Abstract
The purpose of the Big Data to Knowledge (BD2K) initiative is to develop methods for discovering new knowledge from large amounts of data. However, if the resulting knowledge is so large that it resists comprehension, referred to here as Big Knowledge (BK), how can it be used properly and creatively? We call this secondary challenge, Big Knowledge to Use (BK2U). Without a high-level mental representation of the kinds of knowledge in a BK knowledgebase, effective or innovative use of the knowledge may be limited. We describe summarization and visualization techniques that capture the big picture of a BK knowledgebase, possibly created from Big Data. In this research, we distinguish between assertion BK and rule-based BK and demonstrate the usefulness of summarization and visualization techniques of assertion BK for clinical phenotyping. As an example, we illustrate how a summary of many intracranial bleeding concepts can improve phenotyping, compared to the traditional approach. We also demonstrate the usefulness of summarization and visualization techniques of rule-based BK for drug–drug interaction discovery.
Keywords: Big Data, Big Knowledge, drug–drug interactions, clinical phenotyping, summarization of knowledge, visualization of knowledge
Introduction
The purpose of the Big Data to Knowledge (BD2K) initiative is to develop technologies for extracting new knowledge hidden within huge amounts of data.1 However, if the resulting knowledge is stored in a knowledgebase that contains many small units of knowledge, the number of which is so large that it resists easy comprehension, how can the new knowledge be used properly and creatively? When a knowledgebase reaches a size such that its contents cannot be easily comprehended by humans, we call it Big Knowledge (BK). We note that there exist many forms of knowledge. However, the BD2K Request for Proposals did not specify in what format knowledge should be produced. A review of the 11 Centers of Excellence for Big Data Computing2 described on the National Institutes of Health (NIH) webpage entitled "BD2K Centers" does not show any unifying theme.
Looking at a formal notion of knowledge representation as taught in artificial intelligence courses, there are many varieties of knowledge, first differentiated by the symbolic versus numeric distinction, as expressed, for example, in systems of theorems versus neural networks. Among symbolic knowledge representation formats, predicate logic, frame systems, semantic networks, and rule systems from the official production system (OPS) family have been popular in the history of artificial intelligence.3
Our notion of knowledge is in line with the output that is produced by certain branches of data mining, such as Agrawal et al.'s association rules,4 and with the knowledge stored in knowledge representation systems, such as KL-ONE, CLASSIC, LOOM, and the many other members of the Description Logic family that are organized around an IS-A hierarchy.5 Description Logic research has supported the development of the Semantic Web.6 These knowledge representations have in common that basic units of knowledge can be expressed in relatively simple formats of a short length. The complexity arises out of the large number of knowledge units and out of the manner in which they interact with each other, constituting complex networks.
Thus, in this work, we concentrate on the forms of knowledge that pose a human comprehension challenge not owing to the complexity of the elements, but owing to the size and structure of the combined knowledge. Without some high-level mental representation of the kinds of knowledge in a repository of BK, effective or innovative use of this knowledge may be limited.7 In this paper, we address this secondary challenge: how to enable humans to use BK, which we call the Big Knowledge to Use (BK2U) problem. We illustrate the abstract notions of “comprehension” and “kinds of knowledge” in Figures 1 and 2.
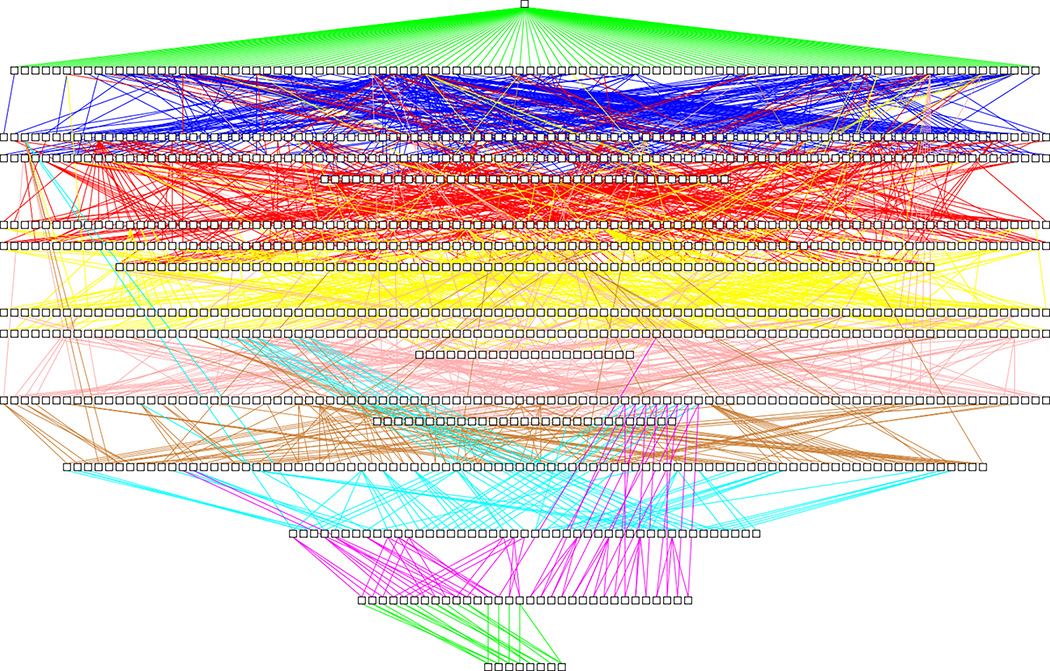
Figure 1.
The 1139 concepts from SNOMED CT’s Bleeding subhierarchy (about 1% of the large Clinical finding hierarchy24), drawn as white boxes organized into levels according to their distance from the root. Only IS-A relationships are shown as lines (colored according to the level of the child concept). The extreme connectedness of SNOMED CT’s concept hierarchy is reflected in the dense mass of lines. Some lines span more than five levels of the hierarchy.
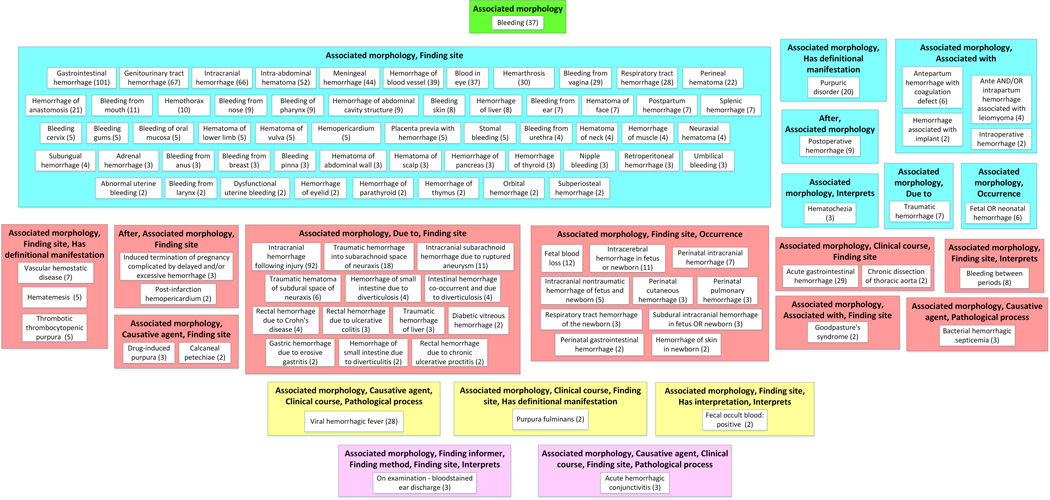
Figure 2.
Big picture of Bleeding subhierarchy: partial-area subtaxonomy for 1139 concepts in Figure 1. It contains 22 areas and 105 partial areas after removing 192 singleton partial-areas, which do not contribute to the big picture.
Large, complex knowledgebases (e.g., triple stores, terminologies, ontologies, and different kinds of rule sets) are found in many domains and are critical to many applications. For example, the Google Knowledge Graph8 is used to provide semantic search results, and DBpedia,9 an ontology derived by mining Wikipedia, contains information on over 4.5 million entities. In the biomedical domain, terminologies are used for encoding of knowledge and interoperability between systems.10, 11 For example, the Gene Ontology (GO) tool12 is used in the annotation of13 genetics findings. In addition, the National Centers for Biomedical Computing (NCBO) BioPortal repository14 contains over 8.1 million biomedical concepts from over 500 ontologies and the Unified Medical Language System (UMLS)15 currently contains over 9 million medical terms.
We distinguish between two kinds of BK: assertion BK and rule-based BK (rule BK). Basic units of assertion BK consist of triples of two concepts and one relationship. Formally, this is written as (concept1, relationship, concept2), for example, (smoking, causes, cancer), (lung, IS-A, body-part), or (cancer, IS-A, genetic-disease). Large repositories of such triples exist in the form of standardized medical terminologies.
A rule may be described, for example, by a Description Logic axiom. We are not addressing the BK2U problem for rules in general, as there is a wide variety of rule formats. Rather, we are limiting ourselves to the simple and practically important rules such as (Condition1, Condition2, Consequence), implying that if the two conditions are fulfilled, then the consequence occurs. The two conditions, and sometimes even the consequence, may be represented as terminological concepts.
Returning to assertion BK, the triple format (concept1, relationship, concept2) is used in the Semantic Web (e.g., in the Resource Description Framework (RDF) format16) and in many medical terminologies (e.g., the NCI thesaurus (NCIt),17 SNOMED CT,18 NDF-RT,19 and LOINC20), which are used in many applications.10, 11
The same unique concept (e.g., cancer) will be a part of many triples, resulting in a complicated network structure. Network structures are by their nature hard to understand, because, among other reasons, they have no intuitive linear format that one can “read from beginning to end.” Thus, we will need to solve the follow-up to the BD2K problem: how to make this knowledge usable (i.e., the BK2U problem).
The complexity of assertion BK and the difficulty humans face when trying to comprehend such content is demonstrated visually in Figure 1, which shows the tiny Bleeding subhierarchy (1139 concepts) taken from the SNOMED CT assertion BK system of over 300,000 concepts. Zooming in does not solve the comprehension problem, because connected elements often end up far (many screens) apart from each other. Figure 1 shows connection lines that span six levels of the BK hierarchy. Extensive experience has shown that utilizing a terminology visualization tool such as Jambalaya21 does not help in most cases, since displays are also overwhelming in Jambalaya. The complexity of Figure 1 illustrates the limitations of the human mental capacity to comprehend assertion BK.
Comprehension of the Bleeding subhierarchy of SNOMED CT, with 1139 concepts (January 2016 release), would include acquiring knowledge about the major kinds of bleeding-related disorders in this subhierarchy and how many concepts belong to each of these disorders. However, most terminology visualization tools only offer a view that displays a concept and its close neighborhood. Therefore, most of the terminology is hidden from the user except for a small window that shows relatively few concepts at a time. No familiar terminology tool, except for the ones we have created, enables a user to acquire knowledge about the major kinds of concepts (e.g., major kinds of bleeding) inside such a subhierarchy. As mentioned previously, without obtaining the big picture of a knowledgebase, one cannot expect the user to utilize the knowledge in a creative or innovative way.7
In contrast, summarizing the Bleeding hierarchy from Figure 1 (using the summary visualization of Figure 2) the user can easily see the major kinds of Bleeding and the number of concepts of each kind, including Gastrointestinal hemorrhage (101 concepts), Genitourinary tract hemorrhage (67), Intracranial hemorrhage (66), Intra-abdominal hematoma (62), Blood in eye (37), Respiratory tract hemorrhage (28), Intracranial hemorrhage following injury (92), Fetal blood loss (12), and others. Figure 2 provides the big picture of the Bleeding subhierarchy. In this paper, we demonstrate the details of summarization and visualization with a smaller subhierarchy within the Bleeding subhierarchy. We will then illustrate the usefulness of these techniques for clinical phenotyping. As an example, we will show how a summary of many intracranial bleeding concepts has the potential to improve phenotyping versus the traditional approach.
Rules in the above rule BK format cover, for example, aspects of the drug–drug interaction (DDI) problem. An example of a DDI rule would be (Aspirin, Warfarin, “Increase the effect of the latter drug”). This rule follows the format (DrugConcept1, DrugConcept2, ClinicalConsequence). Large repositories of such DDI rules exist. For example, First Databank Inc. (www.fdbhealth.com/) maintains a knowledgebase of DDI rules.
Conceptual knowledge that is indicative of deeper levels of comprehension is associated with superior performance across a range of tasks. We have developed summarization and visualization methods to support such comprehension, which are illustrated with improving phenotyping for assertion BK and with DDI discovery for rule-based BK. In the discussion section below, we further illustrate the differences between the BD2K and BK2U challenges.
Summarization approaches for BK2U
In general, the typical way humans comprehend complex knowledge is by summaries, abstraction, and visual representations. Abstracts summarize scientific papers. A table of contents summarizes a book. Road maps abstract satellite photos. The diagram of a cancer cell in a medical text book is not a photograph, but a visual abstraction of the photograph created by a trained artist. What about summaries of Big Data and BK? Creating summaries for data is easier than for knowledge, owing to the linear order of most data repositories. For example, for a drug formulary, a summary may be that “there are 30 antibiotics, 50 analgesics, 10 antifungals” in it. This summary, based on creating, naming, and enumerating groups of similar drugs, using appropriate criteria of similarity, provides a compact view of the formulary. We have designed techniques for summarizing, visualizing, and abstracting the content of BK to support BK2U.
In previous studies,22–34 different assertion BK summaries were successfully applied to terminology quality assurance (QA). For a framework offering support for scaling QA to large families of similar ontologies, see Ochs et al.35 Summaries and their visualizations have been algorithmically constructed for SNOMED CT,18 NCIt,17 NDF-RT,19 GO,12 OCRe,36 SDO,37 DDI,38 and CanCo.39 Our software systems (e.g., the Biomedical Layout Utility for SNOMED CT (BLUSNO)40 and the Ontology Abstraction Framework (OAF)41) generate visual displays of the summaries of such terminologies. We used these displays for QA as well as for obtaining the big picture of these terminologies.42–44 In this paper, we describe summarization and visualization methods (and their implementations) for SNOMED CT in support of clinical phenotyping (assertion BK) and for pharmacovigilance studies (rule BK).
Abstraction networks
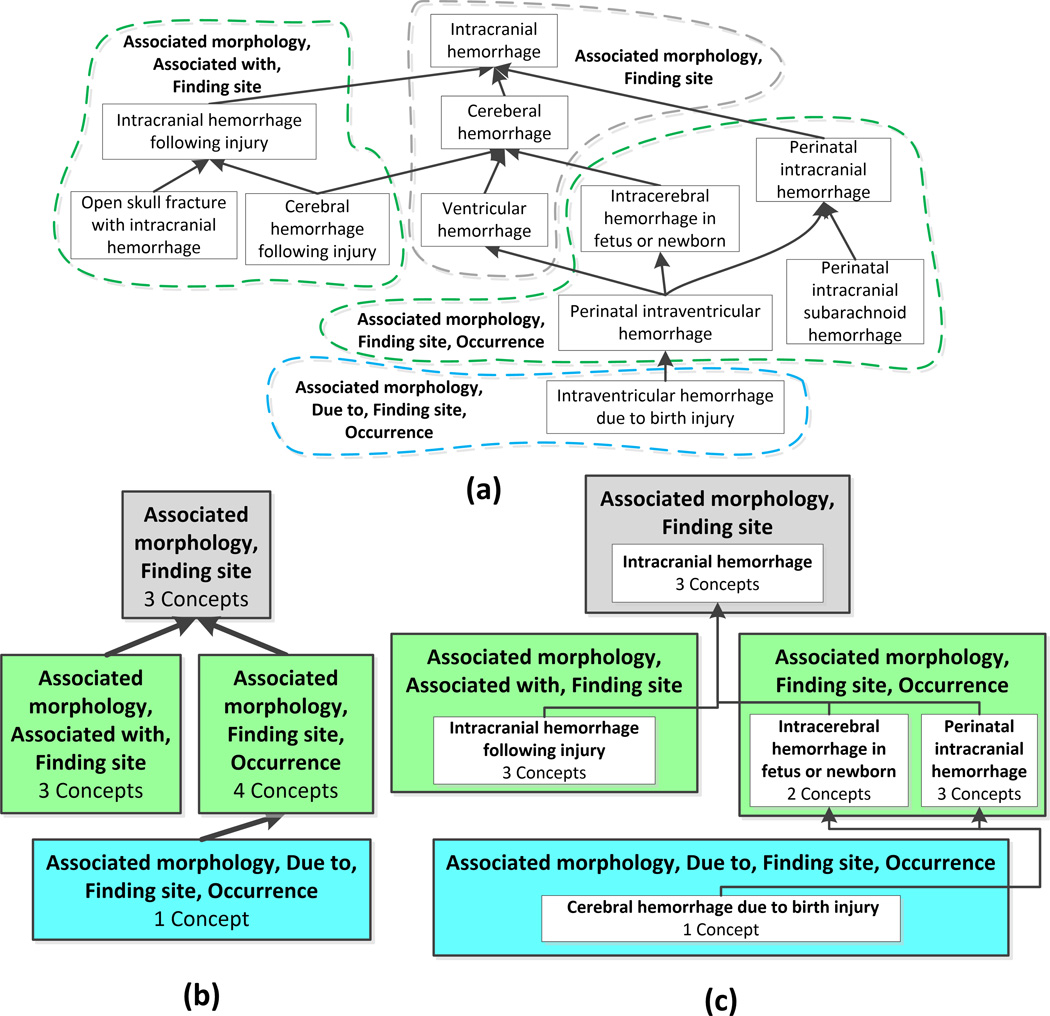
An abstraction network (AbN) of a network-structured medical ontology or terminology is itself a network consisting of nodes. Each node, appearing as a box in a diagram, summarizes several similar concepts of the original medical terminology, and the exact nature of such a node depends on the type of AbN. In previous studies we have developed various kinds of AbNs that summarize different aspects of a terminology.45 Nodes are connected by hierarchical child-of links that are derived from the IS-A relationships in the terminology. A detailed methodology that shows how an AbN is constructed is given in Figure 3 (Fig. 3B summarizes Fig. 3A).
Figure 3.
(A) Excerpt of 11 concepts from the SNOMED CT Intracranial hemorrhage subhierarchy. (B) Area taxonomy for A. (C) Partial-area taxonomy for A. The area taxonomy and partial-area taxonomy are constructed as follows: each area summarizes all concepts with the same set of outgoing relationship types. An area taxonomy is an abstraction network composed of area nodes connected by child-of links based on the IS-A relationships in the ontology. A root of an area is a concept that has no parents in its area (e.g., Perinatal intracranial hemorrhage). An area may have multiple roots. A partial-area consists of a root concept and all of its descendants in its area. Partial-areas summarize semantically similar concepts within each area, since all are descendants of the same root.
We now demonstrate the derivation of an ontology summary, called a partial-area taxonomy. Figure 3A shows an excerpt of 11 concepts from the SNOMED CT Intracranial hemorrhage subhierarchy. IS-A relationships appear as upward arrows, and concepts with the exact same set of outgoing relationships are grouped into dashed bubbles that are labeled with their sets of relationships (e.g., the concepts Intracranial hemorrhage, Cerebral hemorrhage, and Ventricular hemorrhage all have the relationships Associated morphology and Finding site). Thus, they appear together in one dashed bubble.
We define an area as the set of all concepts in an ontology that are modeled using the same set of relationships (“structurally similar concepts”). We define an area taxonomy as an AbN where the nodes represent areas. Thus, the area taxonomy summarizes sets of structurally similar concepts in an ontology. In Figure 3B the area taxonomy for the concepts in Fig. 3A is displayed. Areas appear as colored boxes in the AbN diagram, each named by the common relationship(s). Each area box displays the number of concepts summarized by the area, and areas are organized into color-coded levels according to their numbers of relationships. The particular colors do not carry meaning.
Next, we refine the area taxonomy into the partial-area taxonomy. A root concept is a concept in an area that has no parent concepts in its area (e.g., Perinatal intracranial hemorrhage in Figure 3). An area may have one or more root concepts. We define a partial-area as a set of concepts consisting of a root concept and all of its descendant concepts within the same area. Since all of the concepts in a partial-area are refinements of the root concept, we refer to these concepts as semantically similar, and we name the partial-area after its root. We define a partial-area taxonomy as an AbN consisting of nodes that represent partial-areas. A partial-area taxonomy summarizes sets of structurally (via areas) and semantically (via partial-areas) similar concepts. In Figure 3C, the partial-area taxonomy for the concepts in Fig. 3A is shown. Partial-areas are displayed as white boxes within areas. Each partial-area is labeled with its root concept name and the number of concepts summarized by the partial-area. The partial-area taxonomy provides a refined summary of the original ontology excerpt and explicitly shows the partial-areas within each area.
As shown by Wang et al.,30,46 partial-areas overlap when a concept is a descendant of more than one root in an area. For example, Perinatal interventricular hemorrhage is a descendant of the roots Intracerebral hemorrhage in fetus or newborn and Perinatal intracranial hemorrhage. Thus, this concept is counted twice, since it is summarized by two partial-areas in Fig. 3C, one for each root.
When we apply the partial-area taxonomy derivation method to the Bleeding subhierarchy of SNOMED CT (Fig. 1), we obtain the Bleeding partial-area taxonomy. Figure 2 provides the big picture of the Bleeding subhierarchy from Figure 1. The figure shows the taxonomy after omitting the 192 partial-areas containing only one concept (so called “singletons”), because they do not contribute to the big picture. The taxonomy in Figure 2 consists of 22 areas and 105 partial-areas, summarizing the 1139 concepts of Figure 1. The difference is striking. It is impossible to gain any understanding of the content of the Bleeding subhierarchy from Figure 1. However, in Fig. 2, the main concepts of SNOMED CT dealing with Bleeding are clearly recognizable, and capture the major subjects of Bleeding, as was demonstrated above in the introduction.
Applications of BK
Application of assertion BK: clinical phenotyping using a partial-area taxonomy for intracranial bleeding
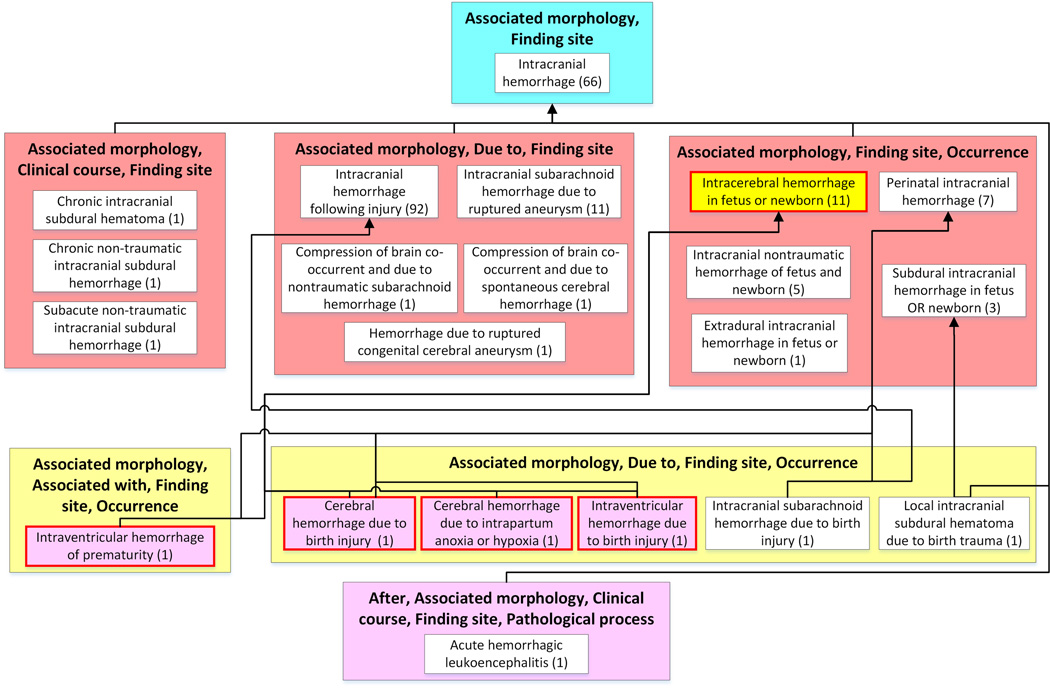
To illustrate the use of summarization for phenotyping, we constructed a partial-area subtaxonomy limited to a subhierarchy rooted at the concept Intracranial hemorrhage. (A subtaxonomy is a taxonomy created for a subhierarchy of a terminology, rooted at a chosen concept24). Figure 4 shows this subtaxonomy, which summarizes 203 concepts (18% of the 1139 concepts in Figure 1) and provides a big picture of the Intracranial hemorrhage subhierarchy. By reviewing the 21 partial-areas of this taxonomy, a user can quickly determine the major types of Intracranial hemorrhages.
Figure 4.
The Intracranial bleeding taxonomy. The seven areas (colored boxes) and 21 partial-areas (white boxes) summarize the 203 types of Intracranial bleeding findings represented as concepts in SNOMED CT. The summary provided by this partial-area taxonomy is demonstrated by the following list of larger partial-areas: Intracranial hemorrhage following injury (92 concepts), Intracranial hemorrhage (63 concepts), Intracranial subarachnoid hemorrhage due to rupture aneurysm (11 concepts), and Intracerebral hemorrhage in fetus or newborn (11 concept). These larger partial-areas identify the major types of intracranial bleeding that are modeled in SNOMED CT, along with their structures, providing the big picture of the Intracranial bleeding subject.
Clinical phenotyping is the process of identifying patients’ diseases, treatments, and response information from electronic health records (EHRs).47 Developing scalable and accurate phenotyping methods is a fundamental challenge in EHR-based studies.48, 49 Various types of clinical information encoded in different terminologies are often used when defining phenotypes (e.g., a disease phenotyping algorithm often considers patients’ ICD-9 or SNOMED CT codes, representing concepts extracted from clinical notes, lab tests, drugs, and procedures).
We use assertion BK summarization to decide on appropriate concepts that should be included when defining a given phenotype. For example, a user searching the EHR for non-injury intracerebral hemorrhages in fetuses or newborn children could start with the SNOMED CT concept Intracerebral hemorrhage in fetus or newborn (marked yellow in Figure 4). However, using only the concept itself will miss occurrences in EHRs of many important relevant concepts, such as Intraventricular hemorrhage of prematurity, and yield a low recall of the phenotyping algorithm. Therefore, a typical approach is to navigate through the hierarchy of the terminology and expand the query by including all of its children or all descendants in the EHR search. HoweverNevertheless, such expanded queries with descendants often introduce irrelevant concepts (e.g., Cerebral hemorrhage due to birth injury in this example), causing a low precision of the phenotyping algorithm. On the other hand, using only all of the children will miss some relevant descendants and result in a low recall.
We propose using BK summarization techniques to resolve the tradeoff between recall and precision of phenotyping algorithms. In the summary-based approach, a user would select one relevant summary group (e.g., the partial-area rooted at the desired concept in a partial-area taxonomy) and the EHR would be queried with the concepts of this partial-area.
Other groups of concepts that summarize topics that refer to different facets of the same finding, expressed by an extra relationship or an extra parent (e.g., Hemorrhages caused by injuries) would not be included in the EHR search. For example, to query for non-injury intracerebral hemorrhages in fetuses or newborns, one could select the Intracerebral hemorrhage in fetus or newborn(11) partial-area (highlighted in yellow) in the Intracranial hemorrhage subtaxonomy as shown in Figure 4. This partial-area summarizes eight children and two grandchildren of the root. All 11 concepts in the partial-area share a common structure and semantics, and, thus, all of them fit the desired query. On the other hand, the partial-area Intracerebral hemorrhage in fetus or newborn(11) has four child partial-areas (highlighted in pink) that introduce a different structure into the hierarchy, which captures, for example, the cause of the hemorrhage with a due to relationship. For example, the child partial-area Cerebral hemorrhage due to birth injury(1) has a due to relationship with the target Birth trauma of fetus. This concept should not be included in the EHR search. Hence, the group of concepts offered by the partial-area taxonomy methodology better captures the proper SNOMED CT concepts that describe the phenotype and has the potential to improve both recall and precision of the phenotyping algorithm, in comparison to current phenotyping techniques. In future research, we will conduct a quantitative study to assess the hypothesis that summarization using AbNs better supports phenotyping algorithms than the current techniques (e.g., including in the search children or descendants of a chosen concept).
Application of rule-based BK: DDI discovery
An example of the use of rule BK can be found in the area of DDIs, which are a particularly important type of adverse drug reaction (ADR)50–53 that can cause excessive responses or altered toxicity.54 For instance, cerivastatin caused 31 cases of fatal rhabdomyolysis prior to its withdrawal in June 2001; the combination cerivastatin–gemfibrozil was implicated in 12 of the 31 deaths55 because gemfibrozil caused elevated blood levels of the statin, resulting in a higher risk of myopathy and rhabdomyolysis. The risk of adverse DDIs increases exponentially for each additional medication.56–60
To summarize rule BK, we associate the different elements of each rule with one or more concepts from a terminology. For example, given DDIs of the form (DrugConcept1, DrugConcept2, ClinicalConsequence), each DrugConcept1 and DrugConcept2 element is coded as a concept in the NDF-RT,19 the National Drug File–Reference Terminology provided by the Veterans Health Administration. The clinical consequence can be coded using an ADR terminology (e.g., the First Databank Medical Lexicon, FML). However, clinical consequence concepts tend to be complex and need to be organized around more basic terminology concepts (e.g., Bleeding24) that occur in clinical terminologies such as SNOMED CT. The transformation from the compound FML concepts to the more basic SNOMED CT concepts will require the extensive use of post-coordination mappings61, 62 provided by First Databank.
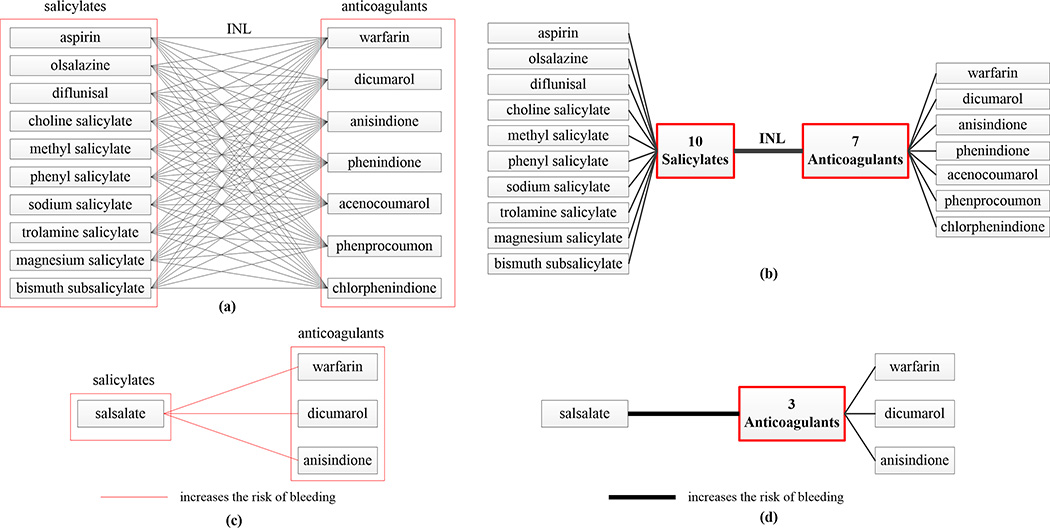
Once each element of a rule is associated with a terminology concept, assertion BK summaries can be constructed to support the summarization of the rule BK. Figure 5A demonstrates a summary of DDI knowledge. There are 10 × 7 = 70 DDIs between the 10 salicylates on the left and the seven anticoagulants on the right in FDB’s DDI knowledgebase, shown explicitly in Figure 5A. The 70 DDI rules between individual drugs can be summarized by one single rule between the nodes representing the two sets of drug concepts (Fig. 5B).
Figure 5.
(A) Illustration of a DDI rule summary. There are 10×7 = 70 DDIs between the 10 salicylates on the left and the seven anticoagulants on the right in FDB’s DDI knowledgebase. (B) The 70 DDI rules between individual drugs are summarized by one rule between the nodes representing the two sets, respectively. INL, increases the effect of latter drug. (C) Three new candidate DDIs not appearing in FDB’s DDI knowledgebase, between the Salicylate Salsalate on the left and the three anticoagulants on the right. (D) They are summarized by one rule between Salsalate and a node representing the three anticoagulants.
The technique, described by Ochs et al.,63 utilizes summarization of drugs by Chemical Ingredient concepts that are derived from the Chemical Ingredient hierarchy of the NDF-RT19 and summarization of DDIs to support the discovery of potentially missing DDI rules in the FDB knowledgebase. For example, there are 18 drugs summarized by the Salicylates classification in the summarization of the NDF-RT that was reported by Ochs et al.63 We reviewed other DDI sources, such as drugs.com (www.drugs.com) for DDIs between additional Salicylates listed in the NDF-RT and Anticoagulants, beyond the drug pairs listed in Figure 5A that are considered DDIs in the FDB knowledgebase. Figure 5C, which presents the findings, and Figure 5D, which summarizes them, both show three new candidate DDIs not appearing in FDB’s DDI knowledgebase between salsalate and three anticoagulants (dicumarol, warfarin, and anisindione) in Figure 5A. In an analysis by JKU, it was found that, while those pairs have the potential to be DDIs, the reason why FDB did not include them in their knowledgebase is that in these cases the drug formulation has a low potential for interaction. One has to realize that, as a leading pharmacological knowledge company, FDB’s DDI knowledge is widely used by pharmacies, doctors, and hospitals to aid in decisions about preventing patients from taking drugs prescribed because of their conditions; these are critical clinical decisions that sometimes involve issues of life or death. Other DDI sources, such as drugs.com, which are not used in this way in the healthcare industry, are thus more lenient in including questionable pairs of drugs as DDIs.
Next, we performed another study to examine several pairs of families of drugs, known to have DDIs, in search of drug pairs with potential DDIs that are not listed in the FDB knowledgebase. For each such pair, one family is a chemical classification, while the other is a pharmaceutical classification. Typically, the NDF-RT contains more drugs under the chemical classification than does the FDB DDI knowledgebase. We explored drugs.com for DDIs for those additional drugs from the NDF-RT, looking for interactions with the drugs that are classified by the corresponding pharmaceutical classification.
Table 1 reports the details of this study for seven pairs of families. Column 2 lists the DDI family pairs (A, B) as given in the FDB DDI knowledgebase. For all seven pairs, A is a chemical ingredient family and B is a pharmaceutical family. Column 3 represents the number of ingredients in Family A (e.g., Sulfonamides) in FDB. Column 4 shows the number of ingredients in Family B (e.g., Antidiabetics, Oral) in FDB. Column 5 gives the number of drug DDI (A, B) pairs in FDB, which is the product of the number in Column 3 and the number in Column 4. Column 6 represents the number of ingredients in Family A in the NDF-RT; column 7 shows the number of ingredients in Family A in both NDF-RT and FDB; and column 8 shows the total number of potential DDIs found in sources other than FDB’s knowledgebase for the specific (A, B) drug pairs.
Table 1.
Potential DDI findings for seven pairs of drug families
| 2 | 3 | 4 | 5 | 6 | 7 | 8 | |
|---|---|---|---|---|---|---|---|
| DDI family pair (A, B) | # of ingredients in Family A in FDB |
# of ingredients in Family B in FDB |
# of actual FDB DDIs |
# of ingredients in Family A in NDF-RT |
# of ingredients in Family A in both FDB and NDF-RT |
# of potential DDIs found in other sources |
|
| 1 | (Sulfonamides; Antidiabetics, Oral) | 6 | 8 | 48 | 52 | 4 | 93 |
| 2 | (Sulfonamides, Anticoagulants) | 22 | 6 | 132 | 52 | 15 | 21 |
| 3 | (NSAIDs, ACE inhibitors or ARBs) | 68 | 24 | 1632 | 43 | 21 | 73 |
| 4 | (NSAIDs, Loop diuretics) | 70 | 7 | 490 | 43 | 21 | 16 |
| 5 | (NSAIDs, Beta blockers) | 12 | 27 | 324 | 43 | 7 | 81 |
| 6 | (Phenothiazines, Narcotics) | 6 | 11 | 66 | 32 | 5 | 73 |
| 7 | (Benzodiazepines, Macrolide antibiotics) |
4 | 5 | 20 | 22 | 3 | 37 |
| Total: | n/a | n/a | 2712 | n/a | n/a | 394 |
For example, the first pair is (Sulfonamides, Antidiabetics, Oral). In FDB the DDIs between six sulfonamides (Family A) and eight antidiabetics (Family B) result in the clinical effect referred to as increased effect of the latter drug (INL). However, in the NDF-RT there are 52 drugs classified under Sulfonamides. When pairing the additional (i.e., not in FDB’s knowledgebase) 48 A drugs with those eight B drugs, 93 pairs of drugs were found in the other sources as DDI pairs between Family A and Family B. An analysis of these 93 pairs revealed that, for (A, B) pairs, the interaction between the two drugs consists of a “protein binding displacement” mechanism that applies only for sulfonamide antibiotics, which describes the six sulfonamides listed in the FDB DDI knowledgebase. This mechanism does not apply to the remaining sulfonamides found in the NDF-RT. One outcome from this study is that FDB will change, in its DDI knowledgebase, the name of this Family A from “Sulfonamides” to “Sulfonamide Antibiotics,” which describes it more accurately.
Out of the 73 potential DDIs (Table 1, line 3) for the family pair (NSAID, ACE inhibitor or ARBS) found in other sources, 16 potential DDIs for the nonsteroidal anti-inflammatory compound (NSAID) diflunisal with 16 different ACE inhibitors should be added to the FDB knowledgebase. As shown in Table 1 (line 5), eight potential DDIs for the NSAID diclofenac with various beta blockers were found, which should be considered for addition to FDB’s knowledgebase. Similarly for its salt form, diclofenac potassium, nine DDIs were found to be missing from FDB’s knowledgebase. However, the clinical studies reported in drugs.com for these pairs did not prove the DDIs to be at the stricter level required for inclusion by FDB. For the two NSAIDs meclofenamate and mefenamic acid, which are currently not available in the U.S. market, eight and 11 DDIs, respectively, were found with various beta blockers and should be added to FDB’s knowledgebase in the case that these drugs are made available for sale.
The DDI family pair in line 6 of Table 1 is known to cause many DDI alerts with a low severity level (i.e., it is a prime example of a combination that causes alert fatigue). Thus, line-6 interactions will be recommended for removal from FDB’s DDI knowledgebase. A detailed pharmacological analysis of these various families of drugs is in progress.
To summarize the results of this study, out of 394 potential DDI drug pairs found in other sources, 80 (20.3%) were approved for inclusion in FDB’s DDI knowledgebase. Another 19 (4.8%) drug pairs will be added if their drugs are made available in the United States. The following additional outcomes of this study do not relate to the potential DDI pairs of column 8, but to the actual FDB DDI pairs of column 5 in Table 1. One such important outcome was that 66 DDI drug pairs for the family pair (phenotiazines, narcotics) were removed from the FDB DDI knowledgebase, since they almost always cause false alerts. Even more interestingly, a deeper analysis of the FDB DDI drug pairs from three family pairs (rows 3, 4, and 5 of Table 1) showed that the issue was not the interaction of one drug with another drug, but the interaction between one drug and the disease that is present when the other drug is used. These interactions will be removed from the FDB DDI knowledgebase, since the ADR is between a drug and a disease, rather than an interaction between two drugs. The relevant ADR knowledge will be placed in the proper FDB ADR knowledgebase. The total number of DDI pairs removed is 1632 + 490 + 324 = 2446. Hence, beyond the additional DDI pairs, the present study led to important changes in the storage of DDI and ADR knowledge in the FDB knowledgebase.
Discussion
In this section, we discuss the differences between the notion of Big Data and the notion of BK, the differences between the challenges of BD2K and BK2U, and the limitations of the BK2U challenge. The purpose of BD2K is to extract knowledge that is hidden in large repositories of data, which can appear in various formats (e.g., tables, text, images). These repositories may require special storage and access techniques and may contain, for example, numeric measurements of experimental results, tables of information derived from various kinds of sensors, scholarly papers describing genomic experiments, and free text annotations in the EHRs of patients in a hospital.
The various techniques used for extracting the knowledge are typically automatic and are fit for processing the large repositories. Furthermore, the knowledge extracted varies widely in its nature and formats (e.g., algorithms, mathematical formulas and models, rules of various complexities and formats). There are ongoing efforts by many research groups to address the BD2K challenge. However, this is not the problem we are addressing with BK2U. Rather, we are addressing a related problem: how to enable the effective utilization of big knowledgebases by humans, whether or not the knowledgebases were created as part of the BD2K challenge.
Furthermore, we are not addressing the BK2U challenge for knowledge in general. For many forms of knowledge, some expertise and/or training are required to be able to use them, and only the proper professionals will be able to do so. We are concerned with the case that knowledge is expressed by many small units that are simple by themselves, but overwhelming when combined into one knowledgebase. The difficulty for effective utilization of such knowledgebases lies in the fact that the many knowledge units constitute a complex network structure. As a result, the orientation into the content and structure of such a knowledgebase poses a mental challenge to humans. How large does a knowledgebase have to be in order to be considered BK? This should be the topic of a future cognitive study, but the practical challenge arises as soon as orientation poses a difficulty to humans who need to work with a knowledgebase. Comprehension capacity is limited, akin to other mental limitations of humans (e.g., being able to remember only about seven items in short-term memory64). It remains for future research to quantify the parameters of a terminology that makes orientation difficult.
Figure 1 showed an example of 1139 concepts from SNOMED CT’s Bleeding subhierarchy. Our approach for addressing the comprehension challenge consists of developing summarization techniques and visualizing the big picture reflected by the partial-area taxonomy (without singletons) shown in Fig. 2. Owing to the different purposes of the two challenges and the different representations of knowledge and data, the techniques developed in the BD2K projects are improper for handling the BK2U challenge, which involves the development of knowledge summaries of large knowledgebases rather than extracting knowledge out of Big Data repositories.
Conclusions
BD2K will only lead to limited results unless the implied problem of BK2U is also solved. We have described strategies based on summarization and visualization to support the comprehension of BK, both for assertions and for rules. We have also indicated how to use these methods for phenotyping (assertion BK) and for DDI discovery (rule BK).
Acknowledgments
Research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under Award Number R01CA190779. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Conflicts of interest
The authors declare no conflicts of interest.
References
- 1.Margolis R, Derr L, Dunn M, et al. The National Institutes of Health's Big Data to Knowledge (BD2K) initiative: capitalizing on biomedical big data. Journal of the American Medical Informatics Association. 2014;21:957–958. doi: 10.1136/amiajnl-2014-002974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. [Accessed 29 June 2016];BD2K Centers. https://datascience.nih.gov/bd2k/funded-programs/centers.
- 3.Brachman RJ, Levesque HJ. Readings in knowledge representation. Morgan Kaufmann Publishers Inc.; [Google Scholar]
- 4.Agrawal R, Imieliński T, Swami A. Mining association rules between sets of items in large databases. Acm sigmod record. 1993;22:207–216. [Google Scholar]
- 5.Baader F, Calvanese D, McGuinness DL, et al. The Description Logic Handbook: Theory, Implementation and Applications. Cambridge University Press; 2010. [Google Scholar]
- 6.Antoniou G, Harmelen FV. A semantic web primer. MIT Press; 2004. [Google Scholar]
- 7.Patel VL, Kaufman DR, Arocha JF. Conceptual change in the biomedical and health sciences domain. Advances in instructional psychology. 2000;5:329–392. [Google Scholar]
- 8.Google. [Accessed 27 June 2016];Google: Inside Search: The Knowledge Graph. 2012 https://www.google.com/intl/es419/insidesearch/features/search/knowledge.html.
- 9.Bizer C, Lehmann J, Kobilarov G, et al. DBpedia-A crystallization point for the Web of Data. Web Semantics: science, services and agents on the world wide web. 2009;7:154–165. [Google Scholar]
- 10.Rubin DL, Shah NH, Noy NF. Biomedical ontologies: a functional perspective. Briefings in Bioinformatics. 2008;9:75–90. doi: 10.1093/bib/bbm059. [DOI] [PubMed] [Google Scholar]
- 11.Bodenreider O. Biomedical ontologies in action: role in knowledge management, data integration and decision support. Yearbook Med Inform. 2008:67–79. [PMC free article] [PubMed] [Google Scholar]
- 12.Ashburner M, Ball CA, Blake JA, et al. Gene Ontology: tool for the unification of biology. Nature Genetics. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Consortium TGO. Gene Ontology annotations and resources. Nucleic acids research. 2013:D530–D535. doi: 10.1093/nar/gks1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Whetzel PL, Noy NF, Sham NH, et al. BioPortal: Enhanced Functionality via New Web services from the National Center for Biomedical Ontology to Access and Use Ontologies in Software Applications. Nucleic Acids Research (NAR) 2011;39:W541–W545. doi: 10.1093/nar/gkr469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bodenreider O. The Unified Medical Language System (UMLS): integrating biomedical terminology. Nucleic acids research. 2004;32:D267–D270. doi: 10.1093/nar/gkh061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. [Accessed 10 February 2015];RDF/XML Syntax Specification. 2004 http://www.w3.org/TR/REC-rdf-syntax/
- 17.Fragoso G, de Coronado S, Haber M, et al. Overview and utilization of the NCI thesaurus. Comp Funct Genomics. 2004;5:648–654. doi: 10.1002/cfg.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stearns MQ, Price C, Spackman KA, et al. SNOMED clinical terms: overview of the development process and project status. Proc AMIA Annu Symp. 2001:662–666. [PMC free article] [PubMed] [Google Scholar]
- 19.Brown SH, Elkin PL, Rosenbloom ST, et al. VA National Drug File Reference Terminology: a cross-institutional content coverage study. Medinfo. 2004;11 [PubMed] [Google Scholar]
- 20.McDonald CJ, Huff SM, Suico JG, et al. LOINC, a universal standard for identifying laboratory observations: a 5-year update. Clin Chem. 2003;49:624–633. doi: 10.1373/49.4.624. [DOI] [PubMed] [Google Scholar]
- 21.Storey M-A, Musen M, Silva J, et al. Jambalaya: Interactive visualization to enhance ontology authoring and knowledge acquisition in Protégé. Workshop on Interactive Tools for Knowledge Capture. 2001 [Google Scholar]
- 22.Ochs C, Perl Y, Halper M, et al. Gene Ontology Summarization to Support Visualization and Quality Assurance. BICoB. 2015:167–174. [Google Scholar]
- 23.Ochs C, Geller J, Perl Y, et al. A Tribal Abstraction Network for SNOMED CT Hierarchies without Attribute Relationships. J Am Med Inform Assoc. 2014;22:628–639. doi: 10.1136/amiajnl-2014-003173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ochs C, Geller J, Perl Y, et al. Scalable Quality Assurance for Large SNOMED CT Hierarchies Using Subject-based Subtaxonomies. J Am Med Inform Assoc. 2014;22:507–518. doi: 10.1136/amiajnl-2014-003151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ochs C, He Z, Perl Y, et al. Choosing the Granularity of Abstraction Networks for Orientation and Quality Assurance of the Sleep Domain Ontology; Proceedings of the 4th International Conference on Biomedical Ontology; 2013. pp. 84–89. [Google Scholar]
- 26.Ochs C, Perl Y, Geller J, et al. Scalability of Abstraction-Network-Based Quality Assurance to Large SNOMED Hierarchies. AMIA Annu Symp Proc. 2013:1071–1080. [PMC free article] [PubMed] [Google Scholar]
- 27.Ochs C, Agrawal A, Perl Y, et al. Deriving an Abstraction Network to Support Quality Assurance in OCRe. AMIA Annu Symp Proc. 2012:681–689. [PMC free article] [PubMed] [Google Scholar]
- 28.Halper M, Wang Y, Min H, et al. Analysis of error concentrations in SNOMED. AMIA Annu Symp Proc. 2007:314–318. [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Y, Halper M, Min H, et al. Structural methodologies for auditing SNOMED. J Biomed Inform. 2007;40:561–581. doi: 10.1016/j.jbi.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 30.Wang Y, Halper M, Wei D, et al. Auditing complex concepts of SNOMED using a refined hierarchical abstraction network. J Biomed Inform. 2012;45:1–14. doi: 10.1016/j.jbi.2011.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Min H, Perl Y, Chen Y, et al. Auditing as part of the terminology design life cycle. J Am Med Inform Assoc. 2006;13:676–690. doi: 10.1197/jamia.M2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.He Z, Ochs C, Agrawal A, et al. A Family-Based Framework for Supporting Quality Assurance of Biomedical Ontologies in BioPortal. Proc AMIA Annu Symp. 2013:581–590. [PMC free article] [PubMed] [Google Scholar]
- 33.He Z, Ochs C, Soldatova L, et al. Auditing Redundant Import in Reuse of a Top Level Ontology for the Drug Discovery Investigations Ontology. VDOS. 2013 [Google Scholar]
- 34.Gu H, Perl Y, Geller J, et al. Representing the UMLS as an object-oriented database: modeling issues and advantages. J Am Med Inform Assoc. 2000;7:66–80. doi: 10.1136/jamia.2000.0070066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ochs C, He Z, Zheng L, et al. Utilizing a structural meta-ontology for family-based quality assurance of the BioPortal ontologies. J Biomed Inform. 2016;61:63–76. doi: 10.1016/j.jbi.2016.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sim I, Carini S, Tu S, et al. The human studies database project: federating human studies design data using the ontology of clinical research. AMIA Summits Transl Sci Proc. 2010:51–55. [PMC free article] [PubMed] [Google Scholar]
- 37.Arabandi S, Ogbuji C, Redline S, et al. Developing a Sleep Domain Ontology. AMIA Clinical Research Informatics Summit. 2010 [Google Scholar]
- 38.Da Q, King R, Hopkins A, et al. An ontology for description of drug discovery investigations. J. Integrative Bioinformatics. 2010;7:126–139. doi: 10.2390/biecoll-jib-2010-126. [DOI] [PubMed] [Google Scholar]
- 39.Zeginis D, Hasnain A, Loutas N, et al. A collaborative methodology for developing a semantic model for interlinking Cancer Chemoprevention linked-data sources. Semantic Web. 2014;5:127–142. [Google Scholar]
- 40.Geller J, Ochs C, Perl Y, et al. New Abstraction Networks and a New Visualization Tool in Support of Auditing the SNOMED CT Content. AMIA Annu Symp Proc. 2012:237–246. [PMC free article] [PubMed] [Google Scholar]
- 41.Ochs C, Geller J, Perl Y, et al. A Unified Software Framework for Deriving, Visualizing, and Exploring Abstraction Networks for Ontologies. J Biomed Inform. 2016;62:90–105. doi: 10.1016/j.jbi.2016.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ochs C, Perl Y, Geller J, et al. Using Aggregate Taxonomies to Summarize SNOMED CT Evolution. International Workshop on Biomedical and Health Informatics. 2015:1008–1015. [Google Scholar]
- 43.Ochs C, Case JT, Perl Y. Tracking the Remodeling of SNOMED CT’s Bacterial Infectious Diseases. AMIA Annu Symp Proc. 2016 (accepted for publication) [PMC free article] [PubMed] [Google Scholar]
- 44.Zheng L, Perl Y, Geller J, et al. How to Summarize Big Knowledge Subjects. ICBO. 2016 (poster accepted for publication) [Google Scholar]
- 45.Halper M, Gu H, Perl Y, et al. Abstraction Networks for Terminologies: Supporting Management of “Big Knowledge”. Artificial intelligence in medicine. 2015;64:1–16. doi: 10.1016/j.artmed.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Y, Halper M, Wei D, et al. Abstraction of complex concepts with a refined partial-area taxonomy of SNOMED. J Biomed Inform. 2012;45:15–29. doi: 10.1016/j.jbi.2011.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kohane IS. Using electronic health records to drive discovery in disease genomics. Nat Rev Genet. 2011;12:417–428. doi: 10.1038/nrg2999. [DOI] [PubMed] [Google Scholar]
- 48.Kho AN, Rasmussen LV, Connolly JJ, et al. Practical challenges in integrating genomic data into the electronic health record. Genetics in Medicine. 2013;15:772–778. doi: 10.1038/gim.2013.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hripcsak G, Albers DJ. Next-generation phenotyping of electronic health records. Journal of the American Medical Informatics Association. 2013;20:117–121. doi: 10.1136/amiajnl-2012-001145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pirmohamed M. Drug interactions of clinical importance. London: Chapman & Hall; 1998. [Google Scholar]
- 51.Kuhlmann J, Muck W. Clinical-pharmacological strategies to assess drug interaction potential during drug development. Drug Saf. 2001;24:715–725. doi: 10.2165/00002018-200124100-00001. [DOI] [PubMed] [Google Scholar]
- 52.Grymonpre RE, Mitenko PA, Sitar DS, et al. Drug-associated hospital admissions in older medical patients. Journal of the American Geriatrics Society. 1988;36:1092–1098. doi: 10.1111/j.1532-5415.1988.tb04395.x. [DOI] [PubMed] [Google Scholar]
- 53.Jankel CA, Speedie SM. Detecting drug interactions: a review of the literature. DICP : the annals of pharmacotherapy. 1990;24:982–989. doi: 10.1177/106002809002401014. [DOI] [PubMed] [Google Scholar]
- 54.Scripture CD, Figg WD. Drug interactions in cancer therapy. Nature reviews. Cancer. 2006;6:546–558. doi: 10.1038/nrc1887. [DOI] [PubMed] [Google Scholar]
- 55.Staffa JA, Chang J, Green L. Cerivastatin and reports of fatal rhabdomyolysis. The New England journal of medicine. 2002;346:539–540. doi: 10.1056/NEJM200202143460721. [DOI] [PubMed] [Google Scholar]
- 56.Zhan C, Correa-de-Araujo R, Bierman AS, et al. Suboptimal prescribing in elderly outpatients: potentially harmful drug-drug and drug-disease combinations. Journal of the American Geriatrics Society. 2005;53:262–267. doi: 10.1111/j.1532-5415.2005.53112.x. [DOI] [PubMed] [Google Scholar]
- 57.Malone DC, Hutchins DS, Haupert H, et al. Assessment of potential drug-drug interactions with a prescription claims database. American journal of health-system pharmacy : AJHP : official journal of the American Society of Health-System Pharmacists. 2005;62:1983–1991. doi: 10.2146/ajhp040567. [DOI] [PubMed] [Google Scholar]
- 58.Janchawee B, Owatranporn T, Mahatthanatrakul W, et al. Clinical drug interactions in outpatients of a university hospital in Thailand. Journal of clinical pharmacy and therapeutics. 2005;30:583–590. doi: 10.1111/j.1365-2710.2005.00688.x. [DOI] [PubMed] [Google Scholar]
- 59.Janchawee B, Wongpoowarak W, Owatranporn T, et al. Pharmacoepidemiologic study of potential drug interactions in outpatients of a university hospital in Thailand. Journal of clinical pharmacy and therapeutics. 2005;30:13–20. doi: 10.1111/j.1365-2710.2004.00598.x. [DOI] [PubMed] [Google Scholar]
- 60.Aparasu R, Baer R, Aparasu A. Clinically important potential drug-drug interactions in outpatient settings. Research in social & administrative pharmacy : RSAP. 2007;3:426–437. doi: 10.1016/j.sapharm.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 61.Pathak J, Wang J, Kashyap S, et al. Mapping clinical phenotype data elements to standardized metadata repositories and controlled terminologies: the eMERGE Network experience. J Am Med Inform Assoc. 2011;18:376–386. doi: 10.1136/amiajnl-2010-000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.SNOMED CT User Guide. http://www.nlm.nih.gov/research/umls/licensedcontent/snomedctarchive.html.
- 63.Ochs C, Zheng L, Perl Y, et al. Drug-drug Interaction Discovery Using Abstraction Networks for "National Drug File – Reference Terminology" Chemical Ingredients. AMIA Annu Symp Proc. 2015:973–982. [PMC free article] [PubMed] [Google Scholar]
- 64.Miller GA. The magical number seven, plus or minus two: Some limits on our capacity for processing information. Psychological review. 1956;63:81. [PubMed] [Google Scholar]