Abstract
The carcinogenic precursor benzo[a]pyrene (BP), a polycyclic aromatic hydrocarbon, is released into the environment through the incomplete combustion of hydrocarbons. Metabolism of BP in the human body yields a potent alkylating agent (benzo[a]pyrene diol epoxide, BPDE) that reacts with guanine (G) in DNA to form an adduct implicated in cancer initiation. We report that the α-hemolysin (αHL) nanopore platform can be used to detect a BPDE adduct to G in synthetic oligodeoxynucleotides. Translocation of a 41-mer poly-2′-deoxycytidine strand with a centrally located BPDE adduct to G through αHL in 1 M KCl produces a unique multi-level current signature allowing the adduct to be detected. This readily distinguishable current modulation was observed when the BPDE-adducted DNA strand translocated from either the 5′ or 3′ directions. This study suggests that BPDE adducts and other large aromatic biomarkers can be detected with αHL, presenting opportunities for the monitoring, quantification, and sequencing of mutagenic compounds from cellular DNA samples.
Keywords: ion channel, DNA adduct, benzo[a]pyrene, alpha hemolysin, biomarker
1. Introduction
Polycyclic aromatic hydrocarbons (PAHs) emitted into the environment by the incomplete combustion of coal, crude oil, and gasoline were reported to have carcinogenic properties in humans as early as 1876 [1,2]; in 1930, the PAH benzo[a]pyrene (BP) was identified as the carcinogen in these substances. Workers in tar distilleries, fossil fuel processing, and road paving are exposed to high levels of BP, as are smokers and consumers of grilled meats [3–6]. Exposure to BP has been shown to increase susceptibility to lung and colon cancers [7].
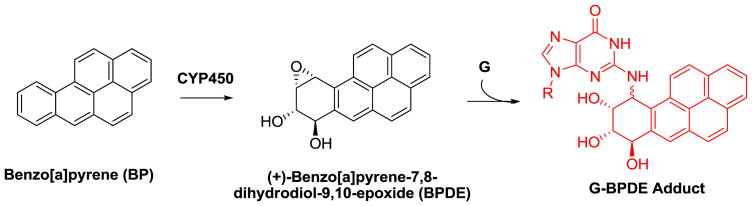
Cellular studies have demonstrated that one of the principal pathways through which BP is removed from the body is via cytochrome p450s (CYP450), yielding the final product benzo[a]pyrene diol epoxide (BPDE, figure 1). BPDE exists in four isomeric forms with the (+)-anti-7α,8β-dihydroxy-9α,10α-epoxy-7,8,9,10-tetrahydrobenzo[a]pyrene (BPDE) isomer being the predominant one observed from enzymatic studies [8,9]. BPDE is electrophilic and susceptible to nucleophilic attack from DNA, where the base guanine (G) is a chief site for adduction of BPDE, yielding a stable adduct at the N2 position (G-BPDE, figure 1). Because the epoxide ring-opening can occur by either SN1 or SN2 mechanisms, two diastereomers of G-BPDE are formed (figure 1), leading to different structural perturbations of the DNA double helix. Moreover, mutations at specific G residues in the TP53 gene are responsible for the mutagenic properties of BPDE that lead to lung cancer [10–12]. Therefore, identification of these adducts and their locations in the genome are critical to addressing an individual’s susceptibility to cancers caused by BPDE.
Figure 1.
Benzo[a]pyrene metabolism leading to guanine adducts in DNA.
Several methods have been developed for quantification of BPDE adducts in genomes [13–15]. The most commonly used methods include [32P]-postlabeling [16], the enzyme-linked immunosorbent assay (ELISA) [17], liquid chromatography coupled to mass spectrometry (LC-MS) [18], capillary electrophoresis MS [16], or HPLC coupled with a fluorescence detector [19]. Analysis of BPDE adducts in DNA by these methods requires exhaustive nuclease digestion of the DNA sample to the nucleoside monomers. There are two major drawbacks with this step: (1) digestion of these adducts in DNA to the nucleoside monomers is often incomplete because the lesion is not a good substrate for nucleases, and (2) digestion of the DNA causes all sequence information to be lost [20]. Methods for quantification of G-BPDE adducts by LC-MS have identified this lesion to exist at a concentration of <10 adducts/108 nucleotides in the human genome [20,21], and a method that can directly analyze these adducts in the genome would be advantageous for quantification of BPDE adducts. In addition, a single-molecule method would have the added advantage of addressing the question of the distribution of adducts and identifying any hotspots for adduct formation.
A powerful strategy for analyzing DNA is achieved by electrophoretically driving single-stranded DNA through the α-hemolysin (αHL) nanopore [22,23]. Studies with this nanopore have demonstrated the potential for sequencing the four DNA bases [24], epigenetic markers [25], damage to DNA resulting from oxidation [26–28] or deamination [29,30], photochemical damage [27], and base release that yields abasic sites [29,31].
Herein, we demonstrate that the nanopore ion channel method can be applied to the direct detection of a G-BPDE adduct. In these studies, short (4-mer) and long (41-mer) synthetic DNA oligomers with a centrally located BPDE adduct were electrophoretically driven through the αHL nanopore while monitoring the current fluctuations and event times. These studies demonstrate that the BPDE adduct is capable of passing through the pore while producing a current blockage signature characteristic of the biomarker. These observations represent the initial step toward applying the nanopore method for the detection and quantification, and ultimately for reading the sequence, in which BPDE adducts reside in the genome.
2. Experimental Section
Caution: All PAHs are potentially carcinogenic and should be handled in accordance with NIH Guidelines for the Use of Chemical Carcinogens.
Chemicals and Materials for Preparation of BPDE-DNA adduct
All DNA strands were synthesized from commercially available phosphoramidites by the DNA/peptide core facility at the University of Utah. (±)-Benzo[a]pyrene-7α,8β-dihydrodiol-9α,10α-epoxide was purchased from MRIGlobal and used as received. All other chemicals were used without further purification.
Preparation of BPDE-DNA adduct
DNA samples were purified by ion-exchange HPLC prior to their use via the following method: solvent A = 10% CH3CN, 90% ddH2O; B = 1 M NaCl in 10% CH3CN 90% ddH2O, 25 mM Tris pH 8; flow rate = 1 mL/min while monitoring the absorbance at 260 nm. The method was initiated at 15% B followed by a linear increase to 100% B over 30 min. Synthesis of the BPDE adducted DNA strands were carried out according to a literature protocol [32]. Briefly, the BPDE stock solution was made by dissolving BPDE in 19:1 THF and 1.5% aqueous triethylamine. Reactions were performed in 100-μL aliquots in Eppendorf tubes containing 2 mM DNA and 1 mM BPDE stock solution in 25 mM Tris, 1.5% aqueous triethylamine, 200 mM NaCl all at pH 9.2. The reaction was carried out overnight in the dark at 37°C. The reaction mixture was neutralized by adding 3 mL of 20 mM PBS buffer (pH 7.5) before purification. Products were purified by ion-exchange HPLC running the same solvent system as reported above. All isomeric products were collected together and analyzed by ESI-MS to give the following result: 41-mer BPDE calcd mass = 12136.9, expt mass = 12139.2. Reaction yields were ~5%.
Glass nanopore membrane (GNM) and bilayer formation for ion channel recording
The method for fabrication of a conical shaped nanopore in a thin glass capillary membrane has been reported previously [33]. The nanopores used for these studies had an orifice with a 300 to 600 nm radius. Silanization of the glass surface was achieved with 2% (v:v) 3-cyanopropyldimethylchlorosilane in CH3CN for 6 h at room temperature to introduce a hydrophobic surface to which the lipid bilayer could form. Two Ag/AgCl electrodes were placed in solution on the inside (trans) and outside (cis) of the capillary. The electrolyte solution was comprised of either 1 M KCl or 3 M NaCl in 10 mM PBS pH 7.4. Current-time (i-t) recordings were performed using a custom built high-impedance, and low-noise system (Electronic BioSciences Inc., San Diego, CA). The lipid bilayer was formed with 1,2-diphytanoyl-sn-glycero-3-phosphochline across the silanized GNM; bilayer formation was indicated by a resistance increase from ~10 MΩ to ~100 GΩ. A gas-tight syringe was used to apply a pressure of 20–40 mmHg to the inside of the GNM capillary that facilitated protein insertion into the lipid bilayer [34]. Wild type αHL was reconstituted from the monomer peptide added to the cis side of the GNM (0.2 μL of a 1 mg/mL solution). Formation of a properly functioning nanopore was determined by an Io at 120 mV of 122 pA or 244 pA at 25 °C in 1 M KCl or 3 M NaCl, respectively. Ion channel measurements were performed at 120, 140, 160 and 180 mV (trans vs. cis), while recording the data with a 100 kHz low-pass filter and at a 500 kHz data acquisition rate. All experiments were performed at 25.0 ± 0.5 °C.
Data Analysis
Events were extracted using QUB 2.0.0.29 and data were analyzed using OriginPro 9.1 and software donated by Electronic BioSciences Inc. (San Diego, CA). The i-t traces presented were re-filtered to 50 kHz for presentation purposes unless stated otherwise.
3. Results and Discussion
Ion channel measurements
Two DNA oligomers were chosen for study, a 4-mer and a 41-mer, with the sequences 5′-CCGC-3′ and 5′-C20-G-C20-3′, respectively. These oligomers were allowed to react with (±)-benzo[a]pyrene-7α,8β-dihydrodiol-9α,10α-epoxide following a literature protocol to yield an adduct at G [32]. The presence of a single G ensured that only one adduct was formed per strand; hereafter, the adducted oligomers are referred to as 4-mer BPDE and 41-mer BPDE. A single αHL ion channel was inserted into a lipid bilayer spanning a glass nanopore membrane [35]. The DNA analyte was added to the cis side of the channel in a buffered (25 mM PBS, pH 7.4) 1 M KCl or 3 M NaCl solution. A voltage was applied to electrophoretically drive the DNA from the cis to trans side of αHL, while monitoring the ion current as a function of time.
Previous studies conducted in our laboratories have monitored translocation of DNA strands modified by a broad range of molecular adducts through the nanopore [29,31]. In these studies, some adducts were found to be too large to translocate through the central constriction of αHL (1.4 nm in diameter) [37]. Due to the size and hydrophobic nature of the BPDE adduct, a short 4-mer BPDE strand was chosen for our initial experiments to determine if the adduct was too large to pass through the narrow constriction zone. The short modified strand was advantageous because of its ease of synthesis and characterization (figure S1), and this simplified study provided the basis for understanding how the BPDE adduct interacts with the αHL channel.
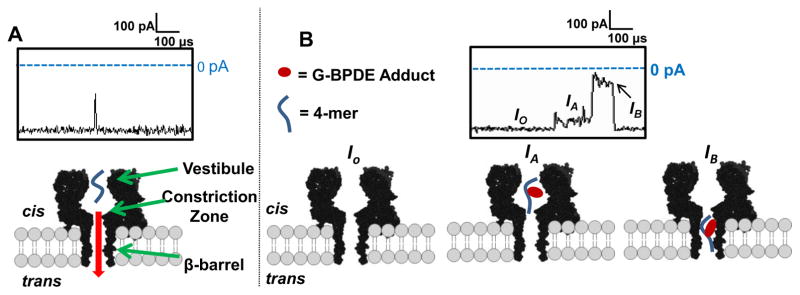
A comparison of ion current vs. time (i-t) recordings for the unmodified 4-mer and the 4-mer BPDE DNA oligomers recorded at 180 mV (trans vs. cis) in 1 M KCl solution is shown in figure 2. Based on previous studies, the anticipated residence time for the C-rich 4-mer strand in the αHL nanopore is predicted to be ~4–8 μs, and events of >50% blockage to the current in this time range were measured [38]. Translocation of the 4-mer BPDE oligomer resulted in longer (>10 μs) events and exhibited unique current patterns that were not observed for the unmodified 4-mer strand (figure 2B). All events initiated with a decrease in the open channel current (figure 2B, Io) to a mid-level current blockage (IA) that was centered at ~10% Io and lasted 10–200 μs. Next, the events progressed to a noisy deep-level current blockage (IB) that was centered at ~75% Io and lasted from 10 to 100 μs (figure 2B). All events returned to Io (figure 2B) without the appearance of another mid-level current suggesting the oligomer moved through the β-barrel and exited the trans side of the pore [38].
Figure 2.
Proposed model for translocation of a 4-mer and 4-mer BPDE adduct through the αHL nanopore. (A) Representative i-t trace for the 4-mer (5′-CCGC-3′) strand, (B) representative i-t trace for a 4-mer BPDE adducted oligomer. All data were recorded at 180 mV (trans vs. cis) in 1 M KCl at 25.0 ± 0.5°C with a 100 kHz low-pass filter and 500 kHz data acquisition rate.
Based on the i-t traces above, we propose the following model to describe how the 4-mer BPDE strand translocates through the αHL pore. The initial mid-level current, IA, is established when the 4-mer BPDE strand enters the vestibule of αHL from the cis side (figure 2B, IA). Next, the BPDE strand enters the β-barrel and the current drops to a deep blockage level (figure 2B, IB), during which the 4-mer BPDE strand is driven through the narrow β-barrel (d ~1.4 nm) to the trans side of αHL. The sharp return to Io, and not back to the mid-level current IA indicates that the 4-mer BPDE exited the trans side of the channel and does not return through the cis opening, because exit from the cis opening would give a second mid-level current (IA). This promising result suggests that BPDE adducts can be detected in longer oligomer model systems that would occupy the full length of the channel while the adduct interacts with the protein walls.
In the second study, a 41-mer poly-2′-deoxycytidine (poly-C) strand with a centrally located G was synthesized and allowed to react with BPDE to give an adduct yield of ~5%. After HPLC purification, reinjection of the adduct sample established that it contained 30% unreacted DNA. Therefore, when analyzing the 41-mer translocation data, events shorter than 50 μs were attributed to unreacted starting material (tmax = 44 μs at 120 mV) and discarded. Analysis of the 41-mer strand with a BPDE adduct (2 μM) was conducted in buffered solutions (25 mM PBS, pH 7.4) containing 1 M KCl and 3 M NaCl. Typical i-t translocation events that are characteristic of the unique pattern observed for the DNA-BPDE adduct in 1 M KCl electrolyte are shown in figure 3, and data collected in 3 M NaCl are presented in figure S3.
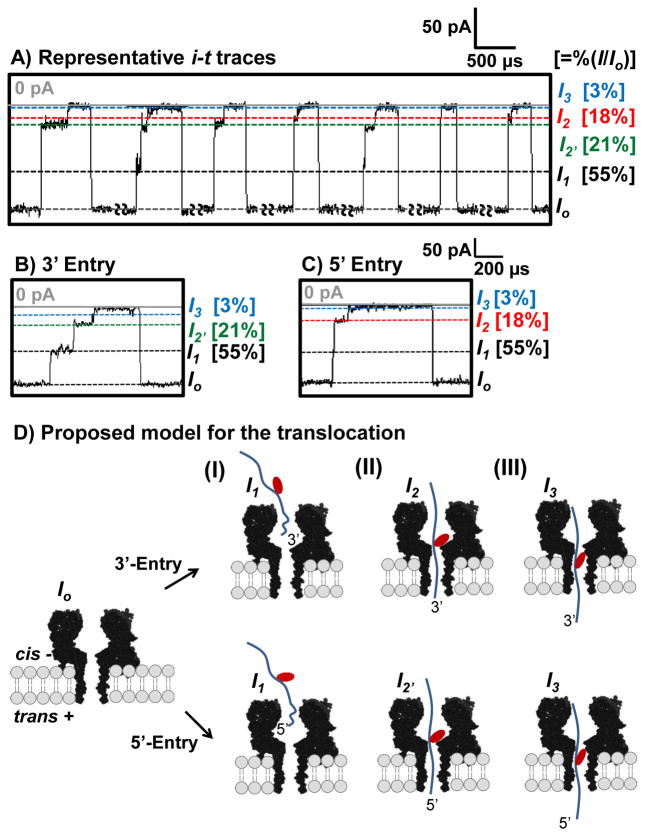
Figure 3.
Event types detected during translocation of the 41-mer BPDE sample. (A) Representative i-t traces for translocation of the 41-mer BPDE sample, (B) blowup of a 3′-entry event, and (C) blowup of a 5′-entry event. The data were recorded at 180 mV (trans vs. cis) at 25.0 ± 0.5°C. The data were refiltered to 50 kHz. Results from measurements are presented as percent ratio of the blockage current vs. open channel current %(I/Io). The i t traces for events >50 μs were analyzed. Long open channel current segments (20 - 500 ms) were manually removed, as indicated on the i-t trace. A relatively low capture rate (~70 events/s) was observed due to the low concentration (2 μM) of the 41-mer BPDE studied.(D) Proposed model for the translocation of a 41-mer BPDE adduct through αHL. (I) DNA enters from the cis side of the channel by threading either the 3′ or 5′ tail. (II) The BPDE adduct becomes caught at the 1.4 nm central constriction that gives rise to the deep blockage in the ion current recorded that marks the presence of the BPDE adduct. (III)The DNA translocates through the β-barrel.
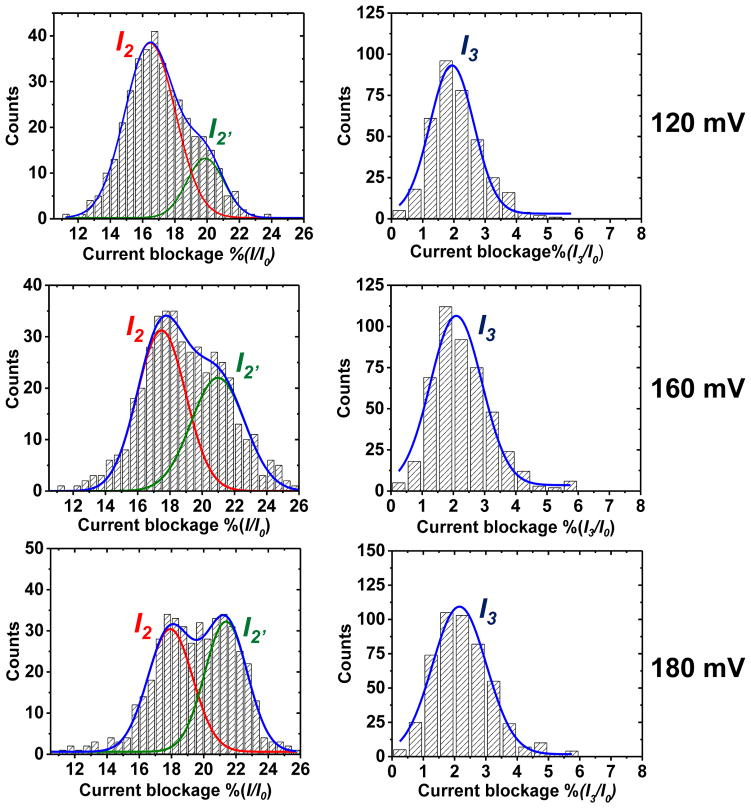
Translocation events for the 41-mer BPDE are initiated when the open channel current is reduced to a shallow shoulder level (I1) that has a %(I1/Io) = 55 ± 5% (figure 3). The entry of either the 3′ or 5′ end of the strand into the vestibule leads to varying lengths of the tail in the vestibule resulting in a shallow and broad distribution in current levels. Currents of this magnitude were previously described to result from partial entry of the DNA strand into the channel [35], similar to our results. Next, the event transitions to a deep level blockage current with values of I2 measured as 18 ± 2% and 21 ± 2% (figure 3B and C). Based on previous studies of poly-C translocation, these two current distributions represent the entry directionally (5′ or 3′) into the β-barrel [39,40]. Current histograms for the I2 currents can be deconvoluted into two pseudo-Gaussian distributions that are centered at the currents for 5′ and 3′ entry (figure 4). In these studies, the 3′ events with lower residual current dominate at lower voltage, and as the voltage is increased, the higher residual current events corresponding to 5′ entry increased. We propose that the %(I2/Io) current represents the bulky BPDE adduct being captured at the central constriction of αHL (figure 3 (D)), The adduct strand then remains caught in this position until a conformation feasible for overcoming the energy barrier and entering into the β-barrel is found. After the strand progresses through the β-barrel the current returns to the open channel value.
Figure 4.
Current histograms for the step-current levels monitored for the 41-mer BPDE events. (A) Plots of frequency distributions for the I2 and I2′ current levels. (B) Plots of frequency distributions for the I3 current levels. The data were collected at 120, 160 and 180 mV (trans vs. cis) in 1 M KCl at 25.0 ± 0.5°C and plotted with a bin size of 0.5 pA. Population distributions represent 400–450 events.
Events then advance to a final current level, I3, in which nearly all of the ion flux is attenuated (%(I3/Io) = ~3%). After the exit of the BPDE adduct, the ion flow returns to the open-channel current indicating translocation because exit through the cis opening would mirror the entry current pattern with a mid-level current blockage. This observation, along with the inverse correlation of time with increased voltage (table 1), supports the hypothesis that the 41-mer BPDE adduct translocates through αHL.
Table 1.
Time constants measured for the 41-mer standard and 41-mer BPDE strand vs. voltage.
| Voltage (mV)a | 41-mer BPDE 5′ & 3′ Events | 41-mer Standard |
|---|---|---|
| τ (μs)b | t max (μs)c | |
| 120 mV | 780 ± 15 | 44.1 ± 0.5 |
| 160 mV | 640 ± 20 | 21.0 ± 0.5 |
| 180 mV | 555 ± 15 | 18.5 ± 0.8 |
Voltage was measured trans vs. cis at 25 °C.
The reported time constant was determined by fitting the frequency vs. time histogram to a single-exponential decay function.
The reported time constant was determined by fitting the frequency vs. time histogram to a Gaussian function (figure S4).
The observed current blocking is consistent with the adduct causing a steric restriction to the movement of the strand through the pore that nearly blocks the ion flux. The distribution in the event time can be modeled by an exponential decay time distribution (figure S4) with a time constant τ for steps 2 and 3. These events were consistently longer (780 μs at 120 mV and 555 μs at 180 mV) than those observed for the standard (44 μs at 120 mV and 18 μs at 180 mV), suggesting that bulky BPDE adducts significantly slows translocation of the DNA strand through αHL (table 1).
Interestingly, 3′-entry and 5′-entry events both give the characteristic ion current, I3, for the BPDE adduct (figure 3B and C). In previous studies from our laboratories, an 18-crown-6 adduct also gave a characteristic low residual current (6–7%) [29]; however, these adducts were only detectable upon 5′-entry of the strand. In contrast, the BPDE adduct yields the current signature upon both 3′ and 5′ entry, which most likely occurs because of the larger size and rigidity of this adduct. Detection of BPDE adducts from both 3′ and 5′ entry will be very advantageous for its detection by the αHL nanopore.
4. Conclusions
In the current work, we set out to determine if BPDE adducts to G in DNA could be detected using the wild-type αHL nanopore. We demonstrate that a 41-mer DNA strand translocates through αHL and yields a characteristic three-step ion-current signature. We anticipate that the approach we outline here will be useful in the study of other lesions created by the adduction of polycyclic organic compounds, such as aflatoxin [40] and ochratoxin [42]. We anticipate that this result will provide the groundwork for future studies that aim to detect, quantify, and eventually sequence this lesion from cellular DNA sources.
Supplementary Material
Acknowledgments
The authors thank Electronic BioSciences Inc. (San Diego, CA) for donating instruments and software and the National Institutes of Health (GM093099) for funding this research.
References
- 1.Toyooka T, Ibuki Y. DNA damage induced by coexposure to PAHs and light. Environ Toxicol Pharmacol. 2007;23:256–263. doi: 10.1016/j.etap.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 2.Baird WM, Hooven LA, Mahadevan B. Carcinogenic polycyclic aromatic hydrocarbon-DNA adducts and mechanism of action. Environ Mol Mutagen. 2005;45:106–114. doi: 10.1002/em.20095. [DOI] [PubMed] [Google Scholar]
- 3.Phillips DH. Fifty years of benzo(a)pyrene. Nature. 1983;303:468–472. doi: 10.1038/303468a0. [DOI] [PubMed] [Google Scholar]
- 4.Armstrong B, Hutchinson E, Unwin J, Fletcher T. Lung cancer risk after exposure to polycyclic aromatic hydrocarbons: a review and meta-analysis. Environ Health Perspect. 2004;112:970–978. doi: 10.1289/ehp.6895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boffetta P, Jourenkova N, Gustavsson P. Cancer risk from occupational and environmental exposure to polycyclic aromatic hydrocarbons. Cancer causes control. 1997;8:444–472. doi: 10.1023/a:1018465507029. [DOI] [PubMed] [Google Scholar]
- 6.Brookes P, Lawley PD. Evidence for the binding of polynuclear aromatic hydrocarbons to the nucleic acids of mouse skin: relation between carcinogenic power of hydrocarbons and their binding to deoxyribonucleic acid. Nature. 1964;202:781–784. doi: 10.1038/202781a0. [DOI] [PubMed] [Google Scholar]
- 7.Mastrangelo G, Fadda E, Marzia V. Polycyclic aromatic hydrocarbons and cancer in man. Environ Health Perspect. 1996;104:1166–1170. doi: 10.1289/ehp.961041166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang SK, McCourt DW, Roller PP, Gelboin HV. Enzymatic conversion of benzo(a)pyrene leading predominantly to the diol-epoxide r-7,t-8-dihydroxy-t-9,10-oxy-7,8,9,10-tetrahydrobenzo(a)pyrene through a single enantiomer of r-7, t-8-dihydroxy-7,8-dihydrobenzo(a)pyrene. Proc Natl Acad Sci. 1976;73:2594–2598. doi: 10.1073/pnas.73.8.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kapitulnik J, Wislocki PG, Levin W, Yagi H, Jerina DM, Conney AH. Tumorigenicity studies with diol-epoxides of benzo(a)pyrene which indicate that (±)-trans-7beta,8alpha±-dihydroxy-9alpha,10alpha-epoxy-7,8,9,10-tetrahydrobenzo(a)pyrene Is an ultimate carcinogen in newborn mice. Cancer Res. 1978;38:354–358. [PubMed] [Google Scholar]
- 10.Pfeifer GP, Besaratinia A. Mutational spectra of human cancer. Human Genetics. 2009;125:493–506. doi: 10.1007/s00439-009-0657-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Denissenko MF, Pao A, Tang M, Pfeifer GP. Preferential formation of benzo[a]pyrene adducts at lung cancer mutational hospots in P53. Science. 1996;274:430–432. doi: 10.1126/science.274.5286.430. [DOI] [PubMed] [Google Scholar]
- 12.Jiang H, Shen Y-M, Quinn AM, Penning TM. Competing roles of cytochrome P450 1A1/1B1 and aldo-keto reductase 1A1 in the metabolic activation of (±)-7,8-dihydroxy-7,8-dihydro-benzo[a]pyrene in human bronchoalveolar cell extracts. Chem Res Toxicol. 2005;18:365–374. doi: 10.1021/tx0497245. [DOI] [PubMed] [Google Scholar]
- 13.Godschalk RW, Van Schooten FJ, Bartsch H. A critical evaluation of DNA adducts as biological markers for human exposure to polycyclic aromatic compounds. J Biochem Mol Biol. 2003;36:1–11. doi: 10.5483/bmbrep.2003.36.1.001. [DOI] [PubMed] [Google Scholar]
- 14.Manchester DK, Weston A, Choi JS, Trivers GE, Fennessey PV, Quintana E, Farmer PB, Mann DL, Harris CC. Detection of benzo[a]pyrene diol epoxide-DNA adducts in human placenta. Proc Natl Acad Sci US A. 1988;85:9243–9247. doi: 10.1073/pnas.85.23.9243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Azbal CC, Skipper PL, Yu MC, London SJ, Dasari RR, Tannenbaum SR. Quantification of (7S,8R)-dihydroxy-(9R,10S)-epoxy-7,8,9,10-tetrahydrobenzo[a]pyrene adducts in human serum albumin by laser-induced fluorescence: Implications for the in vivo metabolism of benzo[a]pyrene. Cancer Epidemiol, Biomarkers Prev. 2000;9:733–739. [PubMed] [Google Scholar]
- 16.Gelboin HV. Benzo[alpha]pyrene metabolism, activation and carcinogenesis: role and regulation of mixed-function oxidases and related enzymes. Physiol Rev. 1980;60:1107–1166. doi: 10.1152/physrev.1980.60.4.1107. [DOI] [PubMed] [Google Scholar]
- 17.Sram RJ, Binkova B. Molecular epidemiology studies on occupational and environmental exposure to mutagens and carcinogens, 1997–1999. Environ Health Perspect. 2000;108:57–70. doi: 10.1289/ehp.108-1637778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen YL, Wang CJ, Wu KY. Analysis of N7-(benzo[a]pyrene-6-yl)guanine in urine using two-step solid-phase extraction and isotope dilution with liquid chromatography/tandem mass spectrometry. Rapid Commun Mass Spectrom. 2005;19:893–898. doi: 10.1002/rcm.1868. [DOI] [PubMed] [Google Scholar]
- 19.Gennaro LA, Vadhanam M, Gupta RC, Vouros P. Selective digestion and novel cleanup techniques for detection of benzo[a]pyrene diol epoxide-DNA adducts by capillary electrophoresis/mass spectrometry. Rapid Commun Mass Spectrom. 2004;18:1541–1547. doi: 10.1002/rcm.1516. [DOI] [PubMed] [Google Scholar]
- 20.Tretyakova N, Villalta PW, Kotapati S. Mass spectrometry of structurally modified DNA. Chem Rev. 2013;113:2395–2436. doi: 10.1021/cr300391r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Singh R, Gaskell M, Le Pla RC, Kaur B, Azim-Araghi A, Roach J, Koukouves G, Souliotis VL, Kyrtopoulos SA, Farmer PB. Detection and quantitation of benzo[a]pyrene-derived DNA adducts in mouse liver by liquid Chromatography-Tandem Mass Spectrometry: Comparison with 32P-Postlabeling. Chem Res Toxicol. 2006;19:868–878. doi: 10.1021/tx060011r. [DOI] [PubMed] [Google Scholar]
- 22.Branton D, Deamer DW, Marziali A, Bayley H, Benner SA, Butler T, Di Ventra M, Garaj S, Hibbs A, Huang X, Jovanovich SB, Krstic PS, Lindsay S, Ling SL, Mastrangelo CH, Meller A, Oliver JS, Pershin YV, Ramsey JM, Riehn R, Soni GV, Tabard-Cossa V, Wanunu M, Wiggin M, Schloss JS. The potential and challenges of nanopore sequencing. Nat Biotechnol. 2008;26:1146–1153. doi: 10.1038/nbt.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stoloff DH, Wanunu M. Recent trends in nanopores for biotechnology. Curr Opin Biotechnol. 2013;24:699–704. doi: 10.1016/j.copbio.2012.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stoddart D, Heron A, Mikhailova E, Maglia G, Bayley H. Single-nucleotide discrimination in immobilized DNA oligonucleotides with a biological nanopore. Proc Natl Acad Sci USA. 2009;106:7702–7707. doi: 10.1073/pnas.0901054106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wallace EVB, Stoddart D, Heron AJ, Mikhailova E, Maglia G, Donohoe TJ, Bayley H. Identification of epigenetic DNA modifications with a protein nanopore. Chem Commun. 2010;46:8195–8197. doi: 10.1039/c0cc02864a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schibel AE, An N, Jin Q, Fleming AM, Burrows CJ, White HS. Nanopore detection of 8-oxo-7,8-dihydro-2′-deoxyguanosine in immobilized single-stranded DNA via adduct formation to the DNA damage site. J Am Chem Soc. 2010;132:17992–17995. doi: 10.1021/ja109501x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wolna AH, Fleming AM, An N, He L, White HS, Burrows CJ. Electrical Current Signatures of DNA Base Modifications in Single Molecules Immobilized in the α-Hemolysin Ion Channel. Isr J Chem. 2013;53:417–430. doi: 10.1002/ijch.201300022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wolna AH, Fleming AM, Burrows CJ. Single-molecule detection of a guanine(C8)-thymine(N3) cross-link using ion channel recording. J Phys Org Chem. 2014;27:247–251. doi: 10.1002/poc.3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.An N, Fleming AM, White HS, Burrows CJ. Crown ether-electrolyte interactions permit nanopore detection of individual DNA abasic sites in single molecules. Proc Natl Acad Sci USA. 2012;109:11504–11509. doi: 10.1073/pnas.1201669109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jin Q, Fleming AM, Johnson RP, Ding Y, Burrows CJ, White HS. Base-excision repair activity of uracil-DNA glycosylase monitored using the latch zone of alpha-hemolysin. J Am Chem Soc. 2013;135:19347–19353. doi: 10.1021/ja410615d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.An N, White HS, Burrows CJ. Modulation of the current signatures of DNA abasic site adducts in the [small alpha]-hemolysin ion channel. Chem Commun. 2012;48:11410–11412. doi: 10.1039/c2cc36366f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pirogov N, Shafirovich V, Kolbanovskiy A, Solntsev K, Courtney SA, Amin S, Geacintov NE. Role of Hydrophobic Effects in the Reaction of a Polynuclear aromatic diol epoxide with oligodeoxynucleotides in aqueous solutions. Chem Res Toxicol. 1998;11:381–388. doi: 10.1021/tx980006q. [DOI] [PubMed] [Google Scholar]
- 33.Zhang B, Galusha J, Shiozawa P, Wang G, Bergren A, Jones R, White R, Ervin E, Cauley C, White H. Bench-top method for fabricating glass-sealed nanodisk electrodes, glass nanpore electrodes, and glass nanopore membranes of controlled size. Anal Chem. 2007;79:4778–4787. doi: 10.1021/ac070609j. [DOI] [PubMed] [Google Scholar]
- 34.Schibel AEP, Heider EC, Harris JM, White HS. Fluorescence microscopy of the pressure-dependent structure of lipid bilayers suspended across conical nanopores. J Am Chem Soc. 2011;133:7810–15. doi: 10.1021/ja1117182. [DOI] [PubMed] [Google Scholar]
- 35.Akeson M, Branton D, Kasianowicz JJ, Brandin E, Deamer DW. Microsecond time-scale discrimination among polycytidylic acid, polyadenylic acid, and polyuridylic acid as homopolymers or as segments within single RNA molecules. Biophys J. 1999;77:3227–33. doi: 10.1016/S0006-3495(99)77153-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.White RJ, Ervin EN, Yang T, Chen X, Daniel S, Cremer PS, White HS. Single ion-channel recordings using glass nanopore membranes. J Am Chem Soc. 2007;129:11766–11775. doi: 10.1021/ja073174q. [DOI] [PubMed] [Google Scholar]
- 37.Song L, Hobaugh M, Shustak C, Cheley S, Bayley H, Gouaux J. Structure of straphylococcal α-hemolysin, a heptameric transmembrane pore. Science. 1996;274:1859–1866. doi: 10.1126/science.274.5294.1859. [DOI] [PubMed] [Google Scholar]
- 38.Kawano R, Schibel A, Cauley C, White H. Controlling the translocation of single-stranded DNA through α-hemolysin ion channels using viscosity. Langmuir. 2009;25:1233–1237. doi: 10.1021/la803556p. [DOI] [PubMed] [Google Scholar]
- 39.Butler TZ, Gundlach JH, Troll MA. Determination of RNA orientation during translocation through a biological nanopore. Biophys J. 2006;90:190–199. doi: 10.1529/biophysj.105.068957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.An N, Fleming AM, White HS, Burrows CJ. Crown ether-electrolyte interactions permit nanopore detection of individual DNA abasic sites in single molecules. Proc Natl Acad Sci U S A. 2012;109:11504–11509. doi: 10.1073/pnas.1201669109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kensler TW, Roebuck BD, Wogan GN, Groopman JD. Aflatoxin: a 50-year odyssey of mechanistic and translational toxicology. Toxicol Sci. 2011;120:S28–48. doi: 10.1093/toxsci/kfq283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Akman SA, Adams M, Case D, Park G, Manderville RA. Mutagenicity of ochratoxin A and its hydroquinone metabolite in the SupF gene of the mutation reporter plasmid Ps189. Toxins (Basel) 2012;4:267–280. doi: 10.3390/toxins4040267. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.