Abstract
Linkage studies of complex genetic diseases have been largely replaced by Genome-Wide Association studies (GWAS), due in part to limited success in complex trait discovery. However, recent interest in rare and low-frequency variants motivates reexamination of family-based methods. In this study we investigated the performance of two-point linkage analysis for over 1.6 million SNPs combined with single variant association analysis to identify high impact variants which are both strongly linked and associated with cardiometabolic traits in up to 1 414 Hispanics from the Insulin Resistance Atherosclerosis Family Study (IRASFS). Evaluation of all 50 phenotypes yielded 83 557 000 LOD scores with 9 214 LOD scores ≥ 3.0, 845 ≥ 4.0, and 89 ≥ 5.0, with a maximal LOD score of 6.49 (rs12956744 in the LAMA1 gene for TNFα receptor 2). Twenty-seven variants were associated with p < 0.005 as well as having a LOD score > 4, including variants in the NFIB gene under a linkage peak with TNFα receptor 2 levels on chromosome 9. Linkage regions of interest included a broad peak (31Mb) on chromosome 1q with acute insulin response (max LOD = 5.37). This region was previously documented with type 2 diabetes in family-based studies, providing support for the validity of these results. Overall, we have demonstrated the utility of two-point linkage and association in comprehensive genome-wide array-based SNP genotypes.
Keywords: linkage analysis, cardiometabolic, acute insulin response, Hispanic
Introduction
Family-based linkage analysis has largely been supplanted by genome-wide association studies, often using unrelated samples, following the limited success of linkage when applied to complex traits. Family-based analyses, however, have inherent strengths which complement other approaches for identification of contributors to complex phenotypes1,2. Such analyses may be especially applicable to identifying low frequency (minor allele frequency [MAF] 0.01–0.05) to rare (MAF < 0.01) alleles with high impact3–8. We have implemented approaches in parallel which utilize simple two-point linkage analysis and conventional association analysis to search for genetic variants with meaningful contributions to phenotypic variance of traits. Two-point linkage analysis considers each variant independently, unlike multipoint analysis which integrates the information from multiple variants simultaneously. Therefore, two-point linkage does not have the same issues with inflation due to linkage disequilibrium between markers and can be used to test putatively impactful variants for linkage directly. The combined two-point linkage and association approach has the advantage of being able to directly align SNP results for the two analyses, pinpointing variants which show evidence of both linkage and association at the single SNP level. In prior studies, this has been applied to exome chip data, thus focusing on coding variants9 and characteristics of a functional SNP10.
Evaluation of association in the context of linkage has an extensive history11–13, with association typically utilized to determine whether genetic variants residing under the linkage peak explain the observed signal. We have observed that instances of strong linkage and association together at a single locus (e.g. APOE with ApoB levels, CETP with HDL levels, ADIPOQ with adiponectin levels)9,10 represent variants or loci which have a striking impact on phenotype, reflected as explanation of a high proportion of the variance of the trait (3–60%). We have also observed this across a range of minor allele frequencies (1–45%), indicating that this approach can be informative for a full range of genetic variation. Other groups have utilized combined metrics of linkage and association to identify variants with large impact11; however, that is a project currently undergoing evaluation separate from these analyses.
Here we have investigated the performance of these approaches in a contemporary genetic dataset consisting of comprehensive genome-wide and exome chip data encompassing 1.6 million SNPs in 90 Hispanic families from the Insulin Resistance Atherosclerosis Family Study (IRASFS). Based on our prior work and recent evidence for the existence of high impact non-coding variants14, we hypothesize this family-based method is applicable to the search for such variants.
Materials and Methods
Samples and Phenotype Data
The samples used in this study are from the Hispanic cohorts of the Insulin Resistance and Atherosclerosis Family Study (IRASFS)15. Briefly, subjects were ascertained on the basis of large family size in San Luis Valley, Colorado and San Antonio, Texas. The sample consisted of 1 425 individuals from 90 families, who were extensively phenotyped, including a frequently sampled intravenous glucose test (FSIGT), measures of blood lipids and inflammatory markers, anthropomorphic measures, as well as fat deposition measures by computed tomography (CT) and dual X-ray absorptiometry (DXA) scans. IRB approval was obtained at all clinical and analysis sites, and all participants provided informed consent.
Genotype Data
SNP genotype data from three genotyping chips were utilized. Illumina OmniExpress and Illumina Omni 1S chips were genotyped as part of the Genetics Underlying Diabetes in Hispanics (GUARDIAN) Consortium (N = 1034 and 1038, respectively)16, and the Illumina HumanExome Beadchip was genotyped on a larger subset (N = 1414)9 of the full IRASFS Hispanic cohorts. Genotyping of the Illumina HumanExome BeadChip v1.0 (N = 552) and v1.1 (N = 862) was performed at the Wake Forest Center for Genomics and Personalized Medicine Research, while the Illumina HumanOmniExpress BeadChip and Illumina Omni1S BeadChip were genotyped at the core genotyping laboratory at Cedars-Sinai Medical Center. All genotypes were called separately by genotyping array using GenomeStudio (Illumina, San Diego, CA). Sample and autosomal SNP call rates were ≥0.98 (>0.99 SNP call rates for the OmniExpress and Omni1S chips), and Exome Chip SNPs with poor cluster separation (<0.35) were excluded. All datasets independently underwent Mendelian error checking using PedCheck17 to detect genotypes discordant in families for Mendelian inheritance, with resolution by removing all inconsistent genotypes. The total number of unique SNPs available for analysis following QC was as follows: 81 559 from the Exome Chip, 668 758 from OmniExpress and 920 823 from the Omni1S chip, for a total of 1 671 140 SNPs.
Imputation to the 1000 Genomes integrated reference panel (version 2) was performed using genotypes and samples from the OmniExpress dataset (N = 634K genotypes and 1034 individuals) using SHAPEIT18 for phasing and IMPUTE219 for imputation.
Analyses
SNPs were evaluated for both two-point family-based linkage and single SNP association using Sequential Oligogenic Linkage Analysis Routines (SOLAR)20 separately by genotyping platform. Both analyses used age, sex, body mass index (BMI), and study center as covariates. All phenotypes evaluated were transformed to approximate normality of the residuals if necessary (Supplementary Table 1). Additionally, due to the high impact of a low frequency variant known to influence adiponectin levels in this population3,10, presence of the variant encoding the G45R missense mutation in ADIPOQ (rs200573126) was included as a covariate for analyses involving adiponectin. Visceral adipose tissue area (VAT), visceral to subcutaneous tissue ratio (VSR), waist circumference, and waist-to-hip ratio (WHR) were run both with and without BMI as a covariate. However subcutaneous adipose tissue area (SAT), percent body fat, and body adiposity index (BAI) were not adjusted for BMI. All association analyses included three admixture proportions as covariates. Existing admixture proportion estimates were available from previously genotyped exome chip data; estimates were computed by maximum likelihood estimation of individual ancestries in ADMIXTURE21 assuming five ancestral populations (K = 5) from exome chip-wide SNP data after pruning for linkage disequilibrium (LD) to produce admixture estimates for the greatest number of samples. Of the five variables considered, three variables were selected as representing the variation in these Hispanic samples, as inclusion of additional postulated ancestral populations began isolating individual pedigrees.
For validation of performance, genotypes imputed to the 1000 Genomes panel were also evaluated for linkage (and association) in two regions which were selected for their linkage regions as well as being phenotypically of particular interest to our group: chromosome 1 for acute insulin response to glucose (AIR) and chromosome 7 for insulin sensitivity index (SI). Best guess genotypes from the imputed data were used in the linkage analysis because methods that account for imputation uncertainty have not been developed for linkage. These analyses used the same covariates as previously mentioned.
Results
The goal of this analysis was to test the utility of carrying out a combined linkage and association analysis in a contemporary dataset made up of GWAS (Illumina OmniExpress and Omni1S) and exome chip data encompassing over 1.6 million SNPs. The combined performance was evaluated for a total of 50 quantitative traits from 7 phenotypic groups: Glucose Homeostasis, Adiposity, Lipids, Biomarkers, Hypertension, Liver Enzymes, and Liver Fat, in 90 families from the IRASFS with an average family size of 15.4 individuals. Overall, 83 557 000 LOD scores and association p-values were calculated across the three genotyping sets.
Characteristics of the samples and genotyping are summarized in Table 1. The sample consisted of 1418 individuals from 90 families. Specifically, for the smallest genotyped sample (OmniExpress), sample sizes ranged from 786 (percent body fat) to 1034 (AIR), although larger sample sizes were available for SNPs present on the exome chip (up to 1256 for fibrinogen and ACR). Across all phenotypes, there were 9214 LOD scores greater than or equal to 3, 845 ≥ 4 and 89 ≥ 5. Of the 57 variants with LOD scores greater than 5.0, 27 were linked to TNFα receptor 2 levels, 13 to HDL levels, 5 to AIR, 4 to G45R-adjusted adiponectin levels, and three to BMI-adjusted VAT. While a detailed summary of each trait analysis is impractical, following on our earlier observations9,10, we have initially focused on the patterns visible in linkage analysis followed by relating these results to association analysis results. In this report, we evaluated linkage and association with 50 cardiometabolic phenotypes (see Supplementary Table 1 for complete listing). Selected phenotypes, namely TNFα receptor 2 levels, high density lipoprotein (HDL) levels, AIR, adiponectin levels (adjusted for G45R, a high impact mutation identified previously in these samples3,10), and VAT adjusted for BMI are summarized in Table 1. Overall, 12 phenotypes (from 4 phenotype groups: glucose homeostasis, lipids, adiposity and biomarkers) were represented in this category of LOD > 5.0 results summarized in Table 2, where highest LOD scores are grouped by phenotype and chromosome. A complete summary of LOD scores greater than 5 is presented in Supplementary Table 2.
Table 1.
Demographic characteristics of the IRASFS Hispanic samples with selected phenotypes.
| Characteristic | Exome Chip (81 559 variants) |
Omni Express (668 758 variants) |
Omni 1S (920 823 variants) |
|||
|---|---|---|---|---|---|---|
| Samples1 | 1 414 | 1 034 | 1 038 | |||
| Age (years) | 1 263 | 42.75 (18–81) | 1 034 | 40.63 (18–81) | 1 038 | 40.61 (18–81) |
| % Female | 823 | 58.3 % F | 609 | 58.90% | 612 | 58.90% |
| BMI (kg/m2) | 1 253 | 28.88 (16–58) | 1 027 | 28.28 (16–58) | 1 027 | 28.28 (16–58) |
| % T2D2 | 187 | 13.20% | 0 | 0% | 0 | 0% |
| AIR (pmol*mL-1*min-1) | 1 035 | 761.86 (−80.9–4 313.7) | 1 034 | 760.29 (−80.9–4 313.7) | 1 038 | 759.21 (−80.9–4 313.7) |
| TNFα receptor 2 (ng/mL) | 982 | 7.05 (2.38–30.00) | 821 | 6.79 (2.38–30.00) | 824 | 6.79 (2.38–30.00) |
| Fibrinogen (mg/dL) | 1 256 | 265.74 (113–591) | 1 032 | 259.37 (113–506) | 1 036 | 259.61 (113–506) |
| Cholesterol (mg/dL) | 1 255 | 177.94 (74–348) | 1 031 | 176.12 (74–311) | 1 035 | 176.17 (74–311) |
| HDL (mg/dL) | 1 254 | 43.82 (18–125) | 1 030 | 43.58 (18–100) | 1 034 | 43.60 (18–100) |
| LDL (mg/dL) | 1 242 | 109.17 (31–218) | 1 022 | 109.04 (31–213) | 1 026 | 109.06 (31–213) |
| Triglycerides (mg/dL) | 1 252 | 124.57 (18–836) | 1 030 | 118.30 (18–836) | 1 034 | 118.31 (18–836) |
| ACR (mg/g) | 1 256 | 53.55 (1.63–3 903.92) | 1 032 | 19.63 (1.93–1 459.68) | 1 036 | 19.58 (1.93–1 459.68) |
| Percent Body Fat | 943 | 33.95 (10.10–55.03) | 786 | 33.51 (10.10–51.78) | 789 | 33.52 (10.10–51.78) |
| VAT (cm2) | 1 206 | 114.02 (10.04–382.56) | 994 | 106.56 (10.04–363.34) | 998 | 106.52 (10.04–363.34) |
| VSR | 1 164 | 0.38 (0.07–1.63) | 963 | 0.36 (0.07–1.56) | 967 | 0.36 (0.07–1.56) |
Data presented as mean (range) or percent.
From 90 pedigrees, not entirely overlapping.
at baseline
Table 2.
Summary of linkage results for phenotypes with at least one variant with LOD >4.
| Phenotype | LOD > 5 | LOD > 4 | LOD > 3 |
|---|---|---|---|
| Acute Insulin Response (AIR) | 24 | 180 | 1 335 |
| Insulin Sensitivity Index (SI) | 1 | 17 | 247 |
| Disposition Index (DI) | 8 | 101 | |
| Metabolic Clearance Rate of Insulin (MCRI) | 6 | 100 | |
| Total Cholesterol | 1 | 16 | 269 |
| High Density Lipoprotein (HDL) | 13 | 129 | 1 202 |
| Low Density Lipoprotein (LDL) | 1 | 9 | 191 |
| Apolipoprotein B (ApoB) | 9 | 291 | |
| Triglycerides | 4 | 18 | 151 |
| Systolic Blood Pressure (SBP) | 1 | 48 | |
| Diastolic Blood Pressure (DBP) | 1 | 24 | |
| Albumin/Creatinine Ratio (ACR) | 3 | 169 | |
| Adiponectin (adjusted) | 13 | 96 | 621 |
| C-Reactive Protein (CRP) | 5 | 84 | |
| Fibrinogen | 16 | 341 | |
| TNFα Receptor 2 (TNF2) | 27 | 259 | 2 458 |
| Retinol Binding Protein 4 (RBP4) | 1 | 20 | |
| Body Mass Index (BMI) | 1 | 11 | 100 |
| Body Adiposity Index (BAI) | 4 | 66 | |
| Percent Body Fat | 1 | 18 | 159 |
| Waist Circumference | 2 | 32 | |
| Waist-to-Hip Ratio (WHR) | 1 | 10 | |
| Subcutaneous Adipose Tissue (SAT) | 1 | 7 | 151 |
| Visceral Adipose Tissue (adj. for BMI) | 1 | 63 | |
| Visceral-to-Subcutaneous Ratio (VSR) | 1 | 8 | 138 |
| Visceral-to-Subcutaneous Ratio (VSR; adj. for BMI) | 3 | 9 | 141 |
| Liver Density | 4 | 123 | |
| Inverse Normalized Liver | 2 | 47 | |
| Gamma Glutamyl Transpeptidase (GGT) | 6 | 126 |
Boldface indicates phenotypes with a LOD score >5.
Evaluation of loci with high LOD scores
The overall maximal LOD score of 6.49 was observed with rs12956744 with the biomarker TNFα receptor 2 levels (Table 3; Figure 1a). This SNP is located in intron 1 (nearer the 5′ end) of LAMA1 (laminin subunit alpha-1 gene) on chromosome 18. Of note, three additional intronic variants in LAMA1 were also linked to TNFα receptor 2 levels with LOD > 6, and 9 SNPs overall were linked with LOD > 3 (Table 3). Notably, one SNP (rs28569884) was also associated with TNFα receptor 2 levels (p-value = 5.9×10−4; LOD = 1.06). The variant rs28569884 (in intron 56) is distal to the striking linkage signal (146 kb apart), though there was another LOD score over 4 (rs4395154; LOD = 4.47) just 13 kb away at the 3′ end of the LAMA1 gene (intron 62). LAMA1 is a very large gene, with 63 exons and 245 SNPs analyzed. Of these, 11 (4.4%) had nominally significant association (p-value < 0.05) with TNFα receptor 2 levels. Comparatively, 9 variants had LOD scores greater than 3 (3.7%) and 23 variants had LOD scores greater than 1 (9.4%).
Table 3.
Selected LAMA1 results with TNFα receptor 2 protein levels (LOD>1 and/or P-value <0.01)
| SNP | Chr | Position | Chip | N | MAF | LOD | P-value | Beta Value | Standard Error | Variance |
|---|---|---|---|---|---|---|---|---|---|---|
| rs4395154 | 18 | 6942805 | OmniExpress | 820 | 0.46 | 4.47 | 0.25 | 0.016 | 0.014 | 0.001 |
| rs2016639 | 18 | 6943264 | OmniExpress | 821 | 0.431 | 3.46 | 0.2 | −0.018 | 0.014 | 0.002 |
| rs17439137 | 18 | 6951060 | OmniExpress | 821 | 0.235 | 1.07 | 0.77 | −0.005 | 0.016 | 0 |
| rs8086875 | 18 | 6951710 | Omni1S | 821 | 0.208 | 1.15 | 0.36 | 0.015 | 0.017 | 0.0008 |
| rs8088218 | 18 | 6951971 | Omni1S | 820 | 0.21 | 1.62 | 0.32 | 0.017 | 0.017 | 0.001 |
| rs12454596 | 18 | 6953989 | OmniExpress | 821 | 0.446 | 1.85 | 0.73 | 0.005 | 0.014 | 0.0002 |
| rs949215 | 18 | 6955676 | OmniExpress | 821 | 0.25 | 1.18 | 0.96 | 0.001 | 0.016 | 0 |
| rs28569884 | 18 | 6956111 | Omni1S | 821 | 0.058 | 1.06 | 5.94E-04 | −0.098 | 0.029 | 0.015 |
| rs509497 | 18 | 6957193 | OmniExpress | 821 | 0.393 | 1.29 | 0.04 | 0.028 | 0.014 | 0.005 |
| rs633691 | 18 | 6967089 | OmniExpress | 821 | 0.419 | 3.18 | 0.085 | 0.024 | 0.014 | 0.0044 |
| rs11873205 | 18 | 6979621 | Omni1S | 818 | 0.13 | 1.54 | 0.0072 | −0.055 | 0.021 | 0.0113 |
| rs538815 | 18 | 6982443 | OmniExpress | 821 | 0.202 | 1.69 | 0.5 | −0.011 | 0.017 | 0.0003 |
| rs619106 | 18 | 7011413 | OmniExpress | 821 | 0.291 | 0.03 | 0.042 | −0.032 | 0.015 | 0.009 |
| rs67268419 | 18 | 7013648 | Omni1S | 820 | 0.077 | 1.74 | 0.74 | −0.009 | 0.025 | 0.0006 |
| rs541928 | 18 | 7034932 | Omni1S | 821 | 0.153 | 2.05 | 0.49 | 0.013 | 0.019 | 0 |
| rs7240767 | 18 | 7070642 | OmniExpress | 821 | 0.468 | 0 | 0.029 | −0.03 | 0.014 | 0.0058 |
| rs7228959 | 18 | 7076464 | OmniExpress | 821 | 0.49 | 0 | 0.044 | −0.027 | 0.014 | 0.0047 |
| rs16951199 | 18 | 7080135 | OmniExpress | 815 | 0.068 | 0 | 0.017 | −0.064 | 0.027 | 0.0081 |
| rs11081298 | 18 | 7085706 | Omni1S | 820 | 0.466 | 2.91 | 0.94 | −0.001 | 0.014 | 0.0001 |
| rs12606163 | 18 | 7096977 | OmniExpress | 807 | 0.485 | 4.78 | 0.11 | 0.022 | 0.014 | 0.0038 |
| rs972038 | 18 | 7102036 | Omni1S | 816 | 0.171 | 0.07 | 0.046 | −0.036 | 0.018 | 0.0103 |
| rs12955222 | 18 | 7102427 | OmniExpress | 821 | 0.482 | 4.53 | 0.13 | 0.02 | 0.013 | 0.0038 |
| rs12956744 | 18 | 7102706 | Omni1S | 821 | 0.407 | 6.49 | 0.03 | 0.03 | 0.014 | 0.0071 |
| rs12959835 | 18 | 7103146 | Omni1S | 820 | 0.408 | 6.38 | 0.034 | 0.029 | 0.014 | 0.0068 |
| rs1462780 | 18 | 7105988 | OmniExpress | 820 | 0.019 | 0 | 0.034 | −0.103 | 0.049 | 0.0072 |
| rs34433741 | 18 | 7108999 | Omni1S | 820 | 0.415 | 6.07 | 0.089 | 0.023 | 0.014 | 0.0031 |
| rs4798533 | 18 | 7109571 | Omni1S | 819 | 0.282 | 1.52 | 0.82 | −0.003 | 0.015 | 0.0002 |
| rs12454984 | 18 | 7109652 | Omni1S | 821 | 0.404 | 6.02 | 0.15 | 0.02 | 0.014 | 0.0019 |
| rs984355 | 18 | 7114212 | OmniExpress | 821 | 0.217 | 2.55 | 0.36 | 0.016 | 0.017 | 0 |
Boldface indicates LOD scores > 3 or p-values < 0.05.
Figure 1.
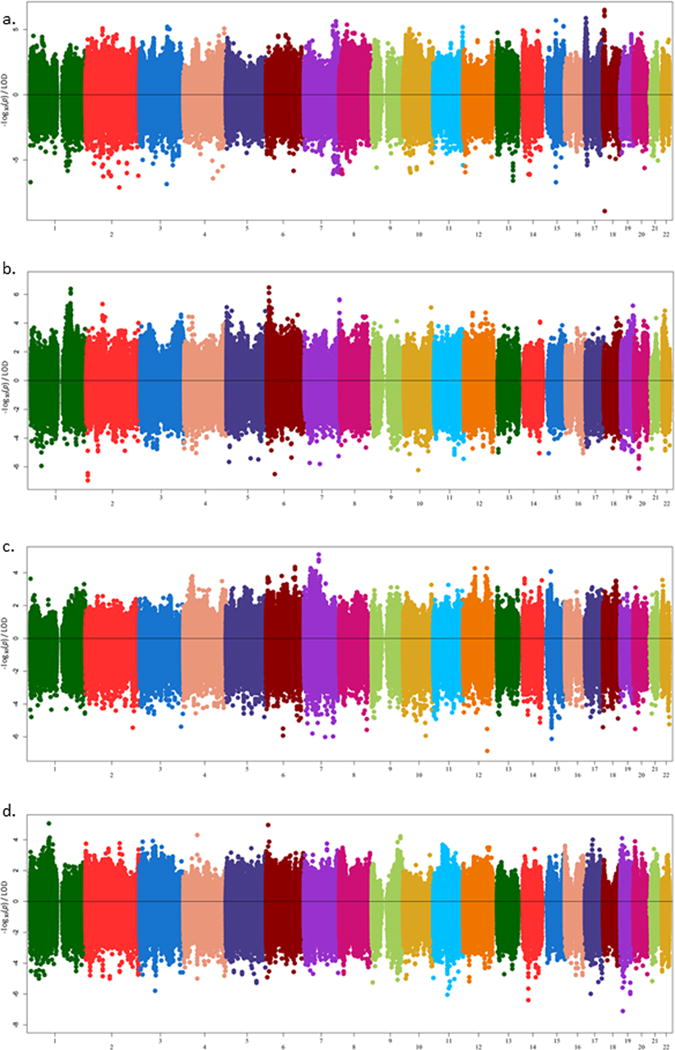
Opposed plots showing LOD scores from the two-point linkage (upper portion) and log-transformed p-values for association (lower portion) results across all arrays for (a.) TNFα receptor 2 levels, (b.) Acute Insulin Response (AIR). (Note the broad linkage peak on Chromosome 1, and the strong linkage also on Chromosome 6), (c.) Insulin Sensitivity Index (SI) (Of particular note are the signals on chromosomes 7 and 12.), and (d.) Low Density Lipoprotein (LDL) levels. (Note the signals on chromosome 4, contributed by LPHN3 and chromosome 19, which represents the APOE locus, evaluated in our previous publication with Apolipoprotein B levels.)
A major focus of our laboratory is identifying genetic contributors to metabolic measures of glucose homeostasis. The top linkage result of LOD = 6.47 (Table 4) for AIR was rs28479408, an intronic variant located in SYCP2L (synaptonemal complex protein 2-like gene) on chromosome 6 (Figure 1b). Although this variant was not associated with AIR (p-value = 0.71), six other SNPs in this gene were also linked (rs4713044, LOD = 6.10; rs12190237, LOD = 5.58; rs12214063, LOD = 3.58; rs1767771, LOD = 3.42; rs2153159, LOD = 3.31; rs1632103, LOD = 3.15) but not associated (p-values > 0.5) (Table 4).
Table 4.
Chromosome 6 AIR linkage peak with linked (LOD>3) and/or associated (p-value <0.05) variants.
| SNP | Chr. | Position | Chip | N | MAF | Gene | LOD | P-value | Beta Value | Standard Error | Variance |
|---|---|---|---|---|---|---|---|---|---|---|---|
| rs12208366 | 6 | 10383410 | Omni1S | 1 034 | 0.146 | 3.43 | 0.578 | 0.39 | 0.701 | 0 | |
| rs480965 | 6 | 10387251 | OmniExpress | 1 033 | 0.142 | 3 | 0.546 | 0.419 | 0.695 | 0 | |
| rs533558 | 6 | 10395572 | OmniExpress | 1 033 | 0.406 | 3.55 | 0.122 | −0.771 | 0.499 | 0.002 | |
| rs79025376 | 6 | 10400618 | Omni1S | 1 033 | 0 | TFAP2A | 0 | 5.06E-03 | −27.514 | 9.816 | 0.008 |
| rs78497087 | 6 | 10471612 | Omni1S | 1 032 | 0.356 | 3.39 | 0.813 | 0.123 | 0.518 | 0 | |
| rs491803 | 6 | 10477438 | Omni1S | 1 033 | 0.331 | 3.31 | 0.885 | 0.075 | 0.521 | 0 | |
| rs9466917 | 6 | 10606584 | Omni1S | 1 033 | 0.492 | GCNT2 | 3.32 | 0.89 | 0.069 | 0.501 | 0 |
| rs3798704 | 6 | 10615268 | Omni1S | 1 034 | 0.494 | GCNT2 | 3.33 | 0.923 | 0.048 | 0.5 | 0 |
| rs1233887 | 6 | 10739432 | OmniExpress | 1 033 | 0.36 | 3.1 | 0.714 | −0.187 | 0.51 | 0 | |
| rs518954 | 6 | 10791859 | OmniExpress | 1 029 | 0.278 | MAK | 3.1 | 0.184 | 0.727 | 0.546 | 0.003 |
| rs12214063 | 6 | 10855738 | Omni1S | 1 032 | 0.213 | SYCP2L | 3.58 | 0.753 | −0.195 | 0.62 | 0 |
| rs1767771 | 6 | 10857646 | Omni1S | 1 034 | 0.473 | SYCP2L | 3.42 | 0.685 | −0.203 | 0.499 | 0 |
| rs1632103 | 6 | 10862649 | Omni1S | 1 034 | 0.478 | SYCP2L | 3.15 | 0.558 | −0.293 | 0.5 | 0 |
| rs2153159 | 6 | 10887932 | Omni1S | 1 033 | 0.36 | SYCP2L | 3.31 | 0.969 | −0.02 | 0.506 | 0 |
| rs4713044 | 6 | 10911282 | OmniExpress | 1 033 | 0.182 | SYCP2L | 6.1 | 0.951 | −0.039 | 0.63 | 0 |
| rs28479408 | 6 | 10912131 | Omni1S | 1 034 | 0.177 | SYCP2L | 6.47 | 0.712 | −0.236 | 0.64 | 0 |
| rs12190237 | 6 | 10922638 | OmniExpress | 1 031 | 0.164 | SYCP2L | 5.58 | 0.775 | 0.188 | 0.66 | 0 |
| rs6457131 | 6 | 11227328 | OmniExpress | 1 029 | 0.207 | NEDD9 | 3.24 | 0.919 | 0.061 | 0.604 | 0 |
| rs55813531 | 6 | 11238023 | Omni1S | 1 031 | 0.185 | NEDD9 | 5.14 | 0.274 | 0.698 | 0.639 | 0.002 |
| rs17496723 | 6 | 11238633 | Omni1S | 1 031 | 0.413 | NEDD9 | 1.2 | 7.89E-03 | −1.323 | 0.498 | 0.004 |
| rs9468690 | 6 | 11239119 | OmniExpress | 1 033 | 0.455 | NEDD9 | 0.86 | 7.86E-03 | −1.316 | 0.495 | 0.005 |
| rs9461574 | 6 | 11239518 | OmniExpress | 1 033 | 0.492 | NEDD9 | 1.94 | 5.77E-03 | −1.354 | 0.49 | 0.006 |
| rs12209631 | 6 | 11242203 | OmniExpress | 1 028 | 0.175 | NEDD9 | 3.08 | 0.0873 | 1.134 | 0.662 | 0.005 |
| rs6908326 | 6 | 11247387 | OmniExpress | 1 033 | 0.204 | NEDD9 | 2.97 | 5.11E-03 | 1.683 | 0.6 | 0.009 |
| rs10947066 | 6 | 11253969 | Omni1S | 1 034 | 0.264 | NEDD9 | 4.34 | 0.0468 | 1.117 | 0.562 | 0.007 |
| rs10947067 | 6 | 11253990 | Omni1S | 1 033 | 0.265 | NEDD9 | 4.25 | 0.0481 | 1.113 | 0.563 | 0.006 |
| rs6457197 | 6 | 11254692 | Omni1S | 1 028 | 0.496 | NEDD9 | 3.72 | 0.0165 | −1.176 | 0.491 | 0.01 |
| rs6457202 | 6 | 11255770 | Omni1S | 1 033 | 0.445 | NEDD9 | 4.29 | 8.71E-03 | 1.324 | 0.505 | 0.013 |
| rs7766626 | 6 | 11256000 | OmniExpress | 1 031 | 0.371 | NEDD9 | 3.73 | 0.0152 | 1.206 | 0.496 | 0.01 |
| rs210903 | 6 | 11724542 | OmniExpress | 1 031 | 0.271 | C6orf105 | 3.93 | 0.954 | −0.032 | 0.561 | 0 |
| rs4713831 | 6 | 11726626 | OmniExpress | 1 014 | 0.298 | C6orf105 | 4.12 | 0.726 | 0.189 | 0.541 | 0 |
| rs210897 | 6 | 11729299 | Omni1S | 1 034 | 0.282 | C6orf105 | 5.49 | 0.893 | 0.075 | 0.557 | 0 |
| rs114551218 | 6 | 11736145 | Omni1S | 1 030 | 0.003 | C6orf105 | 0 | 3.48E-03 | 13.077 | 4.476 | 0.014 |
| rs210890 | 6 | 11740036 | OmniExpress | 1 032 | 0.162 | C6orf105 | 3.13 | 0.552 | 0.4 | 0.673 | 0 |
| rs12204492 | 6 | 11774626 | OmniExpress | 1 032 | 0.424 | C6orf105 | 3.62 | 0.376 | −0.431 | 0.487 | 0.001 |
| rs2235384 | 6 | 11776631 | OmniExpress | 1 031 | 0.205 | C6orf105 | 3.02 | 0.481 | 0.419 | 0.594 | 0 |
Boldface indicates LOD scores > 3 or p-values < 0.05.
Strikingly, chromosome 1 had a broad linkage peak for AIR, with a maximal LOD score of 6.37 (rs2252384) in the region between FAM163A and TOR1AIP2 (located at approximately 179 Mb; 1q25.2; Figure 1b; Table 5). Chromosome 1 has a long history of linkage to diabetes, making this result all the more interesting22–25. Here, variants with LOD scores greater than three spanned much of the proximal q arm of the chromosome, with the most concentrated linkage peak residing between 156Mb and 187 Mb, a region encompassing 357 RefSeq genes (1q22–31.1). Focusing on the peak LOD-1 substantially narrowed the region to a very narrow 1.57 Mb. Of the 343 variants within this region with LOD scores greater than 3, 73 of them had p-values less than 0.05, with a best association signal occurring at rs6426957 (Chr1:165988336; p-value = 6.34×10−4, LOD = 3.09, MAF = 0.441; Supplementary Table 3). Notably, many variants within RASAL2 (RAS protein activator like 2 gene) showed nominal evidence of association (0.05 > p-value > 1.42×10−3) in addition to linkage (N = 45 of 46 linked [LOD>3] SNPs; Tables 5 and 6). LOD scores at this gene ranged from 3.00–5.38.
Table 5.
Broad linkage region on Chromosome 1 with Acute Insulin Response: Variants with LOD >4.5
| SNP | Chr. | Position | Chip | N | MAF | Gene | LOD | P-value | Beta Value | Standard Error | Variance Explained (association) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| rs12047043 | 1 | 164625696 | OmniExpress | 1 029 | 0.225 | AX748175 | 4.95 | 0.16 | 0.832 | 0.594 | 0.005 |
| rs4657367 | 1 | 164627551 | OmniExpress | 1 033 | 0.225 | AX748175 | 4.72 | 0.15 | 0.857 | 0.591 | 0.005 |
| rs4656475 | 1 | 166004063 | Omni1S | 1 032 | 0.14 | Intergenic | 4.66 | 0.38 | 0.635 | 0.721 | 0.001 |
| rs6662013 | 1 | 166042658 | Omni1S | 1 034 | 0.247 | FAM78B | 5.19 | 0.71 | −0.21 | 0.565 | 0 |
| rs6680174 | 1 | 166459849 | OmniExpress | 1 033 | 0.266 | Intergenic | 4.73 | 0.33 | 0.544 | 0.553 | 0.001 |
| rs1476076 | 1 | 167794511 | Omni1S | 1 031 | 0.467 | ADCY10 | 4.74 | 0.81 | −0.113 | 0.48 | 0 |
| rs203849 | 1 | 167849414 | OmniExpress | 1 033 | 0.484 | ADCY10 | 4.62 | 0.43 | −0.395 | 0.5 | 0.002 |
| rs4656148 | 1 | 168179545 | Omni1S | 1 031 | 0.273 | Intergenic | 4.87 | 0.42 | 0.427 | 0.535 | 0 |
| rs11589732 | 1 | 168585289 | OmniExpress | 1 033 | 0.228 | Intergenic | 5.00 | 0.86 | 0.106 | 0.582 | 0 |
| rs7474070 | 1 | 171050589 | OmniExpress | 1 033 | 0.22 | Intergenic | 4.86 | 0.11 | −0.959 | 0.597 | 0.003 |
| rs16863990 | 1 | 171055570 | OmniExpress | 1 032 | 0.193 | Intergenic | 5.09 | 0.15 | −0.929 | 0.644 | 0.003 |
| rs12402693 | 1 | 171057312 | OmniExpress | 1 032 | 0.193 | Intergenic | 5.16 | 0.14 | −0.947 | 0.643 | 0.003 |
| rs12404183 | 1 | 171058946 | OmniExpress | 1 026 | 0.212 | Intergenic | 4.59 | 0.21 | −0.754 | 0.603 | 0.002 |
| rs1800822 | 1 | 171076935 | OmniExpress | 1 029 | 0.201 | FMO3 | 4.62 | 0.3 | −0.637 | 0.613 | 0.002 |
| rs2281002 | 1 | 171080629 | OmniExpress | 1 033 | 0.189 | FMO3 | 4.79 | 0.12 | −1.005 | 0.646 | 0.004 |
| rs909529 | 1 | 171082896 | OmniExpress | 1 033 | 0.201 | FMO3 | 4.72 | 0.078 | −1.103 | 0.624 | 0.004 |
| rs6659102 | 1 | 176535567 | OmniExpress | 1 032 | 0.149 | PAPPA2 | 4.66 | 0.91 | −0.082 | 0.685 | 0 |
| rs7540152 | 1 | 176656255 | OmniExpress | 1 033 | 0.13 | PAPPA2 | 4.53 | 0.73 | 0.256 | 0.731 | 0 |
| rs791031 | 1 | 176667810 | OmniExpress | 1 030 | 0.129 | PAPPA2 | 4.60 | 0.82 | 0.165 | 0.734 | 0 |
| rs11583320 | 1 | 178042145 | OmniExpress | 1 029 | 0.221 | Intergenic | 4.52 | 5.63E-03 | 1.597 | 0.576 | 0.006 |
| rs964993 | 1 | 178062359 | OmniExpress | 1 033 | 0.188 | LOC100302401 | 4.67 | 1.89E-03 | 1.988 | 0.639 | 0.007 |
| rs10913506 | 1 | 178092233 | OmniExpress | 1 033 | 0.186 | RASAL2 | 4.93 | 1.52E-03 | 2.019 | 0.636 | 0.008 |
| rs10798604 | 1 | 178254568 | OmniExpress | 1 029 | 0.174 | RASAL2 | 4.96 | 0.033 | 1.38 | 0.648 | 0.004 |
| rs77603205 | 1 | 178279051 | Omni1S | 1 033 | 0.173 | RASAL2 | 4.52 | 0.021 | 1.504 | 0.652 | 0.005 |
| rs10913550 | 1 | 178408795 | OmniExpress | 1 033 | 0.174 | RASAL2 | 5.16 | 0.027 | 1.435 | 0.65 | 0.004 |
| rs9803679 | 1 | 178410425 | OmniExpress | 1 033 | 0.174 | RASAL2 | 5.21 | 0.027 | 1.435 | 0.65 | 0.004 |
| rs2017349 | 1 | 178419417 | OmniExpress | 1 033 | 0.259 | RASAL2 | 5.38 | 0.07 | 1.034 | 0.57 | 0.004 |
| rs12073428 | 1 | 178427933 | OmniExpress | 1 030 | 0.157 | RASAL2 | 4.98 | 7.40E-03 | 1.829 | 0.682 | 0.006 |
| rs1008495 | 1 | 178458708 | OmniExpress | 1 029 | 0.19 | Intergenic | 4.73 | 0.065 | 1.134 | 0.613 | 0.004 |
| rs2252384 | 1 | 179785891 | OmniExpress | 1 033 | 0.242 | Intergenic | 6.37 | 0.095 | −0.937 | 0.561 | 0.004 |
| rs2794579 | 1 | 179787027 | OmniExpress | 1 033 | 0.243 | Intergenic | 6.12 | 0.09 | −0.965 | 0.568 | 0.004 |
| rs1148821 | 1 | 179795505 | OmniExpress | 1 033 | 0.24 | Intergenic | 6.05 | 0.095 | −0.945 | 0.566 | 0.004 |
| rs2804699 | 1 | 181322837 | Omni1S | 1 026 | 0.351 | Intergenic | 4.91 | 0.49 | 0.353 | 0.515 | 0 |
| rs2804694 | 1 | 181331833 | Omni1S | 1 033 | 0.332 | Intergenic | 4.55 | 0.53 | 0.333 | 0.531 | 0.001 |
Boldface indicates LOD scores > 3 or p-values < 0.05.
Table 6.
Variants with LOD score >4 and p-value <0.005
| SNP | Chr | Position | N | MAF | Trait | Gene | Variant | LOD | P-value | Beta Value | Variance |
|---|---|---|---|---|---|---|---|---|---|---|---|
| rs17109504 | 1 | 83468851 | 965 | 0.2363 | ApoB | .–. | unknown | 4.08 | 3.99E-03 | 0.182 | 0.005 |
| rs10919343 | 1 | 170224982 | 1 032 | 0.205 | AIR | unknown | 4.32 | 0.003 | 1.86 | 0.012 | |
| rs10494510 | 1 | 178074581 | 1 030 | 0.187 | AIR | RASAL2 | intron | 4.08 | 0.002 | 1.98 | 0.007 |
| rs6670912 | 1 | 178082410 | 1 033 | 0.187 | AIR | RASAL2 | intron | 4.28 | 0.0014 | 2.04 | 0.007 |
| rs4440820 | 1 | 178088698 | 1 034 | 0.186 | AIR | RASAL2 | intron | 4.18 | 0.0015 | 2.03 | 0.008 |
| rs12071903 | 1 | 178095804 | 1 034 | 0.187 | AIR | RASAL2 | intron | 4.22 | 0.0014 | 2.041 | 0.007 |
| rs10798597 | 1 | 178108248 | 1 032 | 0.185 | AIR | RASAL2 | intron | 4.01 | 0.0019 | 1.996 | 0.007 |
| rs10157702 | 1 | 178109045 | 1 033 | 0.186 | AIR | RASAL2 | intron | 4.28 | 0.0019 | 1.99 | 0.007 |
| rs10913513 | 1 | 178135941 | 1 034 | 0.186 | AIR | RASAL2 | intron | 4.08 | 0.0018 | 2.002 | 0.007 |
| rs2343249 | 4 | 62419426 | 1 017 | 0.3033 | LDL | LPHN3 | intron | 4.3 | 1.00E-05 | −0.324 | 0.027 |
| rs13245847 | 7 | 38596983 | 821 | 0.431 | TNF2 | AMPH | intron | 4.14 | 5.20E-05 | −0.056 | 0.019 |
| rs723968 | 9 | 14154231 | 820 | 0.2701 | TNF2 | NFIB | intron | 4.11 | 1.28E-03 | −0.05 | 0.012 |
| rs7044402 | 9 | 14157468 | 821 | 0.2966 | TNF2 | NFIB | intron | 4.19 | 9.05E-04 | −0.049 | 0.012 |
| rs16931436 | 9 | 14185939 | 821 | 0.2716 | TNF2 | NFIB | intron | 4.09 | 1.58E-03 | −0.049 | 0.013 |
| rs10756748 | 9 | 16327712 | 1 029 | 0.313 | HDL | unknown | 4.1 | 0.0027 | −0.039 | 0.013 | |
| rs1939523 | 11 | 132599003 | 821 | 0.2954 | TNF2 | OPCML | intron | 4.01 | 3.13E-03 | −0.046 | 0.006 |
| rs73202582 | 12 | 92044537 | 954 | 0.138 | Adiponectin | 0 | unknown | 4.15 | 0.0019 | −0.091 | 0.02 |
| rs9596564 | 13 | 33508797 | 1 029 | 0.2755 | Triglycerides | PDS5B (243392)-KL (81403) | unknown | 4.13 | 4.68E-03 | −0.08 | 0.011 |
| rs11158243 | 14 | 20473910 | 821 | 0.316 | TNF2 | unknown | 4.92 | 0.0037 | −0.046 | 0.014 | |
| rs11643893 | 16 | 16285847 | 784 | 0.425 | Percent Fat | ABCC6 | intron | 4.03 | 0.0034 | −0.891 | 0.018 |
| rs11076039 | 16 | 54450940 | 1 024 | 0.466 | HDL | unknown | 5.43 | 0.0011 | −0.039 | 0.007 | |
| rs11645463 | 16 | 54456353 | 1 030 | 0.47 | HDL | unknown | 5.06 | 0.0049 | −0.033 | 0.004 | |
| rs5882 | 16 | 57016092 | 1 020 | 0.46 | HDL | CETP | Missense V422I | 4.29 | 4.91E-04 | 0.042 | 0.012 |
| rs12602333 | 17 | 10169293 | 821 | 0.1681 | TNF2 | GAS7 (245974)-MYH13 (34889) | unknown | 4.65 | 3.32E-03 | −0.051 | 0.012 |
| rs17745091 | 17 | 52938797 | 785 | 0.498 | Percent Fat | unknown | 5.01 | 1.80E-04 | 1.156 | 0.014 | |
| rs2332308 | 17 | 52944373 | 784 | 0.4802 | Percent Fat | .-TOM1L1 (33678) | unknown | 4.03 | 2.44E-04 | 1.141 | 0.01 |
| rs75500748 | 22 | 48739692 | 819 | 0.093 | TNF2 | unknown | 4.21 | 2.70E-04 | 0.084 | 0.022 |
Additional linkage results of interest include regions on chromosomes 7 and 12 which were linked to insulin sensitivity index (SI). Although these regions did not reach the magnitude seen for TNFα receptor 2 and AIR, the consistency of linkage in the region is compelling. On chromosome 7, the highest LOD score (5.11) was seen with rs1024591, an intergenic SNP over 300kb from the nearest gene (a long intergenic non-coding RNA, LINC01372) (Supplementary Table 4). The linkage signal on chromosome 12 is made up of two distinct peaks (Figure 1c), one at ~53Mb and the second at ~105 Mb (Supplementary Table 5). The LOD scores seen here are not as striking by magnitude (max LOD for each peak 4.27–4.28), but the consistency of LOD scores over 3 into tight peaks is notable (Supplementary Table 5). The first peak consists of 14 variants with LOD scores over 3, from 50.6–54.5Mb, with multiple variants in the KRT8 (keratin 8 gene) and ESPL1 (extra spindle pole bodies like 1, separase) showing evidence for linkage, as well as single variants at the proximal end of the peak in LIMA1 (LIM domain and actin binding 1 gene), DIP2B (disco interacting protein 2 homolog B gene), and SLC4A8 (solute carrier family 4, sodium bicarbonate cotransporter, member 8 gene). There was no evidence for association among linked variants at this linkage peak, though other, unlinked variants in the region showed nominal association (Supplementary Table 5).
The second linkage peak resides from 101–109Mb on chromosome 12, and included 21 linked variants which represented multiple signals from CHST11 (carbohydrate (chondroitin 4) sulfotransferase 11 gene), ACACB (acetyl-CoA carboxylase beta gene), and FOXN4 (forkhead box N4 gene), in addition to intergenic variants and genes implicated by a single variant, such as CMKLR1 (chemerin chemokine-like receptor 1 gene) (Supplementary Table 5). One of these linked variants showed nominal evidence of association, with a p-value of 5.50×10−3 (rs11114094 in SVOP [SV2 related protein gene]; Table 6; Supplementary Tables 3 and 5), although like the prior peak, other unlinked variants in the linkage region also demonstrated evidence of association.
Variants with evidence of both linkage and association
Utilizing the linkage results as a search tool and prioritizing those with any evidence of association identified 1076 variants with p-values less than 0.05 as well as a LOD score greater than or equal to 3 (Supplementary Table 3). Twenty-seven variants were associated with p < 0.005 as well as having a LOD score > 4 (Table 6). NFIB was the primary gene implicated under a linkage peak with TNFα receptor 2 levels on chromosome 9, where there was also evidence of nominal association (p-values on the order of 2×10−4; Figure 1a; Supplementary Table 6). NFIB, which encodes nuclear factor I/B, is represented by 293 SNPs (135 from OmniExpress; 157 from Omni 1S, 1 from exome chip), 289 of which were located in introns. Only one coding variant in this gene was polymorphic from the exome chip dataset, this SNP (rs114558598; I24F) was not linked (LOD = −0.005) or associated (p-value = 0.08). Ten common variants (0.27 < MAF > 0.49) within this gene (all intronic) had LOD scores greater than 3. Overall, 68 NFIB variants had LOD scores greater than 1, and 24 had LOD scores greater than 2.
LPHN3 on chromosome 4 was a strong signal for LDL levels, with two intronic variants being both linked and associated (rs2343249; LOD = 4.30; p-value = 1.00×10−5 and rs9312078, LOD = 3.02; p-value = 8.20×10−5; Table 7; Figure 1d). Both the linkage and association signals were confined to the gene region, with strong LD (r2 > 0.8) between the two top SNPs. There was further support throughout the gene-encoding region for both modest linkage and association with diminishing LD (Supplementary Figure 1). The strongest association result among LOD scores ≥ 3 was with fibrinogen levels; rs1131878 from the OmniExpress chip, LOD = 3.08 and p-value = 1.99×10−6 (Supplementary Table 3). This SNP was located within the UGT2B4 gene, which encodes UDP glucuronosyltransferase 2 family polypeptide B4.
Table 7.
LPHN3 Linkage and Association with LDL levels
| SNP | Chr | Position | Chip | N | MAF | LOD | P-value | Beta Value | Standard Error | Variance |
|---|---|---|---|---|---|---|---|---|---|---|
| rs17828264 | 4 | 62079015 | Omni1S | 1 021 | 0.5 | 1.17 | 0.44 | −0.051 | 0.066 | 0 |
| rs17090416 | 4 | 62098937 | OmniExpress | 1 022 | 0.279 | 1.42 | 0.65 | −0.034 | 0.074 | 0 |
| rs1505682 | 4 | 62111856 | OmniExpress | 1 022 | 0.315 | 1.44 | 0.22 | −0.089 | 0.073 | 0.001 |
| rs1505670 | 4 | 62115243 | Omni1S | 1 021 | 0.475 | 1.26 | 0.69 | −0.027 | 0.067 | 0 |
| rs13140257 | 4 | 62128750 | Omni1S | 999 | 0.321 | 1.66 | 0.037 | −0.152 | 0.073 | 0.003 |
| rs11723103 | 4 | 62128825 | Omni1S | 1 019 | 0.375 | 1.33 | 0.052 | −0.137 | 0.07 | 0.004 |
| rs1505663 | 4 | 62132090 | OmniExpress | 1 022 | 0.229 | 0.15 | 7.90E-03 | 0.213 | 0.08 | 0.003 |
| rs1505664 | 4 | 62132345 | OmniExpress | 1 020 | 0.371 | 1.42 | 0.05 | −0.137 | 0.07 | 0.004 |
| rs67050759 | 4 | 62135455 | Omni1S | 1 019 | 0.496 | 1.49 | 0.12 | −0.105 | 0.068 | 0.003 |
| rs74329144 | 4 | 62136292 | Omni1S | 1 022 | 0.055 | 1.02 | 0.076 | 0.263 | 0.148 | 0.002 |
| rs77082869 | 4 | 62254565 | Omni1S | 1 021 | 0.015 | 0.00 | 1.77E-03 | 0.896 | 0.287 | 0.013 |
| rs10008278 | 4 | 62366666 | OmniExpress | 1 018 | 0.092 | 1.28 | 0.096 | 0.2 | 0.12 | 0.003 |
| rs904243 | 4 | 62406445 | OmniExpress | 1 021 | 0.164 | 0.75 | 6.49E-04 | −0.312 | 0.091 | 0.018 |
| rs7656189 | 4 | 62411676 | OmniExpress | 1 020 | 0.408 | 0.74 | 4.07E-03 | 0.2 | 0.069 | 0.013 |
| rs9312078 | 4 | 62412292 | OmniExpress | 1 015 | 0.331 | 3.02 | 8.20E-05 | −0.282 | 0.071 | 0.022 |
| rs56905501 | 4 | 62413961 | Omni1S | 1 018 | 0.392 | 0.69 | 2.98E-03 | 0.207 | 0.07 | 0.014 |
| rs7688741 | 4 | 62416470 | Omni1S | 1 022 | 0.383 | 1.46 | 2.11E-04 | −0.262 | 0.071 | 0.019 |
| rs2132074 | 4 | 62416499 | OmniExpress | 1 021 | 0.392 | 0.64 | 1.86E-03 | 0.216 | 0.069 | 0.014 |
| rs2343249 | 4 | 62419426 | OmniExpress | 1 017 | 0.303 | 4.30 | 1.00E-05 | −0.324 | 0.073 | 0.027 |
| rs958862 | 4 | 62434848 | OmniExpress | 1 018 | 0.341 | 1.87 | 3.60E-04 | −0.258 | 0.072 | 0.02 |
| rs10018746 | 4 | 62445246 | Omni1S | 1 021 | 0.5 | 0.97 | 4.19E-03 | 0.192 | 0.067 | 0.013 |
| rs11941524 | 4 | 62446484 | Omni1S | 1 022 | 0.5 | 0.86 | 4.17E-03 | 0.192 | 0.067 | 0.013 |
| rs2172802 | 4 | 62453209 | Exome | 1 012 | 0.45 | 0.50 | 6.37E-03 | 0.184 | 0.067 | 0.01 |
| rs17239080 | 4 | 62455462 | OmniExpress | 1 022 | 0.374 | 2.02 | 2.32E-03 | −0.212 | 0.069 | 0.014 |
| rs11131334 | 4 | 62457454 | OmniExpress | 1 017 | 0.379 | 2.11 | 4.84E-03 | −0.195 | 0.069 | 0.011 |
| rs1497901 | 4 | 62461940 | OmniExpress | 1 021 | 0.359 | 2.07 | 1.77E-03 | −0.221 | 0.07 | 0.013 |
| rs2343250 | 4 | 62472682 | Omni1S | 1 022 | 0.36 | 2.09 | 1.59E-03 | −0.224 | 0.071 | 0.013 |
| rs10001410 | 4 | 62474229 | OmniExpress | 1 019 | 0.47 | 0.91 | 3.89E-03 | −0.199 | 0.069 | 0.016 |
| rs1497921 | 4 | 62526281 | OmniExpress | 1 022 | 0.356 | 0.64 | 3.19E-03 | −0.204 | 0.069 | 0.014 |
| rs66614141 | 4 | 62550335 | Omni1S | 1 022 | 0.326 | 1.45 | 1.35E-04 | −0.268 | 0.07 | 0.02 |
| rs6843311 | 4 | 62568688 | OmniExpress | 1 022 | 0.363 | 0.61 | 5.25E-03 | −0.194 | 0.069 | 0.014 |
| rs11734607 | 4 | 62693692 | OmniExpress | 1 021 | 0.453 | 0.24 | 2.44E-03 | 0.204 | 0.067 | 0.015 |
| rs4860106 | 4 | 62850522 | OmniExpress | 1 021 | 0.422 | 1.13 | 0.71 | 0.025 | 0.068 | 0 |
| rs1510921 | 4 | 62895592 | OmniExpress | 1 017 | 0.241 | 0.26 | 4.00E-03 | 0.223 | 0.077 | 0.007 |
| rs6827266 | 4 | 62902162 | Omni1S | 1 020 | 0.437 | 0.08 | 5.00E-03 | 0.188 | 0.067 | 0.004 |
| rs62306380 | 4 | 62908281 | Omni1S | 1 022 | 0.239 | 0.23 | 3.55E-03 | 0.225 | 0.077 | 0.007 |
Boldface indicates LOD scores > 3 or p-values < 0.05.
Discussion
This study evaluated the utility of combining two-point linkage with association analysis in a data set comprised of array-based SNP genotyping totaling 1.6 million non-coding and coding variants in a family-based sample of Hispanics with extensive phenotype information. The goal of the study was to evaluate whether GWAS data in the context of linkage adds insight into the genetic origins of cardiometabolic traits, while utilizing association analysis as a follow up to determine likely candidate loci. This builds upon our prior evaluation of combined linkage and association using exome chip data in this cohort9. Large-scale linkage analysis of SNP genotyping has been uncommon for complex phenotypes recently. To this end, we evaluated 50 phenotypes (46 distinct traits) related to glucose homeostasis, lipids, blood pressure, adiposity, liver fat and enzymes, and biomarkers. Given the breadth of genotypic data and number of phenotypes, the results are extensive, but some noteworthy observations can be made. Broadly speaking, we believe the markedly denser genotypic dataset reveals many insights into the genetic bases of the traits such as TNFα receptor 2, AIR, and SI when compared to our prior study using the more limited data from the exome chip.
Relatively dense genotyping data provides visual evidence of linkage similar to conventional multipoint methods. In addition, while exome chip analysis primarily targets models where functional variants are exonic, the GWAS datasets can potentially address other models such as high impact non-coding variants, especially through linkage analysis. Here we have observed few examples where evidence for both linkage and association are apparent. An example is LPHN3 (Table 7, Supplementary Figure 1), where LOD scores reached 4.30 with a p-value of 1.00×10−5, suggesting a true impact on LDL levels. Given the actual low density of coverage in GWAS datasets which are designed to cover genomic regions through LD relationships, it is unlikely to capture truly causal variants by chance. The ultimate test of whether this approach will be successful will require whole genome sequencing data. Overall, these results incorporating two-point linkage and association analyses can identify meaningful signals that impact cardiometabolic traits, often in the absence of striking association alone. These conclusions are consistent with our prior work9,10 in which we have shown that linkage evidence can be relatively strong, but association evidence only appears when the functional variant is also captured. The latter is unlikely in a GWAS dataset. For these reasons, our main focus was on regions with evidence of linkage based on both the power of linkage methods and the “far-sighted” ability of linkage to identify genetic relationships4–7,9,10.
As noted above, several genomic regions had relatively strong evidence of linkage, but limited association results. Based on our logic, this would suggest the possibility of underlying, as yet unidentified functional variants. Thus, for the strongest linkage with TNF2α receptor levels (LOD = 6.49) we would hypothesize that one or more high impact non-coding variants lie within the linkage region. LAMA1 is similar to LAMA5 which has previously been related to TNFRSF1B expression26, making it plausible for LAMA1 to be related to TNF2α receptor levels.
Analysis of traits of interest to our laboratory (AIR, SI) also resulted in notable linkage peaks. It is tempting to scan these linked regions for biologically relevant genes. Genes located under a broad AIR linkage region on chromosome 1 (Figure 1b, Table 5) included FAM163A, also known as neuroblastoma derived secretory protein (NDSP), TOR1AIP2, and RASAL2. FAM163A (aka NDSP) has been associated in methylation analysis for borderline personality disorder27 with overexpression observed in neuroblastoma28,29. TOR1AIP2 encodes torsin A interacting protein 2, which is involved in the nuclear envelope30,31. Mutations in TOR1AIP1 have been shown to cause muscular dystrophy32. RASAL2 (RAS protein activator like 2) has been implicated as an obesity susceptibility gene in both Chinese33 and Mexican populations34, as well as having a role in the susceptibility of many cancers, including liver35, thyroid36, ovarian37, breast37,38, and lung39.
Genes under the SI linkage peaks also included interesting candidates. On chromosome 12, the most relevant gene with linkage in the distal linkage peak was CMKLR1 (chemerin chemokine-like receptor 1), which is believed to play a role in glucose homeostasis40–42, obesity41,43,44 and diabetes development45. Of note, a strong association signal (p-value = 1×10−7) was also seen within this linkage peak in WSCD2 (WSC domain containing 2; 100Mb from CMKLR1) (Figure 1c).
Additional genes included LIMA1 (LIM domain and actin binding 1, also known as EPLIN and SREPB3), a tumor suppressor; DIP2B (disco interacting protein 2 homolog B), replicated as a susceptibility locus for colorectal cancer46; and SLC4A8, a sodium bicarbonate transporter, which may have a role in regulation of blood pressure with some variants in this gene having been previously implicated47,48. Further, KRT8 (keratin 8, type II) which is overexpressed in human liver disease, resides under the linkage peak on 12q49. The linkage region on chromosome 7 contained only one putative gene, LOC102723427, about which there is no known information.
The most intriguing signal lies in LPHN3 and was both linked and associated with LDL levels at two separate variants. This gene encodes latrophilin 3 (recently renamed as ADGRL350; adhesion G protein-coupled receptor L3), which is related to latrotoxin, the toxin produced by the black widow spider51. There is evidence suggesting a role for latrophilin 3 (among other latrophilins) in binding to fibronectin leucine-rich transmembrane (FLRT) family members, which has been shown to promote the development of glutamatergic synapses52,53. Additionally, genetic variants in LPHN3 have been associated reproducibly with attention deficit hyperactivity disorder (ADHD) and other psychiatric conditions54–56. LPHN3 is also being investigated as a pharmacogenetic target57. Despite the lack of biological evidence directly supporting the link between LPHN3 variants and LDL cholesterol levels, cholesterol is crucially important in the brain, and further study may elucidate a mechanism by which genetic variants in LPHN3 impact plasma LDL levels.
We previously reported CETP (cholesterol ester transfer protein) linkage and association with HDL levels in exome chip data from this Hispanic sample9. Linkage of CETP in this dataset was stronger with LOD scores of up to 5.43, an increase of 1.14 over the previous top signal (Table 6; Supplementary Table 2).The addition of GWAS data implicated additional linked variants (LOD > 5, N = 4) proximal to the coding region, perhaps occluding interpretation of the functional impact of this linkage result.
Here we assessed the impact of SNP density to provide insight into linkage relationships with the conclusion that dense SNP maps do reveal additional insight. We have extended this query further by evaluation of imputed genotype data in regions of particular interest due to evidence of strong linkage with glucose homeostasis-related phenotypes. Three regions were selected based on substantial linkage evidence and a particular interest in glucose homeostasis: chromosome 1 with AIR and chromosomes 7 and 12 with SI. Utilization of imputed data increases the number of markers capturing the region by 22–fold (18 411 directly genotyped markers, 406K imputed markers). The maximal LOD score from the imputed AIR region was 6.45 at rs2252384 (the same SNP implicated in the directly genotyped data; Supplementary Figure 2). The slight increase in LOD score (6.37 to 6.45) can likely be attributed to more complete information following imputation of missing genotypes. For chromosome 7 with SI, a new best SNP rs2530421 had the maximum LOD score of 5.53 (compared to the prior best LOD of 5.11 at rs1024591). The imputed best SNP lies very near the original peak linkage, providing little additional guidance in refining the causal variant(s), given the high degree of correlation between the top linked SNPs (r2 = 0.937). Evaluation of another linked region (chromosome 12 with SI) also showed some limited improvement in linkage signals, but linkage signals were only modestly increased, as could be expected due to the information carried by these imputed markers being wholly derived from the genotyped markers which had already been informative. Thus, inclusion of imputed genotypes marginally improved the maximal LOD scores when evaluated in this small number of examples. However, the improvements did not further refine the regions of interest (Supplementary Figure 2).
In conclusion, we have built upon our previous analysis of combined two-point linkage and association9 and evaluated utility of the approach in a dataset comprised of comprehensive genome-wide array-based SNP genotypes. As seen previously, there were few examples in this data where linkage and association both provided striking evidence at the same locus, which, based on our prior analysis10, would implicate a likely ungentoyped causal variant. However, the GWAS plus exome chip design identified multiple additional regions of linkage which were not seen in exome chip analysis alone. Positive, strong evidence of association with SNPs was not observed, suggesting that functional variants, if they are indeed captured by the linkage signal, have not been identified. To truly test the broad utility of this approach, whole genome sequencing data will be necessary which will incorporate the full spectrum of variant frequencies.
The authors declare no conflicts of interest related to this publication. Supplementary information is available at the Journal of Human Genetics website.
Supplementary Material
Acknowledgments
This work was supported by the grants R01 HG007112 (D.W.B. and C.D.L.) and R01 DK087914 (M.C.Y.N). The GUARDIAN study which contributed the IRASFS GWAS genotypes to this project is supported by grant R01 DK085175 (L.E.W.), and the IRASFS study was supported by HL060944, HL061019, and HL060919. The provision of GWAS genotyping data was supported in part by UL1TR000124 (CTSI), and DK063491 (DRC). The provision of exome chip data was supported in part by the Department of Internal Medicine at University of Michigan, the Doris Duke Medical Foundation, and R01 DK106621 (E.K.S.). Computational support was provided in part by the Center for Public Health Genomics at Wake Forest School of Medicine.
References
- 1.Ott J, Kamatani Y, Lathrop M. Family-based designs for genome-wide association studies. Nat Rev Genet. 2011;12:465–474. doi: 10.1038/nrg2989. [DOI] [PubMed] [Google Scholar]
- 2.Speed D, Balding DJ. Relatedness in the post-genomic era: is it still useful? Nat Rev Genet. 2015;16:33–44. doi: 10.1038/nrg3821. [DOI] [PubMed] [Google Scholar]
- 3.Bowden DW, An SS, Palmer ND, et al. Molecular basis of a linkage peak: exome sequencing and family-based analysis identify a rare genetic variant in the ADIPOQ gene in the IRAS Family Study. Hum Mol Genet. 2010;19:4112–4120. doi: 10.1093/hmg/ddq327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bowden DW. Will Family Studies Return to Prominence in Human Genetics and Genomics? Rare Variants and Linkage Analysis of Complex Traits. Genes & Genomics. 2011:1–8. [Google Scholar]
- 5.Wang Q, Lu Q, Zhao H. A review of study designs and statistical methods for genomic epidemiology studies using next generation sequencing. Frontiers in genetics. 2015;6:149. doi: 10.3389/fgene.2015.00149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Teare MD, Santibanez Koref MF. Linkage analysis and the study of Mendelian disease in the era of whole exome and genome sequencing. Briefings in functional genomics. 2014;13:378–383. doi: 10.1093/bfgp/elu024. [DOI] [PubMed] [Google Scholar]
- 7.Ott J, Wang J, Leal SM. Genetic linkage analysis in the age of whole-genome sequencing. Nat Rev Genet. 2015;16:275–284. doi: 10.1038/nrg3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saad M, Wijsman EM. Power of family-based association designs to detect rare variants in large pedigrees using imputed genotypes. Genet Epidemiol. 2014;38:1–9. doi: 10.1002/gepi.21776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hellwege JN, Palmer ND, Raffield LM, et al. Genome-wide family-based linkage analysis of exome chip variants and cardiometabolic risk. Genet Epidemiol. 2014;38:345–352. doi: 10.1002/gepi.21801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hellwege JN, Palmer ND, Brown WM, et al. Empirical characteristics of family-based linkage to a complex trait: the ADIPOQ region and adiponectin levels. Hum Genet. 2015;134:203–213. doi: 10.1007/s00439-014-1511-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Voruganti VS, Kent JW, Jr, Debnath S, et al. Genome-wide association analysis confirms and extends the association of SLC2A9 with serum uric acid levels to Mexican Americans. Frontiers in genetics. 2013;4:279. doi: 10.3389/fgene.2013.00279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nemesure BB, He Q, Mendell N. Integration of linkage analyses and disease association studies. Genet Epidemiol. 1995;12:653–658. doi: 10.1002/gepi.1370120622. [DOI] [PubMed] [Google Scholar]
- 13.Heo M, Leibel RL, Fontaine KR, et al. A meta-analytic investigation of linkage and association of common leptin receptor (LEPR) polymorphisms with body mass index and waist circumference. International journal of obesity and related metabolic disorders: journal of the International Association for the Study of Obesity. 2002;26:640–646. doi: 10.1038/sj.ijo.0801990. [DOI] [PubMed] [Google Scholar]
- 14.Weedon MN, Cebola I, Patch AM, et al. Recessive mutations in a distal PTF1A enhancer cause isolated pancreatic agenesis. Nat Genet. 2014;46:61–64. doi: 10.1038/ng.2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Henkin L, Bergman RN, Bowden DW, et al. Genetic epidemiology of insulin resistance and visceral adiposity. The IRAS Family Study design and methods. Ann Epidemiol. 2003;13:211–217. doi: 10.1016/s1047-2797(02)00412-x. [DOI] [PubMed] [Google Scholar]
- 16.Palmer ND, Goodarzi MO, Langefeld CD, et al. Genetic Variants Associated with Quantitative Glucose Homeostasis Traits Translate to Type 2 Diabetes in Mexican Americans: The GUARDIAN (Genetics Underlying Diabetes in Hispanics) Consortium. Diabetes. 2014 doi: 10.2337/db14-0732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O’Connell JR, Weeks DE. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet. 1998;63:259–266. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Delaneau O, Marchini J, Zagury JF. A linear complexity phasing method for thousands of genomes. Nat Methods. 2012;9:179–181. doi: 10.1038/nmeth.1785. [DOI] [PubMed] [Google Scholar]
- 19.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet. 1998;62:1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Das SK, Hasstedt SJ, Zhang Z, Elbein SC. Linkage and association mapping of a chromosome 1q21-q24 type 2 diabetes susceptibility locus in northern European Caucasians. Diabetes. 2004;53:492–499. doi: 10.2337/diabetes.53.2.492. [DOI] [PubMed] [Google Scholar]
- 23.Langefeld CD, Wagenknecht LE, Rotter JI, et al. Linkage of the metabolic syndrome to 1q23–q31 in Hispanic families: the Insulin Resistance Atherosclerosis Study Family Study. Diabetes. 2004;53:1170–1174. doi: 10.2337/diabetes.53.4.1170. [DOI] [PubMed] [Google Scholar]
- 24.Wiltshire S, Hattersley AT, Hitman GA, et al. A genomewide scan for loci predisposing to type 2 diabetes in a U.K. population (the Diabetes UK Warren 2 Repository): analysis of 573 pedigrees provides independent replication of a susceptibility locus on chromosome 1q. Am J Hum Genet. 2001;69:553–569. doi: 10.1086/323249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vionnet N, Hani EH, Dupont S, et al. Genomewide search for type 2 diabetes-susceptibility genes in French whites: evidence for a novel susceptibility locus for early-onset diabetes on chromosome 3q27-qter and independent replication of a type 2-diabetes locus on chromosome 1q21–q24. Am J Hum Genet. 2000;67:1470–1480. doi: 10.1086/316887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adair-Kirk TL, Atkinson JJ, Kelley DG, Arch RH, Miner JH, Senior RM. A chemotactic peptide from laminin alpha 5 functions as a regulator of inflammatory immune responses via TNF alpha-mediated signaling. Journal of immunology (Baltimore, Md: 1950) 2005;174:1621–1629. doi: 10.4049/jimmunol.174.3.1621. [DOI] [PubMed] [Google Scholar]
- 27.Prados J, Stenz L, Courtet P, et al. Borderline personality disorder and childhood maltreatment: a genome-wide methylation analysis. Genes, brain, and behavior. 2015;14:177–188. doi: 10.1111/gbb.12197. [DOI] [PubMed] [Google Scholar]
- 28.Vasudevan SA, Russell HV, Okcu MF, et al. Neuroblastoma-derived secretory protein messenger RNA levels correlate with high-risk neuroblastoma. Journal of pediatric surgery. 2007;42:148–152. doi: 10.1016/j.jpedsurg.2006.09.064. [DOI] [PubMed] [Google Scholar]
- 29.Vasudevan SA, Shang X, Chang S, et al. Neuroblastoma-derived secretory protein is a novel secreted factor overexpressed in neuroblastoma. Molecular cancer therapeutics. 2009;8:2478–2489. doi: 10.1158/1535-7163.MCT-08-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goodchild RE, Kim CE, Dauer WT. Loss of the dystonia-associated protein torsinA selectively disrupts the neuronal nuclear envelope. Neuron. 2005;48:923–932. doi: 10.1016/j.neuron.2005.11.010. [DOI] [PubMed] [Google Scholar]
- 31.Vander Heyden AB, Naismith TV, Snapp EL, Hodzic D, Hanson PI. LULL1 retargets TorsinA to the nuclear envelope revealing an activity that is impaired by the DYT1 dystonia mutation. Molecular biology of the cell. 2009;20:2661–2672. doi: 10.1091/mbc.E09-01-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kayman-Kurekci G, Talim B, Korkusuz P, et al. Mutation in TOR1AIP1 encoding LAP1B in a form of muscular dystrophy: a novel gene related to nuclear envelopathies. Neuromuscular disorders: NMD. 2014;24:624–633. doi: 10.1016/j.nmd.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 33.Cheung CY, Tso AW, Cheung BM, et al. Obesity susceptibility genetic variants identified from recent genome-wide association studies: implications in a chinese population. J Clin Endocrinol Metab. 2010;95:1395–1403. doi: 10.1210/jc.2009-1465. [DOI] [PubMed] [Google Scholar]
- 34.Leon-Mimila P, Villamil-Ramirez H, Villalobos-Comparan M, et al. Contribution of common genetic variants to obesity and obesity-related traits in mexican children and adults. PLoS One. 2013;8:e70640. doi: 10.1371/journal.pone.0070640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stefanska B, Cheishvili D, Suderman M, et al. Genome-wide study of hypomethylated and induced genes in patients with liver cancer unravels novel anticancer targets. Clinical cancer research: an official journal of the American Association for Cancer Research. 2014;20:3118–3132. doi: 10.1158/1078-0432.CCR-13-0283. [DOI] [PubMed] [Google Scholar]
- 36.Xu Y, Deng Y, Ji Z, et al. Identification of thyroid carcinoma related genes with mRMR and shortest path approaches. PLoS One. 2014;9:e94022. doi: 10.1371/journal.pone.0094022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Feng M, Bao Y, Li Z, et al. RASAL2 activates RAC1 to promote triple-negative breast cancer progression. J Clin Invest. 2014;124:5291–5304. doi: 10.1172/JCI76711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang Y, Zhao M, Xu H, et al. RASAL2 down-regulation in ovarian cancer promotes epithelial-mesenchymal transition and metastasis. Oncotarget. 2014;5:6734–6745. doi: 10.18632/oncotarget.2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li N, Li S. RASAL2 promotes lung cancer metastasis through epithelial-mesenchymal transition. Biochemical and biophysical research communications. 2014;455:358–362. doi: 10.1016/j.bbrc.2014.11.020. [DOI] [PubMed] [Google Scholar]
- 40.Rourke JL, Muruganandan S, Dranse HJ, McMullen NM, Sinal CJ. Gpr1 is an active chemerin receptor influencing glucose homeostasis in obese mice. The Journal of endocrinology. 2014;222:201–215. doi: 10.1530/JOE-14-0069. [DOI] [PubMed] [Google Scholar]
- 41.Ernst MC, Haidl ID, Zuniga LA, et al. Disruption of the chemokine-like receptor-1 (CMKLR1) gene is associated with reduced adiposity and glucose intolerance. Endocrinology. 2012;153:672–682. doi: 10.1210/en.2011-1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sell H, Laurencikiene J, Taube A, et al. Chemerin is a novel adipocyte-derived factor inducing insulin resistance in primary human skeletal muscle cells. Diabetes. 2009;58:2731–2740. doi: 10.2337/db09-0277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gruben N, Aparicio Vergara M, Kloosterhuis NJ, et al. Chemokine-like receptor 1 deficiency does not affect the development of insulin resistance and nonalcoholic fatty liver disease in mice. PLoS One. 2014;9:e96345. doi: 10.1371/journal.pone.0096345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim SH, Lee SH, Ahn KY, et al. Effect of lifestyle modification on serum chemerin concentration and its association with insulin sensitivity in overweight and obese adults with type 2 diabetes. Clin Endocrinol (Oxf) 2014;80:825–833. doi: 10.1111/cen.12249. [DOI] [PubMed] [Google Scholar]
- 45.Roman AA, Parlee SD, Sinal CJ. Chemerin: a potential endocrine link between obesity and type 2 diabetes. Endocrine. 2012;42:243–251. doi: 10.1007/s12020-012-9698-8. [DOI] [PubMed] [Google Scholar]
- 46.Closa A, Cordero D, Sanz-Pamplona R, et al. Identification of candidate susceptibility genes for colorectal cancer through eQTL analysis. Carcinogenesis. 2014;35:2039–2046. doi: 10.1093/carcin/bgu092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guo L, Liu F, Chen S, et al. Common variants in the Na-coupled bicarbonate transporter genes and salt sensitivity of blood pressure: the GenSalt study. Journal of human hypertension. 2015 doi: 10.1038/jhh.2015.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aalkjaer C, Boedtkjer E, Choi I, Lee S. Cation-coupled bicarbonate transporters. Comprehensive Physiology. 2014;4:1605–1637. doi: 10.1002/cphy.c130005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guldiken N, Usachov V, Levada K, et al. Keratins 8 and 18 are type II acute-phase responsive genes overexpressed in human liver disease. Liver Int. 2015;35:1203–1212. doi: 10.1111/liv.12608. [DOI] [PubMed] [Google Scholar]
- 50.Hamann J, Aust G, Arac D, et al. International Union of Basic and Clinical Pharmacology. XCIV Adhesion G protein-coupled receptors. Pharmacological reviews. 2015;67:338–367. doi: 10.1124/pr.114.009647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Martinez AF, Muenke M, Arcos-Burgos M. From the black widow spider to human behavior: Latrophilins, a relatively unknown class of G protein-coupled receptors, are implicated in psychiatric disorders. Am J Med Genet B Neuropsychiatr Genet. 2011;156b:1–10. doi: 10.1002/ajmg.b.31137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jackson VA, del Toro D, Carrasquero M, et al. Structural basis of latrophilin-FLRT interaction. Structure (London, England: 1993) 2015;23:774–781. doi: 10.1016/j.str.2015.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.O’Sullivan ML, de Wit J, Savas JN, et al. FLRT proteins are endogenous latrophilin ligands and regulate excitatory synapse development. Neuron. 2012;73:903–910. doi: 10.1016/j.neuron.2012.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fallgatter AJ, Ehlis AC, Dresler T, et al. Influence of a latrophilin 3 (LPHN3) risk haplotype on event-related potential measures of cognitive response control in attention-deficit hyperactivity disorder (ADHD) European neuropsychopharmacology: the journal of the European College of Neuropsychopharmacology. 2013;23:458–468. doi: 10.1016/j.euroneuro.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ribases M, Ramos-Quiroga JA, Sanchez-Mora C, et al. Contribution of LPHN3 to the genetic susceptibility to ADHD in adulthood: a replication study. Genes, brain, and behavior. 2011;10:149–157. doi: 10.1111/j.1601-183X.2010.00649.x. [DOI] [PubMed] [Google Scholar]
- 56.Arcos-Burgos M, Jain M, Acosta MT, et al. A common variant of the latrophilin 3 gene, LPHN3, confers susceptibility to ADHD and predicts effectiveness of stimulant medication. Molecular psychiatry. 2010;15:1053–1066. doi: 10.1038/mp.2010.6. [DOI] [PubMed] [Google Scholar]
- 57.Bruxel EM, Salatino-Oliveira A, Akutagava-Martins GC, et al. LPHN3 and attention-deficit/hyperactivity disorder: a susceptibility and pharmacogenetic study. Genes, brain, and behavior. 2015;14:419–427. doi: 10.1111/gbb.12224. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


