Abstract
Avian influenza (AI) viruses circulating in wild birds pose a serious threat to public health. Human and veterinary vaccines against AI subtypes are needed. Here we prepared triple-subtype VLPs that co-localized H5, H7 and H9 antigens derived from H5N1, H7N3 and H9N2 viruses. VLPs also contained influenza N1 neuraminidase and retroviral gag protein. The H5/H7/H9/N1/gag VLPs were prepared using baculovirus expression. Biochemical, functional and antigenic characteristics were determined including hemagglutination and neuraminidase enzyme activities. VLPs were further evaluated in a chicken AI challenge model for safety, immunogenicity and protective efficacy against heterologous AI viruses including H5N2, H7N3 and H9N2 subtypes. All vaccinated birds survived challenges with H5N2 and H7N3 highly pathogenic AI (HPAI) viruses, while all controls died. Immune response was also detectable after challenge with low pathogenicity AI (LPAI) H9N2 virus suggesting that H5/H7/H9/N1/gag VLPs represent a promising approach for the development of broadly protective AI vaccine.
Keywords: Influenza, Avian flu, Vaccine, Recombinant, Virus-Like Particle, VLP
INTRODUCTION
Avian influenza (AI) virus (AIV) belongs to the family Orthomyxoviridae, genus Influenza A virus (type A), and contains a negative-sense, segmented RNA genome. Phylogenetically, there are 18 subtypes of HA that are subdivided into two major antigenic groups (Medina and Garcia-Sastre, 2011; Tong et al., 2013). AIV represents a serious concern for the U.S. and world public health. Humans lack immunity to AIV subtypes, and infections can potentially result in a pandemic. Numerous human infections including avian-origin H5N1, H7N9 and H9N2 subtypes have been documented indicating the potential of these subtypes to adapt to humans and cause life threatening infections (Kang et al., 2009; Xiao et al., 2016). In addition to the pandemic concerns, AI represents a serious threat to the poultry industry and food safety. Wild birds, which are the natural reservoir for the AIV, occasionally transmit the virus to domesticated birds, including chickens, ducks, and turkeys, which are susceptible to AIV. Although most infections in wild birds represent a low-pathogenicity AI (LPAI), infections with the H5 or H7 LPAI viruses can result in the emergence of high-pathogenicity AI (HPAI) viruses through genetic changes of the HA gene. Multiple outbreaks of H5 and H7 HPAI in commercial poultry have been reported in the Americas over the last decade, which renewed interest for AI vaccines (Swayne, 2012). The H9N2 LPAI viruses have also been identified as AIV of concern (Lee et al., 2016).
Together with other measures, vaccines can be an effective measure to prevent AI pandemics, epidemics and epizootics (Kang et al., 2009; Kapczynski and Swayne, 2009; Swayne, 2012). In previous studies, we showed that recombinant influenza virus-like particles (VLPs) protected from pandemic influenza strains including the reconstructed 1918 virus, the 2009 swine-origin pandemic virus, and AI viruses (Kang et al., 2009; Perrone et al., 2009; Pushko et al., 2010). Recombinant VLPs represent inherently safe vaccines that are prepared by using cell culture methods and do not require live AIV for production. Recombinant VLPs comprised of hemagglutinin (HA), neuraminidase (NA) and matrix (M1) proteins have been described (Bright et al., 2007; Galarza et al., 2005; Kang et al., 2009; Perrone et al., 2009; Pushko et al., 2005; Quan et al.; Ross et al., 2009). In some cases, retrovirus gag, such as bovine immunodeficiency virus gag (Bgag), has been used in place of M1 (Kapczynski et al., 2016; Pushko et al., 2016; Tretyakova et al., 2016). Bgag has the advantage of a larger diameter providing more surface area to accommodate multiple HA molecules (Tretyakova et al., 2016). The HA antigen is the major vaccine component, which induces neutralizing antibodies preventing infectious virus from entering cells (Cox et al., 2015; Kang et al., 2009; Pushko et al., 2015). Expression within VLPs increases immunogenicity of the HA antigen (Bright et al., 2007; Pushko et al., 2007). NA is also often included into VLPs because of its role in VLP assembly (Chen et al., 2007) and potential contribution to immunity (Eichelberger and Wan, 2015). Recently, novel design of VLPs was described that allowed co-localization into VLP of the HA proteins derived from several influenza types and subtypes (Kapczynski et al., 2016; Pushko et al., 2011; Tretyakova et al., 2013). This approach was designed to simultaneously elicit specific immunity to multiple influenza subtypes with no requirement for blending individual vaccines. Recombinant VLPs co-localizing three subtypes of HA protected ferrets, a human influenza model, from potentially pandemic viruses of H5, H7 and H9 subtypes (Pushko et al., 2011; Tretyakova et al., 2013).
Here, we prepared H5/H7/H9/N1/gag VLPs containing HA antigens derived from several AIV and evaluated their safety, immunogenicity and efficacy in chickens, a highly sensitive bird model of AI. Immune responses to H5, H7 and H9 antigens and protective efficacy of VLPs after heterologous virus challenges were demonstrated.
MATERIALS AND METHODS
Influenza genes and expression constructs
Influenza H5/H7/H9/N1/gag VLPs were prepared in Spodoptera frugiperda (Sf9) insect cells using a recombinant baculovirus (rBV) expression vector system. VLPs were expressed using rBV containing three HA genes (H5, H7 and H9 subtypes), as well as NA and Bgag genes. Influenza HA gene sequences were derived from A/chicken/West Java Sbg/29/2007 (H5N1 clade 2.1.3), A/turkey/Oregon/1971 (H7N3) and A/turkey/Wisconsin/1/1966 (H9N2) viruses. N1 gene was from H5N1 strain A/chicken/Egypt/121/2012 (clade 2.2.1) (Awad, 2015), while Bgag gene was from BIV R-29 strain retrovirus, GenBank accession number AAA42763.
Three indicated full-length HA genes, as well as NA and Bgag genes were introduced in tandem fashion into the rBV resulting in the vector containing five VLP-relevant genes. Each gene was placed within its own transcriptional cassette that included a polyhedrin promoter upstream from each gene, as described elsewhere (Pushko et al., 2005; Tretyakova et al., 2016). Genes were codon-optimized for high-level expression in Sf9 cells and synthesized (Genscript, Piscataway, NJ). All preparations of rBV were plaque-purified and titrated using standard plaque assay in Sf9 cells.
Expression and characterization of H5/H7/H9 triple-subtype VLP vaccine
To prepare VLP vaccine, Sf9 cells were maintained as suspension cultures in SF900II-SFM insect serum free medium (ThermoFisher Scientific (Thermo), Carlsbad, CA) at 27°C. For production of VLP vaccine, Sf9 cells (2×106 cells/ml) were infected in shaker flasks at a multiplicity of infection (MOI) of 0.1 for approximately 72 h with rBV expressing indicated genes. VLPs were harvested from the growth medium supernatant, clarified using centrifugation and 0.2 μm filtration, concentrated by tangential flow filtration (500 kDa MWCO), and purified by ion exchange chromatography as described elsewhere (Liu et al., 2015). Purified VLPs were further concentrated and purified by ultracentrifugation at 100 000 × g and resuspended in the phosphate buffered saline (PBS). VLPs were characterized including SDS-PAGE and western blot, total protein and HA content, nucleic acid content, functional NA enzyme and hemagglutination activities, as well as particle morphology and size by transmission electron microscopy.
SDS-PAGE was carried out in 4–12% polyacrylamide gels (Thermo) followed by staining with GelCode Blue stain (Pierce, Rockford, IL). Western blots were done using subtype-specific primary antibodies followed by the alkaline phosphatase-conjugated goat IgG (H&L). As primary antibodies, we used mouse anti-H5 (H5N1) and anti-H7 (H7N9) (Immune Tech, New York, NY), as well as chicken anti-H9 (SEPRL, Athens, GA). Total protein in the purified VLPs was determined using Qubit 2.0 fluorometer (Thermo). The HA protein content was determined by gel densitometry of the HA bands using known amounts of reference BSA as a standard.
The nucleic acid content was determined by extracting nucleic acids from the purified VLPs using Trizol LS reagent (Thermo). The extracted nucleic acids were quantitated by using Qubit 2.0 fluorometer using RNA and DNA detection kits. In addition, nucleic acids were treated with either RNAseI or RQ1 DNAse and visualized along with untreated control on the 1% agarose gel in the presence of ethidium bromide.
To determine functional neuraminidase enzyme activity, a fluorescence-based NA assay (NA-Fluor from Thermo) was used with methyl umbelliferone N-acetyl neuraminic acid as a substrate, according to manufacturer’s instructions. Unrelated antigen was used as a negative control, while unrelated H1N1 VLP (A/South Carolina/1/1918) (Perrone et al., 2009) was used a positive control. A standard curve to determine a relative fluorescence unit (R.F.U.) value within the linear range of fluorescence detection was generated using 4-methyl umbelliferone sodium salt (Sigma, St. Louis, MO).
For hemagglutination assay, VLPs were serially diluted in PBS at 2-fold increments in 50 μl volume in a 96-well plate. To each VLP dilution, 50 μl of 1% turkey red blood cell (tRBC) working solution was added as described elsewhere (Tretyakova et al., 2016). Mixtures of VLP s and tRBCs were gently agitated and the plate was incubated at 20°C for 60 min before examination. The titer was calculated as the highest dilution factor that produced a positive reading.
For transmission electron microscopy, purified VLP samples were adsorbed onto a freshly discharged 400 mesh carbon parlodion-coated copper grids, negatively stained with 1% phosphotungstic acid, and visualized on a Hitachi H-7600 transmission electron microscope (Hitachi High Technologies America, Schaumburg, IL).
Vaccinations and challenge
All study protocols were approved by the USDA Institutional Animal Care and Use Committees and all experiments were performed in accordance with the applicable guidelines for the care and use of laboratory animals. H5/H7/H9/N1/gag VLP vaccine was formulated with a commercial adjuvant (SEPPIC, Montanide 70/30, Fairfield, NJ) to contain 1,536 HA units per dose of VLPs (512 HA units of each subtype). Because H5, H7 and H9 genes were expressed from the identical expression cassettes, it is expected that HA subtypes are present at comparable levels in the VLPs, which was confirmed in the previous study (Tretyakova et al., 2016). Specific Pathogen-Free (SPF) chickens (n=30) were vaccinated subcutaneously with 0.2 ml of H5/H7/H9/N1/gag VLP at day 1 of age and 0.5 ml at day 21 of age. Sham-vaccinated birds (n=30) received PBS. Both the control and vaccinated birds were arbitrarily placed into groups of 10 and challenged intranasally (106 EID50 per bird) with one of the following AI isolates at day 35: A/turkey/Minnesota/7172-1/2015 (H5N2 clade 2.3.4.4 HPAI), A/chicken/Jalisco/CPA1/2012 (H7N3 HPAI) and A/chicken/New Jersey/12220/1997 (H9N2 LPAI). After challenges, birds were monitored for clinical signs and mortality daily. HPAI causes rapid progression of disease and death in chickens (Freidl et al., 2014; Kapczynski and Swayne, 2009). If birds were found to be moribund, they were euthanized using AVMA approved methods and counted in the following day after mortality. Because birds rapidly succumbed to HPAI viruses, weight was not measured in these studies, but rather survival. This challenge model was used to demonstrate the effectiveness of the VLP immunization in a system that is similar to natural disease progression with a rapid highly pathogenic virus infection. Serum was taken at day 0 and day 14 post challenge, and oral swabs were taken on days 2 and 4 to measure virus shedding.
Hemagglutination inhibition (HI) assay was performed using standard methods (Pushko et al., 2005). Homologous viruses were used as antigens for HI assay.
Determination of virus shedding
Oropharyngeal and cloacal swabs were collected and kept frozen at −70°C. Viral RNA was extracted using Trizol LS (Invitrogen, Carlsbad, CA) and the MagMAX AI/ND Viral RNA Isolation Kit (Ambion, Austin, TX). Quantitative real time RT-PCR (qRRT-PCR) was performed as previously described (Kapczynski et al., 2016). Briefly, qRRT-PCR targeting the influenza M gene was conducted using AgPath-ID one-step RT-PCR Kit (Ambion) and the ABI 7500 Fast Real-Time PCR system (Applied Biosystems, Carlsbad, CA). For viral quantification, a standard curve was established with viral RNA extracted from the titrated challenge virus. Results were reported as EID50/ml equivalents and the lower limit of detection being 100.9EID50/ml for samples from chickens.
Statistical analysis
Kaplan-Meier survival curves and statistical analyses were done using Prism 5 (GraphPad Co., San Diego, CA). Statistical differences in mean and standard error between HI titers were analyzed using ANOVA. The Mantel-Cox log-rank test was used to compare experimental groups (Prism 5). Lower case letters indicate statistical significance between compared groups. The Student t-test was used was used for pair-wise comparison of virus titers from oral and cloacal swabs. All statistical tests were performed using P < 0.05.
RESULTS
Preparation of VLPs
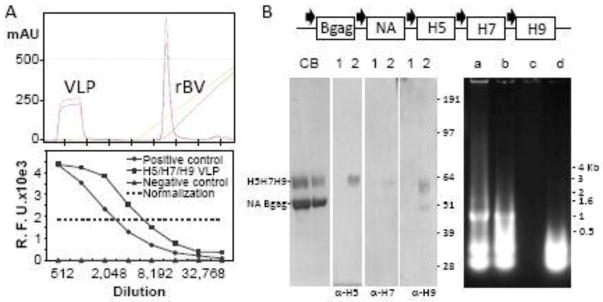
To prepare triple-subtype H5/H7/H9/N1/gag VLPs for vaccinations, HA genes were derived from the three AI viruses, A/chicken/West Java Sbg/29/2007 (H5N1), A/turkey/Oregon/1971 (H7N3) and A/turkey/Wisconsin/1/1966 (H9N2). These viruses represent three subtypes from distinct geographical regions and the time of isolation. In addition to HA genes, we used NA (N1 subtype) and retroviral Bgag as structural components of VLP (Figure 1). NA has been previously shown to be required for assembly and budding of VLPs (Chen et al., 2007); therefore, NA was included into VLPs. Bgag has been previously used for VLP preparation in place of influenza matrix protein (Kapczynski et al., 2016; Pushko et al., 2016; Tretyakova et al., 2016). To prepare VLPs, Sf9 cells were infected with rBV to allow expression and secretion of VLPs containing H5, H7, H9, N1, and Bgag proteins. Cell culture conditions were optimized and the VLPs were harvested from Sf9 culture supernatant. The VLP secretion was monitored by the HA assay. VLPs were separated from rBV by ion exchange chromatography followed by ultracentrifugation. Triple-subtype VLPs migrated in the flow-through fraction during chromatography (Figure 1A, upper panel). The HA proteins in the VLPs were expressed as uncleaved HA0 polypeptides of approximately 62–64 kDa, in agreement with expected sizes of the H5, H7 and H9 proteins of 63.7 kDa, 62.1 kDa, and 62.6 kDa, respectively. The prominent band of ~54 kDa corresponded to the expected molecular weights of the NA (49.0 kDa) and Bgag (53.5 kDa) proteins (Figure 1B, lower left panel).
Figure 1.
Preparation and characterization of VLPs. (A) Upper panel, ion exchange chromatography profile; lower panel, NA enzyme activity. Chromatography chart shows elution volume on X-axis (each tick is 200 ml) and milli-absorbance units (mAU) on Y-axis. NA enzyme activity was determined using a fluorescence-based NA-Fluor assay (Thermo, Carlsbad, CA), with VLP dilutions indicated on the X-axis. R.F.U., relative fluorescence units. Positive control, unrelated H1N1 VLPs (A/South Carolina/1/1918) (Perrone et al., 2009). (B) Upper panel, schematic representation of rBV for expression of H5/H7/H9/N1/gag VLPs. H5, H7 and H9 genes were derived from A/chicken/West Java Sbg/29/2007 (H5N1 clade 2.1.3), A/turkey/Oregon/1971 (H7N3) and A/turkey/Wisconsin/1/1966 (H9N2) viruses, respectively. NA gene was from H5N1 strain A/chicken/Egypt/121/2012 (clade 2.2.1) (Awad, 2015). Bgag indicates BIV retrovirus gag protein. Polyhedrin promoter is indicated with filled arrows. Bottom Left Panel, Coomassie Blue (CB) stained SDS-PAGE and western blot with H5-, H7- and H9-specific antibodies. Lanes: CB, VLP at 5- and 10-fold dilution; 1, negative control VLP; 2, purified VLPs. Right panel, characterization of nucleic acid content in VLPs. Lanes: a, Extracted RNA; b, Extracted RNA incubated with blank buffer; c, Extracted RNA incubated with RNAseI; d, Extracted RNA incubated with RQ1 RNAse-free DNAse.
In addition, we examined the nucleic acid content in the VLPs. The nucleic acids were extracted using Trizol LS and quantitated by fluorometry using RNA- or DNA-specific reagents. The results suggested that extracted nucleic acids from VLPs contained mostly RNA (Table I). This was confirmed by treatment with either RNAse or DNAse. As expected, RNA was undetectable after RNAse treatment, while it was detectable after DNAse treatment using fluorometry (Table I) and agarose gel electrophoresis (Figure 1B, lower right panel), thus confirming that VLPs contain RNA. Notably, RNA bands of ~ 1kB and ~0.5kB were detected in the agarose gel, which disappeared after RNAse treatment. Interestingly enough, these bands converted to the low molecular weight diffuse pattern after incubating with blank buffer or DNAse reaction mixture. One explanation is that RNA extracts from the VLPs as high-molecular weight structures or complexes, which can be disrupted by temperature and other reaction conditions. The nature and origin of the RNA molecules remains to be determined.
Table I.
Analysis of RNA within VLPs by fluorometry.
| Sample | RNA, μg/ml | DNA, μg/ml |
|---|---|---|
| RNA | 370 | 8.2 |
| RNA/blank buffer | 350 | 7.0 |
| RNA/RNAseI | <0.1 | 3.7 |
| RNA/RQ1 DNAse | 222 | 4.7 |
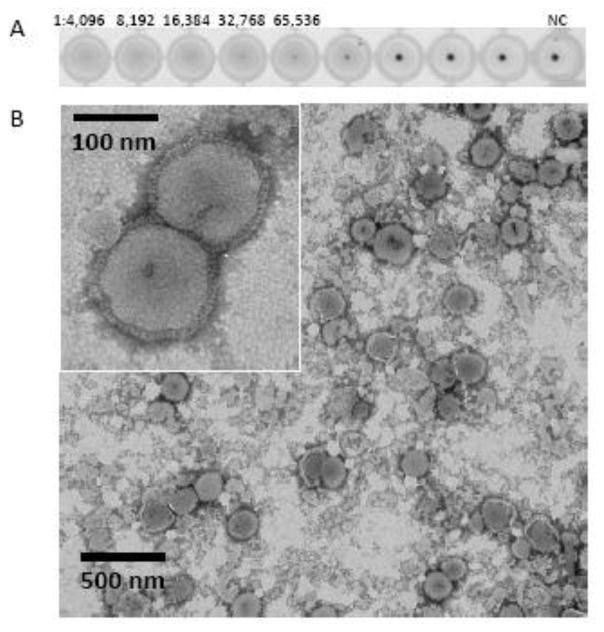
Similar to live influenza virions, VLPs have been previously shown to exhibit functional NA enzyme and hemagglutination activities (Pushko et al., 2005; Tretyakova et al., 2016). Although we did not compare H5/H7/H9/N1/gag VLPs to live virions in the current study, both the NA enzyme and hemagglutination activities of VLPs were evaluated. The NA functional enzyme activity was detected by using fluorescence-based influenza NA assay at more than 8,000-fold dilution of VLPs (Figure 1A, bottom panel). The functional ability of H5/H7/H9/N1/gag VLPs to agglutinate tRBCs was determined by the HA assay (Figure 2A). The hemagglutination activity with the titer of 32,768 per 50 μl of the VLPs (total protein 7 mg/ml, HA content 1 mg/ml) was detected, which confirmed that the HA proteins within VLPs retained their functional stability and tRBC binding activity (Figure 2A). Finally, by negative-staining transmission electron microscopy, the H5/H7/H9/N1/gag VLPs were identified as largely spherical, pleomorphic, enveloped particles approximately 150–200 nm in diameter and containing typical influenza HA spikes protruding from the VLP envelope (Figure 2B).
Figure 2.
Characterization of VLPs by (A) HA assay using tRBC and (B) by negative staining transmission electron microscopy. Purified VLPs were stained using 1% phosphotungstic acid.
Immunogenicity of VLPs
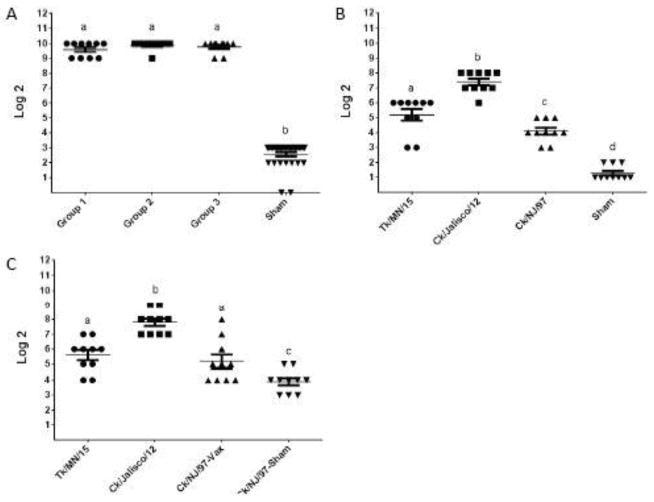
Triple-subtype H5/H7/H9/N1/gag VLPs were formulated with adjuvant for vaccinations. Following vaccination with VLPs, pre-challenge and post-challenge serum antibody levels were determined (Figure 3). Because HA antigen is considered the main protective antigen in people and alone can induce protective response (Cox et al., 2015), we investigated HI responses to H5, H7 and H9. Immunogenicity of N1 has not been determined in this study. Pre-challenge HI titers to the VLP were observed in all three vaccinated groups designated for H5, H7 and H9 challenges (Figure 3A). We also confirmed HI titers in the pre-challenge sera to all three challenge viruses (Figure 3B). The highest HI antibody was detected to A/chicken/Jalisco/CPA1/2012 (H7N3) virus, while the lowest HI titers were detected to A/chicken/New Jersey/12220/1997 (H9N2) virus. As expected, no HI antibody was detected in sham-vaccinated birds (Figure 3A, B).
Figure 3.
Immunogenicity of H5/H7/H9/N1/gag VLPs. X-axis indicates Log2 dilution of sera, y-axis indicated antigen tested. (A) Pre-challenge serum antibody, by HI assay using VLP as antigen. Groups 1, 2, and 3 were vaccinated with VLPs and later challenged with H5, H7 and H9 viruses, respectively. For vaccinations, VLPs were formulated with SEPPIC adjuvant to contain 1,536 HA units per dose (512 HA units of each subtype) and administered to chickens at day 1 and day 21 of age. (B) Pre-challenge serum antibody, by HI assay using indicated virus antigen prepared from A/turkey/Minnesota/7172-1/2015 (H5N2), A/chicken/Jalisco/CPA1/2012 (H7N3) and A/chicken/New Jersey/12220/1997 (H9N2) viruses. (C) Post-challenge serum antibody, by HI assay using indicated virus antigens.
Protective efficacy
The HI data suggested successful seroconvertion after vaccination to the vaccine antigens and to challenge viruses (Figure 3A, B). To test protective efficacy, VLP-vaccinated birds were divided into three groups. Each group of vaccinated birds was challenged with one of three viruses two weeks after vaccination. Similarly, sham-vaccinated birds were divided into three groups, and each group was challenged with one of three viruses two weeks after vaccination. As compared to the VLP vaccine antigens, which were derived from A/chicken/West Java Sbg/29/2007 (H5N1), A/turkey/Oregon/1971 (H7N3) and A/turkey/Wisconsin/1/1966 (H9N2), all three challenge viruses including A/turkey/Minnesota/7172-1/2015 (H5N2), A/chicken/Jalisco/CPA1/2012 (H7N3) and A/chicken/New Jersey/12220/1997 (H9N2) represented heterologous challenges.
Post-challenge HI antibody titers showed no considerable change in pre-challenge and post-challenge HI titers (Figure 3B, C).
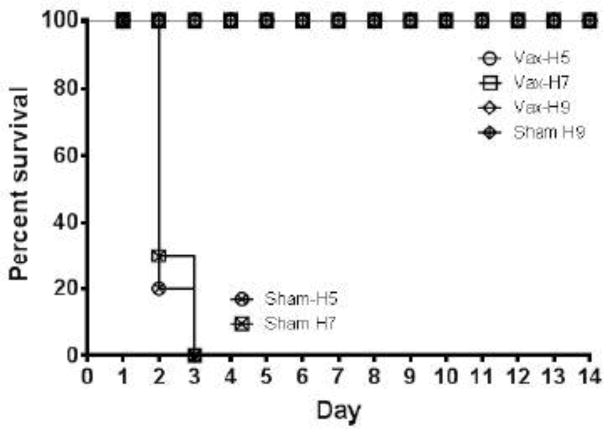
Vaccine protection was measured by survival and reduction in viral shedding in chickens after virus challenges with three indicated viruses. With HPAI viruses, death often occured in the absence of clinical disease. Following virus challenges with 106 EID50 viruses in the H5 HPAI and H7 HPAI challenge experiments, control animals rapidly succumbed to infection and died. In the H5 challenge control group, 8 out of 10 birds died on day 2 post challenge, and the remaining 2 birds died on day 3 postchallenge (Table II). In the H7 control challenge group, 7 out of 10 birds died on day 2, while the remaining 3 birds died on day 3 postchallenge. In contrast, all VLP-vaccinated birds survived H5 and H7 HPAI challenges (Figure 4). Regarding H9 LPAI challenge groups, no deaths were observed in both control and vaccinated groups, as expected for LPAI virus (Table II, Figure 4). Therefore, the main indication of protective effects against H9 LPAI virus challenge was observation of viral shedding as described below.
Table II.
Survival of vaccinated birds after heterologous challenges*.
| Challenge Strain** | Challenge Subtype | Total Birds | Dead | Dead | Survived | Survival |
|---|---|---|---|---|---|---|
| DPI: 2 | DPI: 3 | DPI: 14 | ||||
| Vacc/Ck/NJ | H9 LPAI | 10 | 10 | 10/10 | ||
| Vacc/Jalisco | H7 | 10 | 10 | 10/10 | ||
| Vacc/Tk/MN15 | H5 | 10 | 10 | 10/10 | ||
| Sham-Ck/NJ | H9 LPAI | 10 | 10 | 10/10 | ||
| Sham-Jalisco | H7 | 10 | 7 | 3 | 0 | 0/10 |
| Sham-Tk/MN15 | H5 | 10 | 8 | 2 | 0 | 0/10 |
DPI, days post infection; LPAI, low pathogenicity avian influenza.
Strains included into H5/H7/H9/N1/gag VLP vaccines were the following: A/chicken/West Java Sbg/29/2007 (H5N1), A/turkey/Oregon/1971 (H7N3) and A/turkey/Wisconsin/1/1966 (H9N2).
Challenge strains were the following: A/turkey/Minnesota/7172-1/2015 (H5N2), A/chicken/Jalisco/CPA1/2012 (H7N3) and A/chicken/New Jersey/12220/1997 (H9N2).
Figure 4.
Survival curves of VLP-vaccinated and sham-vaccinated birds after H5 HPAI, H7 HPAI and H9 LPAI virus challenges. Challenges were done using were A/turkey/Minnesota/7172-1/2015 (H5N2), A/chicken/Jalisco/CPA1/2012 (H7N3) and A/chicken/New Jersey/12220/1997 (H9N2) viruses.
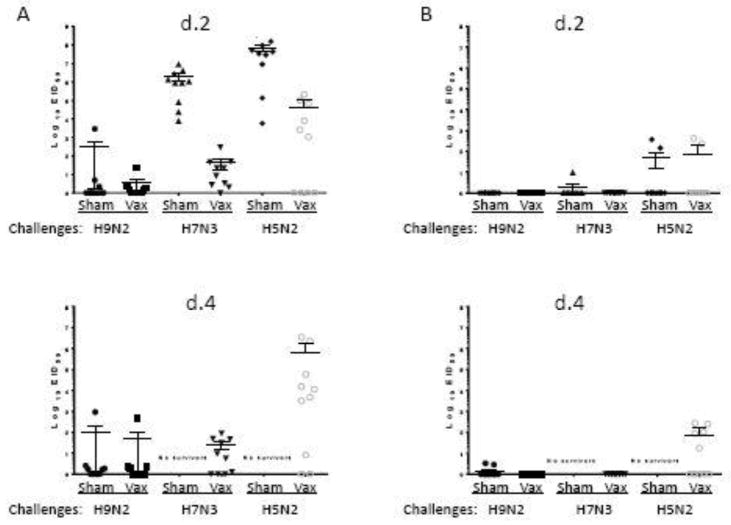
Virus shedding after challenge was measured using oral and cloacal swabs. In all control groups, high titers of H5 and H7 challenge virus were detected in oral swabs on day 2, while in vaccinated birds, lower virus titers were consistently detected. On day 4, while all control birds in H5 and H7 challenge groups died, oral shedding was detectable in vaccinated birds, especially the H5 challenge group. Regarding H9 LPAI challenge, reduction of shedding was detectable in the VLP-vaccinated group after H9 LPAI challenge as compared to H9-challenged sham controls. Examination of postchallenge HI titers in the sham control group that expectedly survived H9N2 LPAI challenge has been carried out, while sham controls in H5 and H7 HPAI challenge groups all died (Figure 3C). After H9N2 virus challenge, sham-vaccinated birds developed lower HI titers than VLP-vaccinated challenged birds suggesting immune response after VLP vaccination and boost after challenge (Figure 3C). However, additional studies are needed to determine protective immune responses against H9 LPAI. Cloacal shedding in all challenge groups was generally low, with elevated titers detected after H5 challenge on days 2 and 4 (Figure 5).
Figure 5.
Virus shedding in chickens after challenges. (A) Oral shedding on days 2 and 4; (B) Cloacal shedding on days 2 and 4. Challenges included A/turkey/Minnesota/7172-1/2015 (H5N2 clade 2.3.4.4), A/chicken/Jalisco/CPA1/2012 (H7N3) and A/chicken/New Jersey/12220/1997 (H9N2) viruses.
Taken together, these data demonstrate that triple-subtype H5/H7/H9/N1/gag VLP immunization induced immunological responses and protection of chickens against heterologous challenges with H5 and H7 HPAI viruses, as well as detectable immune response to heterologous H9 LPAI virus.
DISCUSSION
AI is a viral disease that primarily occurs in many bird species with predominantly asymptomatic LPAI cases, while HPAI variants can result in rapid mortality. AI is also one of the prominent examples of human health risks at the human-animal ecosystem interface. The virus is shed through the intestinal tract of the infected bird that leads to the virus spread. Humans can be exposed to AIV by the contact with infected birds or with contaminated poultry meat. Human infections have been documented for AIV with hemagglutinin subtypes H5, H6, H7, H9 and H10 (Freidl et al., 2014) and most human cases were caused by H5, H7 and H9 viruses (CDC, 2016). The same three subtypes cause large number of infections in wild birds and in commercial poultry species. A major outbreak of Asian-origin HPAI of H5N2, H5N8 and H5N1 viruses occurred during 2015 in the U.S. with more than 50 million birds dead or killed during that time (Ip et al., 2015; Jhung et al., 2015; Lee et al., 2015; Torchetti et al., 2015). The H7N3 AIV has been identified in wild birds throughout the world including H7N3 HPAI virus that cause outbreak in 2012 in Jalisco, Mexico (Kapczynski et al., 2013). Although the index case is unknown, there is a possibility that a wild-bird-origin LPAI virus has mutated into an HPAI variant. Finally, the H9N2 viruses continue to circulate in Africa and Eurasia. Research indicates that at least some H9N2 viruses contain internal genes highly homologous with H10N8 or H7N9 viruses that caused infections in humans and potentially can serve as donors of internal gene complex and lead to new pathogenic viruses (Ye et al., 2016).
Rational use of vaccination can be important measure for prevention and control of HPAI (FAO, 2016). In places where HPAI is endemic, vaccine is used to reduce presence of HPAI viruses in the environment, which reduces human exposure and the likelihood of zoonotic and pandemic influenza, as well as the risk of severe disease in poultry. Considering prevalence of H5, H7 and H9 subtypes, a vaccine that induces immunity to H5, H7 and H9 viruses would be important. Multivalence is a well-known method to prepare broadly protective vaccines and has been used for many viral and bacterial agents including seasonal influenza, HPV, and pneumococcal vaccines. Classic inactivated vaccines represent mixtures of inactivated viruses (Kang et al., 2009; Palese, 2006). However, it is not feasible to make a polyvalent AIV vaccine using current commercial technology, largely due to the costs and biosafety concerns associated with manufacturing of large quantities of multiple HPAI viruses.
Recently, we have shown that recombinant VLPs can co-localize multiple variants of HA in their envelopes and that such multi-HA VLPs induce protective immune responses against multiple homologous strains of influenza (Kapczynski et al., 2016; Pushko et al., 2011). Multi-subtype VLPs can be produced in a single manufacturing process, with no need for individual preparation of each vaccine or blending, which may decrease the cost of vaccine production. Recombinant multi-HA VLPs have inherent safety features, do not dependent on eggs for manufacturing, and provide efficient protection against multiple subtypes and strains. Thus, multi-HA VLPs can represent a promising alternative to blended vaccines in order to prepare multivalent formulations including AI vaccines. In the recent study, we described a recombinant, triple-HA VLP containing HA proteins derived from three distinct clades of H5N1 viruses. The triple-H5 VLP vaccine contained H5 genes from recent H5N1 HPAI isolates A/chicken/Germany/2014 (clade 2.3.4.4), A/chicken/West Java/Subang/29/2007 (clade 2.1.3) and A/chicken/Egypt/121/2012 (clade 2.2.1) (Kapczynski et al., 2016). Vaccination of chickens with these VLPs resulted in induction of serum antibody responses and efficient protection against experimental challenges with three distinct homologous H5 viruses including the recent U.S. H5N8 HPAI isolate. Notably, subtypes within VLPs can be quantitated using standard methods, and expression of VLPs from rBV vectors is stable for at least five passages of rBV (Kapczynski et al., 2016; Tretyakova et al., 2016). However, protective effects of VLPs against heterologous challenges have not been studied yet.
Here, we configured VLPs to express HA proteins derived from H5, H7 and H9 AI viruses and demonstrated that vaccination with H5/H7/H9/N1/gag VLPs induced immune responses capable of protection from experimental challenges with heterologous HPAI viruses. The observed protection was likely because of immune responses to H5, H7 and H9 antigens within VLPs, which were confirmed by HI assays. Contribution of N1 to protection against H5N3, H7N3 and H9N2 challenge viruses is not clear. It has been previously reported that NA immunity is most effective against homologous viruses (Eichelberger and Wan, 2015); therefore protective effect of N1 antigen against H5N3, H7N3 and H9N2 challenge viruses may be limited due to heterologous N2 or N3 in these viruses. However, the potential contribution of N1 to immunity remains to be studied. Regarding H9 LPAI challenge, all birds from the vaccinated and control groups survived, and reduction of shedding was barely detectable in the vaccinated birds. Further studies will be performed to determine to ability of VLPs to protect against H9 LPAI by increasing the H9 virus challenge dose. After challenges with H5 and H7 HPAI viruses, 100% of vaccinated birds survived while all controls died. Shedding was virtually undetectable in the H7 challenge group on day 4, while H5 challenge group still showed residual shedding on day 4. Thus, after H5 heterologous challenge, more time may be required to clear the virus as compared to the homologous challenges observed in our recent study (Kapczynski et al., 2016). Interestingly enough, the H5 of A/chicken/West Java Sbg/29/2007 (H5N1) within H5/H7/H9/N1/gag VLP has only 88% homology with the challenge strain A/turkey/Minnesota/7172-1/2015 (H5N2). The results of the current study suggest that H5/H7/H9/N1/gag VLP vaccine induces protective immunity against heterologous AI virus challenges; however, in order to minimize shedding, vaccine should include the antigens that closely match circulating AI strains. Therefore, in the case when circulating virus is distinct from the vaccine, the H5/H7/H9/N1/gag VLPs can be potentially considered as an emergency vaccine until a specific vaccine is made.
Highlights.
VLPs were prepared that co-localized H5, H7 and H9 subtypes in a VLP envelope.
VLPs were characterized including electron microscopy, HA assay and NA enzyme activity.
Experimental VLP vaccine was evaluated in an avian influenza challenge model.
VLPs induced immune responses against heterologous H5, H7 and H9 virus challenges.
Acknowledgments
Funding
This project was supported in part by grants 2013-33610-21041 (USDA NIFA), 1R01AI111532 (NIH NIAID) and 6040-32000-062-00D (USDA-ARS).
We thank Brian Nickols, Raphael O. Prather, and Michele Exum for their contributions. The authors declare that they have no competing financial interests. The findings and conclusions in this report are those of the authors and do not necessarily reflect the views of the funding agencies.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Awad EST, Gouda E, El-Husseiny MH, Aly MM, Pushko P, Tretyakova I, Arafa ASM. Biochemical and immunogenicity studies on Hemagglutinin protein rescued from H5N1 avian influenza virus like particles. Journal of American Science. 2015;11:1–8. [Google Scholar]
- Bright RA, Carter DM, Daniluk S, Toapanta FR, Ahmad A, Gavrilov V, Massare M, Pushko P, Mytle N, Rowe T, Smith G, Ross TM. Influenza virus-like particles elicit broader immune responses than whole virion inactivated influenza virus or recombinant hemagglutinin. Vaccine. 2007;25:3871–3878. doi: 10.1016/j.vaccine.2007.01.106. [DOI] [PubMed] [Google Scholar]
- CDC. Avian Influenza Current Situation. 2016. [Google Scholar]
- Chen BJ, Leser GP, Morita E, Lamb RA. Influenza virus hemagglutinin and neuraminidase, but not the matrix protein, are required for assembly and budding of plasmid-derived virus-like particles. J Virol. 2007;81:7111–7123. doi: 10.1128/JVI.00361-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox MM, Izikson R, Post P, Dunkle L. Safety, efficacy, and immunogenicity of Flublok in the prevention of seasonal influenza in adults. Ther Adv Vaccines. 2015;3:97–108. doi: 10.1177/2051013615595595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichelberger MC, Wan H. Influenza neuraminidase as a vaccine antigen. Curr Top Microbiol Immunol. 2015;386:275–299. doi: 10.1007/82_2014_398. [DOI] [PubMed] [Google Scholar]
- FAO U. Rational use of vaccination for prevention and control of H5 highly pathogenic avian influenza. 2016. [Google Scholar]
- Freidl GS, Meijer A, de Bruin E, de Nardi M, Munoz O, Capua I, Breed AC, Harris K, Hill A, Kosmider R, Banks J, von Dobschuetz S, Stark K, Wieland B, Stevens K, van der Werf S, Enouf V, van der Meulen K, Van Reeth K, Dauphin G, Koopmans M, Consortium F. Influenza at the animal-human interface: a review of the literature for virological evidence of human infection with swine or avian influenza viruses other than A(H5N1) Euro Surveill. 2014;19 doi: 10.2807/1560-7917.es2014.19.18.20793. [DOI] [PubMed] [Google Scholar]
- Galarza JM, Latham T, Cupo A. Virus-like particle (VLP) vaccine conferred complete protection against a lethal influenza virus challenge. Viral Immunol. 2005;18:244–251. doi: 10.1089/vim.2005.18.244. [DOI] [PubMed] [Google Scholar]
- Ip HS, Torchetti MK, Crespo R, Kohrs P, DeBruyn P, Mansfield KG, Baszler T, Badcoe L, Bodenstein B, Shearn-Bochsler V, Killian ML, Pedersen JC, Hines N, Gidlewski T, DeLiberto T, Sleeman JM. Novel Eurasian highly pathogenic avian influenza A H5 viruses in wild birds, Washington, USA, 2014. Emerg Infect Dis. 2015;21:886–890. doi: 10.3201/eid2105.142020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jhung MA, Nelson DI Centers for Disease C and Prevention. Outbreaks of avian influenza A (H5N2), (H5N8), and (H5N1) among birds--United States, December 2014–January 2015. MMWR Morb Mortal Wkly Rep. 2015;64:111. [PMC free article] [PubMed] [Google Scholar]
- Kang SM, Pushko P, Bright RA, Smith G, Compans RW. Influenza virus-like particles as pandemic vaccines. Curr Top Microbiol Immunol. 2009;333:269–289. doi: 10.1007/978-3-540-92165-3_14. [DOI] [PubMed] [Google Scholar]
- Kapczynski DR, Pantin-Jackwood M, Guzman SG, Ricardez Y, Spackman E, Bertran K, Suarez DL, Swayne DE. Characterization of the 2012 highly pathogenic avian influenza H7N3 virus isolated from poultry in an outbreak in Mexico: pathobiology and vaccine protection. J Virol. 2013;87:9086–9096. doi: 10.1128/JVI.00666-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapczynski DR, Swayne DE. Influenza vaccines for avian species. Curr Top Microbiol Immunol. 2009;333:133–152. doi: 10.1007/978-3-540-92165-3_6. [DOI] [PubMed] [Google Scholar]
- Kapczynski DR, Tumpey TM, Hidajat R, Zsak A, Chrzastek K, Tretyakova I, Pushko P. Vaccination with virus-like particles containing H5 antigens from three H5N1 clades protects chickens from H5N1 and H5N8 influenza viruses. Vaccine. 2016;34:1575–1581. doi: 10.1016/j.vaccine.2016.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DH, Fusaro A, Song CS, Suarez DL, Swayne DE. Poultry vaccination directed evolution of H9N2 low pathogenicity avian influenza viruses in Korea. Virology. 2016;488:225–231. doi: 10.1016/j.virol.2015.11.023. [DOI] [PubMed] [Google Scholar]
- Lee DH, Torchetti MK, Winker K, Ip HS, Song CS, Swayne DE. Intercontinental Spread of Asian-Origin H5N8 to North America through Beringia by Migratory Birds. J Virol. 2015;89:6521–6524. doi: 10.1128/JVI.00728-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YV, Massare MJ, Pearce MB, Sun X, Belser JA, Maines TR, Creager HM, Glenn GM, Pushko P, Smith GE, Tumpey TM. Recombinant virus-like particles elicit protective immunity against avian influenza A(H7N9) virus infection in ferrets. Vaccine. 2015 doi: 10.1016/j.vaccine.2015.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina RA, Garcia-Sastre A. Influenza A viruses: new research developments. Nat Rev Microbiol. 2011;9:590–603. doi: 10.1038/nrmicro2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palese P. Making better influenza virus vaccines? Emerg Infect Dis. 2006;12:61–65. doi: 10.3201/eid1201.051043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrone LA, Ahmad A, Veguilla V, Lu X, Smith G, Katz JM, Pushko P, Tumpey TM. Intranasal vaccination with 1918 influenza virus-like particles protects mice and ferrets from lethal 1918 and H5N1 influenza virus challenge. J Virol. 2009;83:5726–5734. doi: 10.1128/JVI.00207-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pushko P, Kort T, Nathan M, Pearce MB, Smith G, Tumpey TM. Recombinant H1N1 virus-like particle vaccine elicits protective immunity in ferrets against the 2009 pandemic H1N1 influenza virus. Vaccine. 2010;28:4771–4776. doi: 10.1016/j.vaccine.2010.04.093. [DOI] [PubMed] [Google Scholar]
- Pushko P, Pearce MB, Ahmad A, Tretyakova I, Smith G, Belser JA, Tumpey TM. Influenza virus-like particle can accommodate multiple subtypes of hemagglutinin and protect from multiple influenza types and subtypes. Vaccine. 2011;29:5911–5918. doi: 10.1016/j.vaccine.2011.06.068. [DOI] [PubMed] [Google Scholar]
- Pushko P, Pujanauski LM, Sun X, Pearce M, Hidajat R, Kort T, Schwartzman LM, Tretyakova I, Chunqing L, Taubenberger JK, Tumpey TM. Recombinant H7 hemagglutinin forms subviral particles that protect mice and ferrets from challenge with H7N9 influenza virus. Vaccine. 2015 doi: 10.1016/j.vaccine.2015.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pushko P, Sun X, Tretyakova I, Hidajat R, Pulit-Penaloza JA, Belser JA, Maines TR, Tumpey TM. Mono- and quadri-subtype virus-like particles (VLPs) containing H10 subtype elicit protective immunity to H10 influenza in a ferret challenge model. Vaccine. 2016 doi: 10.1016/j.vaccine.2016.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pushko P, Tumpey TM, Bu F, Knell J, Robinson R, Smith G. Influenza virus-like particles comprised of the HA, NA, and M1 proteins of H9N2 influenza virus induce protective immune responses in BALB/c mice. Vaccine. 2005;23:5751–5759. doi: 10.1016/j.vaccine.2005.07.098. [DOI] [PubMed] [Google Scholar]
- Pushko P, Tumpey TM, Van Hoeven N, Belser JA, Robinson R, Nathan M, Smith G, Wright DC, Bright RA. Evaluation of influenza virus-like particles and Novasome adjuvant as candidate vaccine for avian influenza. Vaccine. 2007;25:4283–4290. doi: 10.1016/j.vaccine.2007.02.059. [DOI] [PubMed] [Google Scholar]
- Quan FS, Vunnava A, Compans RW, Kang SM. Virus-like particle vaccine protects against 2009 H1N1 pandemic influenza virus in mice. PloS one. 5:e9161. doi: 10.1371/journal.pone.0009161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross TM, Mahmood K, Crevar CJ, Schneider-Ohrum K, Heaton PM, Bright RA. A trivalent virus-like particle vaccine elicits protective immune responses against seasonal influenza strains in mice and ferrets. PloS one. 2009;4:e6032. doi: 10.1371/journal.pone.0006032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swayne DE. Impact of vaccines and vaccination on global control of avian influenza. Avian Dis. 2012;56:818–828. doi: 10.1637/10183-041012-Review.1. [DOI] [PubMed] [Google Scholar]
- Tong S, Zhu X, Li Y, Shi M, Zhang J, Bourgeois M, Yang H, Chen X, Recuenco S, Gomez J, Chen LM, Johnson A, Tao Y, Dreyfus C, Yu W, McBride R, Carney PJ, Gilbert AT, Chang J, Guo Z, Davis CT, Paulson JC, Stevens J, Rupprecht CE, Holmes EC, Wilson IA, Donis RO. New world bats harbor diverse influenza A viruses. PLoS Pathog. 2013;9:e1003657. doi: 10.1371/journal.ppat.1003657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torchetti MK, Killian ML, Dusek RJ, Pedersen JC, Hines N, Bodenstein B, White CL, Ip HS. Novel H5 Clade 2.3.4.4 Reassortant (H5N1) Virus from a Green-Winged Teal in Washington, USA. Genome Announc. 2015;3 doi: 10.1128/genomeA.00195-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tretyakova I, Hidajat R, Hamilton G, Horn N, Nickols B, Prather RO, Tumpey TM, Pushko P. Preparation of quadri-subtype influenza virus-like particles using bovine immunodeficiency virus gag protein. Virology. 2016;487:163–171. doi: 10.1016/j.virol.2015.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tretyakova I, Pearce MB, Florese R, Tumpey TM, Pushko P. Intranasal vaccination with H5, H7 and H9 hemagglutinins co-localized in a virus-like particle protects ferrets from multiple avian influenza viruses. Virology. 2013;442:67–73. doi: 10.1016/j.virol.2013.03.027. [DOI] [PubMed] [Google Scholar]
- Xiao C, Ma W, Sun N, Huang L, Li Y, Zeng Z, Wen Y, Zhang Z, Li H, Li Q, Yu Y, Zheng Y, Liu S, Hu P, Zhang X, Ning Z, Qi W, Liao M. PB2-588 V promotes the mammalian adaptation of H10N8, H7N9 and H9N2 avian influenza viruses. Sci Rep. 2016;6:19474. doi: 10.1038/srep19474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye G, Liang CH, Hua DG, Song LY, Xiang YG, Guang C, Lan CH, Ping HY. Phylogenetic Analysis and Pathogenicity Assessment of Two Strains of Avian Influenza Virus Subtype H9N2 Isolated from Migratory Birds: High Homology of Internal Genes with Human H10N8 Virus. Front Microbiol. 2016;7:57. doi: 10.3389/fmicb.2016.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]