Abstract
Objective
Several prion amplification systems have been proposed for detection of prions in cerebrospinal fluid (CSF), most recently, the measurements of prion seeding activity with second-generation real-time quaking-induced conversion (RT-QuIC). The objective of this study was to investigate the diagnostic performance of the RT-QuIC prion test in the broad phenotypic spectrum of prion diseases.
Methods
We performed CSF RT-QuIC testing in 2,141 patients who had rapidly progressive neurological disorders, determined diagnostic sensitivity and specificity in 272 cases which were autopsied, and evaluated the impact of mutations and polymorphisms in the PRNP gene, and Type 1 or Type 2 of human prions on diagnostic performance.
Results
The 98.5% diagnostic specificity and 92% sensitivity of CSF RT-QuIC in a blinded retrospective analysis matched the 100% specificity and 95% sensitivity of a blind prospective study. The CSF RT-QuIC differentiated 94% of cases of sporadic Creutzfeldt-Jakob disease (sCJD) MM1 from the sCJD MM2 phenotype, and 80% of sCJD VV2 from sCJD VV1. The mixed prion type 1–2 and cases heterozygous for codon 129 generated intermediate CSF RT-QuIC patterns, while genetic prion diseases revealed distinct profiles for each PRNP gene mutation.
Interpretation
The diagnostic performance of the improved CSF RT-QuIC is superior to surrogate marker tests for prion diseases such as 14-3-3 and Tau proteins and together with PRNP gene sequencing, the test allows the major prion subtypes to be differentiated in vivo. This differentiation facilitates prediction of the clinicopathological phenotype and duration of the disease—two important considerations for envisioned therapeutic interventions.
Keywords: Sporadic and genetic prion diseases, human prions classification, differential diagnostics, cerebrospinal fluid, neurodegeneration
INTRODUCTION
Human prion diseases are highly heterogeneous and invariably fatal neurological disorders. They include Creutzfeldt-Jakob disease (CJD), kuru, Gerstmann-Sträussler-Scheinker (GSS) disease and fatal insomnia (FI)1. Sporadic Creutzfeldt-Jakob disease (sCJD) accounts for 85% of all cases of human prion disease; genetic (gCJD) for 10–15%; and infection from exogenous sources, most frequently iatrogenic (iCJD) for <1%1, 2. Prions propagate by a process in which the pathogenic prion protein (PrPSc) replicates exponentially by templating the misfolding of normal cellular prion protein (PrPC)3.
The diversity of prion diseases is further increased by distinct strains of prions that transmit a particular disease phenotype, including incubation time, clinical signs, progression rate, and patterns of PrPSc deposition and neuropathological lesions4–10. Distinct phenotypes of human prions have been classified according to their clinicopathological characteristics, the methionine (M) or valine (V) polymorphism in codon 129 of the prion protein gene PRNP, and the mass (21 or 19 kDa, as Type 1 or Type 2, respectively) of the deglycosylated, protease-resistant PrPSc fragment1, 11.
Apart from brain biopsy, which with more sensitive detection of PrPSc by conformation-dependent immunoassay (CDI) can reach 100% sensitivity and high specificity12 but carries inherent risks13, there is no disease-specific antemortem diagnostic test for sCJD; consequently a definitive diagnosis of prion disease relies on postmortem detection of PrPSc in brain tissue1, 12, 14. Although technological advancements such as FLAIR, diffusion-weighted imaging (DWI), and apparent diffusion coefficient (ADC) improved both the negative and positive predictive value of brain MRI and became important tools in differential diagnostics of human prion diseases, the origin of detected patterns and relationship to deposits of pathogenic PrPSc remain unclear1, 12, 15, 16. Moreover, surrogate markers that have been proposed for use in diagnostic tests for prion diseases—including the 14-3-3, S-100, and tau proteins in cerebrospinal fluid (CSF)— are released into the CSF as a result of any acute neuronal damage17–22 and thus have low diagnostic specificity, often with contradictory indications in the same patient17, 20, 23.
These difficulties prompted us and other researchers to search for methods having high analytical sensitivity and specificity, which could detect prions directly in tissues and body fluids, and which could therefore be an accurate antemortem diagnostic test for prion diseases. Several techniques were introduced that exploit the self-replicating (seeding) power of misfolded pathogenic PrPSc for detection24–26. The real-time quaking-induced conversion test (RT-QuIC) uses recombinant prion protein (recPrP) as a substrate to amplify very small amounts of PrPSc seed in CSF of CJD patients to detectable levels27, 28. The recent use of shorter fragments of prion protein as a substrate, and replacement of Western blot29 or conformation-dependent immunoassay (CDI)30 detection of the conversion reaction product with real-time monitoring of the amplification time course by a fluorescent dye, Thioflavin T, resulted in a faster and more sensitive second-generation RT-QuIC31. This improved technique paved the way for higher diagnostic throughput in detecting human prion diseases using CSF. Here we describe the diagnostic specificity and sensitivity of human prion detection with the second-generation CSF RT-QuIC in a blinded retrospective, as well as prospective analysis of a cohort of patients suffering from diverse rapidly progressive neurodegenerative disorders and suspected of having prion disease. We compare the results with detailed neuropathological and genetic assessments and analyze the impact of disease duration, phenotype, molecular characteristics of human prions, and polymorphism of codon 129 of the PRNP gene on CSF RT-QuIC in sporadic and genetic prion diseases.
MATERIALS AND METHODS
Ethics statement
All procedures were performed under protocols approved by the Institutional Review Board at Case Western Reserve University. In all cases, written informed consent for research was obtained from the patient or legal guardian and the material used had appropriate ethical approval for use in this project. All patient data and samples were coded and handled according to NIH guidelines to protect patient identities.
Patients and clinical evaluations
As part of the National Prion Disease Pathology Surveillance Center (NPDPSC) protocol, medical record information is requested from clinicians who refer subjects to the Autopsy Program. Clinical records of prion disease cases were reviewed by a clinician familiar with prion disease (BSA) and non-prion disease cases were reviewed by BSA and CF. Demographic data were collected using a standardized abstraction instrument and included approximate month of disease onset as documented in the medical record, gender, date of birth, date of death, clinical symptoms during the disease course, and diagnostic study results. Clinical symptoms pertaining to updated diagnostic criteria for sCJD were included15. Akinetic mutism was omitted due to its end-stage manifestation and commonality among cases. Brain MRIs were considered positive if the neuroradiologist’s reading fulfilled the criteria as previously defined27. Age was calculated as age at the time of death. Data were entered into a database and analyzed via SPSS 22 (IBM Corp, Armonk, NY).
The criteria for inclusion were (1) availability of a clinical diagnosis of CJD according to WHO criteria15, 32–34 and clearly determined and dated initial symptoms upon neurological examination to ascertain the disease duration; (2) methionine or valine at codon 129 of the human prion protein (PrP) gene (PRNP); (3) unequivocal classification of Type 1, Type 2, or Type 1–2 sPrPSc sCJD according to Western blot (WB) pattern; (4) unequivocal classification of pathology as definite Type 1, Type 2, or mixed Type 1–2 at the NPDPSC in Cleveland, OH.
In all cases in which neuropathology, Western blots, and PRNP gene sequencing of brain tissue excluded prion disease, we used for final diagnosis standard clinical and neuropathology protocols we published previously23, 35. The criteria for diagnosis of rapidly progressive Alzheimer disease (AD)35 were as follows: (1) probable clinical diagnosis of AD35, 36; (2) absent autosomal dominant pattern of dementia; (3) unequivocal classification as AD after detailed neuropathology and immunohistochemistry of tau proteins and amyloid beta using the current National Institute of Aging – Alzheimer's Association guidelines for neuropathological diagnosis of AD. That also included assessments of other comorbidities that could contribute to cognitive decline – including Lewy body/alpha-synuclein deposits, vascular parenchymal injury, and hippocampal sclerosis (especially when accompanied by TDP-43 pathology)35, 37
Classification of cases, brain samples, and PRNP gene sequencing
DNA was extracted from frozen brain tissues in all cases, and genotypic analysis of the PRNP coding region was performed as described12, 38, 39. On the basis of diagnostic pathology, immunohistochemistry, and WB examination of 2 or 3 brain regions (including frontal, occipital and cerebellum cortices) with mAb 3F4, the pathogenic PrPSc was classified as described previously1, 30, 40, 41.
Coronal sections of human brain tissues were obtained at autopsy and stored at −80°C. Three 200–350 mg cuts of frontal (superior and more posterior middle gyri) cortex were taken from each brain and used for molecular analyses. The other symmetric cerebral hemisphere was fixed in formalin and used for neuropathological classification of prion disease using histological and immunohistochemical analysis of samples from 16 anatomical areas and NPDPSC’s standard protocols1, 42, 43. In a case of equivocal classification of sCJD subtype between pathology and Western blots, we based the classification on the molecular characteristics of PrPSc on Western blots developed with a panel of Type 1 and Type 2-specific antibodies as described previously1, 44, 45. These criteria allowed the classification of all cases.
CSF samples
Cerebrospinal fluid (CSF) samples were referred to the NPDPSC from US medical institutions that had patients with suspected CJD or other prion diseases. All CSF samples were shipped on dry ice, tested for tTau and 14-3-3 proteins, and stored at −80°C. In the retrospective study, those with an autopsy-confirmed diagnosis of prion disease or an autopsy-confirmed non-prion disease diagnosis were selected for second-generation RT-QuIC analysis and cases were blinded.
Controls
Each RT-QuIC plate contained a positive control run in quadruplicate, a negative control run in octuplicate and up to twenty-one CSF test samples. Positive control wells were seeded with 2 µL of sCJD MM1 brain homogenate diluted 5×10−6 with 0.1% SDS/N2 supplement (Thermo Fisher, Waltham, MA) in PBS, and brought to a final reaction volume of 100 µL with 13 µL of PBS. Negative control wells were seeded with 15 µL of PBS. Testing wells were seeded with 15 µL of neat CSF from patients with an autopsy-confirmed diagnosis of CJD or an autopsy-confirmed non-prion disease diagnosis.
Preparation of recombinant prion protein
N-terminally truncated Syrian hamster recombinant prion protein [SHarPrP(90–231)] was prepared as previously described31, 46 with minor modifications. Briefly, recombinant protein purification was performed using an AKTA Prime Plus system (GE Life Sciences, Marlborough, MA) with the addition of a column prewashing step. In this step, denaturing buffer [100mM sodium phosphate (pH 8.0), 10 mM TRIS, 6 M Guanidine-HCl] was passed through the column at 2.3 mL/min for 30 min prior to protein gradient refolding. Protein concentration was determined using a NanoDrop Lite Spectrophotometer (Thermo Scientific, Wilmington, DE) with absorbance measured at 280 nm. Purity of SHaPrP(90–231) was ≥95%, as estimated by Coomassie staining after SDS-PAGE and by immunoblotting. Analytical sensitivity and bioactivity of purified SHarPrP(90–231) were tested via the second-generation RT-QuIC assay.
Second-generation RT-QuIC
The second-generation RT-QuIC assays31 were performed as previously described, with minor modifications. The RT-QuIC reaction mix was prepared as follows: 10mM phosphate buffer (pH 7.4), 300 mM NaCl, 0.1 mg/mL SHarPrP(23–231), 10 µM thioflavin T (ThT), 1 mM ethylenediaminetetraacteic acid tetrasodium salt (EDTA) and 0.002% sodium dodecyl sulfate (SDS). The 85 µL of reaction mix was loaded into each plate well and seeded with 15 µL of neat CSF for a final reaction volume of 100 µL. The incubation and real time fluorescence monitoring was performed using the FLUOstar Omega plate reader (BMG LABTECH GmbH, Ortenberg, Germany) set to the following parameters: 55°C incubation, 60 hr reaction time, 60 s shake/60 s rest cycles with ThT fluorescence measurements taken every 45 min.
RT-QuIC data analysis
To condense the large amount of raw RT-QuIC data for analysis, the average maximum ThT relative fluorescence units (rfu) of quadruple positive control (Pmax), individual CSF sample quadruples (Smax), and no seed control octuplicates (Nmax) were calculated. To compensate for variations in baselines between individual samples and relative fluorescence readings between plate readers, a baseline correction was performed in which the Nmax value was subtracted from both Pmax and Smax values. Baseline-corrected Smax values were then normalized as a percentage of Pmax. Normalized values were calculated by dividing baseline-corrected Smax by baseline-corrected Pmax and multiplying by 100. The samples were concluded positive if more than one well in the first round or 2 wells total, in first and repeat rounds, were positive and exceeded the diagnostic cutoff calculated as a mean ± 4*SD of all autopsy prion-negative cases.
Tolerance of second-generation RT-QuIC to blood contamination
A human blood sample with a known red blood cell (RBC) count was serially diluted with PBS. Eight dilutions ranging from 300 to 38,400 cells/µL were aliquoted into 1 mL aliquots and stored at −80°C. A 1 mL aliquot of each dilution was removed from the freezer, allowed to thaw and diluted 1:1 with a known RT-QuIC positive and negative CSF sample. The final concentration of RBC in spiked CSF samples ranged from 150 to 19,600 cells/µL. Blood-spiked CSF samples were spun at 2,000 × g for 2 min, loaded onto a 96-well plate and their absorbance read at 540 nm in a FLUOstar plate reader (BMG LABTECH). The seeding activity of each blood-spiked CSF was assessed via RT-QuIC and the maximum allowed tolerance determined.
14-3-3 and total Tau analyses
Surrogate markers for prion disease, 14-3-3, and tau protein, were measured by WB and ELISA respectively, as previously described20.
Conformation-dependent immunoassay (CDI)
CDI was used to quantify the amount of PrPSc present in our sCJD(MM1) brain homogenate control and was performed as previously described30, 41.
Statistical analysis
The sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV) and efficiency of each marker were obtained. The combination of different CSF RT-QuIC data and 14-3-3 or tTau protein results were tested by two different approaches: (i) positive if both results of CSF RT-QuIC and a second test were positive, and a negative result when either or both results of CSF RT-QuIC and a second test were negative; (ii) positive if combined either or both CSF RT-QuIC and a second test were positive, and a negative result if both results of CSF RT-QuIC and 14-3-3 were negative. We investigated the effect of the following demographic and laboratory variables on survival: sex, age at onset, and disease duration; and compared the parameters for the whole group and different phenotypic subgroups with ANOVA or Fisher exact test. Cumulative survival curves were constructed by the Kaplan–Meier method and compared using the Mantel-Cox and Breslow methods. For each type of PrPSc and PRNP codon 129 polymorphism, we report descriptive statistics and each variable was compared with ANOVA.
RESULTS
Analytical sensitivity of second-generation RT-QuIC for human brain prions
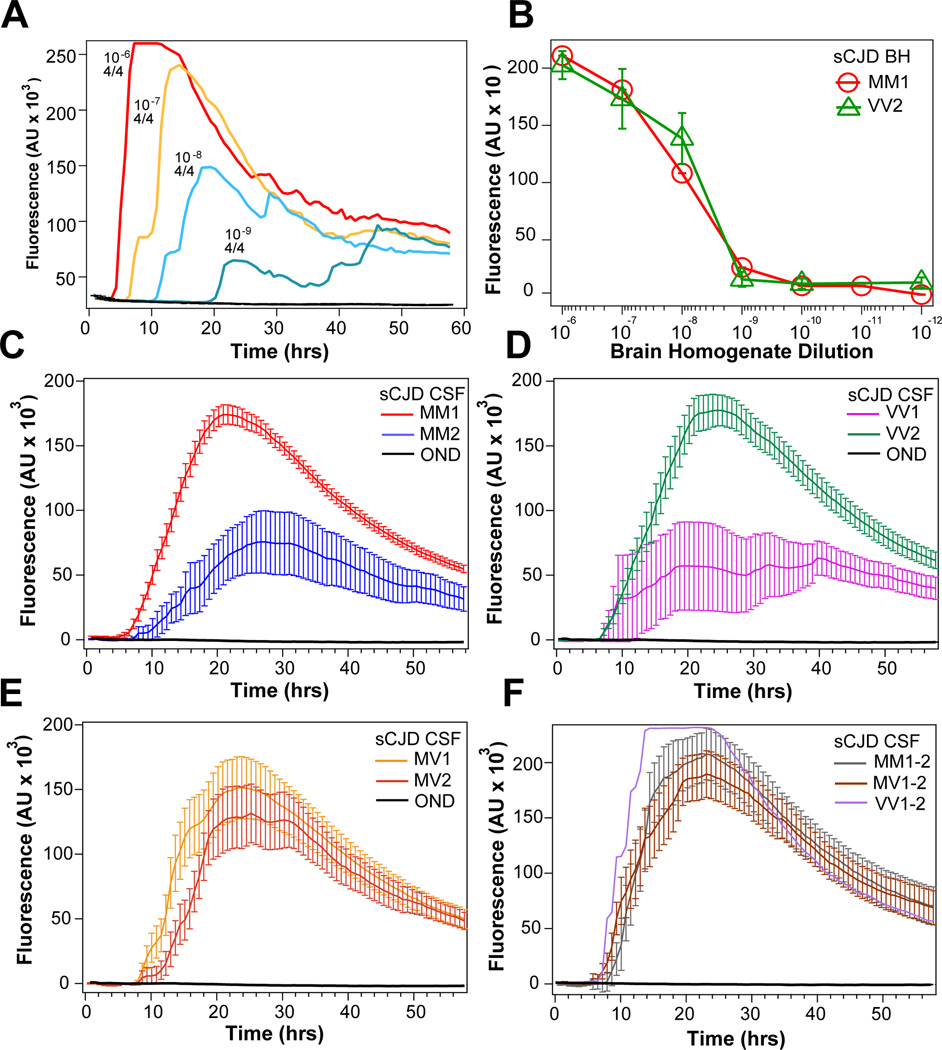
The analytical sensitivity of second-generation RT-QuIC was determined by serial dilution of brain homogenates prepared from three typical sCJD MM1 brain samples, three sCJD VV2 brain samples, and control cases diagnosed with other neurological diseases (Figure 1A and 1B). The raw fluorescence kinetic data and dilutions of a typical sCJD MM1 case shown in Figure 1A demonstrate the quantitative RT-QuIC response with progressively diminishing fluorescence signal and extending lag phase. Cumulatively, the second-generation RT-QuIC test demonstrates analytical sensitivity (Figure 1B) with detection limit for MM1 and VV2 sCJD prions at 109-fold dilution of respective brain homogenates Comparison to PrPSc levels measured in the initial 10% homogenate with CDI indicated that the endpoint sensitivity of second-generation RT-QuIC is ~3 ag of PrPSc per plate well, corresponding to 10–40 prion particles, assuming that the smallest human prion particles with the highest replication potency is composed of 20–78 monomers of PrPSc 30. In contrast, uniformly flat responses were seen with the unseeded reactions and those reactions seeded with brain homogenates of other neurological diseases.
Figure 1.
Analytical sensitivity and seeding response of second-generation CSF RT-QuIC to major human prions. (A) Serial dilution of brain homogenate of a typical sCJD MM1 case monitored in real time by thioflavin T fluorescence in second-generation RT-QuIC. The numbers above RT-QuIC curves denote the dilution and number of positive wells in four wells of the assay. (B) End point RT-QuIC sensitivity determined by serial dilutions of brain homogenate from three cases of sCJD MM1 and sCJD VV2 and monitored with second-generation RT-QuIC (red circles and green triangles). (C) Cumulative plot of second-generation CSF RT-QuIC data obtained in sCJD MM1 (n = 90) and in sCJD MM2 (n = 10). (D) Cumulative plot of CSF RT-QuIC data obtained in sCJD VV1 (n = 7; outlier with no remaining CSF for retesting has been eliminated) and in sCJD VV2 (n = 25). (E) Cumulative plot of CSF RT-QuIC data obtained in sCJD MV1 (n = 15) and in sCJD MV2 (n = 9). (F) Cumulative plot of CSF RT-QuIC data obtained in sCJD MM1–2 (n = 6), sCJD MV1–2 (n = 8), and in sCJD VV1–2 (n = 1). The curves are average ± S.D. of Thioflavin T florescence intensity at a given time point.
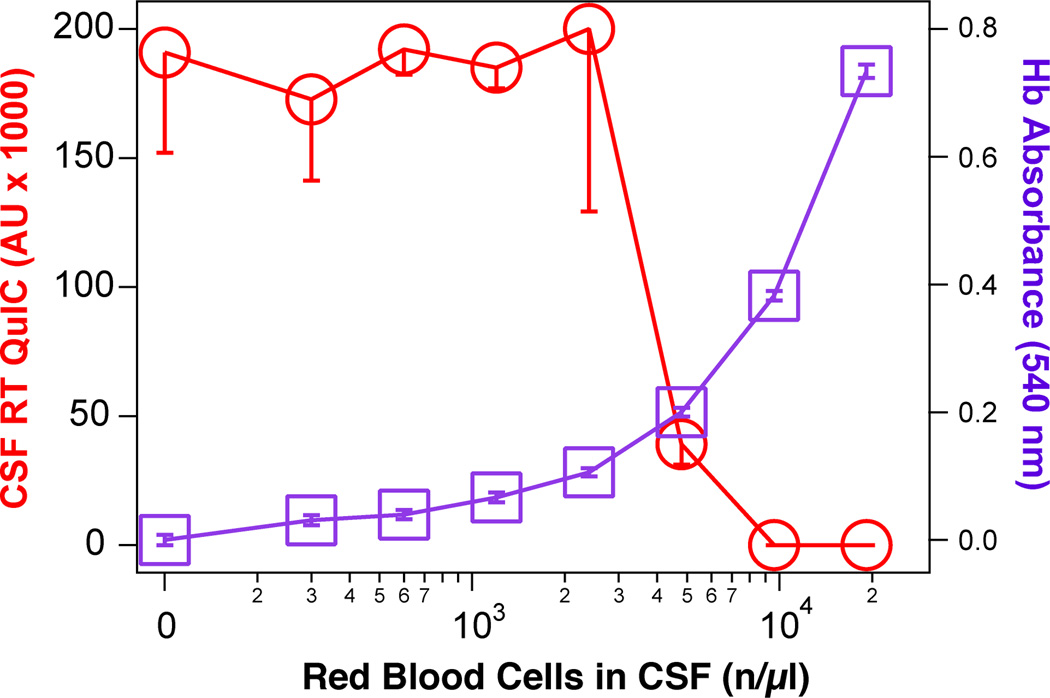
We investigated the known inhibitory effect47 of blood contamination on second-generation CSF RT-QuIC by serially diluting blood into RT-QuIC-positive CSF samples obtained from patients with variable sCJD subtypes: sCJD VV2 (n = 3), sCJD MM1 (n = 3), sCJD MV1 (n = 2), MV1–2 (n = 1), and gCJD with the E200K mutation (n = 1). We observed a uniform inhibitory effect above 9,600 red blood cells (RBC)/µl corresponding, after freezing and thawing, to the 540 nm absorbance of released hemoglobin equal to 0.389. Based on these experiments, we established a threshold (mean – 3×SD) for maximum acceptable blood contamination at hemoglobin absorbance 0.339 corresponding to 8,400 RBC/µl (Figure 2). These criteria excluded 58 (2.7%) out of 2,141 tested CSFs from evaluation.
Figure 2.
Effect of blood contamination on second generation CSF RT QuIC. Human blood was diluted into positive CSF samples received from patients with variable sCJD subtypes: sCJD VV2 (n = 3), sCJD MM1 (n = 3), sCJD MV1 (n = 2), MV1–2 (n = 1), gCJD with the E200K mutation (n = 1), and the second generation CSF RT QuIC was performed as described. Red blood cells (RBC) were counted before disruption by freezing and thawing, followed by in-plate absorbance reading for hemoglobin at 540nm. The curves are average ± S.D. of Thioflavin T florescence intensity in CSF RT QuIC (red circles), and hemoglobin absorbance (purple squares) at a given RBC count.
Diagnostic performance of second-generation CSF RT-QuIC
Autopsy and detailed neuropathological assessment was performed on 272 cases of a total of 2,141 second-generation CSF RT-QuIC tests referred to NPDPSC for evaluation of patients with rapidly progressive neurodegenerative disorders. In the first retrospective RT-QuIC cohort, we blindly tested CJD cases that represented expected frequencies of different prion disease subtypes in the general population1. The samples were defined a priori as RT QuIC positive or negative before unblinding and review of neuropathology. Other rapidly progressive neurological disorders, most frequently rapidly progressive Alzheimer disease, were used as control cases23, 48. The samples that were previously tested for 14-3-3 and tTau proteins were blinded and tested with second-generation CSF RT-QuIC. The demographics, clinical and neuropathological classification of the retrospective cohort are summarized in Table 1. The neuropathologically verified RT-QuIC positive cases were associated with an older age (P = 0.002), shorter disease course (P = 0.014), and a higher frequency of ataxia (P = 0.034). The RT-QuIC was more frequently negative in sCJD MM2 (P = 0.024) and we observed no correlation of the RT-QuIC result with the origin —sporadic or genetic—of prion disease, codon 129 polymorphism alone, gender, EEG, and MRI.
Table 1.
Demographics, CSF data, neuropathological classification, and clinical profiles of retrospective cohort.
| Neuropathological Classification | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Prion Disease Origin |
Sporadic | Genetic | ||||||||||
| Classification | Unit | MM1 | MV1 | MV2 | VV1 | VV2 | MM2-C | sFI | gCJD | GSS | FFI | Total |
| n (%) |
61 (48.4) |
10 (7.9) |
9 (7.1) |
8 (6.3) |
15 (11.9) |
7 (5.6) |
1 (0.8) |
12 (9.5) |
2 (1.6) |
1 (0.8) |
126 (100) |
|
| Age at death | yrs (SD) |
67.7 (9.8) |
70.4 (6.4) |
68.3 (5.0) |
61.9 (15.8) |
63.6 (9.7) |
64.9 (20.8) |
60.0 (0.0) |
57.5 (8.4) |
60.0 (11.3) |
61.0 (0.0) |
65.8 (10.9) |
| Duration | mo (SD) |
3.4 (2.7) |
9.4 (0.0) |
14.2 (9.6) |
6.9 (2.6) |
6.4 (3.6) |
12.5 (11.7) |
13.0 (0.0) |
3.5 (2.4) |
30.9 (0.1) |
9.4 (0.0) |
6.4 (6.9) |
| Male | n (%) |
28 (45.9) |
4 (40.0) |
6 (66.7) |
5 (62.5) |
8 (66.7) |
4 (50.0) |
1 (100.0) |
8 (66.7) |
1 (50.0) |
1 (100.0) |
66 (52.4) |
| 14-3-3 positive | n (%) |
58 (95.1) |
9 (90.0) |
1 (11.1) |
6 (75.0) |
15 (100.0) |
3 (42.9) |
0 (0.0) |
10 (83.3) |
1 (50.0) |
0 (0.0) |
103 (81.7) |
|
tTau positive (>1150 pg/ml) |
n (%) |
58 (95.1) |
9 (90.0) |
7 (77.8) |
8 (100.0) |
15 (100.0) |
4 (57.1) |
0 (0.0) |
11 (91.7) |
1 (50.0) |
1 (100.0) |
114 (90.5) |
| tTau | pg/ml (SD) |
5071 (3971) |
6827 (4588) |
2368 (1245) |
4177 (2276) |
3886 (825) |
2035 (1445) |
125 (0.0) |
6256 (5429) |
2974 (3780) |
4536 (0.0) |
4687 (3758) |
|
RT-QuIC positive |
n (%) |
58 (95.1) |
10 (100.0) |
8 (88.9) |
6 (75.0) |
15 (100.0) |
5 (71.4) |
0 (0.0) |
12 (100.0) |
1 (50.0) |
1 (100.0) |
116 (92.1) |
|
PSWCs on EEG (% with EEG results) |
n (%) |
14 (37.8) |
1 (12.5) |
0 (0.0) |
0 (0.0) |
0 (0.0) |
1 (50.0) |
- | 4 (66.7) |
- | 0 (0.0) |
20 (29.4) |
|
Positive MRI (% with MRI results) |
n (%) |
36 (76.6) |
7 (77.8) |
4 (66.7) |
7 (87.5) |
8 (80.0) |
5 (71.4) |
1 (100.0) |
10 (83.3) |
0 (0.0) |
0 (0.0) |
78 (76.5) |
|
Dementia (% with clinical history) |
n (%) |
52 (96.3) |
8 (88.9) |
6 (100.0) |
8 (100.0) |
11 (100.0) |
8 (100.0) |
1 (100.0) |
11 (91.7) |
2 (100.0) |
1 (100.0) |
108 (96.4) |
|
Visual (% with clinical history) |
n (%) |
16 (30.2) |
3 (33.3) |
2 (33.3) |
2 (25.0) |
3 (27.3) |
3 (37.5) |
0 (0.0) |
2 (16.7) |
0 (0.0) |
0 (0.0) |
31 (27.9) |
|
Ataxia (% with clinical history) |
n (%) |
34 (64.2) |
4 (44.4) |
4 (66.7) |
5 (62.5) |
10 (90.9) |
5 (62.5) |
0 (0.0) |
11 (91.7) |
2 (100.0) |
1 (100.0) |
76 (68.5) |
|
Myoclonus (% with clinical history) |
n (%) |
24 (45.3) |
3 (33.3) |
0 (0.0) |
6 (75.0) |
2 (18.2) |
2 (25.0) |
0 (0.0) |
6 (50.0) |
1 (50.0) |
1 (100.0) |
45 (40.5) |
|
Pyramidal Symptoms (% with clinical history) |
n (%) |
11 (20.8) |
2 (22.2) |
3 (50.0) |
5 (62.5) |
3 (27.3) |
2 (25.0) |
0 (0.0) |
5 (41.7) |
1 (50.0) |
0 (0.0) |
32 (28.8) |
|
Extrapyramidal Symptoms (% with clinical history) |
n (%) |
11 (20.8) |
6 (66.7) |
3 (50.0) |
4 (50.0) |
1 (9.1) |
1 (12.5) |
0 (0.0) |
3 (25.0) |
1 (50.0) |
0 (0.0) |
30 (27.0) |
To evaluate the second-generation CSF RT-QuIC performance during routine CSF testing, we blindly tested all samples sent to NPDPSC with RT-QuIC and in parallel for 14-3-3 and tTau proteins. All data were decoded and evaluated after autopsy, neuropathological assessment, and PRNP gene sequencing. Comparing the retrospective and prospective prion cohort (Table 2), the data indicate a greater preponderance of rapidly progressive prion diseases with shorter illness duration and 23% were mixed PrPSc Type 1–2 cases that were not incorporated into the retrospective study.
Table 2.
Demographics, CSF data, neuropathological classification, and clinical profiles of prospective cohort.
| Neuropathological Classification | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Prion Disease Origin |
Sporadic | Genetic | |||||||||
| Classification | Unit | MM1 | MV1 | VV2 | MM2-C | MM1-2 | MV1-2 | VV1-2 | sFI | gCJD | Total |
| n (%) |
29 (44.6) |
5 (7.7) |
11 (16.9) |
2 (3.1) |
6 (9.2) |
8 (12.3) |
1 (1.5) |
1 (1.5) |
2 (3.1) |
65 | |
| Age at death | yrs (SD) |
69.3 (7.0) |
64.4 (9.3) |
64.5 (8.0) |
66 (4.2) |
74.7 (11.9) |
66.6 (5.2) |
52 (0.0) |
61 (0.0) |
62 (0.0) |
67.5 (8.1) |
| Duration | mo (SD) |
3.4 (2.9) |
3.6 (3.2) |
6.1 (3.2) |
8.5 (0.0) |
2.9 (0.9) |
11.3 (8.9) |
28.9 (0.0) |
7.5 (0.0) |
1.4 (1.1) |
5.5 (5.7) |
| Male | n (%) |
14 (48.3) |
4 (80.0) |
8 (72.7) |
2 (100.0) |
1 (16.7) |
4 (50.0) |
0 (0.0) |
1 (100.0) |
2 (100.0) |
36 (55.4) |
| 14-3-3 positive | n (%) |
25 (86.2) |
2 (40.0) |
11 (100.0) |
1 (50.0) |
6 (100.0) |
5 (62.5) |
1 (100.0) |
0 (0.0) |
2 (100.0) |
53 (81.5) |
|
tTau positive (>1150 pg/ml) |
n (%) |
28 (96.6) |
4 (80.0) |
11 (100.0) |
2 (100.0) |
6 (100.0) |
8 (100.0) |
1 (100.0) |
0 (0.0) |
2 (100.0) |
62 (95.4) |
| tTau | pg/ml (SD) |
6278 (4079) |
3048 (3009) |
8651 (2001) |
4681 (5991) |
5314 (2461) |
3908 (2088) |
6793 (0.0) |
1837 (0.0) |
7372 (462) |
6067 (3528) |
|
RT-QuIC positive |
n (%) |
28 (96.6) |
4 (80.0) |
11 (100.0) |
2 (100.0) |
6 (100.0) |
8 (100.0) |
1 (100.0) |
0 (0.0) |
2 (100.0) |
62 (95.4) |
|
PSWCs on EEG (% with EEG results) |
n (%) |
0 (0.0) |
1 (33.3) |
0 (0.0) |
- | 1 (33.3) |
0 (0.0) |
0 (0.0) |
0 (0.0) |
0 (0.0) |
2 (6.3) |
|
Positive MRI (% with MRI results) |
n (%) |
14 (87.5) |
3 (60.0) |
7 (77.8) |
2 (100) |
4 (80.0) |
4 (57.1) |
1 (100) |
0 (0.0) |
1 (100) |
36 (76.6) |
|
Dementia (% with clinical history) |
n (%) |
20 (95.2) |
5 (100) |
9 (100) |
2 (100) |
5 (100) |
8 (100) |
1 (100) |
1 (100) |
1 (100) |
52 (98.1) |
|
Visual (% with clinical history) |
n (%) |
6 (31.6) |
1 (25.0) |
4 (44.4) |
1 (50.0) |
2 (40.0) |
1 (14.3) |
1 (100) |
0 (0.0) |
0 (0.0) |
16 (32.7) |
|
Ataxia (% with clinical history) |
n (%) |
12 (66.7) |
1 (33.3) |
9 (100) |
1 (50.0) |
2 (40.0) |
6 (85.7) |
1 (100) |
0 (0.0) |
0 (0.0) |
32 (68.1) |
|
Myoclonus (% with clinical history) |
n (%) |
10 (55.6) |
2 (66.7) |
3 (37.5) |
1 (50.0) |
1 (20.0) |
2 (28.6) |
1 (100) |
1 (100) |
0 (0.0) |
21 (45.7) |
|
Pyramidal Symptoms (% with clinical history) |
n (%) |
9 (47.4) |
0 (0.0) |
2 (25.0) |
1 (50.0) |
2 (40.0) |
3 (42.9) |
0 (0.0) |
1 (100) |
0 (0.0) |
18 (38.3) |
|
Extrapyramidal Symptoms (% with clinical history) |
n (%) |
3 (16.7) |
1 (33.3) |
1 (12.5) |
0 (0.0) |
1 (20.0) |
3 (42.9) |
0 (0.0) |
0 (0.0) |
0 (0.0) |
9 (19.6) |
The general diagnostic performance of second-generation CSF RT-QuIC, in the retrospective cohort, is superior to both traditional 14-3-3 and tTau surrogate tests (Table 3). The 98.5% diagnostic specificity in all prion disease forms is due to one case which repeatedly tested positive in CSF RT-QuIC, but which after the neuropathology was found to be Lewy body dementia (LBD). However, after precipitating prions from brain homogenate with PTA followed by WBs and CDI30, 41, we observed low, but above treshhold levels of PrPSc that may have escaped detection by immunohistochemistry12. Whether this case represents a false positive, or a true positive prion disease carrier with LBD comorbidity will require bioassay. Cumulatively, the increase in diagnostic specificity is particularly clinically relevant in the differential diagnostic clinical category of rapidly progressive dementias, because 42% are malignant forms of Alzheimer disease (Table 4). The prospectively determined diagnostic performance of second-generation CSF RT-QuIC maintains 100% specificity and 95% sensitivity for prion diseases of sporadic and genetic origin, with 100% and 82% positive and negative predictive value, respectively. The diagnostic specificity and sensitivity of both 14-3-3 and tTau protein were lower than CSF RT-QuIC, with 14-3-3 protein being the least reliable indicator of prion disease in all statistical parameters (Table 5 and for individual genetic cases Table 6). Combining the prospective CSF RT-QuIC data with the results of 14-3-3 or tTau decreased the sensitivity and specificity for CSF RT-QuIC + 14-3-3 protein to 82% and 43%, respectively, and for CSF RT-QuIC + tTau to 92% and 72%, respectively.
Table 3.
Retrospective neuropathological validation of RT QuIC.
| Neuropathological Assesment | ||||||
|---|---|---|---|---|---|---|
| CSF | Unit | Prion Negative |
Prion Positive |
sCJD | gCJD | |
| RT QuIC | n | 67 | 126 | 111 | 15 | |
| Specificity | % | 98.5 | 98.5 | 98.5 | ||
| Sensitivity | % | 92.1 | 91.9 | 93.3 | ||
| PPV | % | 99.1 | 99.0 | 93.3 | ||
| NPV | % | 86.8 | 88.0 | 98.5 | ||
| 14-3-3 | ||||||
| Specificity | % | 62.7 | 62.7 | 62.7 | ||
| Sensitivity | % | 81.7 | 82.9 | 73.3 | ||
| PPV | % | 81.1 | 79.3 | 31.4 | ||
| NPV | % | 82.4 | 87.5 | 93.3 | ||
| tTau≥1150 pg/ml | ||||||
| Specificity | % | 46.3 | 46.3 | 46.3 | ||
| Sensitivity | % | 90.5 | 91.0 | 86.7 | ||
| PPV | % | 76.0 | 73.7 | 26.5 | ||
| NPV | % | 72.1 | 75.6 | 93.9 | ||
Table 4.
Final neuropathological diagnosis of cases with rapidly progressive other neurological disorders (OND) clinically suspected from prion disorder.
| Neuropathological diagnosis | N | % |
|---|---|---|
| Alzheimer disease (AD) | 34 | 41.5 |
| Multi-infarct dementia | 7 | 8.5 |
| AD with multi-infarcts | 6 | 7.3 |
| AD with Lewy bodies | 5 | 6.1 |
| CNS Lymphoma | 4 | 4.9 |
| Frontotemporal lobar degeneration | 4 | 4.9 |
| Lewy body dementia | 4 | 4.9 |
| Leucodystrophy | 4 | 4.9 |
| Primary angiitis | 2 | 2.4 |
| Progressive multifocal leucoencephalopathy | 2 | 2.4 |
| Coccidiomycosis | 1 | 1.2 |
| Diffuse leucoencephalopathy with axonal spheroids | 1 | 1.2 |
| Encephalitis | 1 | 1.2 |
| Focal necrotizing encephalopathy | 1 | 1.2 |
| Global ischemic encephalopathy | 1 | 1.2 |
| Granulomatous amoebic encephalitis | 1 | 1.2 |
| Hemorrhagic stroke | 1 | 1.2 |
| Leptomeningeal carcinomatosis | 1 | 1.2 |
| Malignant glioma | 1 | 1.2 |
| Wernicke-Korsakoff syndrome with olivary gliosis | 1 | 1.2 |
| Undetermined | 1 | 1.2 |
| Total | 82 | 100 |
Table 5.
Prospective neuropathological validation of CSF RT-QuIC. All statistical conclusions are based on the separate evaluation of each CSF test. The brackets indicate weak CSF data statistics due to the limited number gCJD cases.
| Neuropathological Assessment | ||||||
|---|---|---|---|---|---|---|
| CSF | Unit | Prion Negative |
Prion Positive |
sCJD | gCJD | |
| RT QuIC | n | 14 | 65 | 63 | 2 | |
| Specificity | % | 100.0 | 100.0 | [100.0] | ||
| Sensitivity | % | 95.4 | 95.2 | [100.0] | ||
| PPV | % | 100.0 | 100.0 | [100.0] | ||
| NPV | % | 82.4 | 82.4 | [100.0] | ||
| 14-3-3 | ||||||
| Specificity | % | 42.9 | 42.9 | [42.9] | ||
| Sensitivity | % | 81.5 | 81.0 | [100.0] | ||
| PPV | % | 86.9 | 86.4 | [20.0] | ||
| NPV | % | 50.0 | 50.0 | [100.0] | ||
| tTau > 1150 pg/ml | ||||||
| Specificity | % | 71.4 | 71.4 | [71.4] | ||
| Sensitivity | % | 95.4 | 95.2 | [100.0] | ||
| PPV | % | 93.0 | 93.8 | [33.3] | ||
| NPV | % | 76.9 | 76.9 | [100.0] | ||
Table 6.
Neuropathological classification, PRNP gene sequencing, and CSF data in individual patients with genetic prion diseases.
| Classification |
PRNP gene mutation, codon 129, and chromosomal configuration |
RT-QuIC | 14-3-3 | Tau >1150 pg/ml |
Tau (pg/ml) |
|---|---|---|---|---|---|
| gCJD | p.E200K-p.129M (p.129M/M) | + | − | + | 2576 |
| gCJD | p.E200K-p.129M (p.129M/M) | + | + | + | 2942 |
| gCJD | p.E200K-p.129M (p.129M/M) | + | + | + | 4564 |
| gCJD | p.E200K-p.129M (p.129M/M) | + | + | + | 3811 |
| gCJD | p.E200K-p.129M (p.129M/M) | + | + | + | 7698 |
| gCJD | p.E200K-p.129M (p.129M/M) | + | + | + | 7045 |
| gCJD | p.E200K-p.129M (p.129M/M) | + | + | + | 7698 |
| gCJD | p.E200K-p.129M (p.129M/M) | + | + | + | 7045 |
| gCJD | p.E200K-p.129M (p.129M/M) | + | + | + | 6627 |
| gCJD | p.E200K-p.129V (p.129M/V) | + | + | + | 20360 |
| gCJD | p.E200K-p.129M (p.129M/V) | + | − | − | 991 |
| gCJD | p.E200K-p.129V(p.129V/V) | + | + | + | 4033 |
| gCJD | p.V210I-p.129M (p.129M/M) | + | + | + | 4180 |
| gCJD | p.V210I-p.129M (p.129M/M) | + | + | + | 8468 |
| FFI | p.D178N-p.129M (p.129M/M) | + | − | + | 4536 |
| GSS | p.A117V-p.129V (p.129V/V) | − | − | − | 301 |
| GSS | p.P102L-p.1296M (p.129M/M) | + | + | + | 5647 |
Prion type differentiation with second-generation CSF RT-QuIC
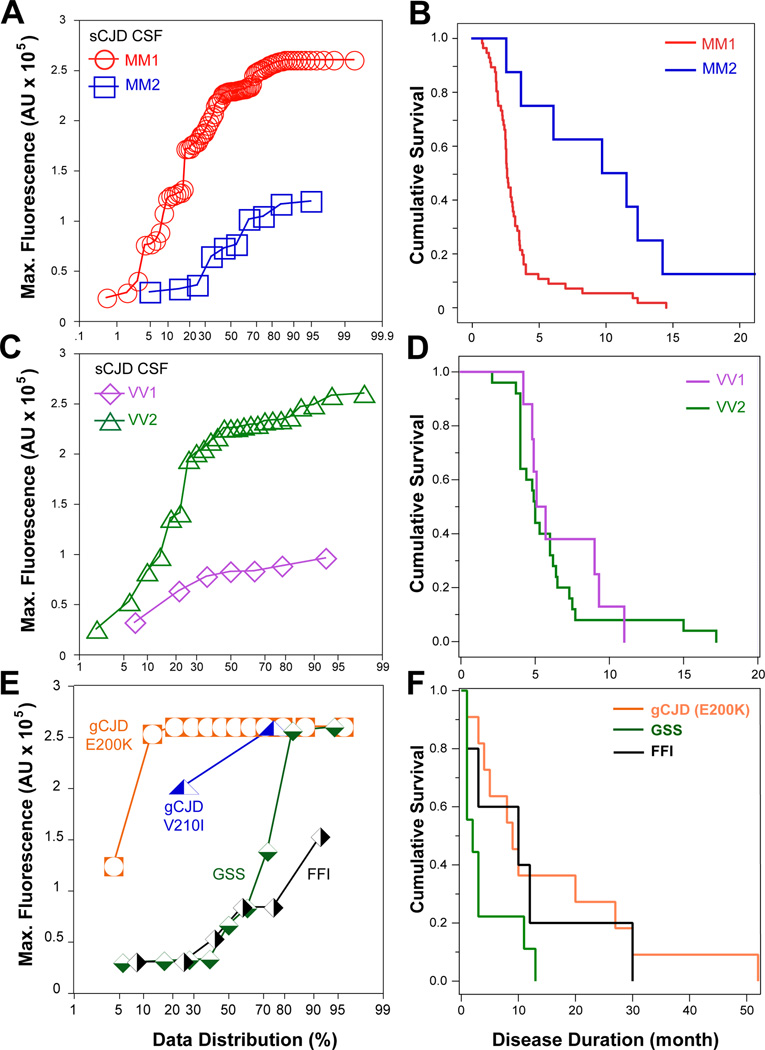
Cumulative plots of raw data revealed significant differences in the second-generation CSF RT-QuIC patterns, in both maximum fluorescence and lag interval, obtained in different subtypes of sCJD cases (Figure 1C–F). All the sCJD MM1 cases showed a significantly shorter lag phase and higher maximum fluorescence values than those in sCJD MM2 cases (P < 0.0001) (Figure 1C). In contrast, the sCJD VV1 cases showed significantly lower fluorescence intensity and an extended lag phase (P = 0.0011) (Figure 1D). Neither parameter differentiated CSF RT-QuIC of heterozygous sCJD MV1 from MV2 and the average fluorescence intensity was intermediate (Figure 1E). Interestingly, the cases with mixed Type 1–2 PrPSc in the same or different brain locations generated patterns of CSF RT-QuIC similar to sCJD MM1, reminiscent of the previous observation in PMCA, in which Type 1 prions progressively won the replication competition with the more slowly replicating Type 2 prions (Figure 1F)40. With the cutoff value for maximum fluorescence intensity of sCJD MM2 set at mean + 3SD, the individual CSF RT-QuIC data allowed differentiation of sCJD MM1 from sCJD MM2 with 95% probability. The higher RT-QuIC seeding potency of sCJD MM1 cases correlated with only a 2.7-month median cumulative survival and contrasted significantly with the median 9.7-month survival of MM2 cases (P = 0.015) (Figure 3A and 3B). Similarly, the CSF RT-QuIC fluorescence intensity differentiated sCJD VV1 from sCJD VV2 with 80% probability (Figure 3C and 3D). The previously reported survival data indicated the significantly longer survival of sCJD VV1 compared to sCJD VV2 cases1, 49, but this trend was not significant in our subset due to the small number of cases. Cases of gCJD, GSS, and FFI had clearly different seeding effects in CSF RT-QuIC that were unique for each mutation. While gCJD with E200K mutation tested in CSF RT-QuIC was invariably highly positive, the CSF RT-QuIC seeding activity varied widely with each specific mutation in GSS and in FFI, and these data correlate with the broad variation in disease duration (Figure 3E and 3F).
Figure 3.
Differentiation of major human prion subtypes based on seeding potency in second-generation CSF RT-QuIC and relationship with the disease progression rate. (A) Individual data distribution of maximum CSF second-generation RT-QuIC fluorescence obtained in sCJD MM1 (n = 90) and sCJD MM2 (n = 10). (B) Cumulative survival curves of sCJD cases plotted in Figure 3A. (C) Cumulative plot of CSF RT-QuIC data obtained in sCJD VV1 (n = 7; outlier with no remaining CSF for retesting is not plotted) and in sCJD VV2 (n = 25). (D) Cumulative survival curves of sCJD cases plotted in Figure 3C (E) Cumulative plot of CSF RT-QuIC data obtained in genetic prion disease (E200K, n = 12; V210I, n = 2), GSS (n = 9), and FFI (n = 6) (F) Cumulative survival curves of the genetic prion cases plotted in Figure 3E.
DISCUSSION
Neuropathological evaluation of the cases from this retrospective and prospective study show that second-generation RT-QuIC analysis of CSF samples from patients with suspected prion diseases can detect prions with a specificity and sensitivity of 98.5–100% and 92–95%, respectively. The high diagnostic specificity compares well with the results of the earlier version of RT-QuIC47, 50, 51. However, high analytical sensitivity with a detection limit of 3 ag of pathogenic PrPSc, corresponding to 10–40 human prion particles, leads to enhanced diagnostic sensitivity and reproducibility. Consequently, second-generation RT-QuIC missed only 5–8% of prion cases compared to 11–23% in the first generation assays31, 47, 50–52.
The results from this study show that the second-generation CSF RT-QuIC has, in a standard surveillance setting, higher sensitivity and specificity to CSF 14-3-3 (monitored here with WBs) and tTau proteins. Combining the CSF results into a two-component panel did not increase diagnostic sensitivity, but significantly decreased the specificity of the final conclusion. However, further studies are required to assess the complementary role, if any, which CSF 14-3-3 or tTau, on the one hand, and CSF second-generation CSF RT-QuIC, on the other, may play in the differential clinical diagnosis of neurodegenerative diseases. The observed lower specificity of 14-3-3 protein and higher specificity of tTau in the prospective cohort may reflect higher proportion of rapidly progressive non-prion cases in this cohort, the trend already noticed in an earlier study53. However, to draw a definitive conclusion would require expanding the numbers of prospectively followed autopsied cases. Ongoing studies are being pursued and more CSF samples need to be investigated from patients with vCJD, gCJD, VPSPr, and other atypical prion disease so that a better understanding can emerge of how this new test can be used in detection and differentiation of the less common and often atypical prion diseases.
Interestingly, in the retrospective second-generation CSF RT-QuIC study, we found one patient in which the initial neuropathology was concluded to be LBD but which RT-QuIC revealed repeatedly positive CSF for prions. Subsequent reinvestigation of the brain homogenate of this patient after PTA precipitation with CDI and WBs revealed low but above threshold levels of PK-resistant PrPSc. Considering the up to 50-year incubation time of prion diseases and the high analytical sensitivity of second-generation CSF RT-QuIC, we conjectured that this patient was likely a subclinical prion carrier who died due to LBD comorbidity. However, a definitive conclusion on this case would require protein misfolding cyclic amplification or bioassay in transgenic mice expressing human or chimeric PrP12, 25, 40.
The analysis of seeding activity kinetics in this study indicates strong agreement between CSF RT-QuIC data and final neuropathological classification after autopsy, which in turn correlates with the cumulative survival and duration of the disease. These correlations allow for the classification and subtyping of prion disease in the majority of cases while the patient is still alive. The CSF RT-QuIC combined with PRNP gene sequencing data allowed us to differentiate with 95% probability the most aggressive sCJD MM1 from slowly progressive sCJD MM2, and with 80% probability more aggressive sCJD VV2 prions from CJD VV1 prions, usually associated with slow progression. The two negative CSF RT QuIC in cases with sFI and one with GSS are intriguing and may suggest either lower levels of prions, distinct prion seeding potency due to the different conformation of PrPSc, or both. These aspects have to be addressed in parallel investigation of brain tissue to determine the levels and conformational strain characteristics of PrPSc present in each case30, 41, 54. Although the conversion substrate used in CSF RT-QuIC was Syrian hamster PrP(90–231), nevertheless the seeding activities observed in this study are in general agreement with data we obtained previously with RT-QuIC protocol and human PrP substrate, and PMCA protocol with transgenic mice brain homogenates expressing human PrPC monitored with CDI30, 54.
Using advanced biophysical tools, we demonstrated recently that the different molecular characteristics of sCJD prions arise from distinct particle sizes and conformational structure14, 30, 41. Furthermore, our data indicate that phenotypically distinct sCJD prions differ considerably in their structural organization, both at the level of the polypeptide backbone (as indicated by backbone amide H/D exchange data), as well as the quaternary packing arrangements (as indicated by H/D exchange kinetics for histidine side chains)54. Remarkably, the structure of sCJD prions is fundamentally different from laboratory rodent prions, and in contrast to previous observations on yeast and some murine prion strains, their replication rate is primarily determined not by conformational stability but by specific structural features that control the growth rate of prion protein aggregates54. Cumulatively, the second-generation CSF RT-QuIC seems to replicate the conformation-driven seeding aspect of human prion replication remarkably well, and thus accurately predicts the biological characteristics of distinct human prions and establishes the prognosis of the patient. These individualized diagnostic data and prognoses are critical for future therapeutic trials and provide measurable objective criteria for therapeutic efficacy.
Acknowledgments
The authors are grateful to the patients’ families, the CJD Foundation, referring clinicians, and all the members of the National Prion Disease Pathology Surveillance Center for invaluable technical help. We thank Dr. Man-Sun Sy for making available hybridoma clone 8H4, and Dr. Earl Poptic for scaled up antibody production. This work was supported by the Centers for Disease Control and Prevention (grant UR8/CCU515004 to JGS), the National Institutes of Health (grant NS074317 to JGS), the Charles S. Britton Fund (to JGS), the Fondation Alliance Biosecure (to JGS and BC), and the Intramural Research Program of the NIAID, NIH to B.C.
Footnotes
AUTHORS’ CONTRIBUTIONS
A.F., C.H., and J.G.S. were responsible for conception and design of the study. A.F., B.S.A., C.H., X.L., S.Y., Y.C., W.C., J.B., C.F., H.W., P.G., S.Z., A.H., C.T., L.B.S., M.L.C., B.C., and J.G.S. were responsible for acquisition and analysis of data. A.F., B.A., and J.G.S. were responsible for drafting of the manuscript and figures.
POTENTIAL CONFLICTS OF INTEREST
None of the authors have conflicts of interest in connection with this paper.
REFERENCES
- 1.Puoti G, Bizzi A, Forloni G, Safar JG, Tagliavini F, Gambetti P. Sporadic human prion diseases: molecular insights and diagnosis. Lancet Neurol. 2012 Jul;11(7):618–628. doi: 10.1016/S1474-4422(12)70063-7. [DOI] [PubMed] [Google Scholar]
- 2.Prusiner SB. Scrapie prions. Annu Rev Microbiol. 1989;43:345–374. doi: 10.1146/annurev.mi.43.100189.002021. [DOI] [PubMed] [Google Scholar]
- 3.Prusiner SB, Scott MR, DeArmond SJ, Cohen FE. Prion protein biology. Cell. 1998;93:337–348. doi: 10.1016/s0092-8674(00)81163-0. [DOI] [PubMed] [Google Scholar]
- 4.Bruce ME, McBride PA, Farquhar CF. Precise targeting of the pathology of the sialoglycoprotein, PrP, and vacuolar degeneration in mouse scrapie. Neurosci Lett. 1989;102:1–6. doi: 10.1016/0304-3940(89)90298-x. [DOI] [PubMed] [Google Scholar]
- 5.Hecker R, Taraboulos A, Scott M, et al. Replication of distinct scrapie prion isolates is region specific in brains of transgenic mice and hamsters. Genes Dev. 1992;6:1213–1228. doi: 10.1101/gad.6.7.1213. [DOI] [PubMed] [Google Scholar]
- 6.Bessen RA, Marsh RF. Distinct PrP properties suggest the molecular basis of strain variation in transmissible mink encephalopathy. J Virol. 1994;68:7859–7868. doi: 10.1128/jvi.68.12.7859-7868.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Telling GC, Parchi P, DeArmond SJ, et al. Evidence for the conformation of the pathologic isoform of the prion protein enciphering and propagating prion diversity. Science. 1996;274:2079–2082. doi: 10.1126/science.274.5295.2079. [DOI] [PubMed] [Google Scholar]
- 8.Safar J, Wille H, Itri V, et al. Eight prion strains have PrPSc molecules with different conformations. Nat Med. 1998 Oct;4(10):1157–1165. doi: 10.1038/2654. [DOI] [PubMed] [Google Scholar]
- 9.Peretz D, Scott M, Groth D, et al. Strain-specified relative conformational stability of the scrapie prion protein. Protein Sci. 2001;10:854–863. doi: 10.1110/ps.39201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Legname G, Baskakov IV, Nguyen H-OB, et al. Synthetic mammalian prions. Science. 2004;305:673–676. doi: 10.1126/science.1100195. [DOI] [PubMed] [Google Scholar]
- 11.Parchi P, Giese A, Capellari S, et al. Classification of sporadic Creutzfeldt-Jakob disease based on molecular and phenotypic analysis of 300 subjects. Ann Neurol. 1999 Aug;46(2):224–233. [PubMed] [Google Scholar]
- 12.Safar JG, Geschwind MD, Deering C, et al. Diagnosis of human prion disease. Proc Natl Acad Sci USA. 2005 Mar 1;102(9):3501–3506. doi: 10.1073/pnas.0409651102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bai HX, Zou Y, Lee AM, Lancaster E, Yang L. Diagnostic Value and Safety of Brain Biopsy in Patients With Cryptogenic Neurological Disease: A Systematic Review and Meta-analysis of 831 Cases. Neurosurgery. 2015 Aug;77(2):283–295. doi: 10.1227/NEU.0000000000000756. discussion 95. [DOI] [PubMed] [Google Scholar]
- 14.Safar JG. Molecular Mechanisms Encoding Quantitative and Qualitative Traits of Prion Strains. In: Gambetti P, editor. Prions and Diseases. New York: Springer Verlag; 2012. [Google Scholar]
- 15.Zerr I, Kallenberg K, Summers DM, et al. Updated clinical diagnostic criteria for sporadic Creutzfeldt-Jakob disease. Brain. 2009 Oct;132(Pt 10):2659–2668. doi: 10.1093/brain/awp191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Forner SA, Takada LT, Bettcher BM, et al. Comparing CSF biomarkers and brain MRI in the diagnosis of sporadic Creutzfeldt-Jakob disease. Neurol Clin Pract. 2015 Apr;5(2):116–125. doi: 10.1212/CPJ.0000000000000111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Geschwind MD, Martindale J, Miller D, et al. Challenging the clinical utility of the 14-3-3 protein for the diagnosis of sporadic Creutzfeldt-Jakob disease. Arch Neurol. 2003;60:813–816. doi: 10.1001/archneur.60.6.813. [DOI] [PubMed] [Google Scholar]
- 18.Sanchez-Juan P, Green A, Ladogana A, et al. CSF tests in the differential diagnosis of Creutzfeldt-Jakob disease. Neurology. 2006;67:637–643. doi: 10.1212/01.wnl.0000230159.67128.00. [DOI] [PubMed] [Google Scholar]
- 19.Sanchez-Juan P, Sanchez-Valle R, Green A, et al. Influence of timing on CSF tests value for Creutzfeldt-Jakob disease diagnosis. J Neurol. 2007;254:901–906. doi: 10.1007/s00415-006-0472-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hamlin C, Puoti G, Berri S, et al. A comparison of tau and 14-3-3 protein in the diagnosis of Creutzfeldt-Jakob disease. Neurology. 2012 Aug 7;79(6):547–552. doi: 10.1212/WNL.0b013e318263565f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmitz M, Ebert E, Stoeck K, et al. Validation of 14-3-3 Protein as a Marker in Sporadic Creutzfeldt-Jakob Disease Diagnostic. Mol Neurobiol. 2015 May 7; doi: 10.1007/s12035-015-9167-5. [DOI] [PubMed] [Google Scholar]
- 22.Karch A, Hermann P, Ponto C, et al. Cerebrospinal fluid tau levels are a marker for molecular subtype in sporadic Creutzfeldt-Jakob disease. Neurobiol Aging. 2015 May;36(5):1964–1968. doi: 10.1016/j.neurobiolaging.2015.01.021. [DOI] [PubMed] [Google Scholar]
- 23.Chitravas N, Jung RS, Kofskey DM, et al. Treatable neurological disorders misdiagnosed as Creutzfeldt-Jakob disease. Ann Neurol. 2011 Sep;70(3):437–444. doi: 10.1002/ana.22454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Colby DW, Zhang Q, Wang S, et al. Prion detection by an amyloid seeding assay. Proc Natl Acad Sci U S A. 2007 Dec 26;104(52):20914–20919. doi: 10.1073/pnas.0710152105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saborio GP, Permanne B, Soto C. Sensitive detection of pathological prion protein by cyclic amplification of protein misfolding. Nature. 2001;411:810–813. doi: 10.1038/35081095. [DOI] [PubMed] [Google Scholar]
- 26.Atarashi R, Moore RA, Sim VL, et al. Ultrasensitive detection of scrapie prion protein using seeded conversion of recombinant prion protein. Nat Methods. 2007;4:645–650. doi: 10.1038/nmeth1066. [DOI] [PubMed] [Google Scholar]
- 27.Peden AH, McGuire LI, Appleford NE, et al. Sensitive and specific detection of sporadic Creutzfeldt-Jakob disease brain prion protein using real-time quaking-induced conversion. J Gen Virol. 2012 Feb;93(Pt 2):438–449. doi: 10.1099/vir.0.033365-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Orru CD, Wilham JM, Vascellari S, Hughson AG, Caughey B. New generation QuIC assays for prion seeding activity. Prion. 2012 Apr 1;6(2) doi: 10.4161/pri.19430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Atarashi R, Wilham JM, Christensen L, et al. Simplified ultrasensitive prion detection by recombinant PrP conversion with shaking. Nat Methods. 2008 Mar;5(3):211–212. doi: 10.1038/nmeth0308-211. [DOI] [PubMed] [Google Scholar]
- 30.Kim C, Haldiman T, Surewicz K, et al. Small Protease Sensitive Oligomers of PrP(Sc) in Distinct Human Prions Determine Conversion Rate of PrP(C) PLoS Pathog. 2012 Aug;8(8):e1002835. doi: 10.1371/journal.ppat.1002835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Orru CD, Groveman BR, Hughson AG, Zanusso G, Coulthart MB, Caughey B. Rapid and sensitive RT-QuIC detection of human Creutzfeldt-Jakob disease using cerebrospinal fluid. MBio. 2015;6(1) doi: 10.1128/mBio.02451-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.World Health Organization. Geneva: 1999. Mar 23–26, WHO infection control guidelines for transmissible spongiform encephalopathies. [Google Scholar]
- 33.Collins SJ, Sanchez-Juan P, Masters CL, et al. Determinants of diagnostic investigation sensitivities across the clinical spectrum of sporadic Creutzfeldt-Jakob disease. Brain. 2006;129:2278–2287. doi: 10.1093/brain/awl159. [DOI] [PubMed] [Google Scholar]
- 34.Geschwind MD, Shu H, Haman A, Sejvar JJ, Miller BL. Rapidly progressive dementia. Ann Neurol. 2008 Jul;64(1):97–108. doi: 10.1002/ana.21430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cohen ML, Kim C, Haldiman T, et al. Rapidly progressive Alzheimer's disease features distinct structures of amyloid-beta. Brain. 2015 Apr;138(Pt 4):1009–1022. doi: 10.1093/brain/awv006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McKhann GM, Knopman DS, Chertkow H, et al. The diagnosis of dementia due to Alzheimer's disease: recommendations from the National Institute on Aging-Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimers Dement. 2011 May;7(3):263–269. doi: 10.1016/j.jalz.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Josephs KA, Nelson PT. Unlocking the mysteries of TDP-43. Neurology. 2015 Mar 3;84(9):870–871. doi: 10.1212/WNL.0000000000001322. [DOI] [PubMed] [Google Scholar]
- 38.Parchi P, Zou W, Wang W, et al. Genetic influence on the structural variations of the abnormal prion protein. Proc Natl Acad Sci USA. 2000;97:10168–10172. doi: 10.1073/pnas.97.18.10168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parchi P, Castellani R, Capellari S, et al. Molecular basis of phenotypic variability in sporadic Creutzfeldt-Jakob disease. Ann Neurol. 1996;39:767–778. doi: 10.1002/ana.410390613. [DOI] [PubMed] [Google Scholar]
- 40.Haldiman T, Kim C, Cohen Y, et al. Co-existence of distinct prion types enables conformational evolution of human PrPSc by competitive selection. J Biol Chem. 2013 Oct 11;288(41):29846–29861. doi: 10.1074/jbc.M113.500108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim C, Haldiman T, Cohen Y, et al. Protease-Sensitive Conformers in Broad Spectrum of Distinct PrP Structures in Sporadic Creutzfeldt-Jakob Disease Are Indicator of Progression Rate. PLoS Pathog. 2011 Sep;7(9):e1002242. doi: 10.1371/journal.ppat.1002242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim JI, Cali I, Surewicz K, et al. Mammalian prions generated from bacterially expressed prion protein in the absence of any mammalian cofactors. J Biol Chem. 2010 May 7;285(19):14083–14087. doi: 10.1074/jbc.C110.113464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gambetti P, Kong Q, Zou W, Parchi P, Chen SG. Sporadic and familial CJD: classification and characterisation. Br Med Bull. 2003;66:213–239. doi: 10.1093/bmb/66.1.213. [DOI] [PubMed] [Google Scholar]
- 44.Parchi P, de Boni L, Saverioni D, et al. Consensus classification of human prion disease histotypes allows reliable identification of molecular subtypes: an inter-rater study among surveillance centres in Europe and USA. Acta Neuropathol. 2012 Oct;124(4):517–529. doi: 10.1007/s00401-012-1002-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cali I, Castellani R, Alshekhlee A, et al. Co-existence of scrapie prion protein types 1 and 2 in sporadic Creutzfeldt–Jakob disease: its effect on the phenotype and prion-type characteristics. Brain. 2009 Oct;132(10):2643–2658. doi: 10.1093/brain/awp196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wilham JM, Orru CD, Bessen RA, et al. Rapid end-point quantitation of prion seeding activity with sensitivity comparable to bioassays. PLoS Pathog. 2010;6(12):e1001217. doi: 10.1371/journal.ppat.1001217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cramm M, Schmitz M, Karch A, et al. Stability and Reproducibility Underscore Utility of RT-QuIC for Diagnosis of Creutzfeldt-Jakob Disease. Mol Neurobiol. 2016 Apr;53(3):1896–1904. doi: 10.1007/s12035-015-9133-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cohen M, Appleby B, Safar JG. Distinct Prion-Like Strains of Amyloid Beta Implicated in Phenotypic Diversity of Alzheimer Disease. Prion. 2016 Jan;25:0. doi: 10.1080/19336896.2015.1123371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pocchiari M, Puopolo M, Croes EA, et al. Predictors of survival in sporadic Creutzfeldt-Jakob disease and other human transmissible spongiform encephalopathies. Brain. 2004 Oct;127(Pt 10):2348–2359. doi: 10.1093/brain/awh249. [DOI] [PubMed] [Google Scholar]
- 50.McGuire LI, Peden AH, Orru CD, et al. Real time quaking-induced conversion analysis of cerebrospinal fluid in sporadic Creutzfeldt-Jakob disease. Ann Neurol. 2012 Aug;72(2):278–285. doi: 10.1002/ana.23589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cramm M, Schmitz M, Karch A, et al. Characteristic CSF prion seeding efficiency in humans with prion diseases. Mol Neurobiol. 2015 Feb;51(1):396–405. doi: 10.1007/s12035-014-8709-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Atarashi R, Satoh K, Sano K, et al. Ultrasensitive human prion detection in cerebrospinal fluid by real-time quaking-induced conversion. Nat Med. 2011 Feb;17(2):175–178. doi: 10.1038/nm.2294. [DOI] [PubMed] [Google Scholar]
- 53.Stoeck K, Sanchez-Juan P, Gawinecka J, et al. Cerebrospinal fluid biomarker supported diagnosis of Creutzfeldt-Jakob disease and rapid dementias: a longitudinal multicentre study over 10 years. Brain. 2012 Oct;135(Pt 10):3051–3061. doi: 10.1093/brain/aws238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Safar JG, Xiao X, Kabir ME, et al. Structural determinants of phenotypic diversity and replication rate of human prions. PLoS Pathog. 2015 Apr;11(4):e1004832. doi: 10.1371/journal.ppat.1004832. [DOI] [PMC free article] [PubMed] [Google Scholar]