Abstract
GATA transcription factors regulate an array of genes important in cell proliferation and differentiation. Here we report the identification of regulator of G protein signaling 4 (RGS4) as a novel target for GATA-6 transcription factor. Although three sites (a, b, c) within the proximal region of rabbit RGS4 promoter for GATA transcription factors were predicted by bioinformatics analysis, only GATA-a site (16 bp from the core TATA box) is essential for RGS4 transcriptional regulation. RT-PCR analysis demonstrated that only GATA-6 was highly expressed in rabbit colonic smooth muscle cells but GATA-4/6 were expressed in cardiac myocytes and GATA-1/2/3 expressed in blood cells. Adenovirus-mediated expression of GATA-6 but not GATA-1 significantly increased the constitutive and IL-1β-induced mRNA expression of the endogenous RGS4 in colonic smooth muscle cells. IL-1β stimulation induced GATA-6 nuclear translocation and increased GATA-6 binding to RGS4 promoter. These data suggest that GATA factor could affect G protein signaling through regulating RGS4 expression, and GATA signaling may develop as a future therapeutic target for RGS4-related diseases.
Keywords: Regulator of G protein signal, GATA transcription factors, smooth muscle cells, gene regulations, Gastrointestinal tract
Introduction
The GATA family of the transcription factors were named because of their direct binding to the consensus DNA sequence (T/A)GATA(A/G) within the promoters and enhancers of target genes. They are evolutionarily conserved. In vertebrates, six members of the GATA family have been identified. GATA-1, -2 and -3 are mainly associated with haematopoiesis, whereas GATA-4, -5 and -6 are involved in cardiogenesis, neurogenesis and gut development[1–3]. GATA transcription factors play important roles in the proliferation and differentiation of various cell types including smooth muscle cells (SMCs)[4, 5]. The cardiomyocytes express GATA-4 and GATA-6, whereas vascular SMC express only GATA-6. GATA-6 plays a critical role in maintaining the contractile phenotype of vascular SMCs[4, 6]. For example, overexpression of GATA-6 activates the transcription of several contractile proteins and leads to cell cycle arrest in cultured vascular SMCs[6]. GATA-6 expression and its DNA-binding activity are reduced in neointimal vascular SMCs after rat carotid artery injury, while overexpression of GATA-6 limits the neointimal proliferative response[7]. However, SMC-specific GATA-6 knockout or dominant-negative GATA-6 mutant inhibition leads to little or no change in expression of genes encoding contractile proteins in vascular SMCs[6]. Earlier studies demonstrated that GATA-6 was not expressed in visceral SMCs populating the gastrointestinal tract and uterus, while it was expressed in the epithelial cells of the gastrointestinal tract[8]. Later study using immunohistochemistry showed that GATA-6 is expressed in the gut[9]. A recent study showed that GATA-6 is expressed in human bladder SMC[10]. GATA-6 regulates CPI-17 expression and Ca2+ sensitization in bladder smooth muscles[5, 11].
Regulator of G protein signaling (RGS) proteins are a large family of highly diverse, multifunctional signaling proteins. RGS proteins bind to the activated Gα subunit and terminates G protein signaling by boosting the intrinsic GTPase activity of Gα subunit. RGS4 is one of seven members of a classic R4 RGS protein that accelerates the GTPase activity of the Gαi/o and Gαq/11 family members[12, 13]. Extensive studies have demonstrated that RGS4 plays an important role in regulating smooth muscle contraction, cardiomyocyte development, neural plasticity and psychiatric disorders[14, 15]. However, the regulatory mechanisms underlying RGS4 expression and function remain largely unclear. We have recently shown that the pro-inflammatory cytokine, interleukin-1β (IL-1β) induces up-regulation of RGS4 mRNA and protein expression in rabbit colonic SMCs[16]. The increase in RGS4 expression is responsible for IL-1β-induced inhibition of initial Ca2+-dependent smooth muscle contraction[16]. Up-regulation of RGS4 expression by IL-1β is mediated by the canonical IKK2/IκBα/NFκB signaling[17], enhanced by extracellular signal-regulated kinases (ERK1/2) and p38 mitogen-activated protein (MAP) kinase[18], and inhibited by phosphoinositide-3 kinase[18] and JNK pathway[19].
To understand the transcriptional mechanism of RGS4 expression, we have cloned rabbit RGS4 promoter[20] and found that several transcription factors regulate RGS4 promoter activity[17–20]. Sequence analysis predicts three sites for GATA transcription factors within the proximal region[20]. In the present study, we characterized the expression pattern of GATA transcription factors in the colonic SMCs and investigated the role of GATA-6 in regulating RGS4 transcription.
Material and Methods
Reagents, antibodies and adenovirus
IL-1β was obtained from Alexis Biochemicals (San Diego, CA). JNK-II inhibitor (SP600125, Anthra[1,9-cd]pyrazol-6(2H)-one, 1,9-pyrazoloanthrone), and IKK2-IV (IKK2 inhibitor IV, [5-(p-Fluorophenyl)-2-ureido]thiophene-3-carboxamide) were obtained from EMD Chemicals (San Diego, CA) and dissolved in dimethyl sulfoxide (DMSO). Antibodies against GATA-1, GATA-6, GAPDH and β-actin were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Affinity-purified anti-Rgs4 antibody was kindly provided by Dr. Susanne M. Mumby (University of Texas Southwest Medical Center). Adenovirus (Ad) encoding EGFP, GATA-1 and GATA-6 were obtained from Vector Biolabs (Philadelphia, PA). All the other reagents were from Sigma (St. Louis, MO).
SMC culture
New Zealand White rabbit (2~2.5 kg, male) was used following IACUC guidance at Temple University. After euthanization with overdose of euthasol (150 mg/kg, i.v.), the distal colon was removed and placed in HEPES-buffered smooth muscle media. The circular SMCs were cultured as previously described[16, 17, 21].
Deletion and site-directed mutagenesis
The wild-type RGS4 promoter (designed as RGS4-P1) was generated by PCR cloning as described previously[20]. Various deletion constructs of pMluc3-Rgs4-P1 (−1389 to +50) were generated through digestion, blunting and ligation by analyzing and combining the digestion sites within the insert and the backbone vector. The mutants pMluc3-Rgs4-P2(−962 to +50) and pMluc-Rgs4-P3(−1389 to −816) were generated by single digestion with PstI and HindIII respectively followed by ligation. The mutant pMluc-Rgs4-P4(−247 to +50) was generated by double digestion with XhoI/XbaI followed by blunting and ligation.
Potential GATA transcription factor binding sites within Rgs4 promoter were identified by MatInspector (http://www.genomatix.de) and TFSEARCH (http://www.cbrc.jp). Mutation of the binding site for the GATA transcription factors in the reporter vector construct was performed by site-directed mutagenesis using the QuikChange kit (Stratagene). Mutagenic primers as shown in Table 1 led to nucleotide change in entire binding site for transcriptional factor.
Table 1.
Sequences for PCR primers, site-mutagenesis oligonucleotides
| Target name | Direction | sequence |
|---|---|---|
| Probe and mutagenic primer | ||
| GATA-a probe | Sense |
|
| Antisense |
|
|
| GATA-a mutation | Sense |
|
| Antisense |
|
|
| GATA-b mutation | Sense |
|
| Antisense |
|
|
| GATA-c mutation | Sense |
|
| Antisense |
|
|
| Primers for conventional PCR | ||
| GATA-1 | Sense | CTCAATTCAGCAGCCTATTCCT |
| Antisense | CCTGTCCTGTCCCTCCGCCACA | |
| GATA-2 | Sense | GTCTTCTTCAAYCAYCTCGACT |
| Antisense | CCCGTCCAGCCAGGGCAAACCC | |
| GATA-3 | Sense | TCTGGAGGAGGAAYGCYAATGG |
| Antisense | CGGTTTCKGGTCTGGATGCCTT | |
| GATA-4 | Sense | TTCTCAGAAGGCAGAGAGTGTG |
| Antisense | TGGCAGTTGGCACAGGAGAGG | |
| GATA-5 | Sense | TCCGACTTCCTGGAGGAGTTCC |
| Antisense | GAGCGGGCGGTTGACGCCGTTCA | |
| GATA-6 | Sense | GACCAGGAAACGAAAACCTAA |
| Antisense | CCTGAGGCTGTRGRTTGTGTTGT | |
| RGS4 | Sense | TCCCACAGCAAGAAGGACAAA |
| Antisense | TTCGGCCCATTTCTTGACTT | |
| GAPDH | Sense | CGCCTGGAGAAAGCTGCTAA |
| Antisense | CGACCTGGTCCTCGGTGTAG | |
Note: The wild-type GATA site highlighted red color but the mutant GATA sites highlighted green color. The overhang tcga on the 5′-end of the probe was used for end-labeling.
Cell transfection, luciferase reporter assays, electrophoretic mobility shift assay, chromatin immunoprecipitation assay, degenerative RT-PCR and RT-qPCR, Western blot, immunocytochemistry and statistical analysis
All these methods were performed as we previously described[16–21].
Results
GATA factor is critical for RGS4 promoter activity
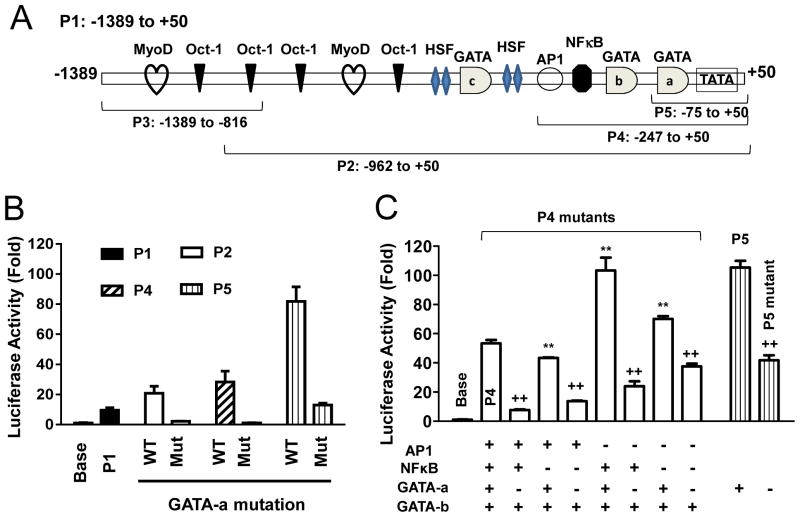
When performing deletion studies on the RGS4 P1 promoter activity using luciferase reporter assay, we identified a proximal promoter region (−247, P4) with a “maximal” promoter activity[20]. Further deletion studies identified the most proximal region (−75, P5) with higher promoter activity (Fig. 1A, 1B). The P5 contains the conserved typical TATA box (−35) that may serve as core promoter and a sole binding site for transcription factor GATA (−51). Site-directed mutation of this GATA binding site (designated GATA-a) P5, P4 and P2 dramatically reduced the promoter activity as compared with corresponding wild-type promoter region (Fig. 1B). These data suggest that the GATA-a response element is critical for RGS4 promoter activity and the binding GATA factor may serve as trans-activator for RGS4 transcription.
Fig. 1. Essential role of GATA-a motif in regulating RGS4 promoter activity in rabbit colonic smooth muscle cells (SMCs).
(A), Diagram showing the location of each promoter mutants and binding sites for predicted transcriptional factors. (B) Removal of promoter regions containing GATA-b and -c sites plus various transcription factor sites increased RGS4 promoter activity. In contrast, site-directed mutagenesis of GATA-a site in P2, P4 and P5 all significantly reduced the corresponding promoter activity. (C) Site-directed mutagenesis of individual or combined binding sites for GATA-a, NFκB and AP1. Cultured colonic SMCs were cotransfected with promoter-less pMlu3 empty vector (Base) or indicated RGS4 promoter vector carrying renilla luciferase and pGL4-CMV vector carrying firefly luciferase (for normalization). After 24 h, the renilla and firefly luciferases were measured separately. The relative fold changes in renilla luciferase activity after normalization by firefly luciferase were expressed as compared with the empty vector. Data represents the mean ± SEM of 3–4 independent experiments. ** P<0.01 indicate statistically significant increase by student’s t test compared with corresponding P1 (B) or P4 (C). ++ p<0.01 indicates significant decrease in GATA-a mutants as compared with corresponding GATA-a non-mutants.
The bioinformatics analysis of the cloned P1 promoter sequence identified two additional potential cis-elements for GATA transcriptional factor designated GATA-b and -c (Fig. 1A). Removal of the regions that contain these two additional GATA sites (GATA-b and -c) plus other transcription factor sites led to the increase of RGS4 promoter activity from 21-fold (P2) or 28-fold (P4) to 82-fold (P5) (Fig. 1B), suggesting that these two additional GATA sites plus other factors may suppress the core promoter activity. To clarify the role of GATA sites and other factors, we chose P4 as a model system because P4 containing AP1, NFκB and GATA-a/b sites (Fig. 1A). As shown in Fig. 1C, mutation of NFκB binding site inhibited while mutation of AP1 binding site enhanced the promoter activity of P4, consistent with our previous observation that NFκB signaling stimulates while AP1 signaling represses the transcription of RGS4[17, 19, 20]. Dual deletion of NFκB and AP1 sites induced a mixed effect on the P4 promoter activity. Again, mutation of GATA-a site in corresponding promoters all reduced the promoter activity, confirming the critical role of GATA-a site as a main trans-activator on RGS4 core promoter.
GATA-a site is highly conserved and contributes to RGS4 promoter activity
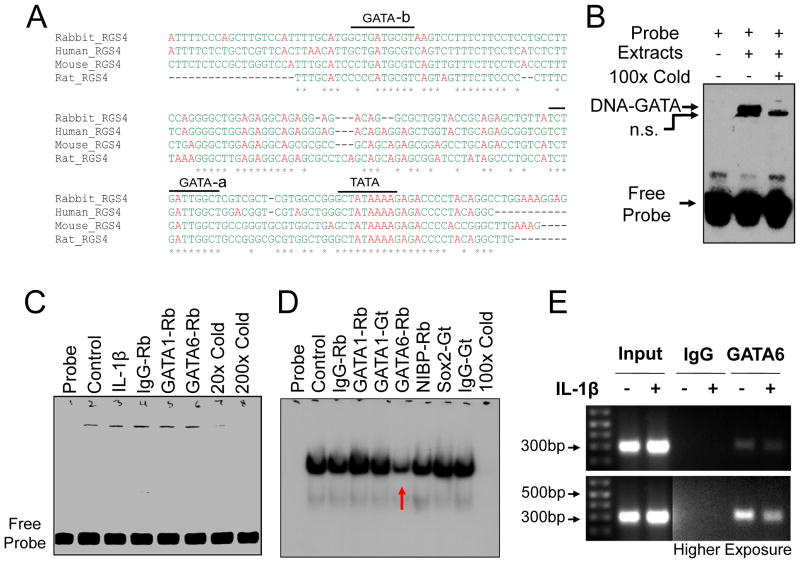
Homology analysis of predicted RGS4 promoter from different species showed that only GATA-a site (TCTGATTGGCT) shares 100% identity (Fig. 2A). To further confirm the binding activity of the conserved GATA-a site with GATA transcription factors, we performed EMSA using biotinylated (Fig. 2B) or radiolabeled (Fig. 2C, D) oligonucleotide probe containing GATA-a motif. The GATA-a DNA-binding complexes were detectable in nuclear extracts from human Hela cells (Fig. 2B, C) and rabbit colonic SMCs (Fig. 2D). The specific binding band of the shifted protein-DNA complex was completely blocked by pretreatment with 20–200× unlabeled GATA-a probe. The specificity of GATA-a binding to GATA-6 but not GATA-1 transcription factor was confirmed by pretreatment with anti-GATA antibody (Fig. 2C, D). The GATA-a binding of the endogenous GATA-6 was corroborated with CHIP-PCR analysis (Fig. 2E). These data suggest that GATA-a site near the core promoter (16 bp from the highly conserved TATA box) is functional in recruiting GATA-6 transcription factor to activate RGS4 transcription.
Fig. 2. GATA-6 binding to the highly conserved GATA-a site within RGS4 promoter.
(A) Multiple alignment from various species by Clustal W method showing the highly-conserved GATA-a site. (B–D) Electrophoretic mobility shift assay of nuclear extracts from human Hela cells (B, C) and rabbit colonic SMCs (D) using biotinylated (B) or radio-labeled (C, D) oligonucleotide probe targeting GATA-a site. The GATA-a-binding band was reduced specifically by pretreatment with rabbit (Rb) anti-GATA-6 antibody (C, D). Additional Rb or goat (Gt) antibodies were used as negative control. n.s. for non-specific band. (E) Chromatin immunoprecipitation assay of serum-starved SMCs after treatment with IL-1β for 3 h. Input indicates the DNA from supernatant after precipitation without IgG.
GATA-6 is highly expressed in rabbit colonic SMCs
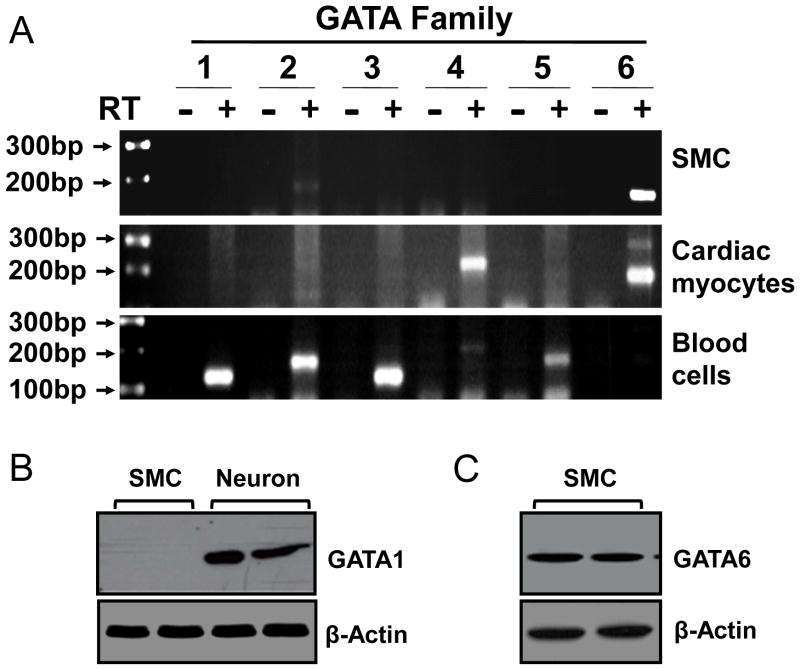
Vascular SMCs express only GATA-6[8]. Previous report using in situ hybridization technique showed the absence of all identified GATA members in mouse gut SMCs[8]. However, immunohistochemical studies demonstrated the presence of GATA-6 in mouse gut SMCs[9]. To determine whether GATA members are expressed in rabbit gut SMCs, we screened the expression pattern of all the family members by performing degenerative RT-PCR using cultured pure SMCs from rabbit colon. The rabbit cardiomyocytes and blood cells were used as positive control for various members of GATA family. Only GATA-6 is highly expressed in colonic SMCs, while GATA-2 is expressed at a lower level (Fig. 3A). The cardiomyocytes expressed GATA-4 and -6 while the blood cells expressed mainly GATA-1 to -3, consistent with previous reports[22, 23]. The expression of GATA-6 but not GATA-1in rabbit colonic SMCs was further corroborated at the protein level by Western blot analysis (Fig. 3B, C). The efficiency of GATA-1 antibody was verified using the enteric neuronal cell line (Fig. 3B).
Fig. 3. Expression of GATA-6 mRNA (A) and protein (B, C) in rabbit colonic SMCs.
(A) Degenerative RT-PCR analysis of 6 GATA transcription factors using total RNA from freshly isolated colonic SMCs, caridiomyocytes and blood cells. (B, C) Western blot analysis with indicated antibodies. Enteric neuronal cell line was used positive control for anti-GATA-1 antibody.
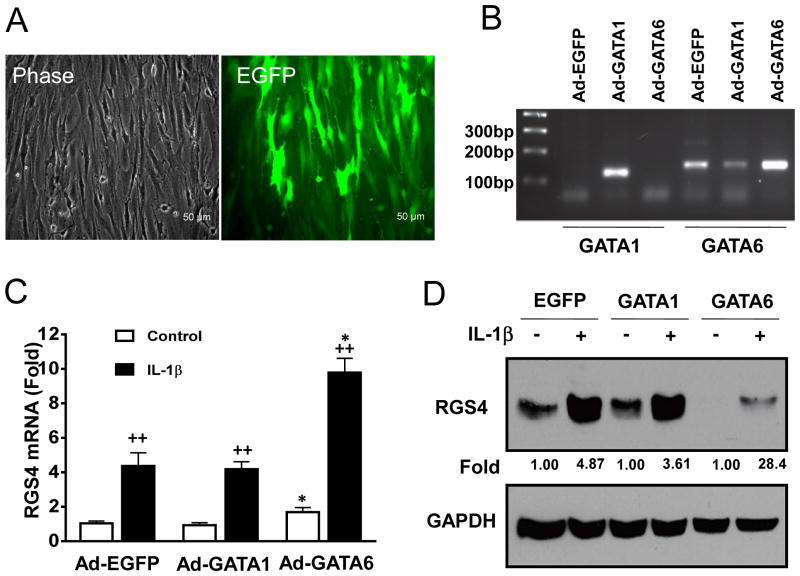
Overexpression of GATA-6 but not GATA-1 increases endogenous RGS4 expression at both mRNA and protein levels in rabbit colonic SMCs
The presence of GATA-6 in SMCs and cardiomyocytes contributes to its function in changing the phenotypes of these cells. As described above, GATA-6 in SMCs binds to the promoter of RGS4 both in vitro and in vivo (Fig. 2B–E). To provide a direct evidence that GATA-6 specifically regulates RGS4 expression, we employed adenovirus-mediated gene delivery system to overexpress GATA-6 in rabbit SMCs and performed RT-qPCR and Western blot analysis. Ad-EGFP virus at 10 MOI achieved over 95% transduction efficiency in rabbit colonic SMCs (Fig. 4A). Overexpression of GATA-6, validated by RT-PCR (Fig. 4B), significantly increased RGS4 transcription (Fig. 4C). In addition, GATA-6 significantly enhanced IL-1β-induced upregulation of RGS4 mRNA and protein expression (Fig. 4C, D). However, overexpression of GATA-1 had no significant effect on the constitutive and IL-1β-induced expression of RGS4 mRNA and protein (Fig. 4C, D), though it moderately inhibited mRNA expression of endogenous GATA-6 (Fig. 4B). These results suggest that GATA-6 expression in SMCs plays an important role in maintaining RGS4 expression in SMCs while GATA-1 is indispensable for RGS4 expression in SMCs.
Fig. 4. Adenovirus (Ad)-mediated overexpression of GATA-6 not GATA-1 in rabbit colonic SMCs significantly increased constitutive and IL-1β-induced RGS4 promoter activity.
(A) Fluorescent micrograph showing the efficiency of Ad-EGFP transduction. (B) RT-PCR analysis validated the expression of Ad-GATA-1 and Ad-GATA-6. Ad-GATA-1 inhibited the mRNA expression of endogenous GATA-6. (C, D) RT-qPCR and Western blot analysis for RGS4 mRNA and protein expression. Rabbit colonic SMCs were infected with indicated Ad for 24 h and treated with or without IL-1β (10 ng/ml) for 3 h before total RNA and protein lysates were prepared. * P<0.05 indicates a statistically significant increase by student’s t test compared with Ad-EGFP control. ++ P<0.01 indicates a significant increase in IL-1β treatment group compared with the control group. The number between panels indicates the relative fold of optical density after IL-1β treatment compared with the corresponding control.
IL-1β stimulation induced GATA-6 nuclear translocation and RGS4 promoter-binding activity
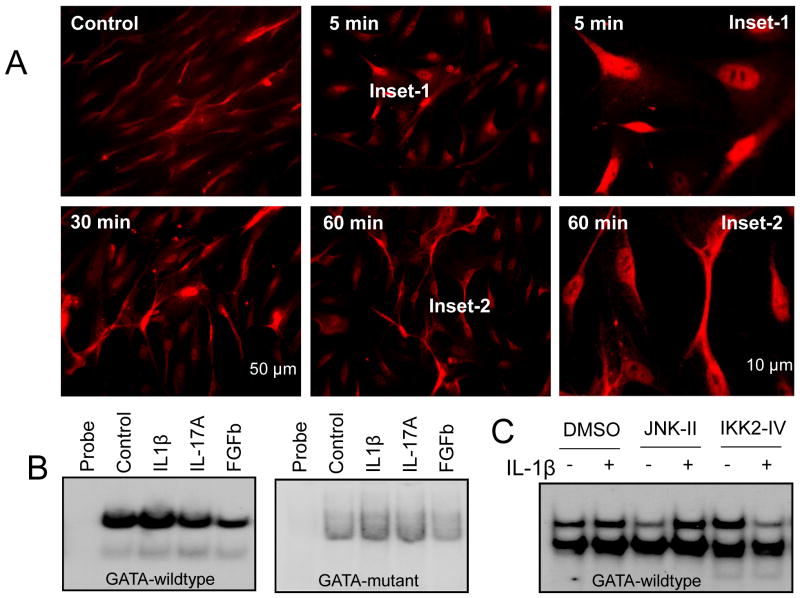
To assess if GATA-6 undergoes nuclear translocation after IL-1β stimulation, we performed immunocytochemistry with GATA-6 antibody at different time point after IL-1β treatment in cultured colonic SMCs. Under physiological condition, GATA-6 was present in the cytoplasm of colonic SMCs. Upon IL-1β stimulation, GATA-6 translocated into nucleus within minutes and recovered partially to cytoplasm at 1 h (Fig. 5A). Treatment for 1 h with IL-1β but not IL-17A increased the binding of GATA-6 to the RGS4 promoter while FGFb reduced GATA-6 binding to RGS4 promoter (Fig. 2C, 5B). To determine the potential effect of IL-1β-stimulated signaling pathways on the RGS4 GATA-a binding activity, we treated the cultured rabbit SMCs with IL-1β in the presence or absence of JNK inhibitor (for AP1 pathway) and IKK2 inhibitor (for NFκB pathway). IL-1β-induced GATA-6 binding to RGS4 promoter was blocked by pretreatment with IKK2 inhibitor but not JNK inhibitor (Fig. 5C). These data suggest that IL-1β-stimulated activation of IKK2/NFκB signaling modulates GATA-6-mediated RGS4 upregulation in SMCs.
Fig. 5. IL-1β induced GATA-6 nuclear translocation and DNA binding to RGS4 promoter in rabbit colonic smooth muscle cells (SMCs).
(A) Immunocytochemistry with goat anti-GATA-6 antibody at indicated time point after IL-1β treatment (10 ng/ml). (B) Electrophoretic mobility shift assay with radiolabeled wildtype or mutant GATA-a probe using nuclear extracts from rabbit colonic SMCs 1 h after treatment with indicated cytokines. (C) The effect of JNK and IKK2 signaling pathway on IL-1β-induced GATA-6 binding to RGS4 promoter. SMCs were pretreated with JNK inhibitor II (10 μM) or IKK2 inhibitor IV (1 μM) for 30 min before IL-1β (10 ng/ml) treatment for 1 h. Electrophoretic mobility shift assay of whole cell lysates with radio-labeled GATA-a wildtype probe was performed.
Discussion
The salient finding in the present study is the identification of RGS4 as a novel target gene for the GATA family of transcription factors. The GATA family executes its functions via regulating the transcription of its target genes. GATA-6 regulates a large array of target genes related to muscle cell differentiation[4–6, 10, 23–29]. For SMC contraction, GATA-6 upregulates contractile proteins[4–6] and caveolin[6, 24, 30]. In the present study, we demonstrated for the first time that GATA-6 is involved in G protein signaling and acts as a critical activator for the transcriptional expression of RGS4. Gut SMC contractility is composed of initial and sustained phases. Our previous studies demonstrated that RGS4 and CPI-17 mediate the initial and sustained contraction of gut SMCs respectively[16, 31]. Upregulation of RGS4 mediates the inhibition of initial contraction induced by IL-1β[16] or colonic inflammation[32]. GATA-6 is highly expressed in gut SMCs, suggesting that GATA-6 may regulate the smooth muscle contraction via affecting RGS4 and perhaps other key targets mediating initial and sustained contraction of gut smooth muscles. For example, GATA-6 upregulates CPI-17 expression in bladder SMCs and thus promotes PKC-mediated signaling and bladder smooth muscle remodeling[5, 11]. Therefore, GATA-6 signaling pathway may be a novel therapeutic target for modulating SMC contractility.
The conserved DNA motif that binds to GATA transcription factor was initially identified in the globin promoter from erythroid cells[33]. Later on, similar GATA motifs have been identified in a huge list of genes important in the proliferation and differentiation of cardiomyocytes and various SMCs [4–6, 10, 23–29, 34]. The family of GATA transcription factors contains 6 members with different pattern of cellular and tissue expression. GATA-6 is expressed during embryogenesis as early as the blastocyst stage at 3.5 days. Mouse GATA-6 knockout is lethal at 5.5 days postcoitum due to a defect in extraembryonic tissue formation[35]. GATA-6 is expressed mainly in heart, muscle, epithelium and all types of smooth muscles in the gastrointestinal tract, respiratory tract, arteries, and urogenital tract[4–6, 10, 23–29, 36–39]. In gastrointestinal tract, previous studies have demonstrated extensive expression of GATA-6 in instestinal epithelium and particularly in proliferative crypt compartment[37–39]. In GATA-6-deficient mice, there are dramatic impairments in the proliferation, migration and lineage maturation of colonic epithelium[40]. However, the expression and function of GATA-6 in gut SMCs remain controversial[8, 9]. We provide convincing evidence that GATA-6 is highly expressed in colonic SMCs and overexpression of GATA-6 but not GATA-1 enhances IL-1β-induced upregulation of RGS4. Such unique expression may contribute to the important role of GATA-6 in regulating gut SMC function. GATA-6 has been shown to maintain the differentiated state of vascular SMCs[41], likely via regulation of contractile protein expression[4]. In gut SMCs, GATA-6 may play a similar role in regulating muscle contractile proteins and cell differentiation.
Within the identified promoter region of rabbit RGS4, there are three sites for GATA factors. Only GATA-a site is highly conserved among different species. Deletion and site-mutagenesis assay validated the essential role of GATA-a site in regulating RGS4 promotor activity. However, the role of GATA-b and GATA-c remains to be determined because they are neighboring with several sites for additional transcription factors such as NFκB, AP1 and HSF, and thus may modulate the coordination of these transcription factors.
In conclusion, we demonstrated that the high level of GATA-6 expression in gut SMCs plays an essential role in regulating the expression of RGS4 via GATA-a cis-element. GATA-6 may act as a novel positive regulator for RGS4 and thus contributes to RGS4-related cellular process and diseases.
Highlights.
GATA-6 is highly expressed in colonic smooth muscle cells.
RGS4 is a novel target for GATA-6 transcription factor.
GATA-a response element is essential to regulate the core promoter of RGS4.
GATA-6 regulates IL-1β-induced RGS4 upregulation.
Acknowledgments
This work was supported by Grant DK075964 (WH) and DK015564 (KSM) from the National Institutes of Diabetes, and Kidney and Digestive Diseases.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Aronson BE, Stapleton KA, Krasinski SD. Role of GATA factors in development, differentiation, and homeostasis of the small intestinal epithelium. Am J Physiol Gastrointest Liver Physiol. 2014;306:G474–490. doi: 10.1152/ajpgi.00119.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morceau F, Schnekenburger M, Dicato M, Diederich M. GATA-1: friends, brothers, and coworkers. Ann N Y Acad Sci. 2004;1030:537–554. doi: 10.1196/annals.1329.064. [DOI] [PubMed] [Google Scholar]
- 3.Charron F, Nemer M. GATA transcription factors and cardiac development. Semin Cell Dev Biol. 1999;10:85–91. doi: 10.1006/scdb.1998.0281. [DOI] [PubMed] [Google Scholar]
- 4.Yin F, Herring BP. GATA-6 can act as a positive or negative regulator of smooth muscle-specific gene expression. J Biol Chem. 2005;280:4745–4752. doi: 10.1074/jbc.M411585200. [DOI] [PubMed] [Google Scholar]
- 5.Boopathi E, Hypolite JA, Zderic SA, Gomes CM, Malkowicz B, Liou HC, Wein AJ, Chacko S. GATA-6 and NF-kappaB activate CPI-17 gene transcription and regulate Ca2+ sensitization of smooth muscle contraction. Mol Cell Biol. 2013;33:1085–1102. doi: 10.1128/MCB.00626-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lepore JJ, Cappola TP, Mericko PA, Morrisey EE, Parmacek MS. GATA-6 regulates genes promoting synthetic functions in vascular smooth muscle cells. Arterioscler Thromb Vasc Biol. 2005;25:309–314. doi: 10.1161/01.ATV.0000152725.76020.3c. [DOI] [PubMed] [Google Scholar]
- 7.Mano T, Luo Z, Malendowicz SL, Evans T, Walsh K. Reversal of GATA-6 downregulation promotes smooth muscle differentiation and inhibits intimal hyperplasia in balloon-injured rat carotid artery. Circ Res. 1999;84:647–654. doi: 10.1161/01.res.84.6.647. [DOI] [PubMed] [Google Scholar]
- 8.Morrisey EE, Ip HS, Lu MM, Parmacek MS. GATA-6: a zinc finger transcription factor that is expressed in multiple cell lineages derived from lateral mesoderm. Dev Biol. 1996;177:309–322. doi: 10.1006/dbio.1996.0165. [DOI] [PubMed] [Google Scholar]
- 9.Nemer G, Nemer M. Transcriptional activation of BMP-4 and regulation of mammalian organogenesis by GATA-4 and -6. Dev Biol. 2003;254:131–148. doi: 10.1016/s0012-1606(02)00026-x. [DOI] [PubMed] [Google Scholar]
- 10.Kanematsu A, Ramachandran A, Adam RM. GATA-6 mediates human bladder smooth muscle differentiation: involvement of a novel enhancer element in regulating alpha-smooth muscle actin gene expression. Am J Physiol Cell Physiol. 2007;293:C1093–1102. doi: 10.1152/ajpcell.00225.2007. [DOI] [PubMed] [Google Scholar]
- 11.Boopathi E, Gomes C, Zderic SA, Malkowicz B, Chakrabarti R, Patel DP, Wein AJ, Chacko S. Mechanical stretch upregulates proteins involved in Ca2+ sensitization in urinary bladder smooth muscle hypertrophy. Am J Physiol Cell Physiol. 2014;307:C542–553. doi: 10.1152/ajpcell.00033.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tokudome T, Kishimoto I, Horio T, Arai Y, Schwenke DO, Hino J, Okano I, Kawano Y, Kohno M, Miyazato M, Nakao K, Kangawa K. Regulator of G-protein signaling subtype 4 mediates antihypertrophic effect of locally secreted natriuretic peptides in the heart. Circulation. 2008;117:2329–2339. doi: 10.1161/CIRCULATIONAHA.107.732990. [DOI] [PubMed] [Google Scholar]
- 13.Druey KM, Sullivan BM, Brown D, Fischer ER, Watson N, Blumer KJ, Gerfen CR, Scheschonka A, Kehrl JH. Expression of GTPase-deficient Gialpha2 results in translocation of cytoplasmic RGS4 to the plasma membrane. J Biol Chem. 1998;273:18405–18410. doi: 10.1074/jbc.273.29.18405. [DOI] [PubMed] [Google Scholar]
- 14.Gerber KJ, Squires KE, Hepler JR. Roles for Regulator of G Protein Signaling Proteins in Synaptic Signaling and Plasticity. Mol Pharmacol. 2016;89:273–286. doi: 10.1124/mol.115.102210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bansal G, Druey KM, Xie Z. R4 RGS proteins: regulation of G-protein signaling and beyond. Pharmacol Ther. 2007;116:473–495. doi: 10.1016/j.pharmthera.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu W, Mahavadi S, Li F, Murthy KS. Upregulation of RGS4 and downregulation of CPI-17 mediate inhibition of colonic muscle contraction by interleukin-1beta. Am J Physiol Cell Physiol. 2007;293:C1991–2000. doi: 10.1152/ajpcell.00300.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu W, Li F, Mahavadi S, Murthy KS. Interleukin-1beta up-regulates RGS4 through the canonical IKK2/IkappaBalpha/NF-kappaB pathway in rabbit colonic smooth muscle. Biochem J. 2008;412:35–43. doi: 10.1042/BJ20080042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu W, Li F, Mahavadi S, Murthy KS. Upregulation of RGS4 expression by IL-1beta in colonic smooth muscle is enhanced by ERK1/2 and p38 MAPK and inhibited by the PI3K/Akt/GSK3beta pathway. Am J Physiol Cell Physiol. 2009;296:C1310–1320. doi: 10.1152/ajpcell.00573.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y, Li F, Liu S, Wang H, Mahavadi S, Murthy KS, Khalili K, Hu W. MEKK1-MKK4-JNK-AP1 pathway negatively regulates Rgs4 expression in colonic smooth muscle cells. PLoS One. 2012;7:e35646. doi: 10.1371/journal.pone.0035646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li F, Murthy KS, Khalili K, Hu W. Cloning and characterization of rabbit Rgs4 promoter in gut smooth muscle. Gene. 2010;451:45–53. doi: 10.1016/j.gene.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li F, Hu DY, Liu S, Mahavadi S, Yen W, Murthy KS, Khalili K, Hu W. RNA-binding protein HuR regulates RGS4 mRNA stability in rabbit colonic smooth muscle cells. Am J Physiol Cell Physiol. 2010;299:C1418–1429. doi: 10.1152/ajpcell.00093.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alexandrovich A, Arno M, Patient RK, Shah AM, Pizzey JA, Brewer AC. Wnt2 is a direct downstream target of GATA6 during early cardiogenesis. Mech Dev. 2006;123:297–311. doi: 10.1016/j.mod.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 23.Lepore JJ, Mericko PA, Cheng L, Lu MM, Morrisey EE, Parmacek MS. GATA-6 regulates semaphorin 3C and is required in cardiac neural crest for cardiovascular morphogenesis. J Clin Invest. 2006;116:929–939. doi: 10.1172/JCI27363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boopathi E, Gomes CM, Goldfarb R, John M, Srinivasan VG, Alanzi J, Malkowicz SB, Kathuria H, Zderic SA, Wein AJ, Chacko S. Transcriptional repression of Caveolin-1 (CAV1) gene expression by GATA-6 in bladder smooth muscle hypertrophy in mice and human beings. Am J Pathol. 2011;178:2236–2251. doi: 10.1016/j.ajpath.2011.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yin F, Hoggatt AM, Zhou J, Herring BP. 130-kDa smooth muscle myosin light chain kinase is transcribed from a CArG-dependent, internal promoter within the mouse mylk gene. Am J Physiol Cell Physiol. 2006;290:C1599–1609. doi: 10.1152/ajpcell.00289.2005. [DOI] [PubMed] [Google Scholar]
- 26.Abe M, Hasegawa K, Wada H, Morimoto T, Yanazume T, Kawamura T, Hirai M, Furukawa Y, Kita T. GATA-6 is involved in PPARgamma-mediated activation of differentiated phenotype in human vascular smooth muscle cells. Arterioscler Thromb Vasc Biol. 2003;23:404–410. doi: 10.1161/01.ATV.0000059405.51042.A0. [DOI] [PubMed] [Google Scholar]
- 27.Wada H, Hasegawa K, Morimoto T, Kakita T, Yanazume T, Abe M, Sasayama S. Calcineurin-GATA-6 pathway is involved in smooth muscle-specific transcription. J Cell Biol. 2002;156:983–991. doi: 10.1083/jcb.200106057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morrisey EE. GATA-6: the proliferation stops here: cell proliferation in glomerular mesangial and vascular smooth muscle cells. Circ Res. 2000;87:638–640. doi: 10.1161/01.res.87.8.638. [DOI] [PubMed] [Google Scholar]
- 29.Perlman H, Suzuki E, Simonson M, Smith RC, Walsh K. GATA-6 induces p21(Cip1) expression and G1 cell cycle arrest. J Biol Chem. 1998;273:13713–13718. doi: 10.1074/jbc.273.22.13713. [DOI] [PubMed] [Google Scholar]
- 30.Fang P, Shi HY, Wu XM, Zhang YH, Zhong YJ, Deng WJ, Zhang YP, Xie M. Targeted inhibition of GATA-6 attenuates airway inflammation and remodeling by regulating caveolin-1 through TLR2/MyD88/NF-kappaB in murine model of asthma. Mol Immunol. 2016;75:144–150. doi: 10.1016/j.molimm.2016.05.017. [DOI] [PubMed] [Google Scholar]
- 31.Huang J, Zhou H, Mahavadi S, Sriwai W, Lyall V, Murthy KS. Signaling pathways mediating gastrointestinal smooth muscle contraction and MLC20 phosphorylation by motilin receptors. Am J Physiol Gastrointest Liver Physiol. 2005;288:G23–31. doi: 10.1152/ajpgi.00305.2004. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Y, Li F, Wang H, Yin C, Huang J, Mahavadi S, Murthy KS, Hu W. Immune/Inflammatory Response and Hypocontractility of Rabbit Colonic Smooth Muscle After TNBS-Induced Colitis. Dig Dis Sci. 2016;61:1925–1940. doi: 10.1007/s10620-016-4078-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gong QH, Stern J, Dean A. Transcriptional role of a conserved GATA-1 site in the human epsilon-globin gene promoter. Mol Cell Biol. 1991;11:2558–2566. doi: 10.1128/mcb.11.5.2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fujiwara T, Yokoyama H, Okitsu Y, Kamata M, Fukuhara N, Onishi Y, Fujimaki S, Takahashi S, Ishizawa K, Bresnick EH, Harigae H. Gene expression profiling identifies HOXB4 as a direct downstream target of GATA-2 in human CD34+ hematopoietic cells. PLoS One. 2012;7:e40959. doi: 10.1371/journal.pone.0040959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koutsourakis M, Langeveld A, Patient R, Beddington R, Grosveld F. The transcription factor GATA6 is essential for early extraembryonic development. Development. 1999;126:723–732. [PubMed] [Google Scholar]
- 36.Yuan X, Xia L, Dong X, Hu S, Zhang Y, Ding F, Liu H, Li L, Wang J. Transcription factors GATA-4 and GATA-6: molecular characterization, expression patterns and possible functions during goose (Anser cygnoides) follicle development. J Reprod Dev. 2014;60:83–91. doi: 10.1262/jrd.2013-080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun L, Wang W, Xiao W, Liang H, Yang Y, Yang H. Angiotensin II induces apoptosis in intestinal epithelial cells through the AT2 receptor, GATA-6 and the Bax pathway. Biochem Biophys Res Commun. 2012;424:663–668. doi: 10.1016/j.bbrc.2012.07.003. [DOI] [PubMed] [Google Scholar]
- 38.Dusing MR, Wiginton DA. Epithelial lineages of the small intestine have unique patterns of GATA expression. J Mol Histol. 2005;36:15–24. doi: 10.1007/s10735-004-2908-9. [DOI] [PubMed] [Google Scholar]
- 39.Gao X, Sedgwick T, Shi YB, Evans T. Distinct functions are implicated for the GATA-4, -5, and -6 transcription factors in the regulation of intestine epithelial cell differentiation. Mol Cell Biol. 1998;18:2901–2911. doi: 10.1128/mcb.18.5.2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Haveri H, Westerholm-Ormio M, Lindfors K, Maki M, Savilahti E, Andersson LC, Heikinheimo M. Transcription factors GATA-4 and GATA-6 in normal and neoplastic human gastrointestinal mucosa. BMC Gastroenterol. 2008;8:9. doi: 10.1186/1471-230X-8-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xie Y, Jin Y, Merenick BL, Ding M, Fetalvero KM, Wagner RJ, Mai A, Gleim S, Tucker DF, Birnbaum MJ, Ballif BA, Luciano AK, Sessa WC, Rzucidlo EM, Powell RJ, Hou L, Zhao H, Hwa J, Yu J, Martin KA. Phosphorylation of GATA-6 is required for vascular smooth muscle cell differentiation after mTORC1 inhibition. Sci Signal. 2015;8:ra44. doi: 10.1126/scisignal.2005482. [DOI] [PMC free article] [PubMed] [Google Scholar]