiliated to the phylogenetic family of the Corynebacterineae (Bou Raad et al. 2010). They possess an atypical cell wall that is composed of a heteropolymer of PG and mycolate-carrying arabinogalactan (AG) cross-linked via trehalose (Tropis et al. 2005) and covalently associated with the outer membrane. Five arabinosyl-transferases are involved in the biosynthesis of AG in Co. glutamicum. AftB catalyses the transfer of Araf onto the arabinan domain of the AG to form terminal β-(1→2)-linked Araf residues. It was shown that in ΔaftB cells, half of the AG mycolylation sites are missing that may facilitate the detachment of membrane patches. These fragments contain mono- and dimycolate of trehalose and PorA/H, the major porin of Co. glutamicum, but lack conventional phospholipids that typify the plasma membrane, suggesting that they are derived from the atypical mycolate outer membrane of the cell envelope wall constituents of mycobacteria.
For M. tuberculosis, the causative agent of TB, the cell envelope provides several glycoconjugates such as glycolipids, lipoglycans and polysaccharides (Torrelles and Schlesinger 2010). In fact, M. tuberculosis can adapt to the human host by decorating its cell envelope molecules with terminal mannosylated (i.e. α-Man-(1→2)-Man) oligosaccharides that resemble the glycoforms of mammalian mannoproteins. These biomolecules engage the mannose receptor (MR) on macrophages during phagocytosis and dictate the intracellular fate of M. tuberculosis by regulating the formation of a unique vesicular compartment in which the bacteria survive. The phylogenetic diversity of M. tuberculosis strains, together with the genetic diversity observed in human populations including those elements that affect macrophage function, may explain the extraordinary evolutionary host adaptation capability of this pathogen (Torrelles and Schlesinger 2010). Recent studies provide evidence that major mannosylated glycoconjugates on the M. tuberculosis cell envelope change as this organism grows in vitro on agar plates. Along with these changes, mannan levels on the outer cell surface also increase significantly over time (Yang et al. 2013). Thus, the impact of interrupting O-mannosylation in the human pathogen M. tuberculosis and in the non-pathogenic saprophyte M. smegmatis by inactivating the respective putative protein mannosyl transferase genes Msmeg_5447 and Rv1002c was analysed. Unexpectedly, while the M. smegmatis phenotype was unaffected by the lack of mannoproteins, the M. tuberculosis mutant showed severely impacted growth in vitro and in cellulo associated with a strong attenuation of its pathogenicity in immunocompromised mice. These data are unique in providing evidence of the biological significance of protein O-mannosylation in mycobacteria (Lommel and Strahl 2009) and demonstrate the crucial contribution of this post-translational protein modification to M. tuberculosis virulence in the host (Liu et al. 2013). Specifically, the 45/47-kDa alanine-proline-rich adhesin (Apa), an immunodominant antigen secreted by M. tuberculosis, is O-mannosylated at multiple sites. Glycosylation of Apa plays a key role in colonisation and invasion of the host cells by M. tuberculosis through interactions of Apa with C-type lectins of the host immune system (Coddeville et al. 2012).
Mycobacterium marinum, a fish pathogen, phylogenetically close to M. tuberculosis, induces a granulomatous response with features similar to those described for M. tuberculosis in humans. O-Mannosylation of M. marinum Apa is required for antigenicity but appears to be dispensable for its immunogenicity and protective efficacy in mice. These results have implications for the development of subunit vaccines using tailored glycoproteins against infectious diseases such as tuberculosis (Nandakumar et al. 2013). Another glycosylated M. tuberculosis protein is the secreted glycoprotein Rv1860, which was shown to inhibit DC-mediated Th1 and Th17 polarisation of T cells and abrogate protective immunity conferred by M. bovis (Satchidanandam et al. 2014). Mycobacterium tuberculosis Rv1860 carries at threonine sites one to three mannose residues linked to each other by α-(1→2) and α-(1→3) glycosidic bonds (Dobos et al. 1996). Based on MS characterisation of Con-A binding proteins, M. tuberculosis codes for at least 41 glycoproteins (González-Zamorano et al. 2009). Recent reports describe interferon-γ secretion by human CD4+ and CD8+ T cells in response to recombinant E. coli-expressed M. tuberculosis Rv1860 protein as well as protection of guinea pigs against a challenge with virulent M. tuberculosis following prime-boost immunisation with a DNA vaccine and poxvirus expressing Rv1860.
The serine-rich repeat (SRR) glycoproteins of Gram-positive bacteria constitute a large family of cell-wall proteins among which there are the surface adhesins; the accessory secretion (Sec) system is a specialised transport system for the export of these SRR glycoproteins (Seepersaud et al. 2012). The system contains two homologues of the general Sec pathway (SecA2 and SecY2) and several other essential proteins (Asp1 to Asp5) that share no homology to proteins of known function (Yen et al. 2013). In Streptococcus gordonii, Asp2 is a bifunctional protein required for the transport of the SRR adhesin GspB and for its correct glycosylation by modulating GlcNAc deposition onto GspB (Bensing, Gibson and Sullam 2004; Zhou and Wu 2009). According to tertiary structure predictions, the carboxyl terminus of Asp2 resembles the catalytic region of numerous enzymes that function through a Ser-Asp-His catalytic triad. In fact, sequence alignment of known Asp2 homologues identified a highly conserved pentapeptide motif (Gly-X-Ser362-X-Gly) typical of most Ser-Asp-His catalytic triads, where Ser forms the reactive residue. The catalytic domain might be responsible for controlling GspB glycosylation, while surrounding regions might be functionally required for glycoprotein transport (Seepersaud et al. 2012).
In S. pneumoniae, the core enzyme GtfA and the coactivator GtfB form an OST complex to O-GlcNAcylate the pneumococcal SRR protein adhesin (PsrP), which is involved in infection and pathogenesis (Lu, Li and Shao 2015). An in vitro glycosylation system enabled mapping of the O-linkages to the serine residues within the first SRR of PsrP. These findings suggest that fusion with an add-on domain might be a universal mechanism for diverse OGTs that recognise varying acceptor proteins/peptides (Shi et al. 2014).
Staphylococcus aureus has been shown to bind to human platelets through a variety of surface molecules, including an SRR adhesin for platelets (SraP), which is a cell-wall-anchored glycoprotein (Zhou and Wu 2009; Li et al. 2014). SecY2, Asp1, Asp2, Asp3 and SecA2 encoded in the SraP operon are required for the efficient transport of glycosylated SraP from the cytoplasm to the bacterial cell surface (Yen et al. 2013). An SraP mutant strain of St. aureus was significantly impaired in its ability to initiate infection compared with the wild type. In fact, it was demonstrated that the predicted OST components GtfA and GtfB transfer GlcNAc-containing oligosaccharides to recombinant SraP. Deletion of either one or both of the OSTs abolished glycosylation of SraP, indicating that both are required for SraP glycosylation in St. aureus (Li et al. 2014; Lu, Li and Shao 2015).
The spores of Ba. anthracis are notorious for their potential in the development of weapons for mass destruction (Tan and Turnbough 2010). Therefore, over recent years, substantial effort has been directed onto the discovery of novel antigens from Ba. anthracis, including the identification and immunochemical evaluation of glycans that might be used for the specific diagnostic detection of this pathogen. Among the carbohydrate structures found on the surface of vegetative Ba. anthracis cells and spores are the cell-wall polysaccharide and multiple copies of an O-linked pentasaccharide that are attached to several sites within the central collagen-like region of BclA (for Bacillus collagen-like protein of anthracis) (Dong et al. 2008). The structure of this oligosaccharide is 2-O-methyl-4-(3-hydroxy-3-methylbutamido)-4,6-dideoxy-β-d-Glcp-(1→3)-α-l-Rhap-(1→3)-α-l-Rhap-(1→2)-l-Rhap-(1→?)-GalNAc (Daubenspeck et al. 2004). The novel terminal sugar 2-O-methyl-4-(3-hydroxy-3-methylbutamido)-4,6-dideoxy-d-glucose was given the trivial name anthrose (Ant). A truncated version of the pentasaccharide, a tetrasaccharide lacking the reducing-end GalNAc residue, was chemically synthesised by several groups and is virtually identical to the material isolated from the Ba. anthracis exosporium, validating the originally proposed structure (Adamo, Saksena and Kováč 2005; Saksena, Adamo and Kováč 2006; Adamo 2014). To produce a conjugate model vaccine, the synthetic tetrasaccharide β-Ant-(1→3)-α-l-Rhap-(1→3)-α-l-Rhap-(1→2)-l-Rhap was covalently linked to a protein carrier (Mehta et al. 2006). Immunisation of animals with the vaccine elicited antibodies that bound specifically to Ba. anthracis spores (Mehta et al. 2006), providing evidence that the oligosaccharide chains are exposed on the spore surface. Since these sugar moieties are highly specific for the spores of Ba. anthracis, they appear to be key biomarkers for the detection of Ba. anthracis spores and promising candidates for the development of novel vaccines targeting anthrax spores (Wang et al. 2007). A detailed reinvestigation of spores of the Ba. cereus group (Ba. cereus, Ba. anthracis and Ba. thuringiensis) revealed species and domain specificity of BclA glycosylation within the Ba. cereus group. The collagen-like regions of both Ba. cereus and Ba. anthracis are similarly substituted by short O-glycans that bear the species-specific deoxyhexose residues anthrose and the newly observed cereose (abbreviated Cro). Besides a Cro-less trisaccharide the wild-type BclA tetrasaccharide has the structure β-Cro-(1→4)-α-3-OMe-Rha-(1→2)-α-Rha-(1→3)-GalNAc-ol (Maes et al. 2016). Moreover, the C-terminal globular domains of BclA from both species are substituted by polysaccharide-like O-glycans whose structures are also species specific. The presence of such polymers on Bacillus spores may have profound impact on spore interaction with biotic and abiotic surfaces.
Recently, evidence was provided that the major spore surface protein BclA of Cl. difficile is glycosylated, in addition to several other proteins of this microorganism (Strong et al. 2014). Biochemical characterisation of glycopeptides corresponding to a putative exosporangial glycoprotein (BclA3) showed that these molecules were modified with multiple HexNAc moieties and, in some cases, capped with novel glycans. The glycosyltransferase gene sgtA (gene CD3350 in strain 630 and CDR3194 in strain R20291), which is located immediately upstream of and cotranscribed with bclA3, was found to be involved in the glycosylation of the spore surface. Reactivity of the spore surface with the anti-β-O-GlcNAc antibody was abolished in the 630 and R20291 glycosyltransferase mutants, while complementation with a wild-type copy of the gene restored β-O-GlcNAc reactivity (Strong et al. 2014).
Recently, a protein O-glycosylation system that is unique in that extracytoplasmic proteins are glycosylated at serine or threonine residues within the specific three-amino acid motif D(S/T)(A/I/L/M/T/V) (Fletcher, Coyne and Comstock 2011) was identified in the Gram-negative intestinal gut symbiont Bac. fragilis. In the course of a computational analysis of the bacterial proteome, 1021 Bac. fragilis candidate glycoproteins were identified. Among those were species predicted to be localised in the inner membrane, a compartment not previously shown to include glycoproteins. In addition, four glycoproteins involved in cell division and chromosomal segregation were predicted. Up to date, all extracytoplasmic Bac. fragilis proteins containing the specific glycosylation motif were found to be glycosylated; it is likely that hundreds of proteins, comprising more than half of the extracytoplasmic proteins of Bac. fragilis, are indeed glycosylated. By engineering glycosylation motifs into a naturally unglycosylated protein, it was possible to bring about site-specific glycosylation at the engineered sites, suggesting that this glycosylation system may have applications for glycoengineering (Fletcher, Coyne and Comstock 2011).
Porphyromonas gingivalis, which is phylogenetically closely related to Bac. fragilis, has been implicated as a major pathogen associated with chronic periodontitis. A proteomic analysis of Po. gingivalis ATCC 33277 revealed the presence of four novel glycoproteins, namely PGN0743, PGN0876, PGN1513 and PGN0729 (Kishi et al. 2012); these are predicted to possess a range of biochemical activities, such as chaperone functions based on partial homology with a cis-trans isomerase (PGN0743), and diverse subcellular localisations. Two of them (PGN0876, PGN1513) contain tetratricopeptide repeat domains known to mediate protein–protein interactions, and PGN0729 encodes the outer membrane protein 41 precursor, which was previously identified as Pgm6 and is a homologue of the OmpA protein in E. coli (Murakami et al. 2002; Dumetz et al. 2007). There is evidence that Po. gingivalis possesses a general mechanism for protein glycosylation and that the novel glycoproteins PGN0743 and PGN0876 may play an important role in bacterial growth and colonisation (Kishi et al. 2012).
Another oral bacterium, the human oropharyngeal pathogen Aggregatibacter (formerly Actinobacillus) actinomycetemcomitans that is a Gram-negative, facultative non-motile, rod-shaped oral commensal synthesises multiple adhesins, including the nonfimbrial extracellular matrix protein adhesin A (EmaA) that is required for collagen binding (Tang and Mintz 2010). Two forms of EmaA were identified, which are suggested to be linked to the O-polysaccharide (O-PS) of the bacterium’s LPS, maintaining collagen-binding activity (Tang et al. 2007). This association was investigated by generating mutants of a Rha biosynthetic enzyme (RmlC; dTDP-4-dehydrorhamnose 3,5-epimerase), the ABC sugar transport protein (Wzt) and the O antigen ligase (WaaL). All three mutants produced reduced amounts of O-PS, and the EmaA monomers in these mutants displayed a modified glycosylation as was suggested by lectin blots using the fucose-specific Lens culinaris agglutinin directed towards the fucose residue present in serotype b O-PS. The ΔrmlC mutant expressing modified EmaA protein demonstrated reduced collagen adhesion, supporting a role of the glycoconjugate in collagen binding. These data provided experimental evidence for the dependence of the post-translational modification of EmaA on the LPS biosynthetic machinery in Ag. actinomycetemcomitans (Tang and Mintz 2010).
While occurrence, biosynthesis and possible functions of glycoproteins are increasingly documented for pathogens, they are not yet widely described for probiotic bacteria. Knowledge of protein glycosylation in these bacteria is pivotal not only for our understanding of their repertoire of glycan-mediated interactions but also for glycoengineering approaches in food-grade microbes like Lactobacillus rhamnosus GG (Lebeer et al. 2012). The peptidoglycan (PG) hydrolase Msp1 of La. rhamnosus is O-glycosylated as evidenced by the identification of a glycopeptide TVETPSSA (amino acids 101–108) bearing hexoses presumably linked to serine residues (Ser106 and Ser107). Obviously, these serine residues of the La. rhamnosus enzyme are glycosylated with ConA-reactive sugars; however, they are absent in the homologous enzyme of several La. casei strains tested, which also failed to bind ConA. Glycosylation is neither required for enzymatic activity of Msp1 nor for Akt activation capacity in epithelial cells, but appears to be important for enzyme stability and protection against proteases (Lebeer et al. 2012).
Recent analyses of the major autolysin Acm2 from La. plantarum strain WCFS1 showed that it undergoes intracellular O-GlcNAcylation (Fredriksen et al. 2012, 2013; Lee et al. 2014). Using biochemical methods, so far 11 Lactobacillus glycoproteins were detected exhibiting in total 49 glycosylation sites for HexNAc. Further, based on a comparative genomic approach, six candidate La. plantarum WCFS1 genes—tagE1 to tagE6—with significant homology to Gtf1, an enzyme involved in the glycosylation of a fimbria-associated protein (Fap1) in S. parasanguinis (Bu et al. 2008), were identified. The candidate genes were targeted by systematic gene deletion, followed by assessment of the consequences on Acm2 glycosylation. sWGA lectin blotting experiments using a panel of eight other La. plantarum strains revealed that protein glycosylation is a common feature in La. plantarum strains. With the establishment of these enzymes as protein glycosyltransferases, renaming was proposed for TagE5 and TagE6 as GtfA and GtfB, respectively (Lee et al. 2014).
S-Linked Glycoproteins
Recently, a new group of glycoconjugates in the form of ribosomal antimicrobial polypeptides, collectively known as bacteriocins, named glycocins (glycosylated bacteriocins) with cysteine-(S-) linked glycans have been identified (Stepper et al. 2011; Wang and van der Donk 2011).
Among them is glycocin F that is secreted by Lactobacillus plantarum KW30; it is modified with an N-acetylhexosamine S-linked to its C-terminal Cys43 residue (Venugopal et al. 2011) in addition to a GlcNAc residue, β-O-linked to Ser18. The latter is essential for bacteriostatic activity of glycocin F in addition to the C-terminus of the pepride being required for its full potency (IC50 2 nM) (Stepper et al. 2011).
Genomic context analysis identified diverse putative glycocins in Firmicutes. One of these, the reputed lantibiotic sublancin 168 of Bacillus subtilis 168 (Paik, Chakicherla and Hansen 1998), was shown to contain a hexose S-linked to Cys22. This residue within the 56-amino acid peptide substrate is selectively modified by a novel S-glycosyltransferase named SunS, which accepts a variety of NDP sugars. It is suggested that SunS recognises an α-helix N-terminal of the SunA cysteine that is present in a flexible linker for glycosylation (Wang and van der Donk 2011). The solution structure of sublancin 168, a 37-amino-acid peptide produced by Ba. subtilis 168, has been solved by NMR spectroscopy; it comprises two α-helices and a well-defined interhelical loop (Garcia De Gonzalo et al. 2014). The two helices span residues 6–16 and 26–35, and the loop region encompasses residues 17–25. The 9-amino-acid loop region contains a β-S-linked glucose moiety attached to Cys22. Interestingly, sugar attachment is not required for sublancin antimicrobial activity as evidenced upon mutation of Cys22. Considering that the sublancin-producing strain Ba. subtilis 168 is susceptible to such mutation also intrinsically, S-glycosylation at Cys22 may constitute an unusual type of self-resistance and, consequently, stability (Wang and van der Donk 2011).
Prokaryotic Surface Layer (S-Layer) Glycoproteins
S-layers are one of the most commonly observed cell-surface structures on different species of nearly every phylogenetic group of prokaryotes. They are formed by self-assembly of monomeric (glyco)proteins into regularly spaced, 2D crystalline arrays with nanometer-scaled periodicity (4–30 nm). Bacteria possess dedicated pathways for the secretion and anchoring of their S-layer to the cell wall, and some Gram-positive species have large S-layer-associated gene families. S-layers have important roles in bacterial growth and survival, and their many functions include the maintenance of cell integrity, enzyme display and, in pathogens and commensals, interaction with the host and its immune system (Sleytr 1978; Sleytr and Messner 1983; Messner and Schäffer 2003; Åvall-Jääskeläinen and Palva 2005; Messner et al. 2010; Hynönen and Palva 2013; Fagan and Fair-weather 2014; Sleytr et al. 2014; Gerbino et al. 2015). Current approaches are exploiting the nanopatterned S-layer matrix for the display of biologically relevant molecules (Schuster and Sleytr 2013; Allievi et al. 2014; Baneyx and Matthaei 2014; Carasi et al. 2014; Turroni et al. 2014; Misra, Basu and Apte 2015; Valero et al. 2015; Zhang et al. 2015).
S-layer protein subunits can undergo glycosylation with glycans of varying complexity (Messner and Sleytr 1991; Messner 1997; Messner and Schäffer 2003; Schäffer and Messner 2004; Messner et al. 2010). Despite the fact that considerable variation exists in the structure and chemistry of prokaryotic cell envelopes, S-layers have apparently coevolved with these diverse structures. In bacteria and archaea, the S-layer lattice assembles on the surface of the rigid wall matrix that is mainly composed of PG or pseudomurein, respectively (Sleytr and Messner 1983). In Gram-positive bacteria, the S-layer has been shown to be attached to PG-bound secondary cell-wall polymers (SCWPs) (Araki and Ito 1989; Mesnage et al. 2000; Sára 2001; Cava et al. 2004; Schäffer and Messner 2005; Pavkov et al. 2008; Messner, Schäffer and Kosma 2013) (vide infra); in Gram-negative bacteria, there are indications for attachement of the S-layer to LPS (Awram and Smit 2001); and in most archaea, S-layers are closely attached to or inserted into the plasma membrane (Sumper 1987; Lechner and Wieland 1989; Abdul Halim et al. 2016).
In this section, we give a general overview on the chemical structure and biosynthesis of both bacterial and archaeal S-layer glycoprotein glycans (Fig. 2) and, then, focus particularly on those organisms that have attracted special attention in the past because of scientific, medical or public implications.
Figure 2.
Schematic depiction of S-layer glycoprotein glycans from Gram-positive and Gram-negative bacteria and archaea and of SCWPs of Gram-positive bacteria. Ac, CH3COO−; CONH2, amidyl-; -COOMe, methyl ester; Me, CH3-; NAc, N-acetyl-; P, phosphate; Pyr, pyruvyl; R1, -COOH; R2, -CONHAc; R3, -CONAc2, S, sulfate;. Sugar symbols:◯, hexose; , hexuronic acid;
, hexuronic acid; , glucose;
, glucose; , N-acetylglucosamine;
, N-acetylglucosamine; , glucuronic acid;
, glucuronic acid; , 3-N-acetylquinovosamine;
, 3-N-acetylquinovosamine; , 2,3-di-N-acetylglucuronic acid;
, 2,3-di-N-acetylglucuronic acid; , 6-sulfoquinovose;
, 6-sulfoquinovose; , 2-amino-6-sulfo-2,6-dideoxy-quinovose;
, 2-amino-6-sulfo-2,6-dideoxy-quinovose; , galactose;
, galactose; , galactofuranose;
, galactofuranose; , N-acetylgalactosamine;
, N-acetylgalactosamine; , galacturonic acid;
, galacturonic acid; , 3-O-methylgalacturonic acid;
, 3-O-methylgalacturonic acid; , mannose;
, mannose; , N-acetylmannosamine;
, N-acetylmannosamine; , mannosaminuronic acid;
, mannosaminuronic acid; , 6-N-amido-Thr mannosaminuronic acid;
, 6-N-amido-Thr mannosaminuronic acid; , l-rhamnose;
, l-rhamnose; , d-rhamnose;
, d-rhamnose; , l-fucose;
, l-fucose; , d-fucose;
, d-fucose; , 3-N-acetylfucosamine;
, 3-N-acetylfucosamine; , iduronic acid;
, iduronic acid; , ribose;
, ribose; , d-glycero-d-manno-heptose;
, d-glycero-d-manno-heptose; , xylose;
, xylose; , digitoxose;
, digitoxose;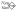 , 5-acetamidino-7-glycerolyl pseudaminic acid;
, 5-acetamidino-7-glycerolyl pseudaminic acid; , N-acetylmuramic acid. Monosaccharide symbols follow the SNFG (Symbol Nomenclature for Glycans) (Appendix-1B 2015). (Mescher and Strominger 1976; Koval and Jarrell 1987; Lechner and Sumper 1987; Sumper et al. 1990; Altman et al. 1991, 1992, 1995, 1996; Baumeister and Lembcke 1992; Christian et al. 1993; Kärcher et al. 1993; Möschl et al. 1993; Bock et al. 1994; Messner et al. 1995; Kosma et al. 1995a,b; Meier-Stauffer et al. 1996; Schäffer et al. 1999a,b, 2000a,b, 2004; Eichler 2000, 2013; Steindl et al. 2002, 2005; Pfoestl et al. 2003; Kählig et al. 2005; Voisin et al. 2005; Steiner et al. 2008; Veith et al. 2009; Peyfoon et al. 2010; Posch et al. 2011; Messner, Schäffer and Kosma 2013; Palmieri et al. 2013; Kandiba and Eichler 2014; Parente et al. 2014; Anzengruber et al. 2014a; Appendix-1B 2015; Lu et al. 2015; Kandiba et al. 2016). (Extended and modified from Eichler 2013. With permission from Nature Publishing Group).
, N-acetylmuramic acid. Monosaccharide symbols follow the SNFG (Symbol Nomenclature for Glycans) (Appendix-1B 2015). (Mescher and Strominger 1976; Koval and Jarrell 1987; Lechner and Sumper 1987; Sumper et al. 1990; Altman et al. 1991, 1992, 1995, 1996; Baumeister and Lembcke 1992; Christian et al. 1993; Kärcher et al. 1993; Möschl et al. 1993; Bock et al. 1994; Messner et al. 1995; Kosma et al. 1995a,b; Meier-Stauffer et al. 1996; Schäffer et al. 1999a,b, 2000a,b, 2004; Eichler 2000, 2013; Steindl et al. 2002, 2005; Pfoestl et al. 2003; Kählig et al. 2005; Voisin et al. 2005; Steiner et al. 2008; Veith et al. 2009; Peyfoon et al. 2010; Posch et al. 2011; Messner, Schäffer and Kosma 2013; Palmieri et al. 2013; Kandiba and Eichler 2014; Parente et al. 2014; Anzengruber et al. 2014a; Appendix-1B 2015; Lu et al. 2015; Kandiba et al. 2016). (Extended and modified from Eichler 2013. With permission from Nature Publishing Group).
Bacterial S-layer glycoproteins
Since the first description of bacterial S-layer glycoproteins on Gram-positive thermophilic clostridia, now renamed Thermoanaerobacter thermohydrosulfuricus (T. thermohydrosulfuricus) L111-69 and Thermoanaerobacterium thermosaccharolyticum (Th. thermosaccharolyticum) D120-70 (Sleytr and Thorne 1976), the number of S-layer glycoprotein-carrying organisms has increased considerably, including Gram-positive isolates such as Geobacillus (formerly Bacillus) stearothermophilus NRS 2004/3a (Steiner et al. 2008), Anoxybacillus (formerly Geobacillus) tepidamans (Kählig et al. 2005), Aneurinibacillus (formerly Bacillus) thermoaerophilus (Kosma et al. 1995b; Pfoestl et al. 2003), different T. thermohydrosulfuricus (Altman et al. 1992; Christian et al. 1993; Bock et al. 1994) and Th. thermosaccharolyticum strains (Altman et al. 1995), Paenibacillus alvei CCM 2051T (Altman et al. 1991; Messner et al. 1995) and different Lactobacillus buchneri strains (Möschl et al. 1993; Anzengruber et al. 2014b), as well as one Gram-negative bacterium—the oral pathogen Tannerella forsythia ATCC 43037 (Posch et al. 2011) (Fig. 2).
The S-layer glycoprotein of Geobacillus stearothermophilus NRS 2004/3a was the first bacterial S-layer protein whose glycosylation was studied in detail (Küpcü et al. 1984). The S-layer was isolated by detergent extraction, purified, and after thorough proteolytic digestion the material was purified by gel permeation chromatography. Colorimetric sugar determination revealed Rha as the predominant constituent (55%) of the glycopeptides and traces of Gal (∼2%). This material was tritium labelled with N-succinimidyl[2,3-3H]propionate and subjected to hydrazinolysis to cleave the glycan off from the labelled glycopeptides (Küpcü et al. 1984). 1H- and 13C NMR spectroscopy revealed two types of glycan chains, one of which, a rhamnan with the repeating unit structure→2)-α-l-Rhap-(1→2)-α-l-Rhap-(1→3)-β-l-Rhap-(l→ (Fig. 2) (Christian et al. 1986), was shown to decorate the 93-kDa surface layer protein SgsE of G. stearothermophilus NRS 2004/3a at least three distinct sites (Thr590, Thr620 and Ser794; (Fig. 2). The other turned out to be a copurified SCWP present in the bacterial cell envelope(Messner et al. 1987; Schäffer et al. 1999a) (vide infra, section ‘SCWP’). The elongated rhamnan chains were subsequently shown to additionally comprise a 2-O-methyl modification of the terminal trisaccharide at the non-reducing end and a core saccharide as a linker to the S-layer protein (Schäffer et al. 1999a). Surprisingly, the rhamnosylated S-layer protein of this bacterium reveals a four-banded pattern on SDS-PAGE. According to our current interpretation, the three high-molecular-mass bands originate from either one, two or three glycosylation sites per S-layer protein, whereas the low-molecular-mass band represents non-glycosylated protein (Steiner et al. 2006). Further, microheterogeneity was assigned to the S-layer glycans, with the most prevalent variation between 12 and 18 trisaccharide repeating units accompanied with the possibility of extension of the already-known→3)-α-l-Rhap-(1→3)-α-l-Rhap-(1→ core by one additional Rha residue. The microheterogeneity of elongated sugar chains in G. stearothermophilus NRS 2004/3a S-layer glycopeptides, containing up to 51 monosaccharide residues at a single O-attachment site on a 12-amino acid peptide backbone, was investigated by Fourier transform ion cyclotron resonance MS (Bindila et al. 2007).
S-layer glycan biosynthesis in G. stearothermophilus NRS 2004/3a is encoded by a polycistronic slg (surface layer glycosylation) gene cluster. To get insight into the glycosylation pathway of the S-layer glycoprotein SgsE, the first step was the characterisation of WsaP from this gene cluster as a UDP-Gal:phosphorylpolyprenol Gal-1-phosphate transferase that primes S-layer glycoprotein glycan biosynthesis in this organism. The enzyme transfers in vitro a galactose-1-phosphate from UDP-Gal to endogenous phosphoryl-polyprenol, with the C-terminal half of WsaP comprising the galactosyltransferase function, in analogy to UDP-Gal:phosphoryl-polyprenol Gal-1-phosphate transferase WbaP from Salmonella enterica (Steiner et al. 2007). Four assigned glycosyltransferases, named WsaC–WsaF, were investigated by a combined biochemical and NMR approach, starting from synthetic octyl-linked saccharide precursors (Messner et al. 2008). Three of the enzymes are rhamnosyl transferases that are responsible for the transfer of l-Rha from a dTDP-β-l-Rha precursor to the nascent S-layer glycan, catalysing the formation of the α1,3- (WsaC and WsaD) and β1,2-linkages (WsaF) (Steiner et al. 2010), present in the adaptor saccharide and in the repeating units of the mature S-layer glycan, respectively. These enzymes work in concert with a multifunctional methylrhamnosyl transferase (WsaE). The N-terminal portion of WsaE is responsible for the S-adenosyl methionine-dependent methylation reaction of the terminal α1,3-lined l-Rha residue, and the central and C-terminal portions are involved in the transfer of l-Rha from dTDP-β-l-Rha to the adaptor saccharide to form the α1,2- and α1,3-linkages during S-layer glycan chain elongation, with the methylation and the glycosylation reactions occurring independently. Characterisation of these enzymes revealed a molecular basis for S-layer glycan biosynthesis in G. stearothermophilus NRS 2004/3a (Steiner et al. 2008).
While protein O-glycosylation at serine, threonine or hydroxyproline residues has been studied in detail in prokaryotes (Jarrell et al. 2014; Varki et al. 2015), only few data are available on O-glycosidic attachment of glycans to the amino acid tyrosine. A bacterial protein tyrosine O-glycosylation system was described for the S-layer glycoproteins of T. thermohydrosulfuricus strains S102-70 (Messner et al. 1992; Christian et al. 1993) and L111-69 (Bock et al. 1994; Messner et al. 1995), Th. thermosaccharolyticum strains D120-70 (Altman et al. 1990) and E207-71 (Altman et al. 1995), and Pa. alvei CCM 2051T (Messner et al. 1995). The latter is a secondary invader of diseased honeybee colonies infected with Melissococcus pluton, the causative agent of European foulbrood (Govan et al. 1998). In Pa. alvei CCM 2051T, a polysaccharide consisting of [→3)-β-d-Galp-(1→4)-[α-d-Glcp-(1→6)]-β-d-ManpNAc-(1→]n repeating units is O-glycosidically linked via an adaptor saccharide with the structure -[GroA-2→OPO2 →4-β-d-ManpNAc-(1→4)]→3)-α-l- Rhap-(1→3)-α-l-Rhap-(1→3)-α-l-Rhap-(1→3)-β-d-Galp-(1→ to specific tyrosine residues of the S-layer protein SpaA (Figs 2 and 3).
Figure 3.
Working model of S-layer glycan biosynthesis in Pa. alvei CCM 2051T. The initial transfer of a Gal residue from UDP-α-d-Gal to a lipid carrier is catalysed by WsfP (A). The adaptor saccharide is formed by the α1,3-linkage of an l-Rha residue from dTDP-β-l-Rha to the linkage sugar d-Gal possibly performed by WsfG, followed by the transfer of two additional α1,3-linked l-Rha residues possibly by the action of WsfF (B). The glycan chain would be elongated by the activity of the aminosugar transferase WsfE and the tripartite transferase WsfC. WsfE may form the β1,4-linkage of a ManNAc residue from UDP-ManNAc to the third rhamnose residue. WsfC putatively adds a single glycerol phosphate from CDP-glycerol to the ManNAc residue of the adaptor oligosaccharide and may form the β1,3-linkage of a ManNAc residue to the third rhamnose residue as well as the β1,4-linkage of a Gal to the ManNAc residues of the repeating units. The glycan chain would be recognised by the carboxy-terminal part of Wzt and exported by the ABC transporter system through the cytoplasmic membrane (C). The transfer of cytoplasmic Glc to the lipid carrier would be carried out by WsfH and, after reorientation, is used at the external face of the cytoplasmic membrane by WsfD for α1,6-linkage of the Glc residues to ManNAc residues of the repeating units (D). The final transfer of the completed S-layer glycan to certain tyrosine residues of the S-layer protein is predicted to occur cosecretionally upon catalysis of the O-OST WsfB (E). Eventually, the mature S-layer glycoprotein would be self-assembled at the cell surface (F). Please note that so far only the WsfP protein has been experimentally verified to perform its predicted role. (Adapted from Zarschler et al. 2010b. With permission from Oxford University Press).
A 24.3-kb slg gene cluster comprising 18 open reading frames (ORF) encodes information for the biosynthesis of nucleotide-activated monosaccharides, glycan assembly and export as well as transfer of the completed polysaccharide chain to the S-layer target by a predicted O-OST (Zarschler et al. 2010b). To study the glycosylation process in detail, all ORFs of the slg gene cluster, except those encoding the nucleotide sugar biosynthesis enzymes and the ABC transporter integral transmembrane proteins (Cuthbertson, Kos and Whitfield 2010) were disrupted by the insertion of the mobile group II intron Ll.LtrB, and S-layer glycoproteins produced in mutant backgrounds were analysed for their glycosylation status by MS. There is evidence that the glycan chain of Pa. alvei CCM 2051T is synthesised in a process comparable to the ABC transporter-dependent pathway of the LPS O-PS biosynthesis (Cuthbertson, Kos and Whitfield 2010). Furthermore, with the protein WsfB, we have identified a good candidate for an OST involved in the formation of the covalent β-d-Galp→Tyr linkage between the glycan chain and the S-layer protein SpaA (Zarschler et al. 2010b). As also evidenced for the S-layer Ser/Thr glycosylation system of G. stearothermophilus NRS 2004/3a, the components encoded in the slg gene cluster work in concert with housekeeping genes such as those required for the production of UDP-Gal, to constitute the full S-layer glycoprotein biosynthesis pathway.
For nanopatterned in vivo cell surface codisplay of peptide and glycan epitopes based on the Pa. alvei CCM 2051T S-layer glycoprotein self-assembly system, a working strategy was developed. The ORF of the S-layer structural gene spaA codes for a protein of 983 amino acids, including a signal peptide of 24 amino acids and the glycosylated S-layer protein SpaA has a the-oretical molecular mass of ~106 kDa. As demonstrated exemplarily with a hexahistidine tag and enhanced green fluorescent protein (eGFP) translationally fused to the S-layer protein, fused epitopes can be efficiently expressed and successfully displayed via the S-layer glycoprotein matrix on the surface of Pa. alvei CCM 2051T cells in vivo as evidenced by immunoblot analysis, immunofluorescence staining and fluorescence microscopy. In contrast, exclusively non-glycosylated chimeric SpaA proteins were displayed, when the S-layer of the glycosylation-deficient wsfP mutant was used as matrix (Zarschler et al. 2010a).
SpaA contains three consecutive S-layer homology (SLH) domains at the N-terminus that are involved in non-covalent anchoring of the glycoprotein via a non-classical, pyruvylated SCWP (vide infra, see chapter ‘SCWP’) to the PG layer of the Pa. alvei cell wall (Schäffer et al. 2000b). The SLH domains contain the amino acid motif TRAE, known to play a key role in cell-wall binding (May et al. 2006), as well as the TVEE and TRAQ variations thereof. The role of the three predicted binding motifs within the SLH domains was analysed by mutating them into TAAA motifs, either individually, pairwise or all of them (Janesch, Messner and Schäffer 2013). Effects were visualised in vivo by homologous expression of chimeras made of the mutated S-layer proteins and eGFP and in an in vitro binding assay using His-tagged SpaA variants and native PG-containing cell-wall sacculi that either contained SCWP or were deprived of it. Experimental data indicated that the TRAE, TVEE and TRAQ motifs are critical for the binding function of SLH domains and two functional motifs are sufficient for cell-wall binding, regardless of the domain location. SLH domains of Pa. alvei CCM 2051T SpaA recognise both SCWP and PG, but cell-wall anchoring is not necessary for SpaA glycosylation. Additionally, we showed that the SLH domains of SpaA are sufficient for in vivo cell-surface display of foreign proteins at the cell surface of Pa. alvei (Janesch, Messner and Schäffer 2013).
In contrast to the previously described long-glycan chain S-layer glycoproteins of thermophilic and mesophilic bacilli, short-glycan chain S-layer glycoproteins are also known, for instance, on the Gram-positive GRAS (generally regarded as safe) organism La. buchneri CD034 (Anzengruber et al. 2014b), or the Gram-negative periodontal pathogen Ta. forsythia ATCC 43037 (Posch et al. 2011).
Based on a previous demonstration of S-layer protein glucosylation in La. buchneri strain 41021/251 (Möschl et al. 1993) and because of general advantages of lactic acid bacteria for applied research, protein glycosylation in this species was investigated in more detail recently. The cell surface of La. buchneri CD034 is completely covered with an oblique S-layer lattice formed by self-assembly of the abundant S-layer protein SlpB (Anzengruber et al. 2014b). Biochemical and MS analyses revealed that SlpB is O-glycosylated at four serine residues within the sequence Ser152-Ala-Ser154-Ser155-Ala-Ser157 with, on average, seven Glc-(α1–6) linked residues, each. Subcellular fractionation of strain CD034 indicated a sequential order of SlpB export and glucosylation as evidenced by lack of glucosylation on cytosolic SlpB. Protein glycosylation analysis was extended to strain La. buchneri NRRL B-30929 (Liu et al. 2011) where an analogous glucosylation scenario was detected, with the S-layer glycoprotein SlpN containing an O-glycosylation motif identical to that of SlpB. This led to the proposal of species-wide S-layer protein O-glucosylation in La. buchneri strains targeted at the sequence motif Ser-Ala-Ser-Ser-Ala-Ser. Search of the La. buchneri genomes for the said glucosylation motif revealed one further ORF, encoding the putative glycosyl-hydrolase LbGH25B and LbGH25N in La. buchneri CD034 and NRRL B-30929, respectively; in fact, there are indications of glycosylation comparable to that of the S-layer proteins (Anzengruber et al. 2014b). The glycosyl hydrolase family 25 domain-containing homologues LbGH25B and LbGH25N from La. buchneri CD034 and NRRL B-30929, respectively, were characterised in detail. Zymogram analysis confirmed hydrolysing activity on bacterial cell walls for both enzymes. Subsequent reversed-phase HPLC and MALDI-TOF MS analysis of the PG breakdown products from La. buchneri strains CD034 and NRRL B-30929 revealed that these enzymes exhibit N-acetylmuramidase activity. Both enzymes were identified as cell-wall-associated proteins due to the binding capability of purified recombinant LbGH25B and LbGH25N to La. buchneri cell walls in vitro. Moreover, similar secondary structures mainly composed of β-sheets and nearly identical thermal stabilities with Tm values around 49°C were found for the two N-acetylmuramidases by far-UV circular dichroism spectroscopy. This work provided for the first time a detailed characterisation of a major N-acetylmuramidase from La. buchneri; this might have implications for other lactic acid bacteria (Anzengruber et al. 2014a).
Also for the Gram-positive pathogen Clostridium difficile that is a major cause of nosocomial diarrhoea (Dingle et al. 2013), there are indications of S-layer protein glycosylation. The cell surface of Cl. difficile is covered by an S-layer encoded by the slpA gene that localises within the cell-wall protein (cwp) gene cluster. For developing safe treatment strategies against Cl. difficile infections, it is important to understand the diversity and evolution of slpA and nearby genes encoding immunodominant cell-surface antigens (Fagan and Fairweather 2014). The whole-genome sequence was determined for 57 Cl. difficile isolates representative of the population structure and different clinical phenotypes. Phylogenetic analyses were performed on their genomic region (>63 kb) spanning the cwp cluster, with genetic diversity across the cwp cluster peaking within slpA, cwp66 (adhesin) and secA2 (secretory translocase). These genes form a 10-kb cassette, of which 12 divergent variants were found. Homologous recombination involving this cassette caused it to associate randomly with the genotype. One cassette contains a novel insertion (~24 kb) that resembles an slg gene cluster. S-layer switching and immune escape could help explain temporal and geographical variation in Cl. difficile epidemiology and may inform genotyping and vaccination strategies (Dingle et al. 2013). Normally, Cl. difficile does not possess a glycosylated S-layer (Qazi et al. 2009), but these variant strains contain a distinct S-layer cassette producing S-layer proteins of reduced polypeptide, as well as a gene cluster encoding dTDP-l-Rha biosynthesis (Giraud and Naismith 2000). Whether these strains do indeed have a glycosylated S-layer and what, if any, phenotype that might confer is currently unknown (Dingle et al. 2013).
The periodontopathic pathogen Ta. forsythia, a Gram-negative, rod-shaped, strict anaerobic, non-pigmented oral bacterium (Haffajee and Socransky 1994; Tanner and Izard 2006), relies on a unique building plan for its S-layer glycoproteins (Lee et al. 2006). Tannerella forsythia is phylogenetically affiliated to the Bacteroidetes and a member of the so-called red complex of oral bacteria (Holt and Ebersole 2005) that is crucially involved in the onset and progression of a set of inflammatory diseases named periodontitis. Periodontitis is the most common inflammatory disease of the gums worldwide and increasingly linked to systemic inflammation, increased risk of stroke, heart attacks and atherosclerosis (Haffajee et al. 2008; Hajishengallis and Lamont 2012). Tannerella forsythia cells are completely covered by a unique square S-layer formed by coassembly of two different glycoproteins in equimolar ratio (Lee et al. 2006; Sekot et al. 2012). The S-layer proteins TfsA and TfsB are both highly O-glycosylated with the equally unique oligosaccharide 4-OMe-β-ManpNAcCONH2 -(1→3)-[Pse5Am7Gra-(2→4)-]-β-ManpNAcA-(1→4)-[4-OMe-α-Galp-(1→2)-]-α-Fucp-(1→4)-[α-Xylp-(1→3)-]-β-GlcpA-(1→3)-[β-Digp-(1→2)-]-α-Galp- (Posch et al. 2011) linked to distinct serine and threonine residues within the Bacteroidetes-wide D(S/T)(A/I/L/M/T/V) glycosylation motif (Coyne et al. 2013) (Fig. 2). The structure of the glycan partially imitates that of host glycoproteins (Stafford et al. 2012), having a terminal sialic acid-like residue (nonulosonic acids) and a terminal fucose (Posch et al. 2011; Megson et al. 2015). The glycobiology of this pathogen, including its repertoire of glycosidases, seems to be key to its physiology and, potentially, pathogenicity (Posch et al. 2012; Douglas et al. 2014). In this context, it could be shown that the terminal trisaccharide motif of this glycan comprising the charged sugar residues acts to modulate DC effector functions to suppress T-helper (Th)17 responses (Settem et al. 2013). In contrast to the wild type, infection with a mutant strain lacking the trisaccharide motif induced a robust Th17 response and reduced periodontal bone loss in mice. These findings demonstrate that surface glycosylation of Ta. forsythia may act to ensure the bacterium’s persistence in the host likely through suppression of Th17 responses. In addition, this data suggest that the bacterium then induces the TLR2—Th2 inflammatory axis that has previously been shown to cause bone destruction. This study provided a biological basis for pathogenesis and opens opportunities in exploiting bacterial glycans as therapeutic targets against periodontitis and a range of other infectious diseases (Settem et al. 2013). Further, Ta. forsythia mutants lacking either the S-layer or glycan assembly and maturation genes display phenotypes involving altered human cell attachment to host cells, biofilm formation and disease progression (Sabet et al. 2003; Honma et al. 2007; Sakakibara et al. 2007).
The Ta. forsythia S-layer glycoproteins belong to a class of so-called CTD (C-terminal domain containing) proteins, with the CTDs on TfsA and TfsB acting as a signal for their translocation across the outer membrane via a Bacteroidetes type IX secretion system (T9SS) (Glew et al. 2012). The genome sequence of Ta. forsythia predicts the presence of the complete set of components for a T9SS in conjunction and a suite of CTD proteins (Tomek et al. 2014). To investigate if T9SS is functional in Ta. forsythia, T9SS-deficient mutants were generated by targeting either TF0955 (putative C-terminal signal peptidase) or TF2327 (PorK orthologue), and the mutants were analysed with respect to secretion, assembly and glycosylation of the S-layer proteins as well as proteolytic processing of the CTD. In either mutant, TfsA and TfsB were incapable of translocation as evidenced by the absence of the S-layer on ultrathin-sectioned bacterial cells using TEM. Despite being entrapped within the periplasm, MS analysis revealed that the S-layer proteins were modified with the complete, mature glycan as found on the secreted proteins, indicating that protein translocation and glycosylation are two independent processes in Ta. forsythia (Tomek et al. 2014). These data are supported by an independent investigation by others (Narita et al. 2014). There, Ta. forsythia mutants deficient in orthologues of the T9SS-encoding genes porK, porT and sov were constructed. All porK, porT and sov single mutants lacked the S-layer and expressed less-glycosylated versions of the S-layer glycoproteins TfsA and TfsB (Narita et al. 2014).
Besides its glycosylated S-layer, outer membrane vesicles (OMVs) of Ta. forsythia enriched in putative glycoproteins were identified as a new addition to the virulence repertoire of Ta. forsythia (Friedrich et al. 2015). The organism was grown anaerobically in serum-free medium and biogenesis of OMVs was analysed by TEM and atomic force microscopy revealing OMVs with a mean diameter of ~100 nm, budding off from the outer membrane while retaining the S-layer. A proteomic analysis of OMVs identified 175 proteins. Of these, 14 exhibited a CTD outer membrane translocation signal that directs them to the cell/vesicle surface, 61 and 53 were localised to the outer membrane and periplasm, respectively, 22 were predicted to be extracellular and 39 to originate from the cytoplasm. A total of 80 proteins contained the Bacteroidales O-glycosylation motif, 18 of which were confirmed as glycoproteins. The inflammatory response elicited by the OMVs was significantly higher than that caused by whole Ta. forsythia cells. This study represents the first characterisation of Ta. forsythia OMVs, their proteomic composition and immunogenic potential (Friedrich et al. 2015), and is complemented by a recent study by others (Veith et al. 2015).
Based on the evidence that the two Bacteroidetes species Ta. forsythia and Bacteroides fragilis both employ a general protein O-glycosylation system and share a common glycosylation sequon, the ability of these organisms to glycosylate a protein native to the other organism was demonstrated (Posch et al. 2013b). Using genetic tools previously developed for Bacteroides species, two Bac. fragilis model glycoproteins were expressed in the fastidious anaerobe Ta. forsythia and the attachment of the known Ta. forsythia O-glycan to these proteins was demonstrated by LC-ESI tandem MS. Likewise, two predominant Ta. forsythia glycoproteins were expressed in Bac. fragilis and glycosylation with the Bac. fragilis O-glycan was confirmed. Compositional and structural similarities between the Ta. forsythia O-glycan and the not yet fully characterised Bac. fragilis O-glycan suggest commonalities in their biosynthesis. These data demonstrate the feasibility of exploiting these organisms for the design of novel glycoproteins (Posch et al. 2013b).
Multidrug-resistant strains of the Gram-negative species Acinetobacter baumannii are increasingly being isolated in hospitals worldwide. Among the virulence factors identified in this pathogen, there is a general protein O-glycosylation system that appears to be important to biofilm formation and virulence, and a capsular polysaccharide that is essential for resistance to complement killing (Lees-Miller et al. 2013). In fact, the [→3)-[β-GlcNAc-(1→6)]-[α-4OAcGlcA-(1→4)-]-α-Gal-(1→6)-β-Glc-(1→3)-β-GalNAc-(1→]n pentasaccharides that decorate the Ac. baumannii glycoproteins are also the building blocks for capsule biosynthesis, with at most two repeat units having been detected on proteins via MS analysis. An O-pentasaccharide biosynthesis gene locus was identified in strain ATCC 17978, with components required for glycan assembly, polymerisation and transport of capsule located within or adjacent to the locus. Mutagenesis of PglC, the initiating glycosyltransferase prevented the synthesis of glycoproteins and capsule, resulting in abnormal biofilm structures and attenuated virulence of Ac. baumannii ATCC 17978 in mice. The discovery of a bifurcated pathway for O-glycosylation and capsule synthesis could not only provide insight into the biology of Ac. baumannii but also identified potential novel candidates for intervention against this emerging pathogen (Lees-Miller et al. 2013). In this context, it should be noted that previous TEM investigations of Acinetobacter sp. strain MJT/F5/199A have shown a presumably square S-layer lattice covering the entire cells (Thornley, Glauert and Sleytr 1973). Outer membrane preparations before and after papain digestion suggested the presence of a glycosylated S-layer protein in that strain that might resemble the O-glycosylated Ac. baumannii ATCC 17978 protein discussed (Lees-Miller et al. 2013).
Very recently, a glycosylated S-layer was shown to be present also on the anamox bacterium candidatus Kuenenia stuttgartiensis (van Teeseling et al. 2014). Anammox bacteria perform anaerobic ammonium oxidation and have a unique compartmentalised cell consisting of three membrane-bound compartments (from inside outwards)—the anammoxosome, riboplasm and paryphoplasm (van Teeseling et al. 2014). The cell envelope of anammox bacteria has been proposed to deviate from typical bacterial cell envelopes by lacking both PG and a conventional outer membrane. In the phylum of the Planctomycetes, for the first time, an S-layer was described on strain candidatus K. stuttgartiensis, providing a new addition to the cell plan of anammox bacteria. This S-layer showed hexagonal symmetry and enrichment of the S-layer from bacterial cells led to a 160-kDa candidate protein, Kustd1514, which has no homology to any known protein and is present in glycosylated form. Antibodies were generated against the glycoprotein and used for immunogold localisation. The antiserum localised Kustd1514 to the S-layer and, thus, confirmed it as the S-layer glycoprotein of candidatus K. stuttgartiensis (van Teeseling et al. 2014).
Secondary cell-wall polymers
In the course of investigations of Gram-positive bacterial S-layer proteins (both non-glycosylated and glycosylated ones), important aspects of a group of non-classical SCWPs, representing cell-wall polysaccharides of moderate molecular mass, have emerged. The identification of these non-classical SCWPs as mediators for non-covalent attachment of S-layers to carbon 6 of muramic acid of the underlying PG meshwork (Figs 2 and 3) underlines their importance for the cell biology of bacteria. Herein we summarise the current knowledge on structural features, cell-wall attachment and interactions of non-classical SCWPs in S-layer-carrying organisms (Araki and Ito 1989; Sára 2001; Cava et al. 2004; Schäffer and Messner 2005; Ferner-Ortner et al. 2007; Pavkov et al. 2008; Messner et al. 2009).
Description of non-classical SCWPs
While a considerable body of knowledge has accumulated concerning structural, biochemical and immunological features of teichoic and teichuronic acids (classical SCWPs) present in cell walls of Gram-positive bacteria (Hancock and Baddiley 1985; Archibald, Hancock and Harwood 1993), it should be kept in mind that additional cell-wall polysaccharides are present in these organisms. Driven by our interest in S-layer glycobiology, we initially coincidentally coisolated a fraction of SCWP with S-layer glycoprotein preparations of different bacteria. Improved purification and separation methods eventually led to the clear assignment of this distinct fraction as a separate class of compounds, i.e. SCWPs, which were shown to account for a substantial proportion (7%–15% by weight) of the bacterial PG (Schäffer and Messner 2005). The investigated SCWP–PG complexes were comprised of intact glycan moieties and, usually, of proportions of PG of variable size, arising from random PG degradation during the preparation procedure (without any acid treatment to prevent undesired removal of acid-labile components) (Messner, Schäffer and Kosma 2013).
Cell-wall polysaccharides of Gram-positive bacteria are generally classified according to their structural characteristics in (i) teichoic acids, (ii) teichuronic acids and (iii) other polysaccharides (neutral or acidic polysaccharides) not classifiable into the two former groups (Araki and Ito 1989). Recent investigations have shown that in S-layer-carrying organisms, teichoic and teichuronic acids as typical representatives of classical SCWPs (Archibald, Hancock and Harwood 1993) are usually not present. Instead, structurally comparable non-classical SCWPs are present. Based on compositional and structural data, we suggested a categorisation of non-classical SCWPs from S-layer-carrying Bacillaceae into three groups (group I to III) (Schäffer and Messner 2005). For each of these, representative structures are given below.
First group
The structure of the glycan portion of the SCWP–PG complex of Pa. alvei CCM 2051T was elucidated to be [(Pyr4,6)-β-d-ManpNAc-(1→4)-β-d-GlcpNAc-(1→3)]n~11 -(Pyr4,6)-β-d-ManpNAc-(1→4)-α-d-GlcpNAc-(1→ (Fig. 2) (Schäffer et al. 2000b). Each repeating-unit disaccharide of this SCWP is substituted with a 4,6-linked pyruvic acid residue, conferring an overall anionic character to the SCWP. Upon prolonged exposure of the polymer to acidic conditions, as occurs for instance upon solvation in D2O during NMR experiments, considerable loss of pyruvate residues can occur. The ManNAc–GlcNAc backbone disaccharide motif corresponds to that frequently observed in certain teichoic acids of other bacilli (Araki and Ito 1989). In the group I non-classical SCWPs, this motif is repeated several times, thus constituting the entire glycan moiety of these SCWPs. Pyruvic acid-containing SCWPs have also been reported for the S-layer-carrying organisms Lysinibacillus (formerly Bacillus) sphaericus CCM 2177 (Ilk et al. 1999) and Ba. anthracis (Mesnage et al. 2000).
In contrast to anionic polymers, neutral polysaccharides possessing an identical backbone motif are found in the PG of other Bacillaceae. The SCWPs of Th. thermosaccharolyticum strains D120-70 (Altman et al. 1990) and E207-71 (Altman et al. 1996) display the commonly encountered →3)-β-d-ManpNAc-(1→4)-β-d-GlcpNAc-(1→4)-β-d-ManpNAc-(1→3)-β-d-GlcpNAc-(1→ motif (Fig. 2) (Altman et al. 1996), which is extended by ribofuranose side chains on alternating ManNAc residues (Altman et al. 1990; Schäffer C, Kählig H, Christian R & Messner P, unpublished results).
Second group
The anionic polymer purified from G. stearothermophilus NRS 2004/3a (Messner et al. 1987; Schäffer et al. 1999a) comprises on average six tetrasaccharide repeating units with the structure →4)-β-d-Manp-2,3-diNAcA-(1→6)-α-d-Glcp-(1→4)-β-d-Manp-2,3-diNAcA-(1→3)-α-d-GlcpNAc(1→ (Fig. 2). It constitutes the SCWP of many G. stearothermophilus wild-type strains and was the first SCWP of an S-layer carrying organism for which the structure was fully elucidated. It covers a mass range of 4000–6000 and substitutes about 20%–25% of the PG muramyl residues (Messner et al. 1987; Schäffer et al. 1999a). Interestingly, analyses of the SCWP of Anoxybacillus (formerly Geobacillus) tepidamans GS5-97T indicated that its backbone structure is reminiscent of that of G. stearothermophilus NRS 2004/3a, but with additional modifications of the carboxyl groups by amide formation at the Manp-2,3-diNAcA residues, turning the anionic character of the SCWP into a neutral one. 31P NMR analysis has demonstrated that the SCWP of Anoxybacillus tepidamans GS5-97T is linked to muramic acid by a common phosphodiester linkage (Steindl et al. 2005).
Third group
In this group, the linkage-region disaccharide (compare with the first group) of teichoic acids is further extended. In addition to linear chains, branched repeating units containing neutral sugars and/or amino sugars have been observed. The charge-neutral SCWP isolated from Aneurinibacillus thermoaerophilus DSM 10155 represents a unique bacterial glycan structure containing repeats of the sequence →3)-α-d-GlcpNAc-(1→3)-β-d-ManpNAc-(1→4)-β-d-GalpNAc-(1→ (Fig. 2) (Steindl et al. 2002) found in the first biantennary SCWP from the domain Bacteria, with the full structure α-d-GlcpNAc-(1→3)-β-d-ManpNAc-(1→4)-β-d-GalpNAc-(1→3)-α-d-GlcpNAc-(1→3)-β-d-ManpNAc-(1→4)-β-d-GalpNAc-(1→3)-α-d-GlcpNAc-(1→4)-[α-d-GlcpNAc-(1→3)-β-d-ManpNAc-(1→4)-β-d-GalpNAc-(1→3)-α-d-GlcpNAc-(1→3)-β-d-ManpNAc-(1→4)-β-d-GalpNAc-(1→3)-α-d-GlcpNAc-(1→3)]-β-d-ManpNAc-(1→3)-α-d-GlcpNAc-(1→3)-β-d-ManpNAc-(1→3)-α-d-GlcpNAc-(1→3)-α-d-GlcpNAc-(1→. The interpretation of the observed NMR data supporting a single branched GalNAc unit need, however, to be treated with caution, since the 13C NMR chemical shifts of the carbon atoms in the branched residue are very similar to those located in the linear chain.
A branched structure was found in the SCWP from G. stearothermophilus PV72/p2, which, however, had undergone treatment with hydrofluoric acid (HF) for cleavage of the linkage to PG. Hence, information on the linkage region as well as the extent of pyruvate substitution and proof for N-deacetylated glycoses is compromised (Petersen et al. 2008). The SCWP contains a pentasaccharide repeating unit of the sequence →4)-[β-d-GlcpNAc-(1→3)]0.3 -β-d-ManpNAcA-(1→4)-β-d-GlcpN/NAc-(1→6)-[4,6-(S)-Pyr-α-d-ManpNAc-(1→4)]-α-d-GlcpNAc-(1→. Similar to the SCWPs of the first group, the (S)-configured pyruvate residues are present on the d-ManpNAc residues, albeit not within the linear backbone chain, but extending from the branching 4,6-disubstituted α-d-GlcpNAc residue.
SCWPs of Ba. anthracis and Ba. cereus may also be classified into the third group. Non-classical SCWPs of Ba. anthracis having approximate molecular masses in the range of 12 000–22 000 mediate binding to the SLH domain of the paracrystalline S-layer proteins Sap and extractable antigen 1 (EA1), respectively (Kern et al. 2010, 2012). The SCWP of several Ba. anthracis strains was released from its covalent linkage to PG by treatment with HF, and the resulting purified fractions were analysed by high-field NMR spectroscopy and MS techniques (Choudhury et al. 2006). The repeating unit of the SCWP comprises the backbone sequence →6)-α-d-GlcpNAc-(1→4)-β-d-ManpNAc-(1→4)-β-d-GlcpNAc-(1→ with α-galactopyranosyl side chains linked to O-3 of the α-d-GlcpNAc and β-d-GlcpNAc units, respectively, and a β-d-Galp residue linked to O-4 of the α-d-GlcpNAc residue. The trisaccharide backbone forms a consensus motif shared by many Ba. anthracis and Ba. cereus strains, respectively. Cross-reactive epitopes are present in these SCWPs as well as epitopes being specific of Ba. anthracis and Ba. cereus, respectively (Choudhury et al. 2006; Leoff et al. 2009). The β-d-galactosyl moiety attached to the non-terminal GlcNAc residue constitutes a unique binding epitope to endolysins from Ba. anthracis bacteriophages (Schuch, Nelson and Fischetti 2002; Mo et al. 2012). In contrast, galactosylation is absent in the SCWP of the avirulent strain Ba. anthracis CDC 648 (Ba684) (Forsberg et al. 2012). This SCWP is tethered via acid-labile phosphodiester bonds to PG. Additionally, pyruvate substitution has been detected in the HF-treated SCWPs from Ba. anthracis Sterne 34F2, 7702 and Ba. cereus strain G9241, 03BB87 and 03BB102 (Forsberg et al. 2011). The extent of pyruvate substitution remains ambiguous, since the cleavage of the SCWP from PG with 48% HF leads to the liberation of the acid-sensitive ketal groups, thus preventing quantification. For the Ba. anthracis SCWP CDC 684, the terminal (S)-pyruvate substituent at O-4 and O-6 of the β-d-ManpNAc residue has been identified by NMR spectroscopy (Forsberg et al. 2012). Earlier work identified the UDP-GlcNAc 2-epimerases GneY (BAS5048) and GneZ (BAS5117) that act as catalysts of ManNAc synthesis, as well as a polysaccharide deacetylase (BAS5051), as factors contributing to SCWP synthesis (Lunderberg et al. 2015). It was shown that tagO (BAS5050), which encodes a UDP-N-acetylglucosamine:undekaprenyl-P N-acetylglucosaminyl 1-P transferase, the enzyme that initiates the synthesis of murein linkage units, is required for Ba. anthracis SCWP synthesis and S-layer assembly. Similar to gneY-gneZ mutants, Ba. anthracis strains lacking tagO cannot maintain cell shape or support vegetative growth. In contrast, mutations in BAS5051 do not affect Ba. anthracis cell shape, vegetative growth, SCWP synthesis or S-layer assembly. These data suggest that TagO-mediated murein linkage unit assembly supports SCWP synthesis and attachment to PG via acid-labile phosphodiester bonds. Further, Ba. anthracis variants unable to synthesise SCWP trisaccharide repeats cannot sustain cell shape and vegetative growth (Lunderberg et al. 2015). A hallmark of S-layer proteins is their C-terminal crystallisation domain that assembles into a crystalline lattice once these polypeptides are deposited on the bacterial surface via association between their N-terminal SLH domains and the SCWP (Huber et al. 2005). In Ba. anthracis, it was shown that slaQ, encoding a small cytoplasmic protein conserved among pathogenic bacilli elaborating S-layers, is required for the efficient secretion and assembly of Sap and EA1. S-layer protein precursors cosediment with SlaQ, and SlaQ appears to facilitate Sap assembly. Purified SlaQ polymerises and when mixed with purified Sap promotes the in vitro formation of tubular S-layer structures. In the current model, SlaQ in conjunction with S-layer secretion factors SecA2 and SlaP promotes localised secretion and S-layer assembly in Ba. anthracis (Nguyen-Mau et al. 2015).
In other Ba. anthracis and Ba. cereus strains, modifications of the basic SCWP structure such as galactosidation and O-acetylation have been reported (Choudhury et al. 2006; Leoff et al. 2008; Forsberg et al. 2011). In addition, environmental conditions and expression levels may play important roles in dictating which of the SCWPs is actually formed. The induction of SCWPs synthesis under specific culture conditions points to the possibility of other, still unknown SCWPs being induced under different environmental conditions, such as the host internal milieu. Changes in the Ba. cereus cell-wall polysaccharide components could be involved in the high adaptability of this bacterium to environmental changes (Candela et al. 2011).
General considerations about non-classical SCWP structures
Comparison of the overall composition of non-classical SCWPs analysed so far revealed that the more complex glycans of the second group exhibit the same alternating order of gluco and manno sugars as glycans from the first group. The glycan chain starts with a GlcNAc residue at the reducing end and ends with Manp-2,3-diNAcA. It is possible that glycan structures of the second group have evolved from the simpler first-group structures by the introduction of residues such as glucose or Manp-2,3-diNAcA through the action of strain-specific enzymes. The same scenario could be imagined for glycans of the third group. However, biosynthesis pathways of greater complexity have to be envisaged and, so far, no experimental evidence is available to support this notion. The linkages between SCWPs and PG are currently being investigated in more detail.
Interactions of SCWPs and S-layers from Bacillaceae
In addition to the general features previously mentioned, novel properties for SCWPs have emerged from research on S-layers (Sleytr 1978; Sleytr et al. 1996). Provision of any kind of selection advantage by the S-layer to a bacterium in its natural environment requires the stable attachment of the S-layer to the bacterial cell surface in vivo. During evolution, Gram-positive bacteria have developed various strategies for displaying proteins on their surface. These strategies include predominantly covalent binding of LPXTG-carrying proteins to PG, but a number of non-covalent binding strategies have also been evolutionary developed (Navarre and Schneewind 1999). Reattachment experiments of isolated S-layer glycoproteins from thermophilic clostridia revealed the non-covalent character of the interaction between the S-layer and PG (Sleytr 1976). For La. buchneri, it was shown very early that hydroxyl groups of a neutral cell-wall polysaccharide are responsible for the attachment of the S-layer protein to the cell wall (Masuda and Kawata 1985).
Possible cell-wall-targeting mechanisms
Recently, the cell-wall-targeting mechanism of S-layer proteins has been investigated in more detail (Mesnage et al. 2000; Sára 2001; Cava et al. 2004; Pavkov et al. 2008; Janesch, Messner and Schäffer 2013). In general, S-layer proteins have two functional regions—a cell-wall-targeting domain, which in most of the organisms thus far investigated is located at the N-terminus, and a C-terminal self-assembly domain. The presence of a cell-wall-targeting domain in S-layer proteins was substantiated by Fujino, Béguin and Aubert (1993) and Lupas et al. (1994), who identified motifs of approximately 55 amino acids, containing 10–15 conserved residues, which were designated SLH (S-layer homology) domains. These SLH domains, usually composed of one to three modules, are means frequently used for targeting proteins to the cell surface. They are found not only in various S-layer proteins but also in many other cell-surface-associated proteins, such as the cellulosomes (Bayer et al. 2004) or other enzymes (Liu et al. 1996).
In the case of Ba. anthracis, the molecular basis of the interaction between SLH domains and SCWP was established by elucidating the binding properties (Mesnage et al. 2000). Both S-layer proteins of Ba. anthracis (EA1 and Sap) possess SLH domains that directly attach the S-layer proteins to PG (Mesnage et al. 1997, 1999). While the overall sequence similarity of SLH domains is rather low, the highly conserved four-amino-acid motif TRAE has been found to play a key role for the binding function to SCWP (May et al. 2006; Kern et al. 2011; Janesch, Messner and Schäffer 2013). The crystal structure recently elucidated of one S-layer protein from Ba. anthracis shows the SLH domains arranged in 3-fold pseudo-symmetry with the TRAE (or similar) motifs so that they would be accessible to the SCWP (Kern et al. 2011). A different binding mechanism was proposed for the S-layer proteins of G. stearothermophilus PV72/p2 (Ries et al. 1997; Sára et al. 1998) and Pa. alvei CCM 2051T (Janesch, Messner and Schäffer 2013). In these organisms, two different binding domains were identified in the N-terminal region of the S-layer proteins, one for SCWP and another for PG. In Thermus thermophilus, a similar binding mechanism was identified between an S-layer outer membrane complex and the cell wall. There is a strong interaction of the SLH domain of the S-layer protein with a pyruvylated component of a highly immunogenic SCWP (Cava et al. 2004).
Originally, the involvement of pyruvate groups in S-layer binding was inferred from observations in Ba. anthracis (Mesnage et al. 2000). It was demonstrated that the pyruvate transferase CsaB is involved in the addition of pyruvate to a PG-associated polysaccharide fraction and that this modification is necessary for the binding of the S-layer via SLH domains. Interestingly, the csaAB operon was found to be present in several bacterial species (Mesnage et al. 2000). Pyruvate was also identified in the SCWP repeating units of Lysinibacillus sphaericus CCM 2177 (Ilk et al. 1999) and Pa. alvei CCM 2051T (Schäffer et al. 2000b) (vide supra), which may be taken as an indication that pyruvate, or more possibly, negative charges of SCWPs, constitute a widespread mechanism for the anchoring of S-layer proteins containing SLH domains to the bacterial cell wall. Besides the anchoring mechanism involving SLH domains, another mechanism that is hypothesised to utilise basic amino acids, present in the cell-wall-targeting region and known for their direct interaction with carbohydrates, may apply for S-layer proteins devoid of SLH domains. In the genus Geobacillus, SLH domains have only been identified on the S-layer protein SbsB of G. stearothermophilus PV72/p2 (Kuen et al. 1997). All other investigated G. stearothermophilus strains possess S-layer proteins devoid of SLH domains (Messner et al. 2010). Interestingly, none of the SCWPs of organisms that possess S-layer proteins lacking SLH domains are modified with pyruvate groups, and some of them even have a net-neutral charge (Schäffer and Messner 2005). These observations support the notion that, in addition to the previously discussed involvement of pyruvate, other mechanisms are involved in the binding of S-layer proteins to the PG (Mesnage et al. 2000; Sára 2001; Cava et al. 2004; Janesch, Messner and Schäffer 2013). The non-conserved character of S-layer-binding mechanisms is supported by the observation that the cell-wall-targeting domain is not necessarily located at the N-terminal region of the S-layer protein. Well-documented examples of C-terminal anchoring domains are the S-layer proteins of La. acidophilus ATCC 4356 (Smit et al. 2001) and La. crispatus (Antikainen et al. 2002).
The diversity observed among different SCWP structures of Bacillaceae follows a general theme that is well known from other bacterial cell-surface structures, such as the serotypes of LPS (Raetz and Whitfield 2002; Whitfield and Trent 2014) and capsular polysaccharides (Sutherland 1999). Presumably, this diversity is responsible for creating microenvironments in which different organisms can survive under unfavourable conditions. Current data indicate that non-classical SCWPs function as mediators for anchoring S-layer (glyco)proteins from Bacillaceae to the bacterial cell wall. The complete elucidation of both the structure and biosynthesis of several non-classical SCWPs will contribute to our general understanding of the various mechanisms underlying the tethering of S-layer (glyco)proteins to the cell surface of Gram-positive bacteria.
Archaeal S-layer glycoproteins
During the past few years, considerable progress has been made in the structural and functional characterisation of carbohydrate-active proteins involved in archaeal S-layer-protein glycosylation. However, only a few new structures of archaeal S-layer glycans have been reported recently (Chaban et al. 2006; Peyfoon et al. 2010; Guan et al. 2012; Kandiba et al. 2016) (Fig. 2); the initial work on S-layer glycosylation of haloarchaea and methanogens is summarised in several excellent publications (Sumper 1987; Lechner and Wieland 1989; Eichler and Adams 2005; Calo, Kaminski and Eichler 2010; Claus and König 2010; Rachel 2010; Jarrell, Jones and Nair 2010; Jarrell et al. 2010; Albers and Meyer 2011; Eichler 2013; Meyer and Albers 2013; Kaminski et al. 2013b; Eichler et al. 2014; Kandiba and Eichler 2014; Kandiba et al. 2016). Typically, the cell envelope architecture in archaea comprises a paracrystalline S-layer situated on top of the cytoplasmic membrane (Sleytr and Messner 1983); in fact, this holds true for the majority of described archaea. Within the Crenarchaea, the S-layer often represents the only cell envelope component, but there are various exceptions from this wall architecture. Besides (glycosylated) S-layers in (hyper)thermophilic Crenarchaea and Euryarchaea as well as in halophilic archaea, there is a great variety of other cell envelope structures such as proteoglycan-like S-layers (halobacteria), glutaminylglycan (natronococci), methanochondroitin (Methanosarcina) or double-layered cell envelopes with pseudomurein (Methanothermus and Methanopyrus) (Klingl 2014).
Concerning halophilic archaea, S-layer glycoproteins from Halobacterium salinarum, Haloferax volcanii, Har. marismortui and Har. japonica have been characterised (Lechner and Sumper 1987; Sumper et al. 1990; Wakai et al. 1997; Calo et al. 2011; Kandiba et al. 2016). Recent molecular analyses of the cell-surface glycoprotein of Hbt. salinarum (Kandiba and Eichler 2015), which was among the first prokaryotic glycoproteins ever described (Mescher and Strominger 1976), show differences in the annotation of sugars compared with previous reports. The structural description in this review is based on the most recent report (Kandiba and Eichler 2015) revealing the S-layer glycoprotein CSG (Lechner and Sumper 1987) to contain two different types of sulfated N-linked saccharides—hexuronic acid-containing, non-repetitive oligosaccharides with the structure (SO3−-)3GlcA3-Glc, N-linked to the Asn residues of the protein via Glc (Wieland, Heitzer and Schaefer 1983) (Fig. 2) and a serially repeated hexuronic acid-containing saccharide with the repeating unit structure [–(H3CO–)GalA–GlcNAc–(SO3−–)(Galf–GalA)–(SO3−–)GalNAc–]n 10–15, N-linked to Asn2 of the S-layer protein CSG via the proximal sulfated GalNAc residue (Paul, Lottspeich and Wieland 1986) (Fig. 2). In addition, approximately 20 O-glycosidically linked neutral disaccharides [Glc–(1→2)–Gal] can be found on CSG of Hbt. salinarum (Lechner and Wieland 1989) (Fig. 2). The archaeal lipid carrier of the lipid-linked precursors of these saccharides is a C60-dolichol rather than the bacterial C55-undekaprenol; the oligosaccharide is bound to this lipid via dolichol monophosphate (Dol-P). Exceptionally, in a total lipid extract from Hbt. salinarum, a tetrasaccharide-charged dolichol was detected in addition to the Dol-P type LLO (Cohen-Rosenzweig et al. 2014). The tetrasaccharide chain was similar but not identical to the N-glycan attached to the Asn-2 position of the Hbt. salinarum S-layer glycoprotein reported previously (Zeitler et al. 1998). Thus, it is possible that both the Dol-PP type LLO and Dol-P LLO are oligosaccharide donors in Hbt. salinarum (Taguchi, Fujinami and Kohda 2016). Sulfation of the saccharides is completed while they are linked to lipid and does not occur after transfer of the saccharides to protein (Lechner and Wieland 1989).
A recent comparative analysis of archaeal LLOs that serve as oligosaccharide donors for Asn glycosylation confirmed the above-mentioned results and showed that in the investigated Euryarchaeota, including Pyrococcus furiosus and Archaeoglobus fulgidus, the oligosaccharide donors were of the Dol-P type whereas crenarchaeotal oligosaccharide donor from species such as Sulfolobus solfataricus and Pyrobaculum calidifontis belong to the Dol-PP type. Further, the Ar. fulgidus cells contain two oligosaccharide donors bearing oligosaccharide moieties with different backbone structures, and the Su. solfataricus cells contain two oligosaccharide donors bearing stereochemically different dolichol chains (Taguchi, Fujinami and Kohda 2016).
In an impressive series of research papers, Eichler and coworkers reinvestigated the structure of the S-layer glycan of Hfx. volcanii and established the complete biosynthesis pathway for an S-layer pentasaccharide Man–(H3CO–)HexA– HexA–HexA–Hex–Asn (Kaminski and Eichler 2010; Kaminski et al. 2010, 2012; Magidovich et al. 2010; Yurist-Doutsch et al. 2010; Cohen-Rosenzweig, Yurist-Doutsch and Eichler 2012; Arbiv et al. 2013; Cohen-Rosenzweig et al. 2014; Kandiba and Eichler 2015). Only recently, this structure of the Hfx. volcanii S-layer glycan was reinvestigated and amended to the novel structure—mannose-(1→2)-[methyl-(O→4)-]-β-GlcA-(1→4)-α-GalA-(1→4)-β-GlcA-(1→4)-β-Glc-(1→N)-Asn (Kandiba et al. 2016) (Fig. 2).
The S-layer glycoprotein forms a hexagonal paracrystalline lattice on the haloarchaeal cell envelope and it was previously believed to be anchored to the cell membrane by the intercalation of a transmembrane segment (Sumper 1987; Sumper et al. 1990). However, previous MS analyses of S-layer proteins have failed to identify peptides at the carboxy-terminus of the protein, which was taken as evidence that the transmembrane segment may not be present in the mature protein (Haft, Payne and Selengut 2012). Moreover, at least a portion of the Hfx. volcanii S-layer glycoprotein has been found to be lipid modified (Kikuchi, Sagami and Ogura 1999; Konrad and Eichler 2002; Kandiba, Guan and Eichler 2013). Comparison of the C-termini of the S-layer protein from both wild-type and ΔartA mutant indeed showed proteolytic processing as evident from the absence of the extreme C-terminal region in the mature wild-type protein. This observation strongly supports the hypothesis that the S-layer glycoprotein is not attached via a C-terminal transmembrane region (which is rather cleaved off), but instead anchored to the lipid bilayer. It is highly likely that ArtA is directly involved in this post-translational processing step (Abdul Halim et al. 2013). Additional experiments revealed a C-terminal tripartite structure of the S-layer glycoprotein, including a highly conserved proline-glycine-phenylalanine (PGF) motif. The S-layer lipid modification requires the PGF motif, is ArtA dependent and underscores the importance of the ArtA-mediated processing and anchoring of the Hfx. volcanii S-layer glycoprotein for growth and maintenance of cell morphology (Abdul Halim et al. 2016).
This archaeal anchoring principle of S-layers clearly contrasts that known for bacterial S-layer proteins, where carbohydrate interactions play a pivotal role in anchoring, either via SCWPs as in Gram-positive bacteria (Mesnage et al. 2000; Sára 2001; Schäffer and Messner 2005; Janesch, Messner and Schäffer 2013) or via rough LPS as suggested for Gram-negative bacteria (Awram and Smit 2001; Posch et al. 2013a).
During these studies, it was discovered that the lipid modification resulted in the formation of two distinct populations of S-layer glycoproteins (Kandiba, Guan and Eichler 2013). These observations are consistent with the S-layer glycoprotein being initially synthesised as an integral membrane protein that subsequently undergoes a processing event in which the extracellular portion of the protein is separated from the membrane-spanning domain and transferred to a ‘waiting’ lipid moiety (Kandiba, Guan and Eichler 2013). In a comparative approach, protein N-glycosylation in Har. marismortui, another haloarchaeon, was investigated (Calo, Guan and Eichler 2011). While both Hfx. volcanii and Har. marismortui decorate their S-layer proteins with the same N-linked pentasaccharide and employ dolichyl phosphate as the lipid glycan carrier, species-specific differences in the two N-glycosylation pathways exist. Haloarcula marismortui first assembles the complete pentasaccharide on dolichyl phosphate and only then transfers the glycan to the target protein. In contrast, Hfx. volcanii initially transfers the first four pentasaccharide subunits from a common dolichyl phosphate carrier to the target protein and subsequently delivers the final pentasaccharide from a distinct dolichyl phosphate to the N-linked tetrasaccharide (Calo, Guan and Eichler 2011). Recent studies of Hfx. volcanii have identified a gene cluster comprised of the agl genes aglB, aglE, aglF, aglG, aglI and aglJ encoding the Agl (archaeal glycosylation) proteins involved in the assembly and attachment of the pentasaccharide to distinct Asn residues of the S-layer protein in this species (Yurist-Doutsch and Eichler 2009). To cope with life in hypersaline environments, halophilic archaeal proteins are enriched in acidic amino acids. Interestingly, deletion of aglD resulted in a reduced growth rate in media with high salinities (3.4 or 4.8 M NaCl), whereas under standard growth conditions (1.7 M NaCl), such a change was not observed. A test of the influence of salinity on archaeal growth was performed by comparing the N-glycosylation of Hfx. volcanii S-layer glycoprotein from cells grown in high (3.4 M NaCl) and low (1.7 M NaCl) salt, as was the glycan bound to dolichyl phosphate and the lipid upon which the N-linked glycan is assembled. In high salt, Asn13 and Asn83 on the S-layer protein are modified by a pentasaccharide (vide supra), while dolichyl phosphate is modified by a tetrasaccharide comprising the first four sugar residues of the S-layer glycan (Kaminski et al. 2013a). When the same targets were considered from cells grown in low salt, substantially less pentasaccharide was detected. Recent efforts have identified Agl5–Agl15 as components of a second Hfx. volcanii N-glycosylation pathway responsible for generating the tetrasaccharide Rha–Hex–Hex–(SO3−–)Hex–Asn (Fig. 2) attached to the S-layer glycoprotein when growth occurs in 1.7 M but not 3.4 M NaCl-containing medium (Eichler et al. 2013; Kaminski et al. 2013a). The same tetrasaccharide modifies S-layer glycoprotein Asn498 in cells grown in low salt, whereas no glycan decorates this residue in cells grown in the high-salt medium. Thus, in response to changes in environmental salinity, Hfx. volcanii not only modulates the N-linked glycans decorating its S-layer glycoprotein but also the sites where this post-translational modification occurs (Guan et al. 2012). This high degree of inherent structural flexibility makes this organism a potent candidate for glycoengineering (Calo, Guan and Eichler 2011).
While genomic analyses point to N-glycosylation as being a common post-translational modification in archaea, to date distinct pathways of archaeal N-glycosylation have only been described for few species (Kandiba and Eichler 2015). With this in mind, the similarities of N-linked glycans decorating glycoproteins in Hfx. volcanii and Hbt. salinarum directed a series of bioinformatic, genetic and biochemical experiments, which were designed to elucidate the Hbt. salinarum pathway responsible for biogenesis of one of the two N-linked oligosaccharides of that species. As in Hfx. volcanii, where agl genes encode proteins responsible for the assembly and attachment of the pentasaccharide to the Asn residues of the target protein, also the Hbt. salinarum genome contains a group of clustered homologous genes. Introduction of these Hbt. salinarum genes into Hfx. volcanii mutant strains deleted of the homologous sequences restored the lost activity. Moreover, transcription of the Hbt. salinarum genes in the native host and in vitro biochemical confirmation of the predicted functions of several of these gene products provided further support for the assignments made according to bioinformatic and genetic experiments, enabling that the first description of the N-glycosylation pathway in Hbt. salinarum was presented (Kandiba and Eichler 2015).
Among halophilic archaea, it was generally assumed that paracrystalline S-layers are assembled only of a single (glyco)protein species. Recently, however, this assumption changed when in Har. hispanica two S-layer glycoproteins, termed Slg1 and Slg2, with a sequence identity of 83%, were identified as constituents of the S-layer (Lu et al. 2015). Based on MS data, the ratio of these glycoproteins was determined to be 1:1. Selective releasing procedures of the decorating glycans from Slg1 and Slg2 allowed discrimination between an N- and an O-linked fraction. In Har. marismotui, Har. japonica, Har. californiae and Har. sinaiiensis, homologous genes corresponding to Slg1/Slg2 and Hvo_2072, the S-layer glycoprotein of Hfx. volcanii, were found (Wakai et al. 1997; Calo et al. 2011). In Har. marismotui and Har. Japonica, expression, regulation and function of the homologous genes of Slg1 and Slg2 are still unknown. In the O-linked glycan fraction containing Slg1 and Slg2, an α-Glc-(1→4)-Gal disaccharide was released at a relatively mild condition (low temperature and short time) compared with the release procedure for the acidic, N-linked trisaccharide. MALDI-TOF MS analyses of the digested glycopeptides obtained from Slg1 and Slg2 revealed that these disaccharides are linked to Thr residues within the DTPE repeats at the C-terminus of Slg2. In Slg1, although there are no DTPE repeats, the corresponding region at the C-terminus of Slg1 is almost completely composed of Asp, Glu, Thr and Pro residues. The Thr residues within this region are very likely modified by the same Glc-Gal disaccharide. The acidic N-linked trisaccharides are linked either to Asn306 in Slg1 or Asn307 in Slg2 (Lu et al. 2015). Thus, the S-layer proteins of Hbt. salinarum as well as Hfx. volcanii can be simultaneously modified by at least two kinds of N-linked glycans (Eichler 2013; Jarrell et al. 2014).
In Har. hispanica, only one N-linked glycan species was detected, which comprises a novel branched trisaccharide containing 2-amino-6-sulfo-2,6-dideoxy-glucose (sulfoquinovosamine) (Fig. 2). The trisaccharide is linked to the S-layer glycoprotein via galactose (Lu et al. 2015). The sulfoquinovose identified in the N-glycan of Har. hispanica is further modified by an amino group at C-2 position. This was the first observation of the naturally occurring form of sulfoquinovosamine, but it was already chemically synthesised, previously (Sacoman and Hollingsworth 2011). As an analogue of glucosamine-6-phosphate, sulfoquinovosamine is an effective inhibitor of the UDP-N-acetylglucosamine synthesis and leads to defects in bacterial or fungal cell-wall biosynthesis. In Har. hispanica, the pathway for biosynthesis and the biological function of this rare amino sugar are still unknown.
Apart from this study, sulfated sugars have been identified in N-linked glycans of several archaeons. In Su. acidocaldarius, the S-layer glycoprotein is glycosylated at multiple sites with chitobiose-linked N-glycans containing 6-sulfoquinovose (Peyfoon et al. 2010). Similarly, in Su. solfataricus P2, 6-sulfoquinovose is present in surface exposed glycoproteins (Palmieri et al. 2013). In Thermoplasma acidophilum, the cell-surface glycoproteins are modified with an N-linked glycan containing 6-sulfofucose (Vinogradov et al. 2012).
Recently, in Hfx. volcanii, the S-layer glycoprotein was found modified at Asn732 by a chitobiose-linked oligosaccharide containing 6-sulfoquinovose (Parente et al. 2014). The structural characterisation of the novel oligosaccharide related a rhomboid protease with the protein glycosylation process. Deletion of the protease gene altered the common S-layer glycoprotein N-glycan structure at the Asn732 site (Parente et al. 2014). Haloferax volcanii is able to grow at salt concentration from 1.7 to 4.8 M NaCl. When growing at high (3.4 M NaCl) and low (1.7 M NaCl) salinity, Hfx. volcanii cells can modify the N-glycans by utilising different N-glycosylation sites on the S-layer glycoprotein (Guan et al. 2012). The reversible changes of protein glycosylation require Hfx. volcanii cells to adapt to the fluctuation of the environmental salinity. If compared with Hfx. volcanii, Har. hispanica needs at least 3.4 M NaCl for decent growth and grows rather poorly at 2.6 M NaCl. Currently, it is not known whether Har. hispanica could adopt to changing environmental salinity and synthesise different N-glycans on the S-layer protein (Lu et al. 2015).
The O-glycan decorating both S-layer glycoproteins of Har. hispancia is an α-Glc-(1→4)-Gal disaccharide. In Hbt. salinarum and Hfx. volcanii, chemical characterisation established a Glc-(1→3)-Gal linkage and a Glc-(1→2)-Gal linkage, respectively (Sumper 1987; Sumper et al. 1990). Although there are differences in sugar linkages, the O-glycan structures seem to be relatively unchanged when compared with the diverse N-glycans found in different haloarchaeal species. The conserved O-glycan structure may indicate a universal function of this Glc-Gal disaccharide in halophilic archaea (Lu et al. 2015).
Recently, by performing for the first time metaproteomics on haloarchaea, genomic variation of S-layer, archaella [ = archaeal flagella; compare with (Jarrell and Albers 2012; Albers and Jarrell 2015); vide infra] and other cell-surface proteins was linked to mechanisms of infection evasion (Tschitschko et al. 2015). In Deep Lake, Antarctica, intergenera gene exchange occurs rampantly within the low complexity, haloarchaea-dominated community, strongly balanced by distinctions in niche adaptation that maintain sympatric speciation. CRISPR defence systems were found to be active, with haloarchaea responding to at least eight distinct types of viruses, including those infecting between genera. Although evasion and defence were evident, both hosts and viruses also may benefit from viruses carrying and expressing host genes, thereby potentially enhancing genetic variation and phenotypic differences within populations (Tschitschko et al. 2015).
The cell surface of the hyperthermophile Methanothermus fervidus is covered by an S-layer glycoprotein with the composition d-3-O-methylmannose, d-Man and d-GalNAc in a molar ratio of 2:3:1, with the first residue partly replaced by 3-O-methylglucose. The heterosaccharide with the proposed structure α-d-3-MeOManp-(1→6)-α-d-3-MeOManp-[(1→2)-α-d-Manp]3-(1→4)-d-GalNAc is linked via GalNAc to asparagines of the peptide moiety (Fig. 2) (Kärcher et al. 1993).
In the course of structural investigations of glycosylated flagellins from methanogenic archaea, Jarrell and coworkers (Koval and Jarrell 1987; Chaban et al. 2009) made the interesting observation that the flagellin and also the glycosylated S-layer protein of Methanococcus voltae were decorated with the same complex N-linked glycan β-ManpNAcA6Thr-(1→4)-β-GlcpNAc3NAcA-(1→3)-β-GlcpNAc-(1→N)-Asn (Fig. 2), where one of the constituting glycan components (β-ManNAcA) having a carbonyl group at C-6 forms an amide bond with the amino group of a threonine residue. Of note is an extension of the trisaccharide, both on the S-layer glycoprotein and the archaellar glycan, with an additional mass of either 220 or 262 Da, when the glycoproteins were prepared from Me. voltae strain PS* (Fig. 2). Searching the Me. voltae genome identified Mv990 and Mv991 as putative glycosyl transferase genes, which are responsible for the mentioned mass addition. Deletions of Mv990 and Mv991 were made, and both resulted in N-glycosylation defects to the archaellin and S-layer proteins (Voisin et al. 2005; Chaban et al. 2009). Using a suite of synthetic and semisynthetic substrates, it was shown that AglK initiates N-linked protein glycosylation in Me. voltae through the formation of α-linked dolichyl monophosphate GlcNAc, which contrasts the polyprenyl diphosphate intermediates that feature in both eukaryotes and bacteria (Weerapana and Imperiali 2006; Varki et al. 2015). Notably, AglK has high-sequence homology to dolichyl phosphate β-glucosyltransferases including Alg5 in eukaryotes (Burda and Aebi 1999), suggesting a common evolutionary origin. The combined action of the first two enzymes, AglK and AglC, affords an α-linked dolichyl monophosphate glycan that serves as a competent substrate for the archaeal OST AglB (Larkin and Imperiali 2011).
The Methanosarcina mazei sheath or S-layer protein MM1976 is one of the most abundant proteins made by the archaeon (Francoleon et al. 2009; Rohlin et al. 2012). ConA-binding enriched cell lysates for this protein, permitting characterisation of low stoichiometry modifications. Bona fide glycosylated proteins, revealed by their oxonium ions in peptide MS/MS spectra, were identified from ConA-eluate as MM0002, MM0716, MM1364 and S-layer protein MM1976. The OST (MM0647) detected in the ConA pull-down experiment is a product of one of three aglB homologues encoded in the Met. mazei genome (MM646, MM0647, MM2210) (Magidovich and Eichler 2009). Its detection makes MM0647 a logical candidate for the AglB OST that links glycans to asparagines on the MM1976 S-layer protein and to other N-linked glycoproteins. In addition, it was found that the hypothetical proteins MM0716 and MM1364 are glycosylated. Glycosylation of MM1364 correlates with homology to known Methanosarcina S-layer proteins (Francoleon et al. 2009; Rohlin et al. 2012) that also bear glycans. To date, no structural information about glycosylated Met. mazei proteins is available (Leon et al. 2015).
Recently, it was shown that the S-layer oligosaccharide of the thermoacidophilic crenarchaeon Su. acidocaldarius consists of a tri-branched hexasaccharide {α-Man-(1→6)-}{α-Man-(1→4)-}-[(HO3S→6)-][β-Glc-(1→4)-]-β-Qui-(1→3)-β-GlcNAc-(1→4)-β-GlcNAc-(1→N)- that is N-glycosidically linked via a chitobiose core to several asparagine sites at the S-layer protein (Baumeister and Lembcke 1992; Veith et al. 2009) (Fig. 2). Interestingly, the same glycan structure was already reported on the Su. acidocaldarius cytochrome b (Zähringer et al. 2000) and on the archaellum (vide infra). Sharing of identical glycan structures on both S-layers and archaella was first reported with the haloarchaeon Hbt. salinarum (Wieland, Paul and Sumper 1985).
The N-glycosylation pattern of cell-surface-exposed proteins of Su. solfataricus P2 (Baumeister and Lembcke 1992; Veith et al. 2009) was analysed by lectin affinity purification, HPAEC-PAD and multiple MS-based techniques (Palmieri et al. 2013). Detailed analysis of SSO1273, one of the most abundant ABC transporters present in the cell-surface fraction of Su. solfataricus, revealed a novel glycan structure composed of a branched sulfated heptasaccharide, Hex4(GlcNAc)2 and sulfoquinovose, where Hex is d-mannose and d-glucose. The same type of N-glycosylation was been found and verified on the S-layer glycoprotein (SSO0389) (Fig. 2) and different proteases, having one monosaccharide unit more than the glycan of the S-layer glycoprotein of Su. acidocaldarius (Peyfoon et al. 2010). This is one of the most complex archaeal glycan structure known today (Palmieri et al. 2013).
Archaeal Surface Appendages
Archaea have evolved fascinating surface structures allowing rapid adaptation to changing environments. Archaeal surface appendages display diverse biological roles such as motility, adhesion, biofilm formation, exchange of genetic material and species-specific interactions and, in turn, increase fitness of the cells (Ng, Chaban and Jarrell 2006; Lassak, Ghosh and Albers 2012).
Archaella
Flagella as motility structures evolved independently in the bacterial and the archaeal kingdom. Since the archaeal archaellum is different from the bacterial flagellum, investigation of structural biochemistry and activities of this motility machinery is of great interest (Ng, Chaban and Jarrell 2006; Jarrell and Albers 2012; Albers and Jarrell 2015). To account for this difference, renaming of the archaeal flagellum to ‘archaellum’ was proposed (Jarrell and Albers 2012; Albers and Jarrell 2015). With increased studies conducted on archaea, it has become clear that the archaellum only functionally appears similar to the bacterial flagellum fulfiling the same swimming function, whereas it structurally resembles a bacterial type IV pilus. Important genes for proper function of the archaella are conserved in the fla operons of the different archaea (Eichler 2000; Ghosh and Albers 2011; Jarrell and Albers 2012), with defects in N-glycosylation influencing the function of the archaellum. In Haloferax volcanii (Abu-Qarn et al. 2007; Tripepi et al. 2012), Methanococcus voltae (Chaban et al. 2006), Me. maripaludis (VanDyke et al. 2009) and Sulfolobus acidocaldarius (Meyer et al. 2011, 2013), for instance, AglB, the archaeal OST is the central enzyme of the N-glycosylation pathway; its deletion results in non-motile cells, lacking any archaella filament.
Previously, an archaellum-related gene cluster was identified in Halobacterium salinarum (Patenge et al. 2001). For the structural characterisation of halobacterial archaella originally Hbt. salinarum strain R1M1 was used (Trachtenberg, Pinnick and Kessel 2000; Cohen-Krausz and Trachtenberg 2002). However, recent work was performed with strain M175, which, under conditions of relatively low ionic strength (0.8 M versus 5 M) and low pH (~2.5 versus ~6.8), forms polyarchaellar bundles at high yield. The archaellum assembles at the proximal end and is constructed from different glycosylated archaellins (Cohen-Krausz and Trachtenberg 2002).
Besides the S-layer glycoproteins of the haloarchaea Hbt. salinarum (Kandiba and Eichler 2015) and Hfx. volcanii (Kandiba et al. 2016), little is known about the glycosylation of other haloarchaeal proteins. Recently, it was demonstrated that the Hfx. volcanii archaellins require archaeal glycosylation (Agl) components involved in S-layer protein glycosylation and that the deletion of any Hfx. volcanii agl gene impairs the baterium’s swimming motility to various extents (Tripepi et al. 2012). A comparison of wild-type Hfx. volcanii and deletion mutants lacking the oligosaccharyltransferase AglB suggests that when the Agl glycosylation pathway is disrupted, cells lack stable archaellins, which consist of the major archaellin, FlgA1, and a minor archaellin, FlgA2. MS analyses of FlgA1 confirm that its three predicted N-glycosylation sites Asn70, Asn115 and Asn172 are modified with covalently linked pentasaccharides having the same mass as that modifying the S-layer glycoprotein, namely mannose-(1→2)-[methyl-(O→4)-]-β-GlcA-(1→4)-α-GalA-(1→4)-β-GlcA-(1→4)-β-Glc-(1→N)-Asn (Kandiba et al. 2016) (Fig. 2). Finally, the replacement of any of three predicted N-glycosylated asparagines of FlgA1 renders cells non-motile, providing direct evidence for the first time that the N-glycosylation of archaellins is critical for motility (Tripepi et al. 2012).
The archaellum of Me. voltae is composed of four structural archaellins FlaA, FlaB1, FlaB2 and FlaB3; they possess a total of 15 potential N-linked (N-X-(S/T)) sequons with 14 of them carrying glycan modifications. The archaellum glycan structure was shown to be a trisaccharide composed of β-ManpNAc3NAcA6Thr-(1→4)-β-GlcpNAc3NAcA-(1→3)-β-GlcpNAc, linked to Asn residues (Voisin et al. 2005). Further, an extension of the archaellar glycan by an additional mass of 220 or 262 Da is possible when the glycoprotein is prepared from Me. voltae strain PS* (vide supra, characterisation of the respective S-layer glycoprotein) (Chaban et al. 2009). In Me. maripaludis, the three archaellins that comprise the archaellum are modified at multiple sites with the N-linked tetrasaccharide Sug-(1→4)-β-ManNAc3NAmA6Thr-(1→4)-β-GlcNAc3NAcA-(1→3)-β-GalNAc-Asn, with the components GalNAc, a diacetylated glucuronic acid (GlcNAc3NAc) and an acetylated and acetamidino-modified mannuronic acid with a threonine substitution in an amid-linkage to carbon 6 of the uronic acid (ManNAc3NAmA6Thr). Further, at the non-reducing end is the novel sugar residue Sug with the structure (5S)-2-acetamido-2,4-dideoxy-5-O-methyl-α-l-erythro-hexos-5-ulo-1,5-pyranose, so far found exclusively in this species. Thus, the Me. voltae GlcNAc-linked trisaccharide is extended in Me. maripaludis by an additional sugar residue and is attached in a different N-linkage via β-GalNAc to Asn residues. The significance of this linkage to archaellins via β-GalNAc rather than β-GlcNAc is not known, but this may be a reflection of the specificity of the respective glycosyltransferases from either strain (Kelly et al. 2009). Several studies were devoted to the analysis of protein functions encoded in the archaellin gene clusters. Successful deletion of genes encoding the three glycosyltransferases and an OST (Stt3p homologue) resulted in archaellins of decreased molecular masses as evidenced by immunoblotting and TEM, indicating partial or completely absent glycan structures (VanDyke et al. 2009; Ding et al. 2013). In an extension of this work, a series of genes in adjacent operons were shown to encode enzymes that complete the pathway for generation of the subsequent sugars of the N-linked tetrasaccharide that modifies archaellins of Me. maripaludis. Among the genes involved in sugar biosynthesis was the putative acetyltransferase gene mmp0350, which appears to attach an acetyl group to the second sugar of the glycan, a diacetylated glucuronic acid (VanDyke et al. 2009; Jarrell, Jones and Nair 2010; Jarrell et al. 2010; Jones et al. 2012). This post-translational modification of archaellins is necessary for archaellum assembly (Siu et al. 2015). Methanococcus maripaludis, the mmp0350-mmp0353 and mmp0357 genes are proposed to be functionally equivalent to the Pseudomonas aeruginosa wbpABEDI genes involved in converting UDP-GlcNAc to UDP-2,3-diacetamido-2,3-dideoxy-d-mannuronic acid, a serotype O5-specific sugar antigen (Westman et al. 2009). Cross-domain complementation of the final step of the P. aeruginosa pathway with mmp0357 supports this hypothesis. This study included a rare example of an archaeal gene functionally replacing a bacterial gene in a complex sugar biosynthesis pathway (Siu et al. 2015). Characterisation of single genes from the cluster mmp1089-mmp1094 and gene annotation and bioinformatic analyses indicated that MMP1090 is a UDP-glucose 4-epimerase, suggesting that the unique terminal sugar of the archaellin N-glycan might be synthesised from UDP-Glc or UDP-GlcNAc with an essential early step in synthesis catalysed by MMP1090 (Ding et al. 2016).
Whereas flagella have been intensively studied (Kearns 2010), the knowledge regarding the archaeal counterpart is mostly restricted to Euryarchaeota rather than crenarchaeal archaella. Therefore, the archaellar assembly system of the crenarchaeal model organism Su. acidocaldarius was investigated in vivo in greater detail (Lassak et al. 2012). Promoter studies and qRT-PCR analyses of the archaella gene cluster provided evidence that the expression of the fla genes was induced by tryptone starvation. Moreover, it was confirmed that a secondary fla promoter is present within the flaB gene that regulates the transcription of downstream genes flaX-J. Markerless in-frame deletions for all fla genes encoded in the fla gene cluster were constructed. Western blot analysis of the fla deletion strains suggested hierarchical protein interactions during archaella assembly. TEM micrographs demonstrated the loss of archaellar assembly coincided with a lack of motility. Thus, all seven fla genes are essential for crenarchaeal archaellum assembly and function.
Recently, the S-layer protein of Su. acidocaldarius was shown to be N-linked with a tri-branched hexasaccharide containing a sulfated sugar called sulfoquinovose (Peyfoon et al. 2010). Concerning the frequently observed convergence of sugar biosynthesis pathways, it was interesting to see that the effect of deletion of agl3, encoding the UDP-sulfoquinovose, did not only result in an altered N-glycan on SlaA but also on the Su. acidocaldarius archaellin FlaB (Meyer et al. 2011). Thus, it was reasoned that deletion of agl16, encoding a glycosyltransferase involved in S-layer N-glycan assembly, might also lead to different migration behaviour of the FlaB protein in SDS-PAGE. MS analyses confirmed that the glycan of the S-layer protein from the agl16 deletion mutant was a pentasaccharide, which was missing a terminal hexose residue; HPLC analyses of the hydrolysed N-glycan indicated that the missing hexose is a glucose residue. A physiological characterisation of the Δagl16 deletion mutant revealed a significant effect on the growth at elevated salt concentrations. At 300 mM NaCl, the doubling time of the Δagl16 mutant was increased 10-fold compared to the background strain. Furthermore, the incomplete glycan structure of the Δagl16 deletion strain affected the assembly and function of the archaellum, as exemplified by semisolid Gelrite plate analysis, in which the motility is decreased in dependence of the N-glycan size. Indeed, both FlaB proteins from the Δagl16 and the Δagl3 deletion strain migrated faster than the wild-type protein, whereas in the complemented strains, the original FlaB wild-type size was restored. As was shown for the S-layer glycoprotein, FlaB in the Δagl16 strain was smaller compared to FlaB in the wild-type and Δagl3 strains. This difference is most likely due to the loss of only one sugar residue on the N-glycan in Δagl16, whereas in Δagl3, the N-glycan is truncated by three sugar residues (Meyer et al. 2011). In Su. acidocaldarius, N-glycosylation mutants, which exhibited reduced N-glycan size, lacking one or three-terminal sugar residues, were severely affected in motility (Meyer et al. 2011, 2013). Interestingly, there are also indications of a physical association between the archaellum and the S-layer glycoprotein in Su. acidocaldarius. FlaF, localised in the archaellum biosynthesis gene cluster, was shown to specifically bind to the S-layer protein, suggesting that its interaction domain is located in the pseudoperiplasm with its N-terminal helix in the membrane. Thus, FlaF may act as the previously unknown archaellum stator protein that anchors the rotating archaellum (Shahapure et al. 2014) to the archaeal cell envelope (Banerjee et al. 2015). Structural comparison with archaella from Su. shibatae B12 and Hbt. salinarum revealed high similarity. This suggests a unique and common underlying symmetry for archaeallar filaments (Cohen-Krausz and Trachtenberg 2008).
The archaella of the euryarchaeon Pyrococcus furiosus are multifunctional cell appendages used for swimming, adhesion to surfaces and formation of cell–cell connections. Archaella were purified by shearing from cells followed by CsCl-gradient centrifugation and were found to consist mainly of an approximately 30-kDa glycoprotein as was demonstrated by SDS-PAGE and glycan staining (Näther-Schindler et al. 2014).
Archaeal pili
In both bacteria and archaea, the biosynthesis of type IV pilus-related structures involves a set of core components (Logan 2006; Makarova, Koonin and Albers 2016). Although in silico analyses showed that most sequenced archaeal genomes encode predicted pilins and conserved pilus biosynthesis components, only recent in vivo analyses of archaeal pili in genetically tractable crenarchaea and euryarchaea revealed archaea-specific type IV pilus functions and biosynthesis components. In the future, studies in a variety of archaeal species will reveal which type IV pilus-like structures are common in all archaea and which are limited only to certain species within this domain (Pohlschroder et al. 2011; Pohlschroder and Esquivel 2015). These structures do not only comprise the widely distributed archaella, but also different highly specialised archaeal pili which may possess different assembly mechanisms, structural aspects and physiological roles (Lassak, Ghosh and Albers 2012; Makarova, Koonin and Albers 2016).
At least two different types of pilus structures have been reported in archaea, including those assembled from type IV pilin-like proteins, as seen in several genera including Hfx. volcanii (Esquivel et al. 2016), Methanococcus and Sulfolobus, and others that are not seemingly related to type IV pili, as in Methanothermobacter thermoautotrophicus (Jarrell, Jones and Nair 2010).
The oligosacharyltransferase (AglB)-dependent N-glycosylation of archaellins is required for archaella assembly in many archaea (Tripepi et al. 2012). However, whether N-glycosylation is required for the assembly and/or function of the structurally related archaeal type IV pili was unknown so far. In a recent study, type IV pilin N-glycans of Hfx. volcanii with diverse roles in pilus biosynthesis, adhesion and microcolony formation have been identified (Esquivel et al. 2016). It was shown that of six adhesion pilins, PilA1 and PilA2, the most abundant pilins in wild-type and ΔaglB strains, are modified under planktonic conditions in an AglB-dependent manner by the same pentasaccharide as was detected on Hfx. volcanii archaellins (mannose-(1→2)-[methyl-(O→4)-]-β-GlcA-(1→4)-α-GalA-(1→4)-β-GlcA-(1→4)-β-Glc-(1→N)-Asn; Fig. 2) (Kandiba et al. 2016). These data revealed that PilA1–PilA4 are N-glycosylated in an AglB-dependent manner and also showed that these pilins inhibit microcolony formation, which is promoted by PilA5 and PilA6 (Esquivel et al. 2016). Non-glycosylated pilins are not able to inhibit this early step in biofilm formation. Since loss of AglB-dependent glycosylation of the archaellins also inhibits Hfx. volcanii archaella biosynthesis (Tripepi et al. 2012), low salt conditions might promote biofilm formation by inhibiting archaella-dependent motility as well as resulting in promotion of microcolony formation, which is dependent on the glycosylation of PilA1–PilA4. This is the first demonstration of a differential effect of glycosylation on pilus assembly and function of paralogous pilins. The growth of wild-type cells in low salt media, a condition that decreases AglB glycosylation, also stimulates microcolony formation and inhibits motility, supporting the hypothesis that N-glycosylation plays an important role in regulating the transition between planktonic to sessile cell states as a response to stress (Esquivel et al. 2016).
A study on Meth. thermoautotrophicus fimbriae/pili indicated that the main structural component is a 16-kDa glycoprotein (Mth60) with no known homologues reported in the databases (Thoma et al. 2008). For the first time in archaea, pili structures were shown to be adhesive surface appendages, and their abundance on the cell surface was greatly enhanced when cells were grown on surfaces rather than in liquid cultures. In this context, it is interesting to note that archaea, like bacteria, predominately exist as biofilms in nature (Thoma et al. 2008).
The type IV pili on the surface of Me. maripaludis are much less numerous than archaella. They are composed of a major structural protein of 17 kDa that is N-glycosylated with the same complex tetrasaccharide as it is present in Me. maripaludis archaella (vide supra), but with an additional hexose attached to the GalNAc linking sugar (Siu et al. 2015).
Sulfolobus acidocaldarius displays three distinct type IV pili-like structures on its surface: the archaellum, the UV-induced pili and the adhesive pili (Henche et al. 2012b; Lassak et al. 2012). To investigate the influence of the diverse set of type IV pili-like structures, deletion mutants lacking the cell-surface appendages were constructed and analysed for their behaviour in attachment assays and during biofilm formation. There is evidence that all three cell-surface appendages play a role in the colonisation of surfaces and only the interplay of all three appendages leads to the observed wild-type biofilm phenotype (Henche et al. 2012b). Type IV pilins and archaellins are N-glycosylated by the archaeal Agl N-glycosylation machinery (Ghosh and Albers 2011). MS analysis of the pilin/archaellin fraction of Su. acidocaldarius identified, among other genes, aapB (Saci2319) as the gene encoding the pilin AapB. Harsh treatment of the isolated pili with phenol and detergents like Triton X-100 did not lead to the disassociation of the pilin subunits. This stability might stem from strong hydrophobic interactions of the pilin subunits and also post-translational modification of the aap pilus by glycosylation. As only AapB was identified in the isolated fraction, it is assumed that this protein is the major pilin of the aap filament. Numerous asparagines are present in the pilins that are possible target residues of the N-glycosylation machinery; the isolated fraction was analysed by TEM analysis, SDS-PAGE and glyco-stained using the Pro-Q Emerald 300 Glycoprotein Gel Stain kit (Henche et al. 2012a). Up to now, no defined pilus glycan structure of Su. acidocaldarius has been reported.
Other archaeal glycoproteins
The glycan structure of Py. furiosus proteins, as determined by MS, is a single oligosaccharide chain composed of two N-acetylhexosamines, two hexoses, one hexuronic acid and two pentoses. The fragmentation pattern of the heptasaccharide suggests the branched structure HexNAc–(Pentose-)HexA)–(Pentose-)Hex–Hex–HexNAc–Asn (Igura et al. 2008). The Py. furiosus linkage unit is probably GalNAc-Asn, as in Hbt. salinarum (Paul, Lottspeich and Wieland 1986).
The N-linked glycoprotein of the hyperthermophilic archaeon Archaeoglobus fulgidus was investigated by MS and revealed a relatively simple N-glycan structure Hex–Hex–dHex–dHex–Hex–Hex with a Hex–Asn linkage unit (Matsumoto et al. 2012). This is rare because the first monosaccharide moiety of most N-glycans contain an N-acetyl group at carbon 2, except for N-glycans from Hbt. salinarum, Hfx volcanii and Har. marismortui (Lechner and Wieland 1989; Calo et al. 2011; Kandiba and Eichler 2014).
Thermoplasma acidophilum is a thermoacidophilic archaeon that grows optimally at pH 2 and 59°C (Vinogradov et al. 2012). This extremophile is remarkable because of the absence of a cell wall or an S-layer. Treating the cells with Triton X-100 at pH 3 allowed the extraction of all of the cell-surface glycoproteins while keeping cells intact. The extracted glycoproteins were partially purified and five glycoproteins were identified by N-terminal sequencing and MS after in-gel tryptic digests. The obtained glycopeptides were positive for carbohydrates in a periodic acid-Schiff staining reaction and had a high content of Asn residues, represented in many Asn-X-Ser/Thr sequons. Structure determination by NMR showed that the carbohydrate portion was represented by two glycans in nearly equal amounts, differing by the presence of one terminal mannose residue. The larger glycan chain consists of eight residues, six hexoses, one heptose and one sugar with an unusual residue mass of 226 Da, which was identified as 6-deoxy-6-C-sulfo-d-galactose (6-C-sulfo-d-fucose). MS analyses of peptides obtained after trypsin and chymotrypsin digestion confirmed the principal structures to be those determined by NMR and identified 14 glycopeptides derived from the main glycoprotein, Ta0280, all containing the Asn-X-Ser/Thr sequons. Thus, T. acidophilum appears to have a general protein N-glycosylation system that targets a number of cell-surface proteins (Vinogradov et al. 2012).
The haloarchaeal pleomorphic virus HRPV-1 infecting Halorubrum sp.strain PV6 and VP4 has a major structural protein that is glycosylated (Kandiba et al. 2012). After β-elimination glycans released from VP4 were analysed by MS and NMR, showing that the major VP4-derived glycan is the pentasaccharide α-5FmLeg-(2→4)-β-(SO3→2)-GlcA-(1→2)-α-Man-(1→4)-β-GlcA-(1→4)–Glc, comprising Glc, GlcA, Man, sulfated GlcA and a terminal 5-N-formyl-legionaminic acid residue (Knirel et al. 2003). The importance of this modified nonulosonic acid (NulO) residue for viral infection was demonstrated upon incubation with N-acetylneuraminic acid. Such treatment reduced progeny virus production by half 4 h post infection. The presence of pentasaccharide precursors on two different VP4-derived peptides bearing the N-glycosylation signal NTT was confirmed by MS (Kandiba et al. 2012). The same sites modified by the native host, Halorubrum sp. strain PV6, were also recognised by the Hfx. volcanii N-glycosylation apparatus, as determined by MS of heterologously expressed VP4. Here, however, the N-linked pentasaccharide was the same as shown to decorate the S-layer glycoprotein in this species. Hence, N-glycosylation of the haloarchaeal viral protein, VP4, is host specific. These results present additional examples of archaeal N-glycosylation diversity and show the ability of archaea to modify heterologously expressed proteins.
Furthermore, Meth. smithii was shown to generate a different NulO, namely pseudaminic acid, although it is not known whether this sugar is used in protein glycosylation (Lewis et al. 2009). The genes for archaeal NulO sugar biosynthesis might have arrived from animals (Lewis et al. 2009) and later emerged in archaea by lateral gene transfer, assuming a major role in the evolutionary history (Kandiba and Eichler 2013).
In this context, it is appropriate to raise the question as to how exchange of genetic material between eukaryotes and prokaryotes and, specifically, bacteria and archaea is facilitated. Acquisition of new genetic material through gene transfer has been shown to be an important feature in the evolution of prokaryotes (Doolittle 1999). Changes in the genetic repertoire occurring through gene acquisition and deletion are major events underlying the emergence and evolution of prokaryotic pathogens (Nelson-Sathi et al. 2015). Thus, the appearance of legionaminic acid in haloarchaea-derived glycans (Kandiba et al. 2012) and pseudaminic acid in Methanobrevibacter (Lewis et al. 2009) is of considerable interest. Members of the domain Archaea have been implicated in the severity of periodontal disease, and a recent report confirms that archaea are present in endodontic infections (Vickerman et al. 2007). A Meth. oralis-like species was detected in one asymptomatic and one symptomatic patient. DNA from root canals of one of these patients showed also the presence of ‘red complex bacteria’ (Holt and Ebersole 2005). These results confirm the presence of archaea in root canals and provide additional insights into the polymicrobial communities in endodontic infections associated with clinical symptoms (Mansfield et al. 2012; Efenberger et al. 2015). Among this group of ‘red complex bacteria’ is also Tannerella forsythia, whose glycosylated S-layer proteins TfsA and TfsB have been characterised recently (Posch et al. 2011) (vide supra and Fig. 2). Very recent unpublished data also show that Ta. forsythia can decorate the S-layer proteins and other cellular proteins, in a strain-dependent manner, either with Pse or Leg (Valentin Friedrich, Bettina Janesch, Markus Windwarder, Matthias Braun, Zoë A. Megson, Evgeny Vinogradov, Ashu Sharma, Friedrich Altmann, Paul Messner, Ian C. Schoenhofen, Christina Schäffer, manuscript in preparation).
Concluding Remarks
The field of prokaryotic glycosylation remains challenging due to its inherent complexity. Our current knowledge summarised in this review represents only a small fraction of the prokaryotic glyco world because all available information originates from cultivable microorganisms only. However, laboratory cultivation conditions in shaking flasks or fermenters do not mirror those encountered in native habitats where most free-living organisms are socialised in biofilms.
Prokaryotic glycoconjugates exhibit an enormous diversity, but their biosynthesis is often based on common themes and pathways. This common ground can enhance the study of the still unexplored glycosylation potential of several prokaryotic organisms but needs to be complemented with efficient analytical methods, since sequence information alone does not adequately predict the glycosylation potential. The increasing availability of microbial whole-genome sequences has expanded the list of putative glycan biosynthetic enzymes from different prokaryotes, and advances in analytical methods have facilitated more comprehensive analyses of glycoproteins.
Although capsular and exopolysaccharides, PG, SCWPs and LPS have been studied for years, many aspects of prokaryotic glycosylation still remain unknown. Research concerning the functional importance of prokaryotic glycoconjugates, in particular of N- and O-linked glycoproteins, is still lagging behind the wealth of information on eukaryotic scenarios. Currently, the main focus lies on the elucidation of prokaryotic glycan structures and on delineating their role in virulence, pathogenesis and microbe–host interactions. Combining prokaryotic genetics with proteomic analysis and structural characterisation of glycans will pave the way for further elucidating the functions of these important molecules and their impact on the organism´s behaviour.
It can be anticipated that the functional importance is more vibrant than currently appreciated. Further, exploitation of new fields for biotechnological and medical applications of the prokaryotic neo-glycoconjugates must be a prime goal in future.
The current knowledge on prokaryotic glycosylation is probably only the tip of the iceberg. However, it is clearer than ever that further elucidation of the enigmatic world of prokaryotic glycoconjugates is worth pursuing, as this can render important insights into microbe–host interactions and increase our understanding of prokaryotic life. Without a doubt, the future of prokaryotic glycobiology looks challenging and exciting.
One sentence summary.
The authors summarise current knowledge of prokaryotic glycobiology, focusing on structures and molecular details of biosynthesis concepts of glycoproteins and secondary cell-wall polymers, including their roles in prokaryotic life and their impact on pathogenicity as well as emerging glycoengineering strategies.
Acknowledgements
Work performed at the Department of NanoBiotechnology, Universität für Bodenkultur Wien, Vienna, Austria, was supported by the Austrian Science Fund FWF, projects P24317-B22, P21954-B22, P27374-B22, P26836-B22, and P24305-B20, to C.S. and P.M.
Abbreviations
- CRISPR
Clustered regularly interspaced short palindromic repeats
- HPLC
high performance liquid chromatography
- LC-ESI
liquid chromatography- electrospray ionisation
- MALDI-TOF
Matrix-assisted laser desorption/ionization-time-of-flight
- NDP
nucleoside diphosphate
- NGT
N-glycosyl transferase
- NMR
nuclear magnetic resonance spectroscopy
- OGT
O-glycosyl transferase
- OST
oligosaccharyl transferase
- O-OST
O-oligosaccharyl transferase
- SDS-PAGE
sodium dodecylsulfate- polyacrylamide gel electrophoresis]
- TEM
transmission electron microscopy
Organisms
Acidovorax avenae, Av. avenae
Acinetobacter baumanii, Ac. baumanii
Actinobacillus pleuropneumoniae, A. pleuropneumoniae
Aeromonas caviae, Ae. caviae
Aeromonas hydrophila, Ae. hydrophila
Aggregatibacter actinomycetemcomitans, Ag. actinomycetemcomitans
Archaeoglobus fulgidus, Ar. fulgidus
Bacillus anthracis, Ba. anthracis
Bacillus cereus, Ba. cereus
Bacillus subtilis, Ba. subtilis
Bacillus thuringiensis, Ba. thuringiensis
Bacteroides fragilis, Bac. fragilis
Burkholderia cenocepacia, B. cenocepacia
Burkholderia cepacia, B. cepacia
Burkholderia thailandensis, B. thailandensis
Campylobacter coli, C. coli
Campylobacter jejuni, C. jejuni
Campylobacter lari, C. lari
Clostridium botulinum, Cl. botulinum
Clostridium difficile, Cl. difficile
Corynebacterium glutamicum, Co. glutamicum
Francisella tularensis, F. tularensis,
Geobacillus stearothermophilus, G. stearothermophilus
Haemophilus influenzae, Ha. influenzae
Haloarcula hispanica, Har. hispanica
Haloarcula japonica, Har. japonica
Haloarcula marismortui, Har. marismortui
Halobacterium salinarum, Hbt. salinarum
Haloferax volcanii, Hfx. volcanii
Helicobacter pullorum, H. pullorum
Helicobacter pylori, H. pylori
Helicobacter canadensis, H. canadensis
Helicobacter winghamensis, H. winghamensis
Kuenenia stuttgartiensis, K. stuttgartiensis
Lactobacillus buchneri, La. buchneri
Lactobacillus acidophilus, La. acidophilus
Lactobacillus crispatus, La. crispatus
Lactobacillus plantarum, La. plantarum
Lactobacillus rhamnosus, La. rhamnosus
Listeria monocytogenes, L. monocytogenes
Methanobrevibacter smithii, Meth. smithii
Methanobrevibacter oralis, Meth. oralis
Methanococcus maripaludis, Me. maripaludis
Methanococcus voltae, Me. voltae
Methanosarcina mazei, Met. mazei
Methanothermobacter thermoautotrophicus, Meth. thermoautotrophicus
Mycobacterium marinum, M. marinum
Mycobacterium smegmatis, M. smegmatis
Mycobacterium tuberculosis, M. tuberculosis
Neisseria gonorrhoeae, N. gonorrhoeae
Neisseria meningitidis, N. meningitidis
Paenibacillus alvei, Pa. alvei
Porphyromonas gingivalis, Po. gingivalis
Pseudomonas aeruginosa, P. aeruginosa
Pseudomonas syringae, P. syringae
Pyrococcus furiosus, Py. furiosus
Staphylococcus aureus, St. aureus
Streptococcus pneumoniae, S. pneumoniae
Sulfolobus acidocaldarius, Su. acidocaldarius
Sulfolobus shibatae, Su. shibatae
Sulfolobus solfataricus, Su. solfataricus
Tannerella forsythia, Ta. forsythia
Thermoanaerobacter thermohydrosulfuricus, T. thermohydrosulfuricus
Thermoanaerobacterium thermosaccharolyticum, Th. thermosaccharolyticum
Thermoplasma acidophilum, T. acidophilum
Footnotes
Conflict of interest. None declared.
References
- Aas FE, Vik Å, Vedde J, et al. Neisseria gonorrhoeae O-linked pilin glycosylation: functional analyses define both the biosynthetic pathway and glycan structure. Mol Microbiol. 2007;65:607–24. doi: 10.1111/j.1365-2958.2007.05806.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdul Halim MF, Karch KR, Zhou Y, et al. Permuting the PGF signature motif blocks both archaeosortase-dependent C-terminal cleavage and prenyl lipid attachment for the Haloferax volcanii S-layer glycoprotein. J Bacteriol. 2016;198:808–15. doi: 10.1128/JB.00849-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdul Halim MF, Pfeiffer F, Zou J, et al. Haloferax volcanii archaeosortase is required for motility, mating, and C-terminal processing of the S-layer glycoprotein. Mol Microbiol. 2013;88:1164–75. doi: 10.1111/mmi.12248. [DOI] [PubMed] [Google Scholar]
- Abu-Qarn M, Yurist-Doutsch S, Giordano A, et al. Haloferax volcanii AglB and AglD are involved in N-glycosylation of the S-layer glycoprotein and proper assembly of the surface layer. J Mol Biol. 2007;374:1224–36. doi: 10.1016/j.jmb.2007.10.042. [DOI] [PubMed] [Google Scholar]
- Adamo R. Glycan surface antigens from Bacillus anthracis as vaccine targets: current status and future perspectives. Expert Rev Vaccines. 2014;13:895–907. doi: 10.1586/14760584.2014.924404. [DOI] [PubMed] [Google Scholar]
- Adamo R, Saksena R, Kováč P. Synthesis of the b anomer of the spacer-equipped tetrasaccharide side chain of the major glycoprotein of the Bacillus anthracis exosporium. Carbohyd Res. 2005;340:2579–82. doi: 10.1016/j.carres.2005.09.015. [DOI] [PubMed] [Google Scholar]
- Albers S-V, Jarrell KF. The archaellum: how Archaea swim. Front Microbiol. 2015;6:23. doi: 10.3389/fmicb.2015.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albers S-V, Meyer BH. The archaeal cell envelope. Nat Rev Microbiol. 2011;9:414–26. doi: 10.1038/nrmicro2576. [DOI] [PubMed] [Google Scholar]
- Alemka A, Nothaft H, Zheng J, et al. N-glycosylation of Campylobacter jejuni surface proteins promotes bacterial fitness. Infect Immun. 2013;81:1674–82. doi: 10.1128/IAI.01370-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allievi MC, Palomino MM, Prado Acosta M, et al. Contribution of S-layer proteins to the mosquitocidal activity of Lysinibacillus sphaericus. PLoS One. 2014;9:e111114. doi: 10.1371/journal.pone.0111114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altman E, Brisson J-R, Gagné SM, et al. Structure of the glycan chain from the surface layer glycoprotein of Clostridium thermohydrosulfuricum L77-66. Biochim Biophys Acta. 1992;1117:71–7. doi: 10.1016/0304-4165(92)90164-p. [DOI] [PubMed] [Google Scholar]
- Altman E, Brisson J-R, Messner P, et al. Chemical characterization of the regularly arranged surface layer glycoprotein of Clostridium thermosaccharolyticum D120-70. Eur J Biochem. 1990;188:73–82. doi: 10.1111/j.1432-1033.1990.tb15373.x. [DOI] [PubMed] [Google Scholar]
- Altman E, Brisson J-R, Messner P, et al. Structure of the glycan chain from the surface layer glycoprotein of Bacillus alvei CCM 2051. Biochem Cell Biol. 1991;69:72–78. doi: 10.1139/o91-010. [DOI] [PubMed] [Google Scholar]
- Altman E, Schäffer C, Brisson J-R, et al. Characterization of the glycan structure of a major glycopeptide from the surface layer glycoprotein of Clostridium thermosaccharolyticum E207-71. Eur J Biochem. 1995;229:308–15. doi: 10.1111/j.1432-1033.1995.tb20470.x. [DOI] [PubMed] [Google Scholar]
- Altman E, Schäffer C, Brisson J-R, et al. Isolation and characterization of an amino sugar-rich glycopeptide from the surface layer glycoprotein of Thermoanaerobacterium thermosaccharolyticum E207-71. Carbohyd Res. 1996;295:245–53. doi: 10.1016/s0008-6215(96)90150-0. [DOI] [PubMed] [Google Scholar]
- Anonsen JH, Vik Å, Børud B, et al. Characterization of a unique tetrasaccharide and distinct glycoproteome in the O-linked protein glycosylation system of Neisseria elongata subsp. glycolytica. J Bacteriol. 2016;198:256–67. doi: 10.1128/JB.00620-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anonsen JH, Vik Å, Egge-Jacobsen W, et al. An extended spectrum of target proteins and modification sites in the general O-linked protein glycosylation system in Neisseria gonorrhoeae. J Proteome Res. 2012;11:5781–93. doi: 10.1021/pr300584x. [DOI] [PubMed] [Google Scholar]
- Antikainen J, Anton L, Sillanpää J, et al. Domains in the S-layer protein CbsA of Lactobacillus crispatus involved in adherence to collagens, laminin and lipoteichoic acids and in self-assembly. Mol Microbiol. 2002;46:381–94. doi: 10.1046/j.1365-2958.2002.03180.x. [DOI] [PubMed] [Google Scholar]
- Anzengruber J, Courtin P, Claes IJJ, et al. Biochemical characterization of the major N-acetylmuramidase from Lactobacillus buchneri. Microbiology. 2014a;160:1807–19. doi: 10.1099/mic.0.078162-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anzengruber J, Pabst M, Neumann L, et al. Protein O-glucosylation in Lactobacillus buchneri. Glycoconj J. 2014b;31:117–31. doi: 10.1007/s10719-013-9505-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appendix-1B. Essentials of glycobiology [Internet]. 3rd edition. Symbol. Nomenclature for Glycans (SNFG) Glycobiology. 2015;25:1323–24. [Google Scholar]
- Apweiler R, Hermjakob H, Sharon N. On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Biochim Biophys Acta. 1999;1473:4–8. doi: 10.1016/s0304-4165(99)00165-8. [DOI] [PubMed] [Google Scholar]
- Araki Y, Ito E. Linkage units in cell walls of gram-positive bacteria. Crit Rev Microbiol. 1989;17:121–35. doi: 10.3109/10408418909105745. [DOI] [PubMed] [Google Scholar]
- Arbiv A, Yurist-Doutsch S, Guan Z, et al. AglQ is a novel component of the Haloferax volcanii N-glycosylation pathway. PLoS One. 2013;8:e81782. doi: 10.1371/journal.pone.0081782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archibald AR, Hancock IC, Harwood CR. Cell wall structure, synthesis and turnover. In: Sonenshein A, Hoch JA, Losick R, editors. Bacillus Subtilis and Other Gram-Positive Bacteria. Washington DC, USA: ASM Press; 1993. pp. 381–410. [Google Scholar]
- Åvall-Jääskeläinen S, Palva A. Lactobacillus surface layers and their applications. FEMS Microbiol Rev. 2005;29:511–29. doi: 10.1016/j.femsre.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Awram P, Smit J. Identification of lipopolysaccharide O antigen synthesis genes required for attachment of the S-layer of Caulobacter crescentus. Microbiology. 2001;147:1451–60. doi: 10.1099/00221287-147-6-1451. [DOI] [PubMed] [Google Scholar]
- Balonova L, Hernychova L, Mann BF, et al. Multimethodological approach to identification of glycoproteins from the proteome of Francisella tularensis, an intracellular microorganism. J Proteome Res. 2010;9:1995–2005. doi: 10.1021/pr9011602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee A, Tsai C-L, Chaudhury P, et al. FlaF is a b-sandwich protein that anchors the archaellum in the archaeal cell envelope by binding the S-layer protein. Structure. 2015;23:863–72. doi: 10.1016/j.str.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee A, Wang R, Supernavage SL, et al. Implications of phase variation of a gene (pgtA) encoding a pilin galactosyl transferase in gonococcal pathogenesis. J Exp Med. 2002;196:147–62. doi: 10.1084/jem.20012022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baneyx F, Matthaei JF. Self-assembled two-dimensional protein arrays in bionanotechnology: from S-layers to designed lattices. Curr Opin Biotechnol. 2014;28:39–45. doi: 10.1016/j.copbio.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartels K-M, Funken H, Knapp A, et al. Glycosylation is required for outer membrane localization of the lectin LecB in Pseudomonas aeruginosa. J Bacteriol. 2011;193:1107–13. doi: 10.1128/JB.01507-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumeister W, Lembcke G. Structural features of archaebacterial cell envelopes. J Bioenerg Biomembr. 1992;24:567–75. doi: 10.1007/BF00762349. [DOI] [PubMed] [Google Scholar]
- Bayer EA, Belaich J-P, Shoham Y, et al. The cellulosomes: multienzyme machines for degradation of plant cell wall polysaccharides. Annu Rev Microbiol. 2004;58:521–54. doi: 10.1146/annurev.micro.57.030502.091022. [DOI] [PubMed] [Google Scholar]
- Bensing BA, Gibson BW, Sullam PM. The Streptococcus gordonii platelet binding protein GspB undergoes glycosylation independently of export. J Bacteriol. 2004;186:638–45. doi: 10.1128/JB.186.3.638-645.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benz I, Schmidt MA. Glycosylation with heptose residues mediated by the aah gene product is essential for adherence of the AIDA-I adhesin. Mol Microbiol. 2001;40:1403–13. doi: 10.1046/j.1365-2958.2001.02487.x. [DOI] [PubMed] [Google Scholar]
- Bindila L, Steiner K, Schäffer C, et al. Sequencing of O-glycopeptides derived from an S-layer glycoprotein of Geobacillus stearothermophilus NRS 2004/3a containing up to 51 monosaccharide residues at a single glycosylation site by Fourier transform ion cyclotron resonance infrared multiphoton dissociation mass spectrometry. Anal Chem. 2007;79:3271–9. doi: 10.1021/ac0617363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock K, Schuster-Kolbe J, Altman E, et al. Primary structure of the O-glycosidically linked glycan chain of the crystalline surface layer glycoprotein of Thermoanaerobacter thermohydrosulfuricus L111-69. Galactosyl tyrosine as a novel linkage unit. J Biol Chem. 1994;269:7137–44. [PubMed] [Google Scholar]
- Børud B, Anonsen JH, Viburiene R, et al. Extended glycan diversity in a bacterial protein glycosylation system linked to allelic polymorphisms and minimal genetic alterations in a glycosyltransferase gene. Mol Microbiol. 2014;94:688–99. doi: 10.1111/mmi.12789. [DOI] [PubMed] [Google Scholar]
- Børud B, Viburiene R, Hartley MD, et al. Genetic and molecular analyses reveal an evolutionary trajectory for glycan synthesis in a bacterial protein glycosylation system. P Natl Acad Sci USA. 2011;108:9643–8. doi: 10.1073/pnas.1103321108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bou Raad R, Méniche X, de Sousa-d’Auria C, et al. A deficiency in arabinogalactan biosynthesis affects Corynebacterium glutamicum mycolate outer membrane stability. J Bacteriol. 2010;192:2691–700. doi: 10.1128/JB.00009-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu S, Li Y, Zhou M, et al. Interaction between two putative glycosyltransferases is required for glycosylation of a serine-rich streptococcal adhesin. J Bacteriol. 2008;190:1256–66. doi: 10.1128/JB.01078-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burda P, Aebi M. The dolichol pathway of N-linked glycosylation. Biochim Biophys Acta. 1999;1426:239–57. doi: 10.1016/s0304-4165(98)00127-5. [DOI] [PubMed] [Google Scholar]
- Calo D, Guan Z, Eichler J. Glyco-engineering in Archaea: differential N-glycosylation of the S-layer glycoprotein in a transformed Haloferax volcanii strain. Microb Biotechnol. 2011;4:461–70. doi: 10.1111/j.1751-7915.2011.00250.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calo D, Guan Z, Naparstek S, et al. Different routes to the same ending: comparing the N-glycosylation processes of Haloferax volcanii and Haloarcula marismortui, two halophilic archaea from the Dead Sea. Mol Microbiol. 2011;81:1166–77. doi: 10.1111/j.1365-2958.2011.07781.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calo D, Kaminski L, Eichler J. Protein glycosylation in Archaea: sweet and extreme. Glycobiology. 2010;20:1065–76. doi: 10.1093/glycob/cwq055. [DOI] [PubMed] [Google Scholar]
- Candela T, Maes E, Garénaux E, et al. Environmental and biofilmdependent changes in a Bacillus cereus secondary cell wall polysaccharide. J Biol Chem. 2011;286:31250–62. doi: 10.1074/jbc.M111.249821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carasi P, Ambrosis NM, De Antoni GL, et al. Adhesion properties of potentially probiotic Lactobacillus kefiri to gastrointestinal mucus. J Dairy Res. 2014;81:16–23. doi: 10.1017/S0022029913000526. [DOI] [PubMed] [Google Scholar]
- Castric P. pilO, a gene required for glycosylation of Pseudomonas aeruginosa 1244 pilin. Microbiology. 1995;141:1247–54. doi: 10.1099/13500872-141-5-1247. [DOI] [PubMed] [Google Scholar]
- Castric P, Cassels FJ, Carlson RW. Structural characterization of the Pseudomonas aeruginosa 1244 pilin glycan. J Biol Chem. 2001;276:26479–85. doi: 10.1074/jbc.M102685200. [DOI] [PubMed] [Google Scholar]
- Cava F, de Pedro MA, Schwarz H, et al. Binding to pyruvylated compounds as an ancestral mechanism to anchor the outer envelope in primitive bacteria. Mol Microbiol. 2004;52:677–90. doi: 10.1111/j.1365-2958.2004.04011.x. [DOI] [PubMed] [Google Scholar]
- Çelik E, Ollis AA, Lasanajak Y, et al. Glycoarrays with engineered phages displaying structurally diverse oligosaccharides enable high-throughput detection of glycan-protein interactions. Biotechnol J. 2015;10:199–209. doi: 10.1002/biot.201400354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaban B, Logan SM, Kelly JF, et al. AglC and AglK are involved in biosynthesis and attachment of diacetylated glucuronic acid to the N-glycan in Methanococcus voltae. J Bacteriol. 2009;191:187–95. doi: 10.1128/JB.00885-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaban B, Voisin S, Kelly J, et al. Identification of genes involved in the biosynthesis and attachment of Methanococcus voltae N-linked glycans: insight into N-linked glycosylation pathways in Archaea. Mol Microbiol. 2006;61:259–68. doi: 10.1111/j.1365-2958.2006.05226.x. [DOI] [PubMed] [Google Scholar]
- Charbonneau M-È, Côté J-P, Haurat MF, et al. A structural motif is the recognition site for a new family of bacterial protein O-glycosyltransferases. Mol Microbiol. 2012;83:894–907. doi: 10.1111/j.1365-2958.2012.07973.x. [DOI] [PubMed] [Google Scholar]
- Che F-S, Nakajima Y, Tanaka N, et al. Flagellin from an incompatible strain of Pseudomonas avenae induces a resistance response in cultured rice cells. J Biol Chem. 2000;275:32347–56. doi: 10.1074/jbc.M004796200. [DOI] [PubMed] [Google Scholar]
- Chen X, Varki A. Advances in the biology and chemistry of sialic acids. ACS Chem Biol. 2010;5:163–76. doi: 10.1021/cb900266r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury B, Leoff C, Saile E, et al. The structure of the major cell wall polysaccharide of Bacillus anthracis is species-specific. J Biol Chem. 2006;281:27932–41. doi: 10.1074/jbc.M605768200. [DOI] [PubMed] [Google Scholar]
- Christian R, Schulz G, Schuster-Kolbe J, et al. Complete structure of the tyrosine-linked saccharide moiety from the surface layer glycoprotein of Clostridium thermohydrosulfuricum S102-70. J Bacteriol. 1993;175:1250–56. doi: 10.1128/jb.175.5.1250-1256.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian R, Schulz G, Unger FM, et al. Structure of a rhamnan from the surface-layer glycoprotein of Bacillus stearothermophilus strain NRS 2004/3a. Carbohyd Res. 1986;150:265–72. doi: 10.1016/0008-6215(86)80021-0. [DOI] [PubMed] [Google Scholar]
- Claus H, König H. Cell envelopes of methanogens. In: König H, Claus H, Varma A, editors. Prokaryotic Cell Wall Compounds—Structure and Biochemistry. Berlin: Springer; 2010. pp. 231–51. [Google Scholar]
- Coddeville B, Wu S-W, Fabre E, et al. Identification of the Mycobacterium marinum Apa antigen O-mannosylation sites reveals important glycosylation variability with the M. tuberculosis Apa homologue. J Proteomics. 2012;75:5695–705. doi: 10.1016/j.jprot.2012.07.017. [DOI] [PubMed] [Google Scholar]
- Cohen-Krausz S, Trachtenberg S. The structure of the archeabacterial flagellar filament of the extreme halophile Halobacterium salinarum R1M1 and its relation to eubacterial flagellar filaments and type IV pili. J Mol Biol. 2002;321:383–95. doi: 10.1016/s0022-2836(02)00616-2. [DOI] [PubMed] [Google Scholar]
- Cohen-Krausz S, Trachtenberg S. The flagellar filament structure of the extreme acidothermophile Sulfolobus shibatae B12 suggests that archaeabacterial flagella have a unique and common symmetry and design. J Mol Biol. 2008;375:1113–24. doi: 10.1016/j.jmb.2007.10.048. [DOI] [PubMed] [Google Scholar]
- Cohen-Rosenzweig C, Guan Z, Shaanan B, et al. Substrate promiscuity: AglB, the archaeal oligosaccharyltransferase, can process a variety of lipid-linked glycans. Appl Environ Microb. 2014;80:486–96. doi: 10.1128/AEM.03191-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen-Rosenzweig C, Yurist-Doutsch S, Eichler J. AglS, a novel component of the Haloferax volcanii N-glycosylation pathway, is a dolichol phosphate-mannose mannosyltransferase. J Bacteriol. 2012;194:6909–16. doi: 10.1128/JB.01716-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corfield AP, Berry M. Glycan variation and evolution in the eukaryotes. Trends Biochem Sci. 2015;40:351–59. doi: 10.1016/j.tibs.2015.04.004. [DOI] [PubMed] [Google Scholar]
- Coyne MJ, Fletcher CM, Chatzidaki-Livanis M, et al. Phylum-wide general protein O-glycosylation system of the Bacteroidetes. Mol Microbiol. 2013;88:772–83. doi: 10.1111/mmi.12220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuccui J, Wren B. Hijacking bacterial glycosylation for the production of glycoconjugates, from vaccines to humanised glycoproteins. J Pharm Pharmacol. 2015;67:338–50. doi: 10.1111/jphp.12321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen TW, Trent MS. A link between the assembly of flagella and lipooligosaccharide of the Gram-negative bacterium Campylobacter jejuni. P Natl Acad Sci USA. 2010;107:5160–5. doi: 10.1073/pnas.0913451107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuthbertson L, Kos V, Whitfield C. ABC transporters involved in export of cell surface glycoconjugates. Microbiol Mol Biol R. 2010;74:341–62. doi: 10.1128/MMBR.00009-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daubenspeck JM, Zeng H, Chen P, et al. Novel oligosaccharide side chains of the collagen-like region of BclA, the major glycoprotein of the Bacillus anthracis exosporium. J Biol Chem. 2004;279:30945–53. doi: 10.1074/jbc.M401613200. [DOI] [PubMed] [Google Scholar]
- Ding Y, Jones GM, Brimacombe C, et al. Identification of a gene involved in the biosynthesis pathway of the terminal sugar of the archaellin N-linked tetrasaccharide in Methanococcus maripaludis. Anton Leeuw. 2016;109:131–48. doi: 10.1007/s10482-015-0615-z. [DOI] [PubMed] [Google Scholar]
- Ding Y, Jones GM, Uchida K, et al. Identification of genes involved in the biosynthesis of the third and fourth sugars of the Methanococcus maripaludis archaellin N-linked tetrasaccharide. J Bacteriol. 2013;195:4094–104. doi: 10.1128/JB.00668-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingle KE, Didelot X, Ansari MA, et al. Recombinational switching of the Clostridium difficile S-layer and a novel glycosylation gene cluster revealed by large-scale whole-genome sequencing. J Infect Dis. 2013;207:675–86. doi: 10.1093/infdis/jis734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobos KM, Khoo K-H, Swiderek KM, et al. Definition of the full extent of glycosylation of the 45-kilodalton glycoprotein of Mycobacterium tuberculosis. J Bacteriol. 1996;178:2498–506. doi: 10.1128/jb.178.9.2498-2506.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doig P, Kinsella N, Guerry P, et al. Characterization of a post-translational modification of Campylobacter flagellin: identification of a sero-specific glycosyl moiety. Mol Microbiol. 1996;19:379–87. doi: 10.1046/j.1365-2958.1996.370890.x. [DOI] [PubMed] [Google Scholar]
- Dong S, McPherson SA, Tan L, et al. Anthrose biosynthetic operon of Bacillus anthracis. J Bacteriol. 2008;190:2350–9. doi: 10.1128/JB.01899-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolittle WF. Phylogenetic classification and the universal tree. Science. 1999;284:2124–9. doi: 10.1126/science.284.5423.2124. [DOI] [PubMed] [Google Scholar]
- Douglas CW, Naylor K, Phansopa C, et al. Physiological adaptations of key oral bacteria. Adv Microb Physiol. 2014;65:257–335. doi: 10.1016/bs.ampbs.2014.08.005. [DOI] [PubMed] [Google Scholar]
- Doyle RJe. Glycomicrobiology. New York: Kluwer Academic/Plenum Publishers; 2000. p. 549. [Google Scholar]
- Dürr C, Nothaft H, Lizak C, et al. The Escherichia coli glycophage display system. Glycobiology. 2010;20:1366–72. doi: 10.1093/glycob/cwq102. [DOI] [PubMed] [Google Scholar]
- Dumetz F, Lapatra SE, Duchaud E, et al. The Flavobacterium psychrophilum OmpA, an outer membrane glycoprotein, induces a humoral response in rainbow trout. J Appl Microbiol. 2007;103:1461–70. doi: 10.1111/j.1365-2672.2007.03359.x. [DOI] [PubMed] [Google Scholar]
- Eckford PD, Sharom FJ. The reconstituted Escherichia coli MsbA protein displays lipid flippase activity. Biochem J. 2010;429:195–203. doi: 10.1042/BJ20100144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efenberger M, Agier J, Pawłowska E, et al. Archaea prevalence in inflamed pulp tissues. Cent Eur J Immunol. 2015;40:194–200. doi: 10.5114/ceji.2015.51358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler J. Novel glycoproteins of the halophilic archaeon Haloferax volcanii. Arch Microbiol. 2000;173:445–8. doi: 10.1007/s002030000152. [DOI] [PubMed] [Google Scholar]
- Eichler J. Extreme sweetness: protein glycosylation in archaea. Nat Rev Microbiol. 2013;11:151–6. doi: 10.1038/nrmicro2957. [DOI] [PubMed] [Google Scholar]
- Eichler J, Adams MWW. Posttranslational protein modification in Archaea. Microbiol Mol Biol R. 2005;69:393–425. doi: 10.1128/MMBR.69.3.393-425.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler J, Arbiv A, Cohen-Rosenzweig C, et al. N-glycosylation in Haloferax volcanii: adjusting the sweetness. Front Microbiol. 2013;4:403. doi: 10.3389/fmicb.2013.00403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler J, Arbiv A, Cohen-Rosenzweig C, et al. Staying in shape: the haloarchaeal cell wall. In: Papke RT, Oren A, editors. Halophiles: Genetics and Genomes. Norwich, UK: Caister Academic Press; 2014. pp. 129–43. [Google Scholar]
- Erhardt M, Namba K, Hughes KT. Bacterial nanomachines: the flagellum and type III injectisome. Cold Spring Harb Perspect Biol. 2010;2:a000299. doi: 10.1101/cshperspect.a000299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esquivel RN, Schulze S, Xu R, et al. Identification of Haloferax volcanii pilin N-glycans with diverse roles in pilus biosynthesis, adhesion, and microcolony formation. J Biol Chem. 2016;291:10602–14. doi: 10.1074/jbc.M115.693556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagan RP, Fairweather NF. Biogenesis and functions of bacterial S-layers. Nat Rev Microbiol. 2014;12:211–22. doi: 10.1038/nrmicro3213. [DOI] [PubMed] [Google Scholar]
- Faridmoayer A, Fentabil MA, Mills DC, et al. Functional characterization of bacterial oligosaccharyltransferases involved in O-linked protein glycosylation. J Bacteriol. 2007;189:8088–98. doi: 10.1128/JB.01318-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferner-Ortner J, Mader C, Ilk N, et al. High-affinity interaction between the S-layer protein SbsC and the secondary cell wall polymer of Geobacillus stearothermophilus ATCC 12980 determined by surface plasmon resonance technology. J Bacteriol. 2007;189:7154–8. doi: 10.1128/JB.00294-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher CM, Coyne MJ, Comstock LE. Theoretical and experimental characterization of the scope of protein O-glycosylation in Bacteroides fragilis. J Biol Chem. 2011;286:3219–26. doi: 10.1074/jbc.M110.194506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsberg LS, Abshire TG, Friedlander A, et al. Localization and structural analysis of a conserved pyruvylated epitope in Bacillus anthracis secondary cell wall polysaccharides and characterization of the galactose-deficient wall polysaccharide from avirulent B. anthracis CDC 684. Glycobiology. 2012;22:1103–17. doi: 10.1093/glycob/cws080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsberg LS, Choudhury B, Leoff C, et al. Secondary cell wall polysaccharides from Bacillus cereus strains G9241, 03BB87 and 03BB102 causing fatal pneumonia share similar glycosyl structures with the polysaccharides from Bacillus anthracis. Glycobiology. 2011;21:934–48. doi: 10.1093/glycob/cwr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsgren E. European foulbrood in honey bees. J Invertebr Pathol. 2010;103(Suppl 1):S5–9. doi: 10.1016/j.jip.2009.06.016. [DOI] [PubMed] [Google Scholar]
- Francoleon DR, Boontheung P, Yang Y, et al. S-layer, surface-accessible, and concanavalin A binding proteins of Methanosarcina acetivorans and Methanosarcina mazei. J Proteome Res. 2009;8:1972–82. doi: 10.1021/pr800923e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredriksen L, Mathiesen G, Moen A, et al. The major autolysin Acm2 from Lactobacillus plantarum undergoes cytoplasmic O-glycosylation. J Bacteriol. 2012;194:325–33. doi: 10.1128/JB.06314-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredriksen L, Moen A, Adzhubei AA, et al. Lactobacillus plantarum WCFS1 O-linked protein glycosylation: an extended spectrum of target proteins and modification sites detected by mass spectrometry. Glycobiology. 2013;23:1439–51. doi: 10.1093/glycob/cwt071. [DOI] [PubMed] [Google Scholar]
- Friedrich V, Gruber C, Nimeth I, et al. Outer membrane vesicles of Tannerella forsythia: biogenesis, composition, and virulence. Mol Oral Microbiol. 2015;30:451–73. doi: 10.1111/omi.12104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujino T, Béguin P, Aubert JP. Organization of a Clostridium thermocellum gene cluster encoding the cellulosomal scaffolding protein CipA and a protein possibly involved in attachment of the cellulosome to the cell surface. J Bacteriol. 1993;175:1891–9. doi: 10.1128/jb.175.7.1891-1899.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabius H-J. The magic of the sugar code. Trends Biochem Sci. 2015;40:341. doi: 10.1016/j.tibs.2015.04.003. [DOI] [PubMed] [Google Scholar]
- Garcia De Gonzalo CV, Zhu L, Oman TJ, et al. NMR structure of the S-linked glycopeptide sublancin 168. ACS Chem Biol. 2014;9:796–801. doi: 10.1021/cb4008106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gawthorne JA, Tan NY, Bailey U-M, et al. Selection against glycosylation sites in potential target proteins of the general HMWC N-glycosyltransferase in Haemophilus influenzae. Biochem Bioph Res Co. 2014;445:633–8. doi: 10.1016/j.bbrc.2014.02.044. [DOI] [PubMed] [Google Scholar]
- Gebhart C, Ielmini MV, Reiz B, et al. Characterization of exogenous bacterial oligosaccharyltransferases in Escherichia coli reveals the potential for O-linked protein glycosylation in Vibrio cholerae and Burkholderia thailandensis. Glycobiology. 2012;22:962–74. doi: 10.1093/glycob/cws059. [DOI] [PubMed] [Google Scholar]
- Gerber S, Lizak C, Michaud G, et al. Mechanism of bacterial oligosaccharyltransferase: in vitro quantification of sequon binding and catalysis. J Biol Chem. 2013;288:8849–61. doi: 10.1074/jbc.M112.445940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerbino E, Carasi P, Mobili P, et al. Role of S-layer proteins in bacteria. World J Microbiol Biot. 2015;31:1877–87. doi: 10.1007/s11274-015-1952-9. [DOI] [PubMed] [Google Scholar]
- Ghosh A, Albers S-V. Assembly and function of the archaeal flagellum. Biochem Soc Trans. 2011;39:64–9. doi: 10.1042/BST0390064. [DOI] [PubMed] [Google Scholar]
- Giltner CL, Nguyen Y, Burrows LL. Type IV pilin proteins: versatile molecular modules. Microbiol Mol Biol R. 2012;76:740–72. doi: 10.1128/MMBR.00035-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraud M-F, Naismith JH. The rhamnose pathway. Curr Opin Struct Biol. 2000;10:687–96. doi: 10.1016/s0959-440x(00)00145-7. [DOI] [PubMed] [Google Scholar]
- Glew MD, Veith PD, Peng B, et al. PG0026 is the C-terminal signal peptidase of a novel secretion system of Porphyromonas gingivalis. J Biol Chem. 2012;287:24605–17. doi: 10.1074/jbc.M112.369223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzélez-Zamorano M, Mendoza-Hernéndez G, Xolalpa W, et al. Mycobacterium tuberculosis glycoproteomics based on ConA-lectin affinity capture of mannosylated proteins. J Proteome Res. 2009;8:721–33. doi: 10.1021/pr800756a. [DOI] [PubMed] [Google Scholar]
- Govan VA, Brozel V, Allsopp MH, et al. A PCR detection method for rapid identification of Melissococcus pluton in honeybee larvae. Appl Environ Microb. 1998;64:1983–5. doi: 10.1128/aem.64.5.1983-1985.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross J, Grass S, Davis AE, et al. The Haemophilus influenzae HMW1 adhesin is a glycoprotein with an unusual N-linked carbohydrate modification. J Biol Chem. 2008;283:26010–5. doi: 10.1074/jbc.M801819200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Z, Naparstek S, Calo D, et al. Protein glycosylation as an adaptive response in Archaea: growth at different salt concentrations leads to alterations in Haloferax volcanii S-layer glycoprotein N-glycosylation. Environ Microbiol. 2012;14:743–53. doi: 10.1111/j.1462-2920.2011.02625.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerry P. Campylobacter flagella: not just for motility. Trends Microbiol. 2007;15:456–61. doi: 10.1016/j.tim.2007.09.006. [DOI] [PubMed] [Google Scholar]
- Guerry P, Ewing CP, Schirm M, et al. Changes in flagellin glycosylation affect Campylobacter autoagglutination and virulence. Mol Microbiol. 2006;60:299–311. doi: 10.1111/j.1365-2958.2006.05100.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerry P, Szymanski CM. Campylobacter sugars sticking out. Trends Microbiol. 2008;16:428–35. doi: 10.1016/j.tim.2008.07.002. [DOI] [PubMed] [Google Scholar]
- Haffajee AD, Socransky SS. Microbial etiological agents of destructive periodontal diseases. Periodontol 2000. 1994;5:78–111. doi: 10.1111/j.1600-0757.1994.tb00020.x. [DOI] [PubMed] [Google Scholar]
- Haffajee AD, Socransky SS, Patel MR, et al. Microbial complexes in supragingival plaque. Oral Microbiol Immunol. 2008;23:196–205. doi: 10.1111/j.1399-302X.2007.00411.x. [DOI] [PubMed] [Google Scholar]
- Haft DH, Payne SH, Selengut JD. Archaeosortases and exosortases are widely distributed systems linking membrane transit with posttranslational modification. J Bacteriol. 2012;194:36–48. doi: 10.1128/JB.06026-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G, Lamont RJ. Beyond the red complex and into more complexity: the polymicrobial synergy and dysbiosis (PSD) model of periodontal disease etiology. Mol Oral Microbiol. 2012;27:409–19. doi: 10.1111/j.2041-1014.2012.00663.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock IC, Baddiley J. Biosynthesis of the bacterial envelope polymers teichoic acid and teichuronic acid. In: Martonosi AN, editor. The Enzymes of Biological Membranes. New York: Plenum Press; 1985. pp. 279–307. [Google Scholar]
- Hanuszkiewicz A, Pittock P, Humphries F, et al. Identification of the flagellin glycosylation system in Burkholderia cenocepacia and the contribution of glycosylated flagellin to evasion of human innate immune responses. J Biol Chem. 2014;289:19231–44. doi: 10.1074/jbc.M114.562603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding CM, Nasr MA, Kinsella RL, et al. Acinetobacter strains carry two functional oligosaccharyltransferases, one devoted exclusively to type IV pilin, and the other one dedicated to O-glycosylation of multiple proteins. Mol Microbiol. 2015;96:1023–41. doi: 10.1111/mmi.12986. [DOI] [PubMed] [Google Scholar]
- Hart GW, Slawson C, Ramirez-Correa G, et al. Cross talk between O-GlcNAcylation and phosphorylation: roles in signaling, transcription, and chronic disease. Annu Rev Biochem. 2011;80:825–58. doi: 10.1146/annurev-biochem-060608-102511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley MD, Morrison MJ, Aas FE, et al. Biochemical characterization of the O-linked glycosylation pathway in Neisseria gonorrhoeae responsible for biosynthesis of protein glycans containing N,N′-diacetylbacillosamine. Biochemistry. 2011;50:4936–48. doi: 10.1021/bi2003372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey H, Kus JV, Tessier L, et al. Pseudomonas aeruginosa d-arabinofuranose biosynthetic pathway and its role in type IV pilus assembly. J Biol Chem. 2011;286:28128–37. doi: 10.1074/jbc.M111.255794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegge FT, Hitchen PG, Aas FE, et al. Unique modifications with phosphocholine and phosphoethanolamine define alternate antigenic forms of Neisseria gonorrhoeae type IV pili. P Natl Acad Sci USA. 2004;101:10798–803. doi: 10.1073/pnas.0402397101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henche A-L, Ghosh A, Yu X, et al. Structure and function of the adhesive type IV pilus of Sulfolobus acidocaldarius. Environ Microbiol. 2012a;14:3188–202. doi: 10.1111/j.1462-2920.2012.02898.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henche AL, Koerdt A, Ghosh A, et al. Influence of cell surface structures on crenarchaeal biofilm formation using a thermostable green fluorescent protein. Environ Microbiol. 2012b;14:779–93. doi: 10.1111/j.1462-2920.2011.02638.x. [DOI] [PubMed] [Google Scholar]
- Hirai H, Takai R, Iwano M, et al. Glycosylation regulates specific induction of rice immune responses by Acidovorax avenae flagellin. J Biol Chem. 2011;286:25519–30. doi: 10.1074/jbc.M111.254029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitchen P, Brzostek J, Panico M, et al. Modification of the Campylobacter jejuni flagellin glycan by the product of the Cj1295 homopolymeric-tract-containing gene. Microbiology. 2010;156:1953–62. doi: 10.1099/mic.0.038091-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt SC, Ebersole JL. Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia: the ‘red complex’, a prototype polybacterial pathogenic consortium in periodontitis. Periodontol 2000. 2005;38:72–122. doi: 10.1111/j.1600-0757.2005.00113.x. [DOI] [PubMed] [Google Scholar]
- Honma K, Inagaki S, Okuda K, et al. Role of a Tannerella forsythia exopolysaccharide synthesis operon in biofilm development. Microb Pathog. 2007;42:156–66. doi: 10.1016/j.micpath.2007.01.003. [DOI] [PubMed] [Google Scholar]
- Hopf PS, Ford RS, Zebian N, et al. Protein glycosylation in Helicobacter pylori: beyond the flagellins? PLoS One. 2011;6:e25722. doi: 10.1371/journal.pone.0025722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber C, Ilk N, Rünzler D, et al. The three S-layer-like homology motifs of the S-layer protein SbpA of Bacillus sphaericus CCM 2177 are not sufficient for binding to the pyruvylated secondary cell wall polymer. Mol Microbiol. 2005;55:197–205. doi: 10.1111/j.1365-2958.2004.04351.x. [DOI] [PubMed] [Google Scholar]
- Hug I, Feldman MF. Analogies and homologies in lipopolysaccharide and glycoprotein biosynthesis in bacteria. Glycobiology. 2011;21:138–51. doi: 10.1093/glycob/cwq148. [DOI] [PubMed] [Google Scholar]
- Hug I, Zheng B, Reiz B, et al. Exploiting bacterial glycosylation machineries for the synthesis of a Lewis antigen-containing glycoprotein. J Biol Chem. 2011;286:37887–94. doi: 10.1074/jbc.M111.287755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynönen U, Palva A. Lactobacillus surface layer proteins: structure, function and applications. Appl Microbiol Biot. 2013;97:5225–43. doi: 10.1007/s00253-013-4962-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igura M, Maita N, Kamishikiryo J, et al. Structure-guided identification of a new catalytic motif of oligosaccharyltransferase. EMBO J. 2008;27:234–43. doi: 10.1038/sj.emboj.7601940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilk N, Kosma P, Puchberger M, et al. Structural and functional analyses of the secondary cell wall polymer of Bacillus sphaericus CCM 2177 that serves as an S-layer-specific anchor. J Bacteriol. 1999;181:7643–6. doi: 10.1128/jb.181.24.7643-7646.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwashkiw JA, Seper A, Weber BS, et al. Identification of a general O-linked protein glycosylation system in Acinetobacter baumannii and its role in virulence and biofilm formation. PLoS Pathog. 2012;8:e1002758. doi: 10.1371/journal.ppat.1002758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwashkiw JA, Vozza NF, Kinsella RL, et al. Pour some sugar on it: the expanding world of bacterial protein O-linked glycosylation. Mol Microbiol. 2013;89:14–28. doi: 10.1111/mmi.12265. [DOI] [PubMed] [Google Scholar]
- Janesch B, Messner P, Schäffer C. Are the surface layer homology domains essential for cell surface display and glycosylation of the S-layer protein from Paenibacillus alvei CCM 2051T? J Bacteriol. 2013;195:565–75. doi: 10.1128/JB.01487-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janesch B, Schirmeister F, Maresch D, et al. Flagellin glycosylation in Paenibacillus alvei CCM 2051T. Glycobiology. 2016;26:74–87. doi: 10.1093/glycob/cwv087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janetzko J, Walker S. The making of a sweet modification: structure and function of O-GlcNAc transferase. J Biol Chem. 2014;289:34424–32. doi: 10.1074/jbc.R114.604405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrell KF, Albers S-V. The archaellum: an old motility structure with a new name. Trends Microbiol. 2012;20:307–12. doi: 10.1016/j.tim.2012.04.007. [DOI] [PubMed] [Google Scholar]
- Jarrell KF, Ding Y, Meyer BH, et al. N-linked glycosylation in Archaea: a structural, functional, and genetic analysis. Microbiol Mol Biol R. 2014;78:304–41. doi: 10.1128/MMBR.00052-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrell KF, Jones GM, Kandiba L, et al. S-layer glycoproteins and flagellins: reporters of archaeal posttranslational modifications. Archaea. 2010;2010:612948. doi: 10.1155/2010/612948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrell KF, Jones GM, Nair DB. Biosynthesis and role of N-linked glycosylation in cell surface structures of Archaea with a focus on flagella and S layers. Int J Microbiol. 2010;2010:470138. doi: 10.1155/2010/470138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings MP, Jen FE-C, Roddam LF, et al. Neisseria gonorrhoeae pilin glycan contributes to CR3 activation during challenge of primary cervical epithelial cells. Cell Microbiol. 2011;13:885–96. doi: 10.1111/j.1462-5822.2011.01586.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings MP, Virji M, Evans D, et al. Identification of a novel gene involved in pilin glycosylation in Neisseria meningitidis. Mol Microbiol. 1998;29:975–84. doi: 10.1046/j.1365-2958.1998.00962.x. [DOI] [PubMed] [Google Scholar]
- Jervis AJ, Butler JA, Lawson AJ, et al. Characterization of the structurally diverse N-linked glycans of Campylobacter species. J Bacteriol. 2012;194:2355–62. doi: 10.1128/JB.00042-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jervis AJ, Langdon R, Hitchen P, et al. Characterization of N-linked protein glycosylation in Helicobacter pullorum. J Bacteriol. 2010;192:5228–36. doi: 10.1128/JB.00211-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannessen C, Koomey M, Børud B. Hypomorphic glycosyl-transferase alleles and recoding at contingency loci influence glycan microheterogeneity in the protein glycosylation system of Neisseria species. J Bacteriol. 2012;194:5034–43. doi: 10.1128/JB.00950-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones GM, Wu J, Ding Y, et al. Identification of genes involved in the acetamidino group modification of the flagellin N-linked glycan of Methanococcus maripaludis. J Bacteriol. 2012;194:2693–702. doi: 10.1128/JB.06686-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan DS, Daubenspeck JM, Laube AH, et al. O-linked protein glycosylation in mycoplasma. Mol Microbiol. 2013;90:1046–53. doi: 10.1111/mmi.12415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josenhans C, Vossebein L, Friedrich S, et al. The neuA/flmD gene cluster of Helicobacter pylori is involved in flagellar biosynthesis and flagellin glycosylation. FEMS Microbiol Lett. 2002;210:165–72. doi: 10.1111/j.1574-6968.2002.tb11176.x. [DOI] [PubMed] [Google Scholar]
- Kählig H, Kolarich D, Zayni S, et al. N-Acetylmuramic acid as capping element of α-d-fucose-containing S-layer glycoprotein glycans from Geobacillus tepidamans GS5-97T. J Biol Chem. 2005;280:20292–9. doi: 10.1074/jbc.M501724200. [DOI] [PubMed] [Google Scholar]
- Kaminski L, Abu-Qarn M, Guan Z, et al. AglJ adds the first sugar of the N-linked pentasaccharide decorating the Haloferax volcanii S-layer glycoprotein. J Bacteriol. 2010;192:5572–9. doi: 10.1128/JB.00705-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminski L, Eichler J. Identification of residues important for the activity of Haloferax volcanii AglD, a component of the archaeal N-glycosylation pathway. Archaea. 2010;2010:315108. doi: 10.1155/2010/315108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminski L, Guan Z, Abu-Qarn M, et al. AglR is required for addition of the final mannose residue of the N-linked glycan decorating the Haloferax volcanii S-layer glycoprotein. Biochim Biophys Acta. 2012;1820:1664–70. doi: 10.1016/j.bbagen.2012.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminski L, Guan Z, Yurist-Doutsch S, et al. Two distinct N-glycosylation pathways process the Haloferax volcanii S-layer glycoprotein upon changes in environmental salinity. mBio. 2013a;4:e00716–13. doi: 10.1128/mBio.00716-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminski L, Naparstek S, Kandiba L, et al. Add salt, add sugar: N-glycosylation in Haloferax volcanii. Biochem Soc Trans. 2013b;41:432–35. doi: 10.1042/BST20120142. [DOI] [PubMed] [Google Scholar]
- Kandiba L, Aitio O, Helin J, et al. Diversity in prokaryotic glycosylation: an archaeal-derived N-linked glycan contains legionaminic acid. Mol Microbiol. 2012;84:578–93. doi: 10.1111/j.1365-2958.2012.08045.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandiba L, Eichler J. Analysis of putative nonulosonic acid biosynthesis pathways in Archaea reveals a complex evolutionary history. FEMS Microbiol Lett. 2013;345:110–20. doi: 10.1111/1574-6968.12193. [DOI] [PubMed] [Google Scholar]
- Kandiba L, Eichler J. Archaeal S-layer glycoproteins: post-translational modification in the face of extremes. Front Microbiol. 2014;5:661. doi: 10.3389/fmicb.2014.00661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandiba L, Eichler J. Deciphering a pathway of Halobacterium salinarum N-glycosylation. Microbiologyopen. 2015;4:28–40. doi: 10.1002/mbo3.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandiba L, Guan Z, Eichler J. Lipid modification gives rise to two distinct Haloferax volcanii S-layer glycoprotein populations. Biochim Biophys Acta. 2013;1828:938–43. doi: 10.1016/j.bbamem.2012.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandiba L, Lin C-W, Aebi M, et al. Structural characterization of the N-linked pentasaccharide decorating glycoproteins of the halophilic archaeon Haloferax volcanii. Glycobiology. 2016;26:745–56. doi: 10.1093/glycob/cww014. [DOI] [PubMed] [Google Scholar]
- Kärcher U, Schröder H, Haslinger E, et al. Primary structure of the heterosaccharide of the surface glycoprotein of Methanothermus fervidus. J Biol Chem. 1993;268:26821–6. [PubMed] [Google Scholar]
- Kawai F, Grass S, Kim Y, et al. Structural insights into the glycosyltransferase activity of the Actinobacillus pleuropneumoniae HMW1C-like protein. J Biol Chem. 2011;286:38546–57. doi: 10.1074/jbc.M111.237602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearns DB. A field guide to bacterial swarming motility. Nat Rev Microbiol. 2010;8:634–44. doi: 10.1038/nrmicro2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly J, Logan SM, Jarrell KF, et al. A novel N-linked flagellar glycan from Methanococcus maripaludis. Carbohydr Res. 2009;344:648–53. doi: 10.1016/j.carres.2009.01.006. [DOI] [PubMed] [Google Scholar]
- Kern J, Ryan C, Faull K, et al. Bacillus anthracis surface-layer proteins assemble by binding to the secondary cell wall polysaccharide in a manner that requires csaB and tagO. J Mol Biol. 2010;401:757–75. doi: 10.1016/j.jmb.2010.06.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kern J, Wilton R, Zhang R, et al. Structure of surface layer homology (SLH) domains from Bacillus anthracis surface array protein. J Biol Chem. 2011;286:26042–9. doi: 10.1074/jbc.M111.248070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kern VJ, Kern JW, Theriot JA, et al. Surface-layer (S-layer) proteins Sap and EA1 govern the binding of the S-layer-associated protein BslO at the cell septa of Bacillus anthracis. J Bacteriol. 2012;194:3833–40. doi: 10.1128/JB.00402-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khemiri A, Naudin B, Franck X, et al. N-glycosidase treatment with 18O labeling and de novo sequencing argues for flagellin FliC glycopolymorphism in Pseudomonas aeruginosa. Anal Bioanal Chem. 2013;405:9835–42. doi: 10.1007/s00216-013-7424-x. [DOI] [PubMed] [Google Scholar]
- Khodai-Kalaki M, Andrade A, Fathy Mohamed Y, et al. Burkholderia cenocepacia lipopolysaccharide modification and flagellin glycosylation affect virulence but not innate immune recognition in plants. MBio. 2015;6:e00679. doi: 10.1128/mBio.00679-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi A, Sagami H, Ogura K. Evidence for covalent attachment of diphytanylglyceryl phosphate to the cell-surface glycoprotein of Halobacterium halobium. J Biol Chem. 1999;274:18011–6. doi: 10.1074/jbc.274.25.18011. [DOI] [PubMed] [Google Scholar]
- Kishi M, Hasegawa Y, Nagano K, et al. Identification and characterization of novel glycoproteins involved in growth and biofilm formation by Porphyromonas gingivalis. Mol Oral Microbiol. 2012;27:458–70. doi: 10.1111/j.2041-1014.2012.00659.x. [DOI] [PubMed] [Google Scholar]
- Klingl A. S-layer and cytoplasmic membrane—exceptions from the typical archaeal cell wall with a focus on double membranes. Front Microbiol. 2014;5:624. doi: 10.3389/fmicb.2014.00624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knirel YA, Rietschel ET, Marre R, et al. The structure of the O-specific chain of Legionella pneumophila serogroup 1 lipopolysaccharide. Eur J Biochem. 1994;221:239–45. doi: 10.1111/j.1432-1033.1994.tb18734.x. [DOI] [PubMed] [Google Scholar]
- Knirel YA, Shashkov AS, Senchenkova SN, et al. Structure of the O-polysaccharide of Aeromonas hydrophila O:34; a case of random O-acetylation of 6-deoxy-l-talose. Carbohyd Res. 2002;337:1381–6. doi: 10.1016/s0008-6215(02)00136-2. [DOI] [PubMed] [Google Scholar]
- Knirel YA, Shashkov AS, Tsvetkov YE, et al. 5,7-diamino-3,5,7,9-tetradeoxynon-2-ulosonic acids in bacterial glycopolymers: chemistry and biochemistry. Adv Carbohyd Chem Bi. 2003;58:371–417. doi: 10.1016/s0065-2318(03)58007-6. [DOI] [PubMed] [Google Scholar]
- Konrad Z, Eichler J. Lipid modification of proteins in Archaea: attachment of a mevalonic acid-based lipid moiety to the surface-layer glycoprotein of Haloferax volcanii follows protein translocation. Biochem J. 2002;366:959–64. doi: 10.1042/BJ20020757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korotkov KV, Sandkvist M, Hol WG. The type II secretion system: biogenesis, molecular architecture and mechanism. Nat Rev Microbiol. 2012;10:336–51. doi: 10.1038/nrmicro2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosma P, Neuninger C, Christian R, et al. Glycan structure of the S-layer glycoprotein of Bacillus sp. L420-91. Glycoconj J. 1995a;12:99–107. doi: 10.1007/BF00731875. [DOI] [PubMed] [Google Scholar]
- Kosma P, Wugeditsch T, Christian R, et al. Glycan structure of a heptose-containing S-layer glycoprotein of Bacillus thermoaerophilus. Glycobiology. 1995b;5:791–6. doi: 10.1093/glycob/5.8.791. Corrigendum: Glycobiology (1996) 6: 5. [DOI] [PubMed] [Google Scholar]
- Koval SF, Jarrell KF. Ultrastructure and biochemistry of the cell wall of Methanococcus voltae. J Bacteriol. 1987;169:1298–306. doi: 10.1128/jb.169.3.1298-1306.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowarik M, Young NM, Numao S, et al. Definition of the bacterial N-glycosylation site consensus sequence. EMBO J. 2006;25:1957–66. doi: 10.1038/sj.emboj.7601087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuen B, Koch A, Asenbauer E, et al. Molecular characterization of the Bacillus stearothermophilus PV72 S-layer gene sbsB induced by oxidative stress. J Bacteriol. 1997;179:1664–70. doi: 10.1128/jb.179.5.1664-1670.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Küpcü Z, März L, Messner P, et al. Evidence for the glycoprotein nature of the crystalline cell wall surface layer of Bacillus stearothermophilus strain NRS 2004/3a. FEBS Lett. 1984;173:185–90. doi: 10.1016/0014-5793(84)81043-1. [DOI] [PubMed] [Google Scholar]
- Kus JV, Kelly J, Tessier L, et al. Modification of Pseudomonas aeruginosa Pa5196 type IV pilins at multiple sites with d-Araf by a novel GT-C family arabinosyltransferase, TfpW. J Bacteriol. 2008;190:7464–78. doi: 10.1128/JB.01075-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin A, Imperiali B. The expanding horizons of asparagine-linked glycosylation. Biochemistry. 2011;50:4411–26. doi: 10.1021/bi200346n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassak K, Ghosh A, Albers S-V. Diversity, assembly and regulation of archaeal type IV pili-like and non-type-IV pili-like surface structures. Res Microbiol. 2012;163:630–44. doi: 10.1016/j.resmic.2012.10.024. [DOI] [PubMed] [Google Scholar]
- Lassak K, Neiner T, Ghosh A, et al. Molecular analysis of the crenarchaeal flagellum. Mol Microbiol. 2012;83:110–24. doi: 10.1111/j.1365-2958.2011.07916.x. [DOI] [PubMed] [Google Scholar]
- Lebeer S, Claes IJJ, Balog CIA, et al. The major secreted protein Msp1/p75 is O-glycosylated in Lactobacillus rhamnosus GG. Microb Cell Fact. 2012;11:15. doi: 10.1186/1475-2859-11-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner J, Sumper M. The primary structure of a procaryotic glycoprotein. Cloning and sequencing of the cell surface glycoprotein gene of halobacteria. J Biol Chem. 1987;262:9724–9. [PubMed] [Google Scholar]
- Lechner J, Wieland F. Structure and biosynthesis of prokaryotic glycoproteins. Annu Rev Biochem. 1989;58:173–94. doi: 10.1146/annurev.bi.58.070189.001133. [DOI] [PubMed] [Google Scholar]
- Lee I-C, van Swam II, Tomita S, et al. GtfA and GtfB are both required for protein O-glycosylation in Lactobacillus plantarum. J Bacteriol. 2014;196:1671–82. doi: 10.1128/JB.01401-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S-W, Sabet M, Um H-S, et al. Identification and characterization of the genes encoding a unique surface (S-) layer of Tannerella forsythia. Gene. 2006;371:102–11. doi: 10.1016/j.gene.2005.11.027. [DOI] [PubMed] [Google Scholar]
- Lees-Miller RG, Iwashkiw JA, Scott NE, et al. A common pathway for O-linked protein-glycosylation and synthesis of capsule in Acinetobacter baumannii. Mol Microbiol. 2013;89:816–30. doi: 10.1111/mmi.12300. [DOI] [PubMed] [Google Scholar]
- Leoff C, Choudhury B, Saile E, et al. Structural elucidation of the nonclassical secondary cell wall polysaccharide from Bacillus cereus ATCC 10987. Comparison with the polysaccharides from Bacillus anthracis and B. cereus type strain ATCC 14579 reveals both unique and common structural features. J Biol Chem. 2008;283:29812–21. doi: 10.1074/jbc.M803234200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leoff C, Saile E, Rauvolfova J, et al. Secondary cell wall polysaccharides of Bacillus anthracis are antigens that contain specific epitopes which cross-react with three pathogenic Bacillus cereus strains that caused severe disease, and other epitopes common to all the Bacillus cereus strains tested. Glycobiology. 2009;19:665–73. doi: 10.1093/glycob/cwp036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leon DR, Ytterberg AJ, Boontheung P, et al. Mining proteomic data to expose protein modifications in Methanosarcina mazei strain Gö1. Front Microbiol. 2015;6:149. doi: 10.3389/fmicb.2015.00149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis AL, Desa N, Hansen EE, et al. Innovations in host and microbial sialic acid biosynthesis revealed by phylogenomic prediction of nonulosonic acid structure. P Natl Acad Sci USA. 2009;106:13552–7. doi: 10.1073/pnas.0902431106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Huang X, Li J, et al. Both GtfA and GtfB are required for SraP glycosylation in Staphylococcus aureus. Curr Microbiol. 2014;69:121–6. doi: 10.1007/s00284-014-0563-2. [DOI] [PubMed] [Google Scholar]
- Lithgow KV, Scott NE, Iwashkiw JA, et al. A general protein O-glycosylation system within the Burkholderia cepacia complex is involved in motility and virulence. Mol Microbiol. 2014;92:116–37. doi: 10.1111/mmi.12540. [DOI] [PubMed] [Google Scholar]
- Liu C-F, Tonini L, Malaga W, et al. Bacterial protein-O-mannosylating enzyme is crucial for virulence of Mycobacterium tuberculosis. P Natl Acad Sci USA. 2013;110:6560–5. doi: 10.1073/pnas.1219704110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Leathers TD, Copeland A, et al. Complete genome sequence of Lactobacillus buchneri NRRL B-30929, a novel strain from a commercial ethanol plant. J Bacteriol. 2011;193:4019–20. doi: 10.1128/JB.05180-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S-Y, Gherardini FC, Matuschek M, et al. Cloning, sequencing, and expression of the gene encoding a large S-layer-associated endoxylanase from Thermoanaerobacterium sp. strain JW/SL-YS 485 in Escherichia coli. J Bacteriol. 1996;178:1539–47. doi: 10.1128/jb.178.6.1539-1547.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lizak C, Gerber S, Numao S, et al. X-ray structure of a bacterial oligosaccharyltransferase. Nature. 2011;474:350–5. doi: 10.1038/nature10151. [DOI] [PubMed] [Google Scholar]
- Logan SM. Flagellar glycosylation - a new component of the motility repertoire? Microbiology. 2006;152:1249–62. doi: 10.1099/mic.0.28735-0. [DOI] [PubMed] [Google Scholar]
- Lomino JV, Naegeli A, Orwenyo J, et al. A two-step enzymatic glycosylation of polypeptides with complex N-glycans. Bioorg Med Chem. 2013;21:2262–70. doi: 10.1016/j.bmc.2013.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lommel M, Strahl S. Protein O-mannosylation: conserved from bacteria to humans. Glycobiology. 2009;19:816–28. doi: 10.1093/glycob/cwp066. [DOI] [PubMed] [Google Scholar]
- Lu H, Lü Y, Ren J, et al. Identification of the S-layer glycoproteins and their covalently linked glycans in the halophilic archaeon Haloarcula hispanica. Glycobiology. 2015;25:1150–62. doi: 10.1093/glycob/cwv050. [DOI] [PubMed] [Google Scholar]
- Lu Q, Li S, Shao F. Sweet talk: protein glycosylation in bacterial interaction with the host. Trends Microbiol. 2015;23:630–41. doi: 10.1016/j.tim.2015.07.003. [DOI] [PubMed] [Google Scholar]
- Lunderberg JM, Liszewski Zilla M, Missiakas D, et al. Bacillus anthracis tagO is required for vegetative growth and secondary cell wall polysaccharide synthesis. J Bacteriol. 2015;197:3511–20. doi: 10.1128/JB.00494-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas A, Engelhardt H, Peters J, et al. Domain structure of the Acetogenium kivui surface layer revealed by electron crystallography and sequence analysis. J Bacteriol. 1994;176:1224–33. doi: 10.1128/jb.176.5.1224-1233.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons JJ, Milner JD, Rosenzweig SD. Glycans instructing immunity: the emerging role of altered glycosylation in clinical immunology. Front Pediatr. 2015;3:54. doi: 10.3389/fped.2015.00054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maes E, Krzewinski F, Garenaux E, et al. Glycosylation of BclA glycoprotein from Bacillus cereus and Bacillus anthracis exosporium is domain-specific. J Biol Chem. 2016;291:9666–77. doi: 10.1074/jbc.M116.718171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magidovich H, Eichler J. Glycosyltransferases and oligosaccharyltransferases in Archaea: putative components of the N-glycosylation pathway in the third domain of life. FEMS Microbiol Lett. 2009;300:122–30. doi: 10.1111/j.1574-6968.2009.01775.x. [DOI] [PubMed] [Google Scholar]
- Magidovich H, Yurist-Doutsch S, Konrad Z, et al. AglP is a S-adenosyl-l-methionine-dependent methyltransferase that participates in the N-glycosylation pathway of Haloferax volcanii. Mol Microbiol. 2010;76:190–9. doi: 10.1111/j.1365-2958.2010.07090.x. [DOI] [PubMed] [Google Scholar]
- Mahdavi J, Pirinccioglu N, Oldfield NJ, et al. A novel O-linked glycan modulates Campylobacter jejuni major outer membrane protein-mediated adhesion to human histo-blood group antigens and chicken colonization. Open Biol. 2014;4:130202. doi: 10.1098/rsob.130202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Koonin EV, Albers SV. Diversity and evolution of type IV pili systems in archaea. Front Microbiol. 2016;7:667. doi: 10.3389/fmicb.2016.00667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally DJ, Aubry AJ, Hui JP, et al. Targeted metabolomics analysis of Campylobacter coli VC167 reveals legionaminic acid derivatives as novel flagellar glycans. J Biol Chem. 2007;282:14463–75. doi: 10.1074/jbc.M611027200. [DOI] [PubMed] [Google Scholar]
- McNally DJ, Hui JPM, Aubry AJ, et al. Functional characterization of the flagellar glycosylation locus in Campylobacter jejuni 81-176 using a focused metabolomics approach. J Biol Chem. 2006;281:18489–98. doi: 10.1074/jbc.M603777200. [DOI] [PubMed] [Google Scholar]
- Manina G, Pasca MR, Buroni S, et al. Decaprenylphosphoryl-β-d-ribose 2′-epimerase from Mycobacterium tuberculosis is a magic drug target. Curr Med Chem. 2010;17:3099–108. doi: 10.2174/092986710791959693. [DOI] [PubMed] [Google Scholar]
- Mansfield JM, Campbell JH, Bhandari AR, et al. Molecular analysis of 16S rRNA genes identifies potentially periodontal pathogenic bacteria and archaea in the plaque of partially erupted third molars. J Oral Maxil Surg. 2012;70:1507–14, e1–6. doi: 10.1016/j.joms.2011.09.049. [DOI] [PubMed] [Google Scholar]
- Masuda K, Kawata T. Reassembly of a regularly arranged protein in the cell wall of Lactobacillus buchneri and its reattachment to cell walls: chemical modification studies. Microbiol Immunol. 1985;29:927–38. [PubMed] [Google Scholar]
- Matsumoto S, Igura M, Nyirenda J, et al. Crystal structure of the C-terminal globular domain of oligosaccharyltransferase from Archaeoglobus fulgidus at 1.75 Å resolution. Biochemistry. 2012;51:4157–66. doi: 10.1021/bi300076u. [DOI] [PubMed] [Google Scholar]
- Matsumoto S, Shimada A, Nyirenda J, et al. Crystal structures of an archaeal oligosaccharyltransferase provide insights into the catalytic cycle of N-linked protein glycosylation. P Natl Acad Sci USA. 2013;110:17868–73. doi: 10.1073/pnas.1309777110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May A, Pusztahelyi T, Hoffmann N, et al. Mutagenesis of conserved charged amino acids in SLH domains of Thermoanaerobacterium thermosulfurigenes EM1 affects attachment to cell wall sacculi. Arch Microbiol. 2006;185:263–9. doi: 10.1007/s00203-006-0092-x. [DOI] [PubMed] [Google Scholar]
- Megson ZA, Koerdt A, Schuster H, et al. Characterization of an α-l-fucosidase from the periodontal pathogen Tannerella forsythia. Virulence. 2015;6:282–92. doi: 10.1080/21505594.2015.1010982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta AS, Saile E, Zhong W, et al. Synthesis and antigenic analysis of the BclA glycoprotein oligosaccharide from the Bacillus anthracis exosporium. Chemistry. 2006;12:9136–49. doi: 10.1002/chem.200601245. [DOI] [PubMed] [Google Scholar]
- Meier-Stauffer K, Busse H-J, Rainey FA, et al. Description of Bacillus thermoaerophilus sp. nov., to include sugar beet isolates and Bacillus brevis ATCC 12990. Int J Syst Bacteriol. 1996;46:532–41. [Google Scholar]
- Merino S, Fulton KM, Twine SM, et al. Aeromonas hydrophila flagella glycosylation: involvement of a lipid carrier. PLoS One. 2014;9:e89630. doi: 10.1371/journal.pone.0089630. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Mescher MF, Strominger JL. Purification and characterization of a prokaryotic glucoprotein from the cell envelope of Halobacterium salinarium. J Biol Chem. 1976;251:2005–14. [PubMed] [Google Scholar]
- Mesnage S, Fontaine T, Mignot T, et al. Bacterial SLH domain proteins are non-covalently anchored to the cell surface via a conserved mechanism involving wall polysaccharide pyruvylation. EMBO J. 2000;19:4473–84. doi: 10.1093/emboj/19.17.4473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesnage S, Tosi-Couture E, Mock M, et al. Molecular characterization of the Bacillus anthracis main S-layer component: evidence that it is the major cell-associated antigen. Mol Microbiol. 1997;23:1147–55. doi: 10.1046/j.1365-2958.1997.2941659.x. [DOI] [PubMed] [Google Scholar]
- Mesnage S, Tosi-Couture E, Mock M, et al. The S-layer homology domain as a means for anchoring heterologous proteins on the cell surface of Bacillus anthracis. J Appl Microbiol. 1999;87:256–60. doi: 10.1046/j.1365-2672.1999.00880.x. [DOI] [PubMed] [Google Scholar]
- Messner P. Bacterial glycoproteins. Glycoconjugate J. 1997;14:3–11. doi: 10.1023/a:1018551228663. [DOI] [PubMed] [Google Scholar]
- Messner P, Christian R, Kolbe J, et al. Analysis of a novel linkage unit of O-linked carbohydrates from the crystalline surface layer glycoprotein of Clostridium thermohydrosulfuricum S102-70. J Bacteriol. 1992;174:2236–40. doi: 10.1128/jb.174.7.2236-2240.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messner P, Christian R, Neuninger C, et al. Similarity of ‘core’ structures in two different glycans of tyrosine-linked eubacterial S-layer glycoproteins. J Bacteriol. 1995;177:2188–93. doi: 10.1128/jb.177.8.2188-2193.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messner P, Egelseer EM, Sleytr UB, et al. Surface layer glycoproteins and ‘non-classical’ secondary cell wall polymers. In: Moran AP, Brennan PJ, Holst O, et al., editors. Microbial Glycobiology: Structures, Relevance and Applications. San Diego, CA: Academic Press-Elsevier; 2009. pp. 109–28. [Google Scholar]
- Messner P, Schäffer C. Prokaryotic glycoproteins. In: Herz W, Falk H, Kirby GW, editors. Progress in the Chemistry of Organic Natural Products. Wien: Springer; 2003. pp. 51–124. [DOI] [PubMed] [Google Scholar]
- Messner P, Schäffer C, Kosma P. Bacterial cell-envelope glycoconjugates. Adv Carbohyd Chem Bi. 2013;69:209–72. doi: 10.1016/B978-0-12-408093-5.00006-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messner P, Schäffer C, Egelseer E-M, et al. Occurrence, structure, chemistry, genetics, morphogenesis, and functions of S-layers. In: König H, Claus H, Varma A, editors. Prokaryotic Cell Wall Compounds—Structure and Biochemistry. Berlin: Springer; 2010. pp. 53–109. [Google Scholar]
- Messner P, Sleytr UB, Christian R, et al. Isolation and structure determination of a diacetamidodideoxyuronic acid-containing glycan chain from the S-layer glycoprotein of Bacillus stearothermophilus NRS 2004/3a. Carbohyd Res. 1987;168:211–8. doi: 10.1016/0008-6215(87)80027-7. [DOI] [PubMed] [Google Scholar]
- Messner P, Sleytr UB. Bacterial surface layer glycoproteins. Glycobiology. 1991;1:545–51. doi: 10.1093/glycob/1.6.545. [DOI] [PubMed] [Google Scholar]
- Messner P, Steiner K, Zarschler K, et al. S-layer nanoglycobiology of bacteria. Carbohyd Res. 2008;343:1934–51. doi: 10.1016/j.carres.2007.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer BH, Albers S-V. Hot and sweet: protein glycosylation in Crenarchaeota. Biochem Soc Trans. 2013;41:384–92. doi: 10.1042/BST20120296. [DOI] [PubMed] [Google Scholar]
- Meyer BH, Peyfoon E, Dietrich C, et al. Agl16, a thermophilic glycosyltransferase mediating the last step of N-glycan biosynthesis in the thermoacidophilic crenarchaeon Sulfolobus acidocaldarius. J Bacteriol. 2013;195:2177–86. doi: 10.1128/JB.00035-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer BH, Zolghadr B, Peyfoon E, et al. Sulfoquinovose synthase - an important enzyme in the N-glycosylation pathway of Sulfolobus acidocaldarius. Mol Microbiol. 2011;82:1150–63. doi: 10.1111/j.1365-2958.2011.07875.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller WL, Matewish MJ, McNally DJ, et al. Flagellin glycosylation in Pseudomonas aeruginosa PAK requires the O-antigen biosynthesis enzyme WbpO. J Biol Chem. 2008;283:3507–18. doi: 10.1074/jbc.M708894200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra CS, Basu B, Apte SK. Surface (S)-layer proteins of Deinococcus radiodurans and their utility as vehicles for surface localization of functional proteins. Biochim Biophys Acta. 2015;1848:3181–7. doi: 10.1016/j.bbamem.2015.09.021. [DOI] [PubMed] [Google Scholar]
- Mo K-F, Li X, Li H, et al. Endolysins of Bacillus anthracis bacteriophages recognize unique carbohydrate epitopes of vegetative cell wall polysaccharides with high affinity and selectivity. J Am Chem Soc. 2012;134:15556–62. doi: 10.1021/ja3069962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morelle W, Canis K, Chirat F, et al. The use of mass spectrometry for the proteomic analysis of glycosylation. Proteomics. 2006;6:3993–4015. doi: 10.1002/pmic.200600129. [DOI] [PubMed] [Google Scholar]
- Morrison MJ, Imperiali B. The renaissance of bacillosamine and its derivatives: pathway characterization and implications in pathogenicity. Biochemistry. 2014;53:624–38. doi: 10.1021/bi401546r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möschl A, Schäffer C, Sleytr UB, et al. Characterization of the S-layer glycoproteins of two lactobacilli. In: Beveridge TJ, Koval SF, editors. Advances in Paracrystalline Bacterial Surface Layers. New York: Plenum Press; 1993. pp. 281–4. [Google Scholar]
- Mukherjee S, Kearns DB. The structure and regulation of flagella in Bacillus subtilis. Annu Rev Genet. 2014;48:319–40. doi: 10.1146/annurev-genet-120213-092406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami Y, Imai M, Nakamura H, et al. Separation of the outer membrane and identification of major outer membrane proteins from Porphyromonas gingivalis. Eur J Oral Sci. 2002;110:157–62. doi: 10.1034/j.1600-0722.2002.11171.x. [DOI] [PubMed] [Google Scholar]
- Murphy TF. The role of bacteria in airway inflammation in exacerbations of chronic obstructive pulmonary disease. Curr Opin Infect Dis. 2006;19:225–30. doi: 10.1097/01.qco.0000224815.89363.15. [DOI] [PubMed] [Google Scholar]
- Musumeci MA, Faridmoayer A, Watanabe Y, et al. Evaluating the role of conserved amino acids in bacterial O-oligosaccharyltransferases by in vivo, in vitro and limited proteolysis assays. Glycobiology. 2014;24:39–50. doi: 10.1093/glycob/cwt087. [DOI] [PubMed] [Google Scholar]
- Musumeci MA, Hug I, Scott NE, et al. In vitro activity of Neisseria meningitidis PglL O-oligosaccharyltransferase with diverse synthetic lipid donors and a UDP-activated sugar. J Biol Chem. 2013;288:10578–87. doi: 10.1074/jbc.M112.432815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naegeli A, Aebi M. Current approaches to engineering N-linked protein glycosylation in Bacteria. Methods Mol Biol. 2015;1321:3–16. doi: 10.1007/978-1-4939-2760-9_1. [DOI] [PubMed] [Google Scholar]
- Naegeli A, Neupert C, Fan Y-Y, et al. Molecular analysis of an alternative N-glycosylation machinery by functional transfer from Actinobacillus pleuropneumoniae to Escherichia coli. J Biol Chem. 2014;289:2170–9. doi: 10.1074/jbc.M113.524462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandakumar S, Kannanganat S, Dobos KM, et al. O-mannosylation of the Mycobacterium tuberculosis adhesin Apa is crucial for T cell antigenicity during infection but is expendable for protection. PLoS Pathog. 2013;9:e1003705. doi: 10.1371/journal.ppat.1003705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita Y, Sato K, Yukitake H, et al. Lack of a surface layer in Tannerella forsythia mutants deficient in the type IX secretion system. Microbiology. 2014;160:2295–303. doi: 10.1099/mic.0.080192-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Näther-Schindler DJ, Schopf S, Bellack A, et al. Pyrococcus furiosus flagella: biochemical and transcriptional analyses identify the newly detected flaB0 gene to encode the major flagellin. Front Microbiol. 2014;5:695. doi: 10.3389/fmicb.2014.00695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarre WW, Schneewind O. Surface proteins of gram-positive bacteria and mechanisms of their targeting to the cell wall envelope. Microbiol Mol Biol R. 1999;63:174–229. doi: 10.1128/mmbr.63.1.174-229.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson-Sathi S, Sousa FL, Röttger M, et al. Origins of major archaeal clades correspond to gene acquisitions from bacteria. Nature. 2015;517:77–80. doi: 10.1038/nature13805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuberger A. Carbohydrates in protein: the carbohydrate component of crystalline egg albumin. Biochem J. 1938;32:1435–51. doi: 10.1042/bj0321435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng SYM, Chaban B, Jarrell KF. Archaeal flagella, bacterial flagella and type IV pili: a comparison of genes and posttranslational modifications. J Mol Microb Biotech. 2006;11:167–91. doi: 10.1159/000094053. [DOI] [PubMed] [Google Scholar]
- Nguyen-Mau S-M, Oh S-Y, Schneewind DI, et al. Bacillus anthracis SlaQ promotes S-layer protein assembly. J Bacteriol. 2015;197:3216–27. doi: 10.1128/JB.00492-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nothaft H, Scott NE, Vinogradov E, et al. Diversity in the protein N-glycosylation pathways within the Campylobacter genus. Mol Cell Proteomics. 2012;11:1203–19. doi: 10.1074/mcp.M112.021519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nothaft H, Szymanski CM. Bacterial protein, N-glycosylation: new perspectives and applications. J Biol Chem. 2013;288:6912–20. doi: 10.1074/jbc.R112.417857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nothaft H, Szymanski CM. Protein glycosylation in bacteria: sweeter than ever. Nat Rev Microbiol. 2010;8:765–78. doi: 10.1038/nrmicro2383. [DOI] [PubMed] [Google Scholar]
- Olivier NB, Chen MM, Behr JR, et al. In vitro biosynthesis of UDP-N,N′-diacetylbacillosamine by enzymes of the Campylobacter jejuni general protein glycosylation system. Biochemistry. 2006;45:13659–69. doi: 10.1021/bi061456h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ollis AA, Chai Y, DeLisa MP. GlycoSNAP: a high-throughput screening methodology for engineering designer glycosylation enzymes. Methods Mol Biol. 2015;1321:37–47. doi: 10.1007/978-1-4939-2760-9_3. [DOI] [PubMed] [Google Scholar]
- Ortega X, Hunt TA, Loutet S, et al. Reconstitution of O-specific lipopolysaccharide expression in Burkholderia cenocepacia strain J2315, which is associated with transmissible infections in patients with cystic fibrosis. J Bacteriol. 2005;187:1324–33. doi: 10.1128/JB.187.4.1324-1333.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paik SH, Chakicherla A, Hansen JN. Identification and characterization of the structural and transporter genes for, and the chemical and biological properties of, sublancin 168, a novel lantibiotic produced by Bacillus subtilis 168. J Biol Chem. 1998;273:23134–42. doi: 10.1074/jbc.273.36.23134. [DOI] [PubMed] [Google Scholar]
- Palmieri G, Balestrieri M, Peter-Katalinić J, et al. Surface-exposed glycoproteins of hyperthermophilic Sulfolobus solfataricus P2 show a common N-glycosylation profile. J Proteome Res. 2013;12:2779–90. doi: 10.1021/pr400123z. [DOI] [PubMed] [Google Scholar]
- Parente J, Casabuono A, Ferrari MC, et al. A rhomboid protease gene deletion affects a novel oligosaccharide, N-linked to the S-layer glycoprotein of Haloferax volcanii. J Biol Chem. 2014;289:11304–17. doi: 10.1074/jbc.M113.546531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker JL, Day-Williams MJ, Tomás JM, et al. Identification of a putative glycosyltransferase responsible for the transfer of pseudaminic acid onto the polar flagellin of Aeromonas caviae Sch 3N. MicrobiologyOpen. 2012;1:149–60. doi: 10.1002/mbo3.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkhill J, Wren BW, Mungall K, et al. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hyper-variable sequences. Nature. 2000;403:665–8. doi: 10.1038/35001088. [DOI] [PubMed] [Google Scholar]
- Patenge N, Berendes A, Engelhardt H, et al. The fla gene cluster is involved in the biogenesis of flagella in Halobacterium salinarum. Mol Microbiol. 2001;41:653–63. doi: 10.1046/j.1365-2958.2001.02542.x. [DOI] [PubMed] [Google Scholar]
- Paul G, Lottspeich F, Wieland F. Asparaginyl-N-acetylgalactosamine. Linkage unit of halobacterial glycosaminoglycan. J Biol Chem. 1986;261:1020–4. [PubMed] [Google Scholar]
- Pavkov T, Egelseer EM, Tesarz M, et al. The structure and binding behavior of the bacterial cell surface layer protein SbsC. Structure. 2008;16:1226–37. doi: 10.1016/j.str.2008.05.012. [DOI] [PubMed] [Google Scholar]
- Perez C, Gerber S, Boilevin J, et al. Structure and mechanism of an active lipid-linked oligosaccharide flippase. Nature. 2015;524:433–8. doi: 10.1038/nature14953. [DOI] [PubMed] [Google Scholar]
- Petersen BO, Sára M, Mader C, et al. Structural characterization of the acid-degraded secondary cell wall polymer of Geobacillus stearothermophilus PV72/p2. Carbohyd Res. 2008;343:1346–58. doi: 10.1016/j.carres.2008.03.029. Corrigendum: Carbohydr Res 373 (2013) 52. [DOI] [PubMed] [Google Scholar]
- Peyfoon E, Meyer B, Hitchen PG, et al. The S-layer glycoprotein of the crenarchaeote Sulfolobus acidocaldarius is glycosylated at multiple sites with chitobiose-linked N-glycans. Archaea. 2010;2010:754101. doi: 10.1155/2010/754101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfoestl A, Hofinger A, Kosma P, et al. Biosynthesis of dTDP-3-acetamido-3,6-dideoxy-α-d-galactose in Aneurinibacillus thermoaerophilus L420-91T. J Biol Chem. 2003;278:26410–7. doi: 10.1074/jbc.M300858200. [DOI] [PubMed] [Google Scholar]
- Pohlschroder M, Esquivel RN. Archaeal type IV pili and their involvement in biofilm formation. Front Microbiol. 2015;6:190. doi: 10.3389/fmicb.2015.00190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohlschroder M, Ghosh A, Tripepi M, et al. Archaeal type IV piluslike structures–evolutionarily conserved prokaryotic surface organelles. Curr Opin Microbiol. 2011;14:357–63. doi: 10.1016/j.mib.2011.03.002. [DOI] [PubMed] [Google Scholar]
- Posch G, Andrukhov O, Vinogradov E, et al. Structure and immunogenicity of the rough-type lipopolysaccharide from the periodontal pathogen Tannerella forsythia. Clin Vaccine Immunol. 2013a;20:945–53. doi: 10.1128/CVI.00139-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posch G, Pabst M, Brecker L, et al. Characterization and scope of S-layer protein O-glycosylation in Tannerella forsythia. J Biol Chem. 2011;286:38714–24. doi: 10.1074/jbc.M111.284893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posch G, Pabst M, Neumann L, et al. ‘Cross-glycosylation’ of proteins in Bacteroidales species. Glycobiology. 2013b;23:568–77. doi: 10.1093/glycob/cws172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posch G, Sekot G, Friedrich V, et al. Glycobiology aspects of the periodontal pathogen Tannerella forsythia. Biomolecules. 2012;2:467–82. doi: 10.3390/biom2040467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power PM, Roddam LF, Dieckelmann M, et al. Genetic characterization of pilin glycosylation in Neisseria meningitidis. Microbiology. 2000;146:967–79. doi: 10.1099/00221287-146-4-967. [DOI] [PubMed] [Google Scholar]
- Power PM, Seib KL, Jennings MP. Pilin glycosylation in Neisseria meningitidis occurs by a similar pathway to wzy-dependent O-antigen biosynthesis in Escherichia coli. Biochem Bioph Res Co. 2006;347:904–8. doi: 10.1016/j.bbrc.2006.06.182. [DOI] [PubMed] [Google Scholar]
- Qazi O, Hitchen P, Tissot B, et al. Mass spectrometric analysis of the S-layer proteins from Clostridium difficile demonstrates the absence of glycosylation. J Mass Spectrom. 2009;44:368–74. doi: 10.1002/jms.1514. [DOI] [PubMed] [Google Scholar]
- Rachel R. Cell envelopes of crenarchaeota and nanoarchaeota. In: König H, Claus H, Varma A, editors. Prokaryotic Cell Wall Compounds—Structure and Biochemistry. Berlin: Springer; 2010. pp. 271–91. [Google Scholar]
- Raetz CRH, Whitfield C. Lipopolysaccharide endotoxins. Annu Rev Biochem. 2002;71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramström O, Yan M. Glyconanomaterials for combating bacterial infections. Chemistry. 2015;21:16310–7. doi: 10.1002/chem.201502842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reunanen J, von Ossowski I, Hendrickx AP, et al. Characterization of the SpaCBA pilus fibers in the probiotic Lactobacillus rhamnosus GG. Appl Environ Microb. 2012;78:2337–44. doi: 10.1128/AEM.07047-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ries W, Hotzy C, Schocher I, et al. Evidence that the N-terminal part of the S-layer protein from Bacillus stearothermophilus PV72/p2 recognizes a secondary cell wall polymer. J Bacteriol. 1997;179:3892–8. doi: 10.1128/jb.179.12.3892-3898.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ristl R, Steiner K, Zarschler K, et al. The S-layer glycomeadding to the sugar coat of bacteria. Int J Microbiol. 2011;2011:127870. doi: 10.1155/2011/127870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohlin L, Leon DR, Kim U, et al. Identification of the major expressed S-layer and cell surface-layer-related proteins in the model methanogenic archaea: Methanosarcina barkeri Fusaro and Methanosarcina acetivorans C2A. Archaea. 2012;2012:873589. doi: 10.1155/2012/873589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz N, Kahne D, Silhavy TJ. Transport of lipopolysaccharide across the cell envelope: the long road of discovery. Nat Rev Microbiol. 2009;7:677–83. doi: 10.1038/nrmicro2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabet M, Lee S-W, Nauman RK, et al. The surface (S-) layer is a virulence factor of Bacteroides forsythus. Microbiology. 2003;149:3617–27. doi: 10.1099/mic.0.26535-0. [DOI] [PubMed] [Google Scholar]
- Sacoman JL, Hollingsworth RI. Synthesis and evaluation of an N-acetylglucosamine biosynthesis inhibitor. Carbohyd Res. 2011;346:2294–9. doi: 10.1016/j.carres.2011.07.004. [DOI] [PubMed] [Google Scholar]
- Sakakibara J, Nagano K, Murakami Y, et al. Loss of adherence ability to human gingival epithelial cells in S-layer protein-deficient mutants of Tannerella forsythensis. Microbiology. 2007;153:866–76. doi: 10.1099/mic.0.29275-0. [DOI] [PubMed] [Google Scholar]
- Saksena R, Adamo R, Kováč P. Synthesis of the tetrasaccharide side chain of the major glycoprotein of the Bacillus anthracis exosporium. Bioorg Med Chem Lett. 2006;16:615–7. doi: 10.1016/j.bmcl.2005.10.056. [DOI] [PubMed] [Google Scholar]
- Sára M. Conserved anchoring mechanisms between crystalline cell surface S-layer proteins and secondary cell wall polymers in Gram-positive bacteria? Trends Microbiol. 2001;9:47–49. doi: 10.1016/s0966-842x(00)01905-3. discussion 49–50. [DOI] [PubMed] [Google Scholar]
- Sára M, Dekitsch C, Mayer HF, et al. Influence of the secondary cell wall polymer on the reassembly, recrystallization, and stability properties of the S-layer protein from Bacillus stearothermophilus PV72/p2. J Bacteriol. 1998;180:4146–53. doi: 10.1128/jb.180.16.4146-4153.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satchidanandam V, Kumar N, Jumani RS, et al. The glycosylated Rv1860 protein of Mycobacterium tuberculosis inhibits dendritic cell mediated TH1 and TH17 polarization of T cells and abrogates protective immunity conferred by BCG. PLoS Pathog. 2014;10:e1004176. doi: 10.1371/journal.ppat.1004176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäffer C, Dietrich K, Unger B, et al. A novel type of carbohydrate-protein linkage region in the tyrosine-bound S-layer glycan of Thermoanaerobacterium thermosaccharolyticum D120-70. Eur J Biochem. 2000a;267:5482–92. doi: 10.1046/j.1432-1327.2000.01610.x. [DOI] [PubMed] [Google Scholar]
- Schäffer C, Franck WL, Scheberl A, et al. Classification of isolates from locations in Austria and Yellowstone National Park as Geobacillus tepidamans sp. nov. Int J Syst Evol Micr. 2004;54:2361–8. doi: 10.1099/ijs.0.63227-0. [DOI] [PubMed] [Google Scholar]
- Schäffer C, Kählig H, Christian R, et al. The diacetamidodideoxyuronic-acid-containing glycan chain of Bacillus stearothermophilus NRS 2004/3a represents the secondary cell-wall polymer of wild-type B. stearothermophilus strains. Microbiology. 1999a;145:1575–83. doi: 10.1099/13500872-145-7-1575. [DOI] [PubMed] [Google Scholar]
- Schäffer C, Messner P. Surface-layer glycoproteins: an example for the diversity of bacterial glycosylation with promising impacts on nanobiotechnology. Glycobiology. 2004;14:31R–42R. doi: 10.1093/glycob/cwh064. [DOI] [PubMed] [Google Scholar]
- Schäffer C, Messner P. The structure of secondary cell wall polymers: how Gram-positive bacteria stick their cell walls together. Microbiology. 2005;151:643–51. doi: 10.1099/mic.0.27749-0. [DOI] [PubMed] [Google Scholar]
- Schäffer C, Müller N, Christian R, et al. Complete glycan structure of the S-layer glycoprotein of Aneurinibacillus thermoaerophilus GS4-97. Glycobiology. 1999b;9:407–14. doi: 10.1093/glycob/9.4.407. [DOI] [PubMed] [Google Scholar]
- Schäffer C, Müller N, Mandal PK, et al. A pyrophosphate bridge links the pyruvate-containing secondary cell wall polymer of Paenibacillus alvei CCM 2051 to muramic acid. Glycoconj J. 2000b;17:681–90. doi: 10.1023/a:1011062302889. [DOI] [PubMed] [Google Scholar]
- Schirm M, Arora SK, Verma A, et al. Structural and genetic characterization of glycosylation of type a flagellin in Pseudomonas aeruginosa. J Bacteriol. 2004a;186:2523–31. doi: 10.1128/JB.186.9.2523-2531.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirm M, Kalmokoff M, Aubry A, et al. Flagellin from Listeria monocytogenes is glycosylated with β-O-linked N-acetylglucosamine. J Bacteriol. 2004b;186:6721–27. doi: 10.1128/JB.186.20.6721-6727.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirm M, Schoenhofen IC, Logan SM, et al. Identification of unusual bacterial glycosylation by tandem mass spectrometry analyses of intact proteins. Anal Chem. 2005;77:7774–82. doi: 10.1021/ac051316y. [DOI] [PubMed] [Google Scholar]
- Schirm M, Soo EC, Aubry AJ, et al. Structural, genetic and functional characterization of the flagellin glycosylation process in Helicobacter pylori. Mol Microbiol. 2003;48:1579–92. doi: 10.1046/j.1365-2958.2003.03527.x. [DOI] [PubMed] [Google Scholar]
- Schoenhofen IC, McNally DJ, Vinogradov E, et al. Functional characterization of dehydratase/aminotransferase pairs from Helicobacter and Campylobacter: enzymes distinguishing the pseudaminic acid and bacillosamine biosynthetic pathways. J Biol Chem. 2006;281:723–32. doi: 10.1074/jbc.M511021200. [DOI] [PubMed] [Google Scholar]
- Schoenhofen IC, Vinogradov E, Whitfield DM, et al. The CMPlegionaminic acid pathway in Campylobacter: biosynthesis involving novel GDP-linked precursors. Glycobiology. 2009;19:715–25. doi: 10.1093/glycob/cwp039. [DOI] [PubMed] [Google Scholar]
- Schuch R, Nelson D, Fischetti VA. A bacteriolytic agent that detects and kills Bacillus anthracis. Nature. 2002;418:884–9. doi: 10.1038/nature01026. [DOI] [PubMed] [Google Scholar]
- Schulz BL, Jen FEC, Power PM, et al. Identification of bacterial protein O-oligosaccharyltransferases and their glycoprotein substrates. PLoS One. 2013;8:e62768. doi: 10.1371/journal.pone.0062768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster B, Sleytr UB. Nanotechnology with S-layer proteins. Methods Mol Biol. 2013;996:153–75. doi: 10.1007/978-1-62703-354-1_9. [DOI] [PubMed] [Google Scholar]
- Schwarz F, Aebi M. Mechanisms and principles of N-linked protein glycosylation. Curr Opin Struct Biol. 2011;21:576–82. doi: 10.1016/j.sbi.2011.08.005. [DOI] [PubMed] [Google Scholar]
- Schwarz F, Fan Y-Y, Schubert M, et al. Cytoplasmic N-glycosyltransferase of Actinobacillus pleuropneumoniae is an inverting enzyme and recognizes the NX(S/T) consensus sequence. J Biol Chem. 2011;286:35267–74. doi: 10.1074/jbc.M111.277160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz F, Huang W, Li C, et al. A combined method for producing homogeneous glycoproteins with eukaryotic N-glycosylation. Nat Chem Biol. 2010;6:264–6. doi: 10.1038/nchembio.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seepersaud R, Bensing BA, Yen YT, et al. The accessory Sec protein Asp2 modulates GlcNAc deposition onto the serine-rich repeat glycoprotein GspB. J Bacteriol. 2012;194:5564–75. doi: 10.1128/JB.01000-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekot G, Posch G, Oh YJ, et al. Analysis of the cell surface layer ultrastructure of the oral pathogen Tannerella forsythia. Arch Microbiol. 2012;194:525–39. doi: 10.1007/s00203-012-0792-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settem RP, Honma K, Nakajima T, et al. A bacterial glycan core linked to surface (S)-layer proteins modulates host immunity through Th17 suppression. Mucosal Immunol. 2013;6:415–26. doi: 10.1038/mi.2012.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahapure R, Driessen RPC, Haurat MF, et al. The archaellum: a rotating type IV pilus. Mol Microbiol. 2014;91:716–23. doi: 10.1111/mmi.12486. [DOI] [PubMed] [Google Scholar]
- Sherlock O, Dobrindt U, Jensen JB, et al. Glycosylation of the self-recognizing Escherichia coli Ag43 autotransporter protein. J Bacteriol. 2006;188:1798–807. doi: 10.1128/JB.188.5.1798-1807.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi W-W, Jiang Y-L, Zhu F, et al. Structure of a novel O-linked Nacetyl-d-glucosamine (O-GlcNAc) transferase, GtfA, reveals insights into the glycosylation of pneumococcal serine-rich repeat adhesins. J Biol Chem. 2014;289:20898–907. doi: 10.1074/jbc.M114.581934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siu S, Robotham A, Logan SM, et al. Evidence that biosynthesis of the second and third sugars of the archaellin tetrasaccharide in the archaeon Methanococcus maripaludis occurs by the same pathway used by Pseudomonas aeruginosa to make a di-N-acetylated sugar. J Bacteriol. 2015;197:1668–80. doi: 10.1128/JB.00040-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleytr UB. Self-assembly of the hexagonally and tetragonally arranged subunits of bacterial surface layers and their reattachment to cell walls. J Ultrastruct Res. 1976;55:360–77. doi: 10.1016/s0022-5320(76)80093-7. [DOI] [PubMed] [Google Scholar]
- Sleytr UB. Regular arrays of macromolecules on bacterial cell walls: structure, chemistry, assembly, and function. Int Rev Cytol. 1978;53:1–62. doi: 10.1016/s0074-7696(08)62240-8. [DOI] [PubMed] [Google Scholar]
- Sleytr UB, Messner P. Crystalline surface layers on bacteria. Annu Rev Microbiol. 1983;37:311–39. doi: 10.1146/annurev.mi.37.100183.001523. [DOI] [PubMed] [Google Scholar]
- Sleytr UB, Messner P, Pum D, et al. Crystalline surface layers on eubacteria and archaeobacteria. In: Sleytr UB, Messner P, Pum D, editors. Crystalline Bacterial Cell Surface Proteins. R.G. Austin, TX: Landes/Academic Press; 1996. pp. 211–25. [Google Scholar]
- Sleytr UB, Schuster B, Egelseer E-M, et al. S-layers: principles and applications. FEMS Microbiol Rev. 2014;38:823–64. doi: 10.1111/1574-6976.12063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleytr UB, Thorne KJI. Chemical characterization of the regularly arranged surface layers of Clostridium thermosaccharolyticum and Clostridium thermohydrosulfuricum. J Bacteriol. 1976;126:377–83. doi: 10.1128/jb.126.1.377-383.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit E, Oling F, Demel R, et al. The S-layer protein of Lactobacillus acidophilus ATCC 4356: identification and characterisation of domains responsible for S-protein assembly and cell wall binding. J Mol Biol. 2001;305:245–57. doi: 10.1006/jmbi.2000.4258. [DOI] [PubMed] [Google Scholar]
- Spiro RG. Protein glycosylation: nature, distribution, enzymaticformation, and disease implications of glycopeptide bonds. Glycobiology. 2002;12:43R–56R. doi: 10.1093/glycob/12.4.43r. [DOI] [PubMed] [Google Scholar]
- St Geme JW, 3rd, Yeo H-J. A prototype two-partner secretion pathway: the Haemophilus influenzae HMW1 and HMW2 adhesin systems. Trends Microbiol. 2009;17:355–60. doi: 10.1016/j.tim.2009.06.002. [DOI] [PubMed] [Google Scholar]
- Stafford G, Roy S, Honma K, et al. Sialic acid, periodontal pathogens and Tannerella forsythia: stick around and enjoy the feast! Mol Oral Microbiol. 2012;27:11–22. doi: 10.1111/j.2041-1014.2011.00630.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steindl C, Schäffer C, Smrečki V, et al. The secondary cell wall polymer of Geobacillus tepidamans GS5-97T: structure of different glycoforms. Carbohyd Res. 2005;340:2290–6. doi: 10.1016/j.carres.2005.07.005. [DOI] [PubMed] [Google Scholar]
- Steindl C, Schäffer C, Wugeditsch T, et al. The first biantennary bacterial secondary cell wall polymer and its influence on S-layer glycoprotein assembly. Biochem J. 2002;368:483–94. doi: 10.1042/BJ20020988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner K, Hagelueken G, Messner P, et al. Structural basis of substrate binding in WsaF, a rhamnosyltransferase from Geobacillus stearothermophilus. J Mol Biol. 2010;397:436–47. doi: 10.1016/j.jmb.2010.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner K, Novotny R, Patel K, et al. Functional characterization of the initiation enzyme of S-layer glycoprotein glycan biosynthesis in Geobacillus stearothermophilus NRS 2004/3a. J Bacteriol. 2007;189:2590–8. doi: 10.1128/JB.01592-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner K, Novotny R, Werz DB, et al. Molecular basis of S-layer glycoprotein glycan biosynthesis in Geobacillus stearothermophilus. J Biol Chem. 2008;283:21120–33. doi: 10.1074/jbc.M801833200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner K, Pohlentz G, Dreisewerd K, et al. New insights into the glycosylation of the surface layer protein SgsE from Geobacillus stearothermophilus NRS 2004/3a. J Bacteriol. 2006;188:7914–21. doi: 10.1128/JB.00802-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepper J, Shastri S, Loo TS, et al. Cysteine S-glycosylation, a new post-translational modification found in glycopeptide bacteriocins. FEBS Lett. 2011;585:645–50. doi: 10.1016/j.febslet.2011.01.023. [DOI] [PubMed] [Google Scholar]
- Stimson E, Virji M, Makepeace K, et al. Meningococcal pilin: a glycoprotein substituted with digalactosyl 2,4-diacetamido-2,4,6-trideoxyhexose. Mol Microbiol. 1995;17:1201–14. doi: 10.1111/j.1365-2958.1995.mmi_17061201.x. [DOI] [PubMed] [Google Scholar]
- Strong PC, Fulton KM, Aubry A, et al. Identification and characterization of glycoproteins on the spore surface of Clostridium difficile. J Bacteriol. 2014;196:2627–37. doi: 10.1128/JB.01469-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumper M. Halobacterial glycoprotein biosynthesis. Biochim Biophys Acta. 1987;906:69–79. doi: 10.1016/0304-4157(87)90005-0. [DOI] [PubMed] [Google Scholar]
- Sumper M, Berg E, Mengele R, et al. Primary structure and glycosylation of the S-layer protein of Haloferax volcanii. J Bacteriol. 1990;172:7111–8. doi: 10.1128/jb.172.12.7111-7118.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland IW. Microbial polysaccharide products. Biotechnol Genet Eng. 1999;16:217–29. doi: 10.1080/02648725.1999.10647976. [DOI] [PubMed] [Google Scholar]
- Szymanski CM, Wren BW. Protein glycosylation in bacterial mucosal pathogens. Nat Rev Microbiol. 2005;3:225–37. doi: 10.1038/nrmicro1100. [DOI] [PubMed] [Google Scholar]
- Szymanski CM, Yao R, Ewing CP, et al. Evidence for a system of general protein glycosylation in Campylobacter jejuni. Mol Microbiol. 1999;32:1022–30. doi: 10.1046/j.1365-2958.1999.01415.x. [DOI] [PubMed] [Google Scholar]
- Tabei SMB, Hitchen PG, Day-Williams MJ, et al. An Aeromonas caviae genomic island is required for both O-antigen lipopolysaccharide biosynthesis and flagellin glycosylation. J Bacteriol. 2009;191:2851–63. doi: 10.1128/JB.01406-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi Y, Fujinami D, Kohda D. Comparative analysis of archaeal lipid-linked oligosaccharides that serve as oligosaccharide donors for Asn glycosylation. J Biol Chem. 2016;291:11042–54. doi: 10.1074/jbc.M115.713156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi K, Ono H, Yoshida M, et al. Flagellin glycans from two pathovars of Pseudomonas syringae contain rhamnose in D and L configurations in different ratios and modified 4-amino-4,6-dideoxyglucose. J Bacteriol. 2007;189:6945–56. doi: 10.1128/JB.00500-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan FYY, Tang CM, Exley RM. Sugar coating: bacterial protein glycosylation and host-microbe interactions. Trends Biochem Sci. 2015;40:342–50. doi: 10.1016/j.tibs.2015.03.016. [DOI] [PubMed] [Google Scholar]
- Tan L, Turnbough CL., Jr Sequence motifs and proteolytic cleavage of the collagen-like glycoprotein BclA required for its attachment to the exosporium of Bacillus anthracis. J Bacteriol. 2010;192:1259–68. doi: 10.1128/JB.01003-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G, Mintz KP. Glycosylation of the collagen adhesin EmaA of Aggregatibacter actinomycetemcomitans is dependent upon the lipopolysaccharide biosynthetic pathway. J Bacteriol. 2010;192:1395–404. doi: 10.1128/JB.01453-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G, Ruiz T, Barrantes-Reynolds R, et al. Molecular heterogeneity of EmaA, an oligomeric autotransporter adhesin of Aggregatibacter (Actinobacillus) actinomycetemcomitans. Microbiology. 2007;153:2447–57. doi: 10.1099/mic.0.2007/005892-0. [DOI] [PubMed] [Google Scholar]
- Tanner ACR, Izard J. Tannerella forsythia, a periodontal pathogen entering the genomic era. Periodontol 2000. 2006;42:88–113. doi: 10.1111/j.1600-0757.2006.00184.x. [DOI] [PubMed] [Google Scholar]
- Tavares-Carreón F, Patel KB, Valvano MA. Burkholderia cenocepacia and Salmonella enterica ArnT proteins that transfer 4-amino-4-deoxy-l-arabinose to lipopolysaccharide share membrane topology and functional amino acids. Sci Rep. 2015;5:10773. doi: 10.1038/srep10773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibault P, Logan SM, Kelly JF, et al. Identification of the carbohydrate moieties and glycosylation motifs in Campylobacter jejuni flagellin. J Biol Chem. 2001;276:34862–70. doi: 10.1074/jbc.M104529200. [DOI] [PubMed] [Google Scholar]
- Thoma C, Frank M, Rachel R, et al. The Mth60 fimbriae of Methanothermobacter thermoautotrophicus are functional adhesins. Environ Microbiol. 2008;10:2785–95. doi: 10.1111/j.1462-2920.2008.01698.x. [DOI] [PubMed] [Google Scholar]
- Thornley MJ, Glauert AM, Sleytr UB. Isolation of outer membranes with an ordered array of surface subunits from Acinetobacter. J Bacteriol. 1973;114:1294–308. doi: 10.1128/jb.114.3.1294-1308.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomek MB, Neumann L, Nimeth I, et al. The S-layer proteins of Tannerella forsythia are secreted via a type IX secretion system that is decoupled from protein O-glycosylation. Mol Oral Microbiol. 2014;29:307–20. doi: 10.1111/omi.12062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrelles JB, Schlesinger LS. Diversity in Mycobacterium tuberculosis mannosylated cell wall determinants impacts adaptation to the host. Tuberculosis (Edinb) 2010;90:84–93. doi: 10.1016/j.tube.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trachtenberg S, Pinnick B, Kessel M. The cell surface glycoprotein layer of the extreme halophile Halobacterium salinarum and its relation to Haloferax volcanii: cryo-electron tomography of freeze-substituted cells and projection studies of negatively stained envelopes. J Struct Biol. 2000;130:10–26. doi: 10.1006/jsbi.2000.4215. [DOI] [PubMed] [Google Scholar]
- Tripepi M, You J, Temel S, et al. N-glycosylation of Haloferax volcanii flagellins requires known Agl proteins and is essential for biosynthesis of stable flagella. J Bacteriol. 2012;194:4876–87. doi: 10.1128/JB.00731-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tropis M, Meniche X, Wolf A, et al. The crucial role of trehalose and structurally related oligosaccharides in the biosynthesis and transfer of mycolic acids in Corynebacterineae. J Biol Chem. 2005;280:26573–85. doi: 10.1074/jbc.M502104200. [DOI] [PubMed] [Google Scholar]
- Tschitschko B, Williams TJ, Allen MA, et al. Antarctic archaeavirus interactions: metaproteome-led analysis of invasion, evasion and adaptation. ISME J. 2015;9:2094–107. doi: 10.1038/ismej.2015.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turroni F, Ventura M, Buttó LF, et al. Molecular dialogue between the human gut microbiota and the host: a Lactobacillus and Bifidobacterium perspective. Cell Mol Life Sci. 2014;71:183–203. doi: 10.1007/s00018-013-1318-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twine SM, Paul CJ, Vinogradov E, et al. Flagellar glycosylation in Clostridium botulinum. FEBS J. 2008;275:4428–44. doi: 10.1111/j.1742-4658.2008.06589.x. [DOI] [PubMed] [Google Scholar]
- Twine SM, Reid CW, Aubry A, et al. Motility and flagellar glycosylation in Clostridium difficile. J Bacteriol. 2009;191:7050–62. doi: 10.1128/JB.00861-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tytgat HLP, Lebeer S. The sweet tooth of bacteria: common themes in bacterial glycoconjugates. Microbiol Mol Biol R. 2014;78:372–417. doi: 10.1128/MMBR.00007-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valderrama-Rincon JD, Fisher AC, Merritt JH, et al. An engineered eukaryotic protein glycosylation pathway in Escherichia coli. Nat Chem Biol. 2012;8:434–6. doi: 10.1038/nchembio.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valero E, Martín M, Gálvez N, et al. Nanopatterning of magnetic CrNi Prussian Blue nanoparticles using a bacterial S-layer as a biotemplate. Inorg Chem. 2015;54:6758–62. doi: 10.1021/acs.inorgchem.5b00555. [DOI] [PubMed] [Google Scholar]
- Valguarnera E, Kinsella RL, Feldman MF. Sugar and spice make bacteria not nice: protein glycosylation and its influence in pathogenesis. J Mol Biol. 2016;428:3206–20. doi: 10.1016/j.jmb.2016.04.013. [DOI] [PubMed] [Google Scholar]
- van Teeseling MCF, de Almeida NM, Klingl A, et al. A new addition to the cell plan of anammox bacteria: ‘Candidatus Kuenenia stuttgartiensis’ has a protein surface layer as the outermost layer of the cell. J Bacteriol. 2014;196:80–9. doi: 10.1128/JB.00988-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanDyke DJ, Wu J, Logan SM, et al. Identification of genes involved in the assembly and attachment of a novel flagellin N-linked tetrasaccharide important for motility in the archaeon Methanococcus maripaludis. Mol Microbiol. 2009;72:633–44. doi: 10.1111/j.1365-2958.2009.06671.x. [DOI] [PubMed] [Google Scholar]
- Varki A, Cummings RD, Esko JD, et al. Essentials of Glycobiology. 3rd edn. Woodbury, NY, USA: Cold Spring Harbor Laboratory Press; 2015. p. 800. [PubMed] [Google Scholar]
- Varki NM, Varki A. Diversity in cell surface sialic acid presentations: implications for biology and disease. Lab Invest. 2007;87:851–7. doi: 10.1038/labinvest.3700656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veith A, Klingl A, Zolghadr B, et al. Acidianus, Sulfolobus and Metallosphaera surface layers: structure, composition and gene expression. Mol Microbiol. 2009;73:58–72. doi: 10.1111/j.1365-2958.2009.06746.x. [DOI] [PubMed] [Google Scholar]
- Veith PD, Chen Y-Y, Chen D, et al. Tannerella forsythia outer membrane vesicles are enriched with substrates of the type IX secretion system and TonB-dependent receptors. J Proteome Res. 2015;14:5355–66. doi: 10.1021/acs.jproteome.5b00878. [DOI] [PubMed] [Google Scholar]
- Venugopal H, Edwards PJB, Schwalbe M, et al. Structural, dynamic, and chemical characterization of a novel S-glycosylated bacteriocin. Biochemistry. 2011;50:2748–55. doi: 10.1021/bi200217u. [DOI] [PubMed] [Google Scholar]
- Verma A, Schirm M, Arora SK, et al. Glycosylation of b-type flagellin of Pseudomonas aeruginosa: structural and genetic basis. J Bacteriol. 2006;188:4395–403. doi: 10.1128/JB.01642-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickerman MM, Brossard KA, Funk DB, et al. Phylogenetic analysis of bacterial and archaeal species in symptomatic and asymptomatic endodontic infections. J Med Microbiol. 2007;56:110–8. doi: 10.1099/jmm.0.46835-0. [DOI] [PubMed] [Google Scholar]
- Vijayakumar S, Merkx-Jacques A, Ratnayake DB, et al. Cj1121c, a novel UDP-4-keto-6-deoxy-GlcNAc C-4 aminotransferase essential for protein glycosylation and virulence in Campylobacter jejuni. J Biol Chem. 2006;281:27733–43. doi: 10.1074/jbc.M511714200. [DOI] [PubMed] [Google Scholar]
- Vinogradov E, Deschatelets L, Lamoureux M, et al. Cell surface glycoproteins from Thermoplasma acidophilum are modified with an N-linked glycan containing 6-C-sulfofucose. Glycobiology. 2012;22:1256–67. doi: 10.1093/glycob/cws094. [DOI] [PubMed] [Google Scholar]
- Voisin S, Houliston RS, Kelly J, et al. Identification and characterization of the unique N-linked glycan common to the flagellins and S-layer glycoprotein of Methanococcus voltae. J Biol Chem. 2005;280:16586–93. doi: 10.1074/jbc.M500329200. [DOI] [PubMed] [Google Scholar]
- Vorwerk H, Huber C, Mohr J, et al. A transferable plasticity region in Campylobacter coli allows isolates of an otherwise nonglycolytic food-borne pathogen to catabolize glucose. Mol Microbiol. 2015;98:809–30. doi: 10.1111/mmi.13159. [DOI] [PubMed] [Google Scholar]
- Wacker M, Linton D, Hitchen PG, et al. N-linked glycosylation in Campylobacter jejuni and its functional transfer into E. coli. Science. 2002;298:1790–3. doi: 10.1126/science.298.5599.1790. [DOI] [PubMed] [Google Scholar]
- Wakai H, Nakamura S, Kawasaki H, et al. Cloning and sequencing of the gene encoding the cell surface glycoprotein of Haloarcula japonica strain TR-1. Extremophiles. 1997;1:29–35. doi: 10.1007/s007920050012. [DOI] [PubMed] [Google Scholar]
- Wang D, Carroll GT, Turro NJ, et al. Photogenerated glycan arrays identify immunogenic sugar moieties of Bacillus anthracis exosporium. Proteomics. 2007;7:180–4. doi: 10.1002/pmic.200600478. [DOI] [PubMed] [Google Scholar]
- Wang H, van der Donk WA. Substrate selectivity of the sublancin S-glycosyltransferase. J Am Chem Soc. 2011;133:16394–7. doi: 10.1021/ja2075168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L-X. The amazing transglycosylation activity of endo-β-Nacetylglucosaminidases. Trends Glycosci Glyc. 2011;23:33–52. doi: 10.4052/tigg.23.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z-x, Deng R-p, Jiang H-W, et al. Global identification of prokaryotic glycoproteins based on an Escherichia coli proteome microarray. PLoS One. 2012;7:e49080. doi: 10.1371/journal.pone.0049080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward A, Reyes CL, Yu J, et al. Flexibility in the ABC transporter MsbA: alternating access with a twist. P Natl Acad Sci USA. 2007;104:19005–10. doi: 10.1073/pnas.0709388104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren MJ, Jennings MP. Identification and characterization of pptA: a gene involved in the phase-variable expression of phosphorylcholine on pili of Neisseria meningitidis. Infect Immun. 2003;71:6892–8. doi: 10.1128/IAI.71.12.6892-6898.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weerapana E, Imperiali B. Asparagine-linked protein glycosylation: from eukaryotic to prokaryotic systems. Glycobiology. 2006;16:91R–101R. doi: 10.1093/glycob/cwj099. [DOI] [PubMed] [Google Scholar]
- Westman EL, McNally DJ, Charchoglyan A, et al. Characterization of WbpB, WbpE, and WbpD and reconstitution of a pathway for the biosynthesis of UDP-2,3-diacetamido-2,3-dideoxy-d-mannuronic acid in Pseudomonas aeruginosa. J Biol Chem. 2009;284:11854–62. doi: 10.1074/jbc.M808583200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield C, Trent MS. Biosynthesis and export of bacterial lipopolysaccharides. Annu Rev Biochem. 2014;83:99–128. doi: 10.1146/annurev-biochem-060713-035600. [DOI] [PubMed] [Google Scholar]
- Wieland F, Heitzer R, Schaefer W. Asparaginylglucose: novel type of carbohydrate linkage. P Natl Acad Sci USA. 1983;80:5470–4. doi: 10.1073/pnas.80.18.5470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieland F, Paul G, Sumper M. Halobacterial flagellins are sulfated glycoproteins. J Biol Chem. 1985;260:15180–5. [PubMed] [Google Scholar]
- Wilhelms M, Fulton KM, Twine SM, et al. Differential glycosylation of polar and lateral flagellins in Aeromonas hydrophila AH-3. J Biol Chem. 2012;287:27851–62. doi: 10.1074/jbc.M112.376525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan A, Wu E, Lennarz WJ. Studies of yeast oligosaccharyl transferase subunits using the split-ubiquitin system: topological features and in vivo interactions. P Natl Acad Sci USA. 2005;102:7121–6. doi: 10.1073/pnas.0502669102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Q, Lennarz WJ. Studies on the function of oligosaccharyl transferase subunits. Stt3p is directly involved in the glycosylation process. J Biol Chem. 2002;277:47692–700. doi: 10.1074/jbc.M208136200. [DOI] [PubMed] [Google Scholar]
- Yang L, Sinha T, Carlson TK, et al. Changes in the major cell envelope components of Mycobacterium tuberculosis during in vitro growth. Glycobiology. 2013;23:926–34. doi: 10.1093/glycob/cwt029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen YT, Cameron TA, Bensing BA, et al. Differential localization of the streptococcal accessory sec components and implications for substrate export. J Bacteriol. 2013;195:682–95. doi: 10.1128/JB.01742-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young NM, Brisson J-R, Kelly J, et al. Structure of the N-linked glycan present on multiple glycoproteins in the Gram negative bacterium, Campylobacter jejuni. J Biol Chem. 2002;277:42530–9. doi: 10.1074/jbc.M206114200. [DOI] [PubMed] [Google Scholar]
- Yurist-Doutsch S, Eichler J. Manual annotation, transcriptional analysis, and protein expression studies reveal novel genes in the agl cluster responsible for N glycosylation in the halophilic archaeon Haloferax volcanii. J Bacteriol. 2009;191:3068–75. doi: 10.1128/JB.01838-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurist-Doutsch S, Magidovich H, Ventura VV, et al. N-glycosylation in Archaea: on the coordinated actions of Haloferax volcanii AglF and AglM. Mol Microbiol. 2010;75:1047–58. doi: 10.1111/j.1365-2958.2009.07045.x. [DOI] [PubMed] [Google Scholar]
- Zähringer U, Moll H, Hettmann T, et al. Cytochrome b558/566 from the archaeon Sulfolobus acidocaldarius has a unique Asn-linked highly branched hexasaccharide chain containing 6-sulfoquinovose. Eur J Biochem. 2000;267:4144–9. doi: 10.1046/j.1432-1327.2000.01446.x. [DOI] [PubMed] [Google Scholar]
- Zampronio CG, Blackwell G, Penn CW, et al. Novel glycosylation sites localized in Campylobacter jejuni flagellin FlaA by liquid chromatography electron capture dissociation tandem mass spectrometry. J Proteome Res. 2011;10:1238–45. doi: 10.1021/pr101021c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarschler K, Janesch B, Kainz B, et al. Cell surface display of chimeric glycoproteins via the S-layer of Paenibacillus alvei. Carbohyd Res. 2010a;345:1422–31. doi: 10.1016/j.carres.2010.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarschler K, Janesch B, Pabst M, et al. Protein tyrosine O-glycosylation–a rather unexplored prokaryotic glycosylation system. Glycobiology. 2010b;20:787–98. doi: 10.1093/glycob/cwq035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitler R, Hochmuth E, Deutzmann R, et al. Exchange of Ser-4 for Val, Leu or Asn in the sequon Asn-Ala-Ser does not prevent N-glycosylation of the cell surface glycoprotein from Halobacterium halobium. Glycobiology. 1998;8:1157–64. doi: 10.1093/glycob/8.12.1157. [DOI] [PubMed] [Google Scholar]
- Zeituni AE, McCaig W, Scisci E, et al. The native 67-kilodalton minor fimbria of Porphyromonas gingivalis is a novel glycoprotein with DC-SIGN-targeting motifs. J Bacteriol. 2010;192:4103–10. doi: 10.1128/JB.00275-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W-M, Wang H-F, Gao K, et al. Lactobacillus reuteri glyceraldehyde-3-phosphate dehydrogenase functions in adhesion to intestinal epithelial cells. Can J Microbiol. 2015;61:373–80. doi: 10.1139/cjm-2014-0734. [DOI] [PubMed] [Google Scholar]
- Zhou M, Wu H. Glycosylation and biogenesis of a family of serine-rich bacterial adhesins. Microbiology. 2009;155:317–27. doi: 10.1099/mic.0.025221-0. [DOI] [PubMed] [Google Scholar]