Abstract
Current preclinical drug testing does not predict some forms of adverse drug reactions in humans. Efforts at improving predictability of drug-induced tissue injury in humans include using stem cell technology to generate human cells for screening for adverse effects of drugs in humans. The advent of induced pluripotent stem cells means that it may ultimately be possible to develop personalised toxicology to determine inter-individual susceptibility to adverse drug reactions. However, the complexity of idiosyncratic drug-induced liver injury (DILI) means that no current single cell model, whether of primary liver tissue origin, from liver cell lines, or derived from stem cells, adequately emulates what is believed to occur during human DILI. Nevertheless, a single cell model of a human hepatocyte which emulates key features of a hepatocyte is likely to be valuable in assessing potential chemical risk; furthermore understanding how to generate a relevant hepatocyte will also be critical to efforts to build complex multicellular models of the liver. Currently, hepatocyte-like cells differentiated from stem cells still fall short of recapitulating the full mature hepatocellular phenotype. Therefore, we convened a number of experts from the areas of preclinical and clinical hepatotoxicity and safety assessment, from industry, academia and regulatory bodies, to specifically explore the application of stem cells in hepatotoxicity safety assessment, and to make recommendations for the way forward. In this short review, we particularly discuss the importance of benchmarking stem cell-derived hepatocyte-like cells to their terminally-differentiated human counterparts using defined phenotyping, to make sure the cells are relevant and comparable between labs, and outline why this process is essential before the cells are introduced into chemical safety assessment.
Keywords: hepatocyte, hepatotoxicity, cardiomyocyte, biomarker, de-differentiation
Prediction of adverse drug reactions in the liver: why it is important, limitations of current in vitro models and how stem cells may prove useful in drug screening
Adverse drug reactions (ADRs) are a significant clinical problem, resulting in considerable patient morbidity and mortality(1) and thus represent a major financial burden on healthcare systems. ADRs also represent a major challenge for the pharmaceutical industry leading to attrition of drugs in development and the withdrawal of drugs post-licensing(2). Amongst different forms of ADRs, the liver is particularly susceptible to drug toxicity; drug-induced liver injury (DILI) is the second highest cause of attrition and accounts for more than 50% of cases of acute liver failure(3).
The principal cause of these high attrition rates is the failure of current preclinical drug testing procedures to effectively predict idiosyncratic DILI in patients(2). This is true for in vitro models and even for in vivo models - a recent study that related the preclinical assessment of drugs with the occurrence of DILI in the clinic showed that between 38% (Medline database: 269 out of 710 compounds) and 51% (EMEA database: 70 out of 137 compounds) of drugs that subsequently caused liver injury in patients were not predicted from animal studies(4). Concerted worldwide efforts are therefore required to improve the assessment of hepatotoxic risk for new compounds. In Europe, the SEURAT (http://www.seurat-1.eu/pages/cluster-projects/scrtox.php) and MIP-DILI (http://www.mip-dili.eu/) consortia, and in the US, DILIN (http://www.dilin.org/) and iSAEC (http://www.saeconsortium.org/) are attempting to address this issue. The clinical manifestation of DILI indicates that it is a multi-dimensional and multi-faceted disease(5). Indeed, the diagnosis of DILI is largely based upon exclusion criteria(5). Although the use of currently available cell lines and primary human hepatocyte models has been able to correctly classify a number of DILI compounds as hepatoxins(6–9), idiosyncratic DILI is inherently difficult to model in the laboratory, and therefore highly unlikely to be predicted by simplistic screening strategies, often based on single-cell models involving cell lines. Many approaches use liver-derived cancer cell lines, e.g. HepG2 and HepaRG, which may have value for identifying drugs lacking a propensity to cause idiosyncratic DILI (90-95% predictability), but perform less well for positive predictions (50-89%)(9–11). Metabolically-competent freshly-isolated, or cryopreserved human primary adult hepatocytes are still considered to be the gold-standard single cell model of DILI. Nevertheless, human hepatocytes are difficult to source, they are also costly and functionally variable (reflecting variation in the human population), they undergo severe stress during the isolation process and, critically, they rapidly lose key functions when cultured in vitro. Moreover, it is important to note that hepatocyte toxicity per se is not the sole cause of hepatotoxicity which, in the intact liver, may involve multiple different cell types including lymphocytes and macrophages. Yet it is reasonable to assume from the work of several groups, over many years, that a metabolically-competent hepatocyte will be an essential component of any model of hepatotoxicity in vitro. Thus, a robust and reproducible metabolically-competent hepatocyte-like cell derived from directly reprogrammed cells, or from pluripotent stem cells, would represent a major step forward for the development of a new generation of in vitro models.
The imperatives of industry and academia are driven by different model requirements. The priority for industry is a cost-effective and scalable high-throughput screening model that has direct input into ‘go/no go’ decision making during drug development, whilst academic scientists are driven by the need to understand hepatic physiology and the mechanistic basis of DILI. Hepatocytes derived from stem cells can, however, be central to both of these objectives. Whilst significant progress towards a functional hepatic phenotype has been made, it is clear that stem-cell-derived hepatocyte-like cells (SC-HLCs) still fall well short of recapitulating the full mature hepatocellular phenotype(12–15).
Because of the importance and likely impact of developments in this field, scientists with expertise in preclinical and clinical hepatotoxicityand complex and novel forms of in vitro cell culture, representing industry, academia and regulatory bodies, assembled at a workshop at the University of Liverpool, under the auspices of the European Partnership for Alternative Approaches to Animal Testing (EPAA) (http://ec.europa.eu/growth/sectors/chemicals/epaa/index_en.htm) and the MRC Centre for Drug Safety Science (https://www.liverpool.ac.uk/drug-safety/). The purpose of the workshop was to specifically explore the application of stem cells in hepatotoxicity safety assessment, and to make recommendations for the way forward. This workshop follows the EPAA/NC3Rs (National Centre for the Replacement, Refinement and Reduction of Animals in Research) (https://www.nc3rs.org.uk/) “Stem Cells in Safety Testing Forum” workshop that took place in 2013, with a mandate to provide a platform for permanent dialogue between research groups, to share experiences, problems, successes and opportunities.
Current challenges in the use of stem cell-derived hepatocytes in the safety assessment of new chemical entities
It is clear from a large number of studies(13, 14, 16–47) (see Table 1) that hepatocytes generated from stem cells are not currently sufficiently mature to emulate an adult primary human hepatocyte, and that these cells are probably closer in phenotype to a fetal hepatocyte(12). Many studies using SC-HLCs purport to demonstrate a hepatocyte-like phenotype but do not actually incorporate a physiologically-relevant benchmark (e.g. freshly-isolated human hepatocytes) and a non-physiologically-relevant benchmark (e.g. HepG2 cells); in addition, often very few markers of the hepatic phenotype are used and studies do not always employ quantitatively-relevant assays (e.g mass spectrometry). Thus, inadequate benchmarking has hampered the field and there is likely significant value in identifying a common framework that might allow end users to readily interpret cell phenotype.
Table 1.
Summary of studies post-2007 of HLC-derivation from human pluripotent stem cells (adapted from Table 1 and Table 2, Kia et al(13) with modification). Note the limited number of Phase 1 and 2 phenotyping markers generally employed in the characterization of the HLCs.
| Reference | Method of stem cell differentiation | Differentiation efficiency % ALB +ve HLCs | PHENOTYPING: Phase I and II enzyme activity | ||||
|---|---|---|---|---|---|---|---|
| Stem cell (cell line) | Culture format | Differentiation factors | % ALB +ve HLCs (assay method) | Enzyme (assay method) | % hPH comparator | Other comparators | |
| Cai et al., 2007(14) | hESC (H1, H9) | Monolayer, EB formation | AF V, AA, ITS, BMP2, FGF4, HGF, OSM, DEX | 70 (ICC) | CYP2B6 (Fluorescence) | ND | hESC |
| Ek et al., 2007(15) | hESC (SA002, SA002.5, SA167) | Monolayer | Proprietary differentiation medium, FGF2 | ND | CYP1A1 (Fluorescence) | 0 | – |
| CYP3A4 (Fluorescence) | 0 | – | |||||
| Söderdahl et al., 2007(16) | hESC (SA001, SA002, SA002.5, AS034, SA121, and SA167) | Monolayer | Proprietary differentiation medium, bFGF | ND | GST (Fluorescence) | 80 | HepG2 |
| Hay et al., 2008(17); | hESC (H1, H9) | Monolayer | AA, Wnt3a | 90 (ICC) | CYP 1A2 (LC-MS-MS) | 24 | hESC |
| Godoy et al., 2015(11); | CYP1A2 (Luminescence) 100 | ||||||
| Cameron et al., 2015(10) | CYP3A4 (Luminescence) 100 | ||||||
| Shiraki et al., 2008(18) | hESC (Khes-1) | Co-culture with M15 cell line | AA, BMP4, bFGF, HGF, DMSO, DEX, Ly294002 | 9 (ICC) | ND | – | – |
| Agarwal et al.,2008(19) | hESC (WA01, WA09) | Monolayer | AA, FGF4, HGF, BSA, OSM, DEX | 67.4 (ICC) | ND | – | – |
| Moore et al., 2009(20) | hESC (H1) | Monolayer, EB formation | AA, Wnt3a, HGF, OSM, DEX | 72.8 (ICC) | CYP 1A2 (Fluorescence) | ND | hESC-derived HLCs in culture media of different components |
| Basma et al., 2009(21) | hESC (H1) | Monolayer, EB formation | AA, FGF2, HGF, DMSO, DEX | 55.5 (ICC) | CYP1A (Fluorescence) | 30 | – |
| CYP3A (LC-MS-MS) | 90 | – | |||||
| Song et al., 2009(22) | hESC (H1), hiPSC (hFb-derived 3U1, 3U2) | Monolayer | AF V, AA, ITS, BMP2, FGF4, OSM, DEX, KGF, B27 | 60 (ICC) | CYP2B6 (Fluorescence) | ND | hiPSC-derived versus hESC-derived HLCs |
| Duan et al., 2010(23) | hESC (H9) | Monolayer | AA, sodium butyrate, BMP2, BMP4, FGF4, HGF DMSO, B27 | 75-90 (ICC, FACS) | CYP1A2 (LC-MS-MS) | 100 | – |
| CYP2C9 (LC-MS-MS) | 60 | – | |||||
| CYP2D6 (LC-MS-MS) | 95 | – | |||||
| CYP3A4 (LC-MS-MS) | 90 | – | |||||
| Synnergren et al., 2010(24) | hESC (SA002, SA167, SA461) | Monolayer | AA, ITS, FGF1, FGF2, BMP2, BMP4, HGF, OSM, DEX | ND | ND | – | – |
| Touboul et al., 2010(25) | hESC (H9) | Monolayer | AA, BMP4,FGF2, FGF4, FGF10, HGF, EGF, retinoic acid, SB431542, Ly294002 | ND | CYP3A (Bioluminescence) | ND | – |
| Brolén et al., 2010(26) | hESC (SA001, SA002, SA002.5, SA167) | Monolayer | AA, BMP2, BMP4, FGF1, FGF2, HGF, OSM, DEX, Wnt3A | ND | CYP1A (LC-MS-MS) | ND | Spontaneously differentiated hESC-derived HLCs, HepG2 |
| CYP2C (LC-MS-MS) | |||||||
| CYP2A (LC-MS-MS) | |||||||
| Ghodsizadeh et al., 2010(27) | hiPSC (hFb-derived) | EB formation | AA, FGF2, HGF, DMSO, DEX | 50 (FACS) | CYP2B6 (Fluorescence) | ND | hiPSC |
| Liu et al., 2010(28) | hESC (WA01, WA09), hiPSC (hPH-derived) | Monolayer | AA, FGF4, HGF, OSM, DEX | ND | CYP1A2 (Bioluminescence) | ND | – |
| CYP3A4 (Bioluminescence) | |||||||
| Si-Tayeb et al., 2010(29) | hESC (H9), hiPSC (hFb-derived) | Monolayer | AA, BMP4, FGF2, OSM, B27 | 80 (FACS) | ND | – | – |
| Sullivan et al., 2010(30) | hiPSC (hFb-derived) | Monolayer | AA, HGF, Wnt3A, DMSO, OSM, hydrocortisone, tryptose phosphate broth, B27 | 70-90 (ICC) | CYP1A2 (Bioluminescence) | ND | – |
| CYP3A4 (Bioluminescence) | |||||||
| Rashid et al., 2010(31) | hiPSC (hFb-derived) | Monolayer | AA, BMP4, FGF2, HGF, OSM, Ly294002, CHIR99021 (GSK-3 inhibitor) | 83 (FACS) | CYP3A4 (Bioluminescence) | ND | hiPSC |
| Zhang et al., 2011(32) | hESC (H9), hiPSC (hFb-derived) | Monolayer, EB formation | AA, BMP2, FGF4, HGF, KGF, OSM, DEX | 60-80 (ICC, FACS) | CYP3A4 (Bioluminescence) | 0.32 | hESC-derived HLCs |
| Bone et al., 2011(33) | hESC (Shef1, Shef3) | Monolayer | FGF4, HGF, OSM, DEX, 1 m (GSK-3 inhibitor) | ND | ND | – | – |
| Yildirimman et al., 2011(34) | hESC (SA002) | Monolayer | Proprietary differentiation medium | ND | CYP1A2 (LC-MS-MS) | 50 | – |
| CYP3A4 (LC-MS-MS) | 50 | – | |||||
| CYP2B6 (LC-MS-MS) | 10 | – | |||||
| CYP2C9 (LC-MS-MS) | 50 | – | |||||
| CYP2C19 (LC-MS-MS) | 50 | – | |||||
| Chen et al., 2012(35) | hESC (H9), hiPSC (hFb-derived, CFB46) | Monolayer | AA, ITS, HGF, Wnt3A, OSM, DMSO, DEX | ND | CYP3A4 (Bioluminescence) | 100 | hiPSC |
| Cayo et al., 2012(36) | hiPSC (FH patient JD fibroblast-derived) | Monolayer | OCT4, SOX2, NANOG, LIN28 | ND | ND | – | – |
| Schwartz et al., 2012(37) | hiPSC (hFb-derived) | Monolayer | AA, BMP4, FGF2, HGF, OSM | 80 (ICC) | ND | – | – |
| Takayama et al., 2012(38) | hES (H9), hiPSC (hFb-derived, MCR5 & 201B7) | Monolayer | AA, SOX17, HEX, BMP4, FGF4, LacZ, HNF4α, HGF, OSM, DEX | ND | CYP3A4 (Fluorescence) | 100 | – |
| CYP2C9 (Fluorescence) | > 10 | – | |||||
| CYP1A2 (Fluorescence) | < 1 | – | |||||
| Choi et al., 2013(39) | hiPSC (derived from AAT deficient patients) | Monolayer | B27, AA, FGF4, HGF, OSM, DEX | ND | CYP3A4 (Bioluminescence) | 80 | – |
| CYP2D6 (Bioluminescence) | 70 | – | |||||
| CYP2C19 (Bioluminescence) | 90 | – | |||||
| CYP1A2 (Bioluminescence) | 90 | – | |||||
| Ramasamy et al., 2013(40) | hESC (H1) | Monolayer & 3D culture in Algimatrix plate | AA, DMSO, HGF, OSM | ND | CYP3A4 (Bioluminescence) | ND | HepG2 |
| Gieseck et al., 2014(41) | hiPSC (hFb-derived) | Monolayer, 3D-single cell or Clump culture in RAFT system | AA, FGF2,BMP4, LY-294002, Hepatozyme-SFM | ND | CYP3A4 for 2D Day 35 (HPLC-MS) | 4 | – |
| CYP3A4 for 3D Day 45 (Bioluminescence) | 25 | ||||||
| Jia et al., 2014(42) | hiPSC (from urine cells of HA patient) | Monolayer, EB formation | AA, FGF4, BMP2, HGF, KGF, OSM, DEX | 64 (FACS) | ND | – | – |
| Avior et al., 2015(43) | hESC (I3) | Monolayer | AA, B27, Wnt3A, HGF, DMSO, DEX, OSM, FGF2, LCA, MK4 | 83 (FACS) | CYP3A4, 1A2 (Fluorescence) | 30 | HepG2, hESC without LCA/MK4 |
| CYP2E1, 2C9 (Fluorescence) | 8 | ||||||
| Chien et al., 2015(44) | hiPSC (from dental pulp stromal cells) | Co-culture with MEF, EB formation | AF V, AA, FGF4, BMP2, HGF, KFG, OSM, DEX, B27, miR122 (delivered by PU-PEI in CHC) | ND | ND | – | – |
Abbreviations: AA, activin A; AF V, albumin fraction V; bFGF, human recombinant basic FGF; BMP, bone morphogenic protein; BSA, bovine serum albumin; CHC, carboxymethyl-hexanoyl chitosan; DEX, dexamethasone; DMSO, dimethyl sulfoxide; EGF, epidermal growth factor; FACS, fluorescence-activated cell sorting; FGF, fibroblast growth factor; GFP, green fluorescent protein; GSK, glycogen synthase kinase; Hepatozyme-SMF, hepatozyme serum free medium; HEX, hematopoietically-expressed homeobox protein; hESC, human embryonic stem cell; hiPSC, human induced pluripotent stem cell; HLCs, hepatocyte-like cells; HGF, hepatocyte growth factor; HNF4alpha, hepatocyte nuclear factor 4 alpha; ICC, immunocytochemistry; ITS, insulin-transferrin-selenium; KGF, keratinocyte growth factor; LacZ, beta-D-glactosidase; Ly294002, phosphoinositide 3-kinase inhibitor; miR122, microRNA 122; ND, not determined; OCT, octamer-binding transcription factor; OSM, oncostatin M; PU-PEI, biodegradable polyurethane-graft-short-branch polyethylenimine; qRT-PCR, quantitative real time polymerase chain reaction; SB431542, inhibitor for activin receptor-like kinase receptors ALK5, ALK4 and ALK7; SOX, sex determining region Y-box; Wnt3a, wingless-type MMTV integration site family, member 3a
Despite the challenges in generating mature hepatocytes, SC-HLCs have recently been shown to retain the cytochrome P450 (CYP) expression profile (specifically CYP2C9 and CYP2D6) of the donor hepatocyte(48, 49), yielding metabolism-specific toxicity for CYP2C9 (benzbromarone) and CYP2D6 (tamoxifen). This is highly relevant as the CYPs are key enzymes of Phase I drug metabolism, that play a key role in the chemical functionalization and eventual elimination of drugs from the body, but which also can yield significant intracellular concentrations of chemically reactive metabolites, leading to cellular and tissue damage of the liver, and therefore DILI (for a review of this area, see Park et al, 2011(50)).
The recent studies outlined above (48, 49) are particularly important as they suggest that modelling some forms of DILI (such as that elicited by benzbromarone or tamoxifen) using stem cell-derived hepatocytes may be possible, and that ultimately the challenges to generating a fully mature HLC will not always be insurmountable.
We consider that there are at least three major challenges to producing mature, physiologically- and pharmacologically-relevant hepatocytes from stem cells:
Stem cell-derived hepatocytes must mimic several years of development in vivo.
Like primary hepatocytes, the stem cell-derived hepatocyte phenotype is unstable currently in culture(51).
At the moment, it is difficult to emulate the complexity of the liver, with its unique blood supply and exposure to relevant concentrations of intestinal products and nutrients in vitro. Development of three-dimensional culture systems that employ co-cultivation of all cell types found in the liver acinus is likely to be required if we are to recapitulate the liver in vitro(51, 52). Following on from this, it is important to remember that a hepatocyte is not a single entity but varies functionally according to the hepatic zone in which it is located. The consequence of this is that some hepatotoxins induce hepatocellular damage in a zone-specific manner and this has not yet begun to be addressed meaningfully in the stem cell field, as we focus our attempts on improving basic functional maturity of the SC-derived cells, but it will need to be considered.
Despite these challenges, there are many promising leads in development, e.g. the discovery of several small molecule inducers of the hepatic phenotype(53), and the finding that microbial-derived secondary metabolites to which immature hepatocytes are likely to be exposed to post-partum may induce a significant increase in maturity. A further paradigm comes from the exploitation of SC-HLCs for demonstration of efficacy; specifically, for the reversal of the hepatic alpha1-antitrypsin-deficient phenotype, shown by Yusa et al(54). This study demonstrated restoration of alpha1-antitrypsin activity was possible on a “sufficiently” mature background, rather than one that was necessarily fully mature and identical to a freshly-isolated adult hepatocyte. Furthermore, a recent study by Ware et al(55) suggests that DILI detection is possible using SC-HLCs in micopatterned co-cultures, in which cells mature to significant levels. It is worth remembering that the hepatocyte exhibits more individual functions (>500) than any of the other ~200 terminally differentiated cell types in the human body. Therefore it is perhaps not surprising that this cell is amongst the most challenging to mature, and we should still continue to explore the utility of hepatocyte-like cells as prototypes rather than await the final “product”.
Lessons learned from the use of stem cell-derived cardiomyocytes in detecting cardiotoxicity
A parallel example, from which lessons can be learned, comes from the use of stem cells in the assessment of drug-induced cardiotoxicity – a primary cause of drug attrition. Cardiotoxicity, specifically QT prolongation, has already been successfully modelled using such cells(56–58). In comparison, there is only very recent evidence that SC-HLCs are able to recapitulate hepatotoxic events(49, 55). The difference between successful application of cardiac models compared with hepatic models may reflect the relative specificity of some forms of drug-induced cardiotoxicity, in contrast with the rather pleiotropic and diverse manifestations of hepatotoxicity, at the molecular, cellular, and tissular level(59). Cardiotoxicity often arises due to drug-induced electrical perturbation of the cell interfering with its contractile function(60). Here, the stem cell-cardiomyocyte model provides advantages over recombinant tumour models. Thus, the impact of drugs that cause simple single ion channel or complex multi-channel perturbation can be related to cardiomyocyte arrhythmias and abnormalities in contractility(61). In hepatotoxicity, however, there are myriad factors required to recapitulate toxicity, especially idiosyncratic toxicity where the immune system is also implicated. This is compounded by inter-individual variation in expression of xenobiotic metabolism and transporter proteins in addition to the chemistry of each drug.
Whilst protocols to differentiate stem cells towards cardiomyocytes generate cells that are not fully mature(61), these cells can recapitulate some facets of the cell phenotype required to produce specific forms of cardiotoxicity. This has prompted major international efforts to search for methods to further mature stem cell cardiomyocytes. Each incremental improvement made towards progressing the compliment of ion channels, regulatory pathways and structural proteins to the complete sets found in adult cells will dramatically increase the utility of stem cell cardiomyocytes. The demonstration that specific toxicological phenotypes can be mimicked by stem cell-derived cardiomyocytes allows the cell model to be considered “fit-for-purpose”. This raises the notion of using stem cell-derived hepatocytes that may be sufficiently mature for a specific toxicological assessment even though the cells may lack the full hepatic functionality with respect to drug metabolism, transporter expression etc. For example, where one or two cytochrome P450s (P450s), some relevant phase II enzymes, such as the glutathione transferases and UDP-glucuronyl transferases, and some Phase III proteins (influx and efflux transporters) are expressed at a set and reproducible % of a “typical” human hepatocyte, this cell may in some cases represent a significant and useful model in understanding specifically drug metabolism and possible metabolism-dependent toxicity.
The Importance of Phenotypic Characterisation
For the field to continue to move forward and develop liver cell models that are useful in prediction and mechanistic understanding of DILI, it is essential that the SC-HLCs are properly benchmarked against currently used and relevant human cells, especially fresh primary human hepatocytes and HepG2 cells (see Table 1 and Figure 1). Moreover, the phenotype of the HLCs must be as reproducible as possible, and they should be fully characterised, particularly with reference to the pharmacological phenotype (using a defined panel of training compounds). It is also important that the cell model can provide a static point of reference that can be used to ascertain if real progress is being made. When assessing novel models of hepatotoxicity it is important to use functional assays employing quantitative mass spectrometry whenever possible, as this is now being routinely employed (48, 62, 63)in order to determine the true phenotype of the model. A global proteomic analysis however may be the most appropriate way to characterise the cells, as this would represent a broad visualisation of the physiological phenotype of the cells. Similarity to freshly-isolated hepatocytes/tissue can be established through proteomics and targeted multiple- reaction-monitoring (MRM)-based mass spectrometric analysis of key proteins, such as CYP450s, transporters and intracellular signalling molecules and metabolic and cellular uptake profiles determined. Developments in mass spectrometric technologies mean that it is now possible to analyse small panels of proteins (for example 10-20 transporters or P450s) using MRM, in order to quantify proteins per cell at an absolute level(64). This would ensure valid comparisons between currently used models and cells, as well as cells that are developed in the future. Given the inherent deficiencies in a transcriptomic-only approach, which are well-illustrated in a recent landmark paper reporting only a 39% correlation between mRNA and protein at a global level(65), measuring mRNA levels is not recommended for cell characterisation purposes.
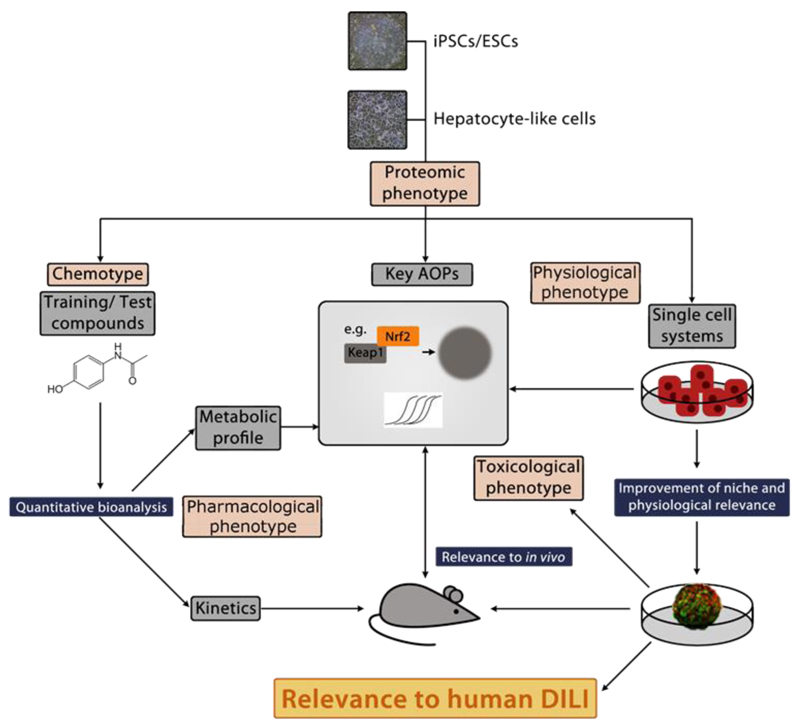
Figure 1. Roadmap for producing stem cell-derived models to improve mechanistic understanding and prediction of human DILI.
The physiological, pharmacological and toxicological characterisation of stem cell-derived hepatocytes is necessary before the cells can be fully utilised. This will include the use of toxicity/stress reporters, and a small panel of well-defined chemicals, thereby defining the toxicological purpose for which each line is suitable. This will position the new cells within a screening toolbox that could be validated for drug/chemical safety evaluation. The use of iPSC lines with drug toxicity-relevant mutations and the use of CRISPR technology to edit genes involved in drug metabolism may also be important in this regard.
Abbreviation: AOP = Adverse Outcome Pathway
As part of a comprehensive assessment of HLC phenotype, recent developments in the field of hepatocyte-selective translatable biomarkers (e.g. miR122(66)) might allow us to translate the response to chemicals between humans, model organisms and cells including SC-HLCs and it is likely that additional novel and selective biomarkers will be identified in the future using models such as SC-HLCs. This is an important area for industry which requires selective and translatable biomarkers of liver injury to monitor potentially hepatotoxic compounds in the clinic.
The recently developed concepts of adverse outcome pathways and points of departure(67) in the field of systems toxicology should also be considered in the context of phenotyping the response to chemical exposure of hepatocyte-like cells that express relevant proteins and pathways. To this end, cells expressing genetic reporters for key adaptive pathways such as Nrf2, PXR andNF-κB will be useful as a means for understanding the earliest events in the biological response to a drug(68–70). However, it is imperative that we develop ways to bridge our findings from these molecular investigations to what actually occurs in DILI in humans –the development of novel bridging biomarkers that allow extrapolation from in vitro test system to man will be invaluable in this endeavour. Another important development in relation to hepatocyte genotype and phenotype in DILI is the derivation of SC-HLCs with specific polymorphisms relevant to drug toxicology. Of particular interest in this regard is the developing use of CRISPR technology in SC-HLCs to edit, for example, genes relevant to drug metabolism and toxicity thereby providing a wild type cell and an almost identical cell with an alteration in drug metabolism and toxicological responses, respectively.
Finally, phenotypic characterisation may be assisted by a better understanding of the mechanisms contributing to de-differentiation or loss of phenotype. Consideration of the cellular complexity of the liver and the functional sophistication of a hepatocyte makes it unsurprising that the maintenance of a fully functional hepatocyte in culture is difficult to achieve(71). The cells have been removed from their neighbouring hepatocytes, disrupting their gap junctions and tight junctions which are important for their phenotype, as well as their juxtaposed non-parenchymal cells, which may also be responsible for the differentiated hepatocyte phenotype(72, 73). Dedifferentiation is not a unique process to the liver; when cardiomyocytes are cultured, they also lose some of their in vivo phenotype, e.g. the t-tubules are lost, glycogen is accumulated and chromatin becomes dispersed in vitro(74). However, the key difference between hepatocytes and myocytes is the importance of the metabolic phenotype with respect to drug toxicity, and it is this function – particularly the phase I CYP450 capacity – that is most rapidly and profoundly depleted(71, 75) - and it is also this function, at a defined proportion of the activity present in human liver, that is essential in any in vitro model of a hepatocyte
One area of research that could have a significant impact on attempts to re-establish a functional hepatocyte from stem cells, is the investigation of the precise cellular mechanisms underlying the de-differentiation process that occurs in hepatocytes once they have been removed from the liver. Whilst the factors driving de-differentiation may not be identical to those that drive differentiation, it is likely that one or more pathways and processes uncovered through research into de-differentiation will be amenable for testing in differentiation experiments. If it is not understood how to maintain the dynamic and sophisticated machinery of a fully mature hepatocyte in vitro, it is likely to be difficult to capture the same phenotype in a stem cell-derived cell grown under similar conditions.
Summary and recommendations
DILI is a complex, multi-dimensional disease, with variable phenotype between individuals, even for a single drug. There is essentially no ideal in vitro or in vivo model that recapitulates all of the potential features of this injury.
The aspiration of the field is a “perfect” mature hepatocyte as it exists in a liver - this has not yet been achieved. Until it is, hepatocyte-like cells with known, quantifiable and reproducible proportions of the function of two widely-used standards, i.e. primary fresh human hepatocytes, and HepG2, will be valuable biological models to explore the physiological, pharmacological and toxicological response of hepatocytes to drug exposure.
These “immature” cells should be explored as models of chemical perturbation using genetic reporters and biomarkers, with continual effort to relate findings to human DILI.
Global proteomic analysis aligned with biological pathway analysis may be the most appropriate way to characterise HLCs – a small targeted panel of proteins will also help to compare cells for key proteins and functions using absolute quantitation by mass spectrometry. Crucially, this will advance the field by avoiding over-reliance on a small panel of liver proteins, such as albumin, that may not be representative of a fully mature and functioning liver cell.
It is likely that niche creation in vitro, deploying enhanced matrices(13) and even 3D bioprinting(76) , and incorporating other cell types such as endothelial cells(76, 77) and Kupffer cells(78) inter alia, will mature and support hepatocyte function.
A small panel of chemical benchmarks will be needed to probe the physiological, pharmacological and toxicological function of the cells, only once they have been properly phenotyped. There is little point in exposing HLCs to chemicals chosen as hepatotoxins in man unless we fully characterise the cells.
Acknowledgments
Financial Support :
CG1, DJA1, NK1, AN1, CP1, RSY1, FZ1 and BKP1 are supported by grants from the Medical Research Council (grant number MR/L006758/1) and the European Community under the Innovative Medicine Initiative project MIP-DILI [grant agreement number 115336].
FB 2 is supported by Stem Cells for Safer Medicines (SC4SM).
CD 4 is supported by EPSRC, BHF, Heart Research UK, Medical Research Council and National Centre for the Replacement, Refinement and Reduction of Animals in Research (NC3Rs).
RJF 5 is a member of the Drug Induced Liver Injury Network (DILIN) a U01 cooperative agreement supported by the National Institute of Diabetes and Digestive and Kidney Diseases (U01DK065184).
NAH 6 is supported by Wellcome Trust in addition to Stem Cells for Safer Medicines (SC4SM).
JR 7 is supported by the Innovative Medicines Initiative Joint Undertaking under grant agreement n° 115439, resources of which are composed of financial contribution from the European Union's Seventh Framework Programme (FP7/2007-2013) and EFPIA companies in kind contribution. This publication reflects only the author’s views and neither the IMI JU nor EFPIA nor the European Commission are liable for any use that may be made of the information contained therein.
MIS 8 is supported by grants from The Swedish Research Council and from the European Community under the Innovative Medicine Initiative project MIP-DILI [grant agreement number 115336].
BvdW 13 is supported by the EC FP7 project DETECTIVE (grant agreement n° 266838), the IMI MIP-DILI project (grant agreement n° 115336), and the Horizon2020 EU-ToxRisk project (grant agreement n° 681002).
List of Abbreviations
- DILI
drug-induced liver injury
- ADR
adverse drug reaction
- SC-HLC
stem-cell-derived hepatocyte-like cell
- EPAA
European Partnership for Alternative Approaches to Animal Testing
- NC3Rs
National Centre for the Replacement, Refinement and Reduction of Animals in Research
- MRM
multiple-reaction-monitoring
- CRISPR
clustered regularly-interspaced short palindromic repeats
- iPSC
induced pluripotent stem cell
Bibliography
- 1.Pirmohamed M, James S, Meakin S, Green C, Scott AK, Walley TJ, Farrar K, et al. Adverse drug reactions as cause of admission to hospital: prospective analysis of 18 820 patients. BMJ. 2004;329:15–19. doi: 10.1136/bmj.329.7456.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Waring MJ, Arrowsmith J, Leach AR, Leeson PD, Mandrell S, Owen RM, Pairaudeau G, et al. An analysis of the attrition of drug candidates from four major pharmaceutical companies. Nat Rev Drug Discov. 2015;14:475–486. doi: 10.1038/nrd4609. [DOI] [PubMed] [Google Scholar]
- 3.Ostapowicz G, Fontana RJ, Schiodt FV, Larson A, Davern TJ, Han SH, McCashland TM, et al. Results of a prospective study of acute liver failure at 17 tertiary care centers in the United States. Ann Intern Med. 2002;137:947–954. doi: 10.7326/0003-4819-137-12-200212170-00007. [DOI] [PubMed] [Google Scholar]
- 4.Spanhaak S, Cook D, Barnes J, Reynolds J. Species Concordance for Liver Injury. BioWisdom Report. 2008 http://bioblog.instem.com/downloads/SIP_Board_Species_Concordance.pdf.
- 5.Verma S, Kaplowitz N. Diagnosis, management and prevention of drug-induced liver injury. Gut. 2009;58:1555–1564. doi: 10.1136/gut.2008.163675. [DOI] [PubMed] [Google Scholar]
- 6.Tolosa L, Pinto S, Donato MT, Lahoz A, Castell JV, O'Connor JE, Gomez-Lechon MJ. Development of a multiparametric cell-based protocol to screen and classify the hepatotoxicity potential of drugs. Toxicol Sci. 2012;127:187–198. doi: 10.1093/toxsci/kfs083. [DOI] [PubMed] [Google Scholar]
- 7.Xu JJ, Henstock PV, Dunn MC, Smith AR, Chabot JR, de Graaf D. Cellular imaging predictions of clinical drug-induced liver injury. Toxicol Sci. 2008;105:97–105. doi: 10.1093/toxsci/kfn109. [DOI] [PubMed] [Google Scholar]
- 8.Tolosa L, Gomez-Lechon MJ, Lopez S, Guzman C, Castell JV, Donato MT, Jover R. Human Upcyte Hepatocytes: Characterization of the Hepatic Phenotype and Evaluation for Acute and Long-Term Hepatotoxicity Routine Testing. Toxicol Sci. 2016 doi: 10.1093/toxsci/kfw078. [DOI] [PubMed] [Google Scholar]
- 9.Khetani SR, Kanchagar C, Ukairo O, Krzyzewski S, Moore A, Shi J, Aoyama S, et al. Use of micropatterned cocultures to detect compounds that cause drug-induced liver injury in humans. Toxicol Sci. 2013;132:107–117. doi: 10.1093/toxsci/kfs326. [DOI] [PubMed] [Google Scholar]
- 10.Gerets HH, Tilmant K, Gerin B, Chanteux H, Depelchin BO, Dhalluin S, Atienzar FA. Characterization of primary human hepatocytes, HepG2 cells, and HepaRG cells at the mRNA level and CYP activity in response to inducers and their predictivity for the detection of human hepatotoxins. Cell Biol Toxicol. 2012;28:69–87. doi: 10.1007/s10565-011-9208-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guillouzo A, Corlu A, Aninat C, Glaise D, Morel F, Guguen-Guillouzo C. The human hepatoma HepaRG cells: a highly differentiated model for studies of liver metabolism and toxicity of xenobiotics. Chem Biol Interact. 2007;168:66–73. doi: 10.1016/j.cbi.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 12.Baxter M, Withey S, Harrison S, Segeritz CP, Zhang F, Atkinson-Dell R, Rowe C, et al. Phenotypic and functional analyses show stem cell-derived hepatocyte-like cells better mimic fetal rather than adult hepatocytes. J Hepatol. 2015;62:581–589. doi: 10.1016/j.jhep.2014.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cameron K, Tan R, Schmidt-Heck W, Campos G, Lyall MJ, Wang Y, Lucendo-Villarin B, et al. Recombinant Laminins Drive the Differentiation and Self-Organization of hESC-Derived Hepatocytes. Stem Cell Reports. 2015;5:1250–1262. doi: 10.1016/j.stemcr.2015.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Godoy P, Schmidt-Heck W, Natarajan K, Lucendo-Villarin B, Szkolnicka D, Asplund A, Bjorquist P, et al. Gene networks and transcription factor motifs defining the differentiation of stem cells into hepatocyte-like cells. J Hepatol. 2015;63:934–942. doi: 10.1016/j.jhep.2015.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu J, Brzeszczynska J, Samuel K, Black J, Palakkan A, Anderson RA, Gallagher R, et al. Efficient episomal reprogramming of blood mononuclear cells and differentiation to hepatocytes with functional drug metabolism. Exp Cell Res. 2015;338:203–213. doi: 10.1016/j.yexcr.2015.08.004. [DOI] [PubMed] [Google Scholar]
- 16.Kia R, Sison RL, Heslop J, Kitteringham NR, Hanley N, Mills JS, Park BK, et al. Stem cell-derived hepatocytes as a predictive model for drug-induced liver injury: are we there yet? Br J Clin Pharmacol. 2013;75:885–896. doi: 10.1111/j.1365-2125.2012.04360.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cai J, Zhao Y, Liu Y, Ye F, Song Z, Qin H, Meng S, et al. Directed differentiation of human embryonic stem cells into functional hepatic cells. Hepatology. 2007;45:1229–1239. doi: 10.1002/hep.21582. [DOI] [PubMed] [Google Scholar]
- 18.Ek M, Soderdahl T, Kuppers-Munther B, Edsbagge J, Andersson TB, Bjorquist P, Cotgreave I, et al. Expression of drug metabolizing enzymes in hepatocyte-like cells derived from human embryonic stem cells. Biochem Pharmacol. 2007;74:496–503. doi: 10.1016/j.bcp.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 19.Soderdahl T, Kuppers-Munther B, Heins N, Edsbagge J, Bjorquist P, Cotgreave I, Jernstrom B. Glutathione transferases in hepatocyte-like cells derived from human embryonic stem cells. Toxicol In Vitro. 2007;21:929–937. doi: 10.1016/j.tiv.2007.01.021. [DOI] [PubMed] [Google Scholar]
- 20.Hay DC, Fletcher J, Payne C, Terrace JD, Gallagher RC, Snoeys J, Black JR, et al. Highly efficient differentiation of hESCs to functional hepatic endoderm requires ActivinA and Wnt3a signaling. Proc Natl Acad Sci U S A. 2008;105:12301–12306. doi: 10.1073/pnas.0806522105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shiraki N, Umeda K, Sakashita N, Takeya M, Kume K, Kume S. Differentiation of mouse and human embryonic stem cells into hepatic lineages. Genes Cells. 2008;13:731–746. doi: 10.1111/j.1365-2443.2008.01201.x. [DOI] [PubMed] [Google Scholar]
- 22.Agarwal S, Holton KL, Lanza R. Efficient differentiation of functional hepatocytes from human embryonic stem cells. Stem Cells. 2008;26:1117–1127. doi: 10.1634/stemcells.2007-1102. [DOI] [PubMed] [Google Scholar]
- 23.Moore RN, Moghe PV. Expedited growth factor-mediated specification of human embryonic stem cells toward the hepatic lineage. Stem Cell Res. 2009;3:51–62. doi: 10.1016/j.scr.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 24.Basma H, Soto-Gutierrez A, Yannam GR, Liu L, Ito R, Yamamoto T, Ellis E, et al. Differentiation and transplantation of human embryonic stem cell-derived hepatocytes. Gastroenterology. 2009;136:990–999. doi: 10.1053/j.gastro.2008.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Song Z, Cai J, Liu Y, Zhao D, Yong J, Duo S, Song X, et al. Efficient generation of hepatocyte-like cells from human induced pluripotent stem cells. Cell Res. 2009;19:1233–1242. doi: 10.1038/cr.2009.107. [DOI] [PubMed] [Google Scholar]
- 26.Duan Y, Ma X, Zou W, Wang C, Bahbahan IS, Ahuja TP, Tolstikov V, et al. Differentiation and characterization of metabolically functioning hepatocytes from human embryonic stem cells. Stem Cells. 2010;28:674–686. doi: 10.1002/stem.315. [DOI] [PubMed] [Google Scholar]
- 27.Synnergren J, Heins N, Brolen G, Eriksson G, Lindahl A, Hyllner J, Olsson B, et al. Transcriptional profiling of human embryonic stem cells differentiating to definitive and primitive endoderm and further toward the hepatic lineage. Stem Cells Dev. 2010;19:961–978. doi: 10.1089/scd.2009.0220. [DOI] [PubMed] [Google Scholar]
- 28.Touboul T, Hannan NR, Corbineau S, Martinez A, Martinet C, Branchereau S, Mainot S, et al. Generation of functional hepatocytes from human embryonic stem cells under chemically defined conditions that recapitulate liver development. Hepatology. 2010;51:1754–1765. doi: 10.1002/hep.23506. [DOI] [PubMed] [Google Scholar]
- 29.Brolen G, Sivertsson L, Bjorquist P, Eriksson G, Ek M, Semb H, Johansson I, et al. Hepatocyte-like cells derived from human embryonic stem cells specifically via definitive endoderm and a progenitor stage. J Biotechnol. 2010;145:284–294. doi: 10.1016/j.jbiotec.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 30.Ghodsizadeh A, Taei A, Totonchi M, Seifinejad A, Gourabi H, Pournasr B, Aghdami N, et al. Generation of liver disease-specific induced pluripotent stem cells along with efficient differentiation to functional hepatocyte-like cells. Stem Cell Rev. 2010;6:622–632. doi: 10.1007/s12015-010-9189-3. [DOI] [PubMed] [Google Scholar]
- 31.Liu H, Ye Z, Kim Y, Sharkis S, Jang YY. Generation of endoderm-derived human induced pluripotent stem cells from primary hepatocytes. Hepatology. 2010;51:1810–1819. doi: 10.1002/hep.23626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Si-Tayeb K, Noto FK, Nagaoka M, Li J, Battle MA, Duris C, North PE, et al. Highly efficient generation of human hepatocyte-like cells from induced pluripotent stem cells. Hepatology. 2010;51:297–305. doi: 10.1002/hep.23354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sullivan GJ, Hay DC, Park IH, Fletcher J, Hannoun Z, Payne CM, Dalgetty D, et al. Generation of functional human hepatic endoderm from human induced pluripotent stem cells. Hepatology. 2010;51:329–335. doi: 10.1002/hep.23335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rashid ST, Corbineau S, Hannan N, Marciniak SJ, Miranda E, Alexander G, Huang-Doran I, et al. Modeling inherited metabolic disorders of the liver using human induced pluripotent stem cells. J Clin Invest. 2010;120:3127–3136. doi: 10.1172/JCI43122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang S, Chen S, Li W, Guo X, Zhao P, Xu J, Chen Y, et al. Rescue of ATP7B function in hepatocyte-like cells from Wilson's disease induced pluripotent stem cells using gene therapy or the chaperone drug curcumin. Hum Mol Genet. 2011;20:3176–3187. doi: 10.1093/hmg/ddr223. [DOI] [PubMed] [Google Scholar]
- 36.Bone HK, Nelson AS, Goldring CE, Tosh D, Welham MJ. A novel chemically directed route for the generation of definitive endoderm from human embryonic stem cells based on inhibition of GSK-3. J Cell Sci. 2011;124:1992–2000. doi: 10.1242/jcs.081679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yildirimman R, Brolen G, Vilardell M, Eriksson G, Synnergren J, Gmuender H, Kamburov A, et al. Human embryonic stem cell derived hepatocyte-like cells as a tool for in vitro hazard assessment of chemical carcinogenicity. Toxicol Sci. 2011;124:278–290. doi: 10.1093/toxsci/kfr225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen YF, Tseng CY, Wang HW, Kuo HC, Yang VW, Lee OK. Rapid generation of mature hepatocyte-like cells from human induced pluripotent stem cells by an efficient three-step protocol. Hepatology. 2012;55:1193–1203. doi: 10.1002/hep.24790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cayo MA, Cai J, DeLaForest A, Noto FK, Nagaoka M, Clark BS, Collery RF, et al. JD induced pluripotent stem cell-derived hepatocytes faithfully recapitulate the pathophysiology of familial hypercholesterolemia. Hepatology. 2012;56:2163–2171. doi: 10.1002/hep.25871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schwartz RE, Trehan K, Andrus L, Sheahan TP, Ploss A, Duncan SA, Rice CM, et al. Modeling hepatitis C virus infection using human induced pluripotent stem cells. Proc Natl Acad Sci U S A. 2012;109:2544–2548. doi: 10.1073/pnas.1121400109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takayama K, Inamura M, Kawabata K, Katayama K, Higuchi M, Tashiro K, Nonaka A, et al. Efficient generation of functional hepatocytes from human embryonic stem cells and induced pluripotent stem cells by HNF4alpha transduction. Mol Ther. 2012;20:127–137. doi: 10.1038/mt.2011.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Choi SM, Kim Y, Shim JS, Park JT, Wang RH, Leach SD, Liu JO, et al. Efficient drug screening and gene correction for treating liver disease using patient-specific stem cells. Hepatology. 2013;57:2458–2468. doi: 10.1002/hep.26237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ramasamy TS, Yu JS, Selden C, Hodgson H, Cui W. Application of three-dimensional culture conditions to human embryonic stem cell-derived definitive endoderm cells enhances hepatocyte differentiation and functionality. Tissue Eng Part A. 2013;19:360–367. doi: 10.1089/ten.tea.2012.0190. [DOI] [PubMed] [Google Scholar]
- 44.Gieseck RL, 3rd, Hannan NR, Bort R, Hanley NA, Drake RA, Cameron GW, Wynn TA, et al. Maturation of induced pluripotent stem cell derived hepatocytes by 3D-culture. PLoS One. 2014;9:e86372. doi: 10.1371/journal.pone.0086372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jia B, Chen S, Zhao Z, Liu P, Cai J, Qin D, Du J, et al. Modeling of hemophilia A using patient-specific induced pluripotent stem cells derived from urine cells. Life Sci. 2014;108:22–29. doi: 10.1016/j.lfs.2014.05.004. [DOI] [PubMed] [Google Scholar]
- 46.Avior Y, Levy G, Zimerman M, Kitsberg D, Schwartz R, Sadeh R, Moussaieff A, et al. Microbial-derived lithocholic acid and vitamin K2 drive the metabolic maturation of pluripotent stem cells-derived and fetal hepatocytes. Hepatology. 2015;62:265–278. doi: 10.1002/hep.27803. [DOI] [PubMed] [Google Scholar]
- 47.Chien Y, Chang YL, Li HY, Larsson M, Wu WW, Chien CS, Wang CY, et al. Synergistic effects of carboxymethyl-hexanoyl chitosan, cationic polyurethane-short branch PEI in miR122 gene delivery: accelerated differentiation of iPSCs into mature hepatocyte-like cells and improved stem cell therapy in a hepatic failure model. Acta Biomater. 2015;13:228–244. doi: 10.1016/j.actbio.2014.11.018. [DOI] [PubMed] [Google Scholar]
- 48.Ulvestad M, Nordell P, Asplund A, Rehnstrom M, Jacobsson S, Holmgren G, Davidson L, et al. Drug metabolizing enzyme and transporter protein profiles of hepatocytes derived from human embryonic and induced pluripotent stem cells. Biochem Pharmacol. 2013;86:691–702. doi: 10.1016/j.bcp.2013.06.029. [DOI] [PubMed] [Google Scholar]
- 49.Takayama K, Morisaki Y, Kuno S, Nagamoto Y, Harada K, Furukawa N, Ohtaka M, et al. Prediction of interindividual differences in hepatic functions and drug sensitivity by using human iPS-derived hepatocytes. Proc Natl Acad Sci U S A. 2014;111:16772–16777. doi: 10.1073/pnas.1413481111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Park BK, Boobis A, Clarke S, Goldring CE, Jones D, Kenna JG, Lambert C, et al. Managing the challenge of chemically reactive metabolites in drug development. Nat Rev Drug Discov. 2011;10:292–306. doi: 10.1038/nrd3408. [DOI] [PubMed] [Google Scholar]
- 51.Berger DR, Ware BR, Davidson MD, Allsup SR, Khetani SR. Enhancing the functional maturity of induced pluripotent stem cell-derived human hepatocytes by controlled presentation of cell-cell interactions in vitro. Hepatology. 2015;61:1370–1381. doi: 10.1002/hep.27621. [DOI] [PubMed] [Google Scholar]
- 52.Davidson MD, Ware BR, Khetani SR. Stem cell-derived liver cells for drug testing and disease modeling. Discov Med. 2015;19:349–358. [PMC free article] [PubMed] [Google Scholar]
- 53.Shan J, Schwartz RE, Ross NT, Logan DJ, Thomas D, Duncan SA, North TE, et al. Identification of small molecules for human hepatocyte expansion and iPS differentiation. Nat Chem Biol. 2013;9:514–520. doi: 10.1038/nchembio.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yusa K, Rashid ST, Strick-Marchand H, Varela I, Liu PQ, Paschon DE, Miranda E, et al. Targeted gene correction of alpha1-antitrypsin deficiency in induced pluripotent stem cells. Nature. 2011;478:391–394. doi: 10.1038/nature10424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ware BR, Berger DR, Khetani SR. Prediction of Drug-Induced Liver Injury in Micropatterned Co-cultures Containing iPSC-Derived Human Hepatocytes. Toxicol Sci. 2015;145:252–262. doi: 10.1093/toxsci/kfv048. [DOI] [PubMed] [Google Scholar]
- 56.Lahti AL, Kujala VJ, Chapman H, Koivisto AP, Pekkanen-Mattila M, Kerkela E, Hyttinen J, et al. Model for long QT syndrome type 2 using human iPS cells demonstrates arrhythmogenic characteristics in cell culture. Dis Model Mech. 2012;5:220–230. doi: 10.1242/dmm.008409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Matsa E, Rajamohan D, Dick E, Young L, Mellor I, Staniforth A, Denning C. Drug evaluation in cardiomyocytes derived from human induced pluripotent stem cells carrying a long QT syndrome type 2 mutation. Eur Heart J. 2011;32:952–962. doi: 10.1093/eurheartj/ehr073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Itzhaki I, Maizels L, Huber I, Gepstein A, Arbel G, Caspi O, Miller L, et al. Modeling of catecholaminergic polymorphic ventricular tachycardia with patient-specific human-induced pluripotent stem cells. J Am Coll Cardiol. 2012;60:990–1000. doi: 10.1016/j.jacc.2012.02.066. [DOI] [PubMed] [Google Scholar]
- 59.Kaplowitz N. Idiosyncratic drug hepatotoxicity. Nat Rev Drug Discov. 2005;4:489–499. doi: 10.1038/nrd1750. [DOI] [PubMed] [Google Scholar]
- 60.Force T, Kolaja KL. Cardiotoxicity of kinase inhibitors: the prediction and translation of preclinical models to clinical outcomes. Nat Rev Drug Discov. 2011;10:111–126. doi: 10.1038/nrd3252. [DOI] [PubMed] [Google Scholar]
- 61.Denning C, Borgdorff V, Crutchley J, Firth KS, George V, Kalra S, Kondrashov A, et al. Cardiomyocytes from human pluripotent stem cells: From laboratory curiosity to industrial biomedical platform. Biochim Biophys Acta. 2015 doi: 10.1016/j.bbamcr.2015.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sengupta S, Johnson BP, Swanson SA, Stewart R, Bradfield CA, Thomson JA. Aggregate culture of human embryonic stem cell-derived hepatocytes in suspension are an improved in vitro model for drug metabolism and toxicity testing. Toxicol Sci. 2014;140:236–245. doi: 10.1093/toxsci/kfu069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ma X, Duan Y, Tschudy-Seney B, Roll G, Behbahan IS, Ahuja TP, Tolstikov V, et al. Highly efficient differentiation of functional hepatocytes from human induced pluripotent stem cells. Stem Cells Transl Med. 2013;2:409–419. doi: 10.5966/sctm.2012-0160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kitteringham NR, Jenkins RE, Lane CS, Elliott VL, Park BK. Multiple reaction monitoring for quantitative biomarker analysis in proteomics and metabolomics. J Chromatogr B Analyt Technol Biomed Life Sci. 2009;877:1229–1239. doi: 10.1016/j.jchromb.2008.11.013. [DOI] [PubMed] [Google Scholar]
- 65.Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, et al. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 66.Starkey Lewis PJ, Dear J, Platt V, Simpson KJ, Craig DG, Antoine DJ, French NS, et al. Circulating microRNAs as potential markers of human drug-induced liver injury. Hepatology. 2011;54:1767–1776. doi: 10.1002/hep.24538. [DOI] [PubMed] [Google Scholar]
- 67.Willett C, Caverly Rae J, Goyak KO, Minsavage G, Westmoreland C, Andersen M, Avigan M, et al. Building shared experience to advance practical application of pathway-based toxicology: liver toxicity mode-of-action. ALTEX. 2014;31:500–519. doi: 10.14573/altex.1401281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Herpers B, Wink S, Fredriksson L, Di Z, Hendriks G, Vrieling H, de Bont H, et al. Activation of the Nrf2 response by intrinsic hepatotoxic drugs correlates with suppression of NF-kappaB activation and sensitizes toward TNFalpha-induced cytotoxicity. Arch Toxicol. 2015 doi: 10.1007/s00204-015-1536-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fredriksson L, Wink S, Herpers B, Benedetti G, Hadi M, de Bont H, Groothuis G, et al. Drug-induced endoplasmic reticulum and oxidative stress responses independently sensitize toward TNFalpha-mediated hepatotoxicity. Toxicol Sci. 2014;140:144–159. doi: 10.1093/toxsci/kfu072. [DOI] [PubMed] [Google Scholar]
- 70.Wink S, Hiemstra S, Huppelschoten S, Danen E, Niemeijer M, Hendriks G, Vrieling H, et al. Quantitative high content imaging of cellular adaptive stress response pathways in toxicity for chemical safety assessment. Chem Res Toxicol. 2014;27:338–355. doi: 10.1021/tx4004038. [DOI] [PubMed] [Google Scholar]
- 71.Elaut G, Henkens T, Papeleu P, Snykers S, Vinken M, Vanhaecke T, Rogiers V. Molecular mechanisms underlying the dedifferentiation process of isolated hepatocytes and their cultures. Curr Drug Metab. 2006;7:629–660. doi: 10.2174/138920006778017759. [DOI] [PubMed] [Google Scholar]
- 72.Bhatia SN, Balis UJ, Yarmush ML, Toner M. Probing heterotypic cell interactions: hepatocyte function in microfabricated co-cultures. J Biomater Sci Polym Ed. 1998;9:1137–1160. doi: 10.1163/156856298x00695. [DOI] [PubMed] [Google Scholar]
- 73.Zinchenko YS, Schrum LW, Clemens M, Coger RN. Hepatocyte and kupffer cells co-cultured on micropatterned surfaces to optimize hepatocyte function. Tissue Eng. 2006;12:751–761. doi: 10.1089/ten.2006.12.751. [DOI] [PubMed] [Google Scholar]
- 74.Ausma J, Borgers M. Dedifferentiation of atrial cardiomyocytes: from in vivo to in vitro. Cardiovasc Res. 2002;55:9–12. doi: 10.1016/s0008-6363(02)00434-0. [DOI] [PubMed] [Google Scholar]
- 75.Rowe C, Gerrard DT, Jenkins R, Berry A, Durkin K, Sundstrom L, Goldring CE, et al. Proteome-wide analyses of human hepatocytes during differentiation and dedifferentiation. Hepatology. 2013;58:799–809. doi: 10.1002/hep.26414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ma X, Qu X, Zhu W, Li YS, Yuan S, Zhang H, Liu J, et al. Deterministically patterned biomimetic human iPSC-derived hepatic model via rapid 3D bioprinting. Proc Natl Acad Sci U S A. 2016 doi: 10.1073/pnas.1524510113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Takebe T, Sekine K, Enomura M, Koike H, Kimura M, Ogaeri T, Zhang RR, et al. Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature. 2013;499:481–484. doi: 10.1038/nature12271. [DOI] [PubMed] [Google Scholar]
- 78.Nguyen TV, Ukairo O, Khetani SR, McVay M, Kanchagar C, Seghezzi W, Ayanoglu G, et al. Establishment of a hepatocyte-kupffer cell coculture model for assessment of proinflammatory cytokine effects on metabolizing enzymes and drug transporters. Drug Metab Dispos. 2015;43:774–785. doi: 10.1124/dmd.114.061317. [DOI] [PubMed] [Google Scholar]