Summary
Background
Statin treatment and variants in the gene encoding HMG-CoA reductase are associated with reductions in both the concentration of LDL cholesterol and the risk of coronary heart disease, but also with modest hyperglycaemia, increased bodyweight, and modestly increased risk of type 2 diabetes, which in no way offsets their substantial benefits. We sought to investigate the associations of LDL cholesterol-lowering PCSK9 variants with type 2 diabetes and related biomarkers to gauge the likely effects of PCSK9 inhibitors on diabetes risk.
Methods
In this mendelian randomisation study, we used data from cohort studies, randomised controlled trials, case control studies, and genetic consortia to estimate associations of PCSK9 genetic variants with LDL cholesterol, fasting blood glucose, HbA1c, fasting insulin, bodyweight, waist-to-hip ratio, BMI, and risk of type 2 diabetes, using a standardised analysis plan, meta-analyses, and weighted gene-centric scores.
Findings
Data were available for more than 550 000 individuals and 51 623 cases of type 2 diabetes. Combined analyses of four independent PCSK9 variants (rs11583680, rs11591147, rs2479409, and rs11206510) scaled to 1 mmol/L lower LDL cholesterol showed associations with increased fasting glucose (0·09 mmol/L, 95% CI 0·02 to 0·15), bodyweight (1·03 kg, 0·24 to 1·82), waist-to-hip ratio (0·006, 0·003 to 0·010), and an odds ratio for type diabetes of 1·29 (1·11 to 1·50). Based on the collected data, we did not identify associations with HbA1c (0·03%, −0·01 to 0·08), fasting insulin (0·00%, −0·06 to 0·07), and BMI (0·11 kg/m2, −0·09 to 0·30).
Interpretation
PCSK9 variants associated with lower LDL cholesterol were also associated with circulating higher fasting glucose concentration, bodyweight, and waist-to-hip ratio, and an increased risk of type 2 diabetes. In trials of PCSK9 inhibitor drugs, investigators should carefully assess these safety outcomes and quantify the risks and benefits of PCSK9 inhibitor treatment, as was previously done for statins.
Funding
British Heart Foundation, and University College London Hospitals NHS Foundation Trust (UCLH) National Institute for Health Research (NIHR) Biomedical Research Centre.
Introduction
The benefit of statins in reducing LDL cholesterol and coronary heart disease (CHD) risk is well established. More recently, and only after completion of numerous randomised controlled trials, was it discovered that statins increase risk of type 2 diabetes,1, 2 although this effect is modest and greatly outweighed by the benefits of this drug class. Genetic studies based on common variants in the gene encoding the target of statins, HMG-CoA reductase (HMGCR), suggest the effect is mechanism-based (ie, on-target).3 Genetic studies assessing the effects of variants in a broader range of genes suggest a more general link between lower LDL cholesterol and higher risk of type 2 diabetes.4, 5 Consistent with this finding, patients with autosomal dominant familial hypercholesterolaemia caused by mutations in the LDL receptor and apolipoprotein B genes are 50% less likely to be diagnosed with type 2 diabetes compared with their unaffected relatives.6
Research in context.
Evidence before this study
We searched PubMed for “pcsk9[All Fields] AND (“antagonists and inhibitors”[Subheading] OR (“antagonists”[All Fields] AND “inhibitors”[All Fields]) OR “antagonists and inhibitors”[All Fields] OR “inhibitors”[All Fields]) AND (“diabetes mellitus”[MeSH Terms] OR (“diabetes”[All Fields] AND “mellitus”[All Fields]) OR “diabetes mellitus”[All Fields])” for articles published up to Oct 8, 2016, to identify studies that assessed treatment with PCSK9 inhibitors or carriage of genetic variants in PCSK9 in relation to diabetes. This search identified 17 studies, two of which presented novel, yet contrasting findings in relation to genetic variants in PCSK9 and glycaemic status.
Randomised trials of treatment with statins and carriage of corresponding genetic variants in HMGCR that lower LDL cholesterol both show and increase in the risk of type 2 diabetes. More recently, genetic predisposition to lower LDL cholesterol concentrations has been linked to an increased risk of diabetes, suggesting that dysglycaemia might be a consequence of lowering LDL cholesterol in general. Whether lowering of LDL cholesterol by PCSK9 inhibitors results in increased risk of diabetes is currently unknown. Clinical trials of PCSK9 inhibitors to assess their effect on cardiovascular outcomes are ongoing, but reliable evidence for a possible association between PCSK9 inhibition and risk of diabetes could take longer to accrue.
Added value of this study
Mendelian randomisation is an established approach that uses randomly allocated variants in the encoding gene to infer mechanism-based efficacy and safety outcomes from pharmacological perturbation of a drug target. We used four genetic variants in PCSK9 in more than 550 000 individuals (including about 50 000 diabetes cases) and showed that PCSK9 genetic variants associated with lower LDL cholesterol concentrations were associated with increased concentration of fasting glucose, bodyweight, and risk of diabetes. This finding adds robust new evidence to previous research that identified weak associations of PCSK9 with risk of diabetes.
Implications of all the available evidence
Similar to statin therapy, treatment with PCSK9 inhibitors is likely to increase the risk of diabetes. Patients treated with PCSK9 inhibitors should be carefully monitored for dysglycaemia, including within ongoing and future clinical trials.
Gain-of-function mutations in PCSK9, the gene encoding proprotein convertase subtilisin/kexin type 9, also cause familial hypercholesterolaemia,7 whereas loss-of-function mutations in the same gene lower LDL cholesterol and protect against CHD.8 Consequently, monoclonal antibodies inhibiting PCSK9 have been developed9 and are effective in lowering LDL cholesterol by 50–70%,10 with preliminary evidence suggesting that this effect might be associated with reduced risk of myocardial infarction and all-cause mortality.9 Although large phase 3 trials to assess the effects of PCSK9 monoclonal antibodies on cardiovascular events are underway, conclusive evidence for the specific effect of PCSK9 inhibition on risk of type 2 diabetes from individual randomised controlled trials or meta-analyses might not emerge for some time.
We used the principle of mendelian randomisation as a tool for drug target validation, whereby common variants in a gene that encodes a drug target, through effects on expression or activity, are used to predict the on-target effect of pharmacological modification of the same target.3, 11, 12 We investigated associations of common genetic variants in PCSK9 with markers of glycaemia, bodyweight, and risk of type 2 diabetes to assess the potential on-target effects of PCSK9 inhibition on these traits. Although results of a recent study provided evidence of an association of a single nucleotide polymorphism (SNP) in PCSK9 with type 2 diabetes risk,13 our aim was to confirm the type 2 diabetes risk-increasing effect of PCSK9 variation and explore potential biological mechanisms that might explain this effect. To do this we used four SNPs in the PCSK9 locus collected in 50 studies supplemented with data from large genetic consortia.
Methods
Genetic variant selection
We selected four SNPs in or near PCSK9 on the basis of a strong association with LDL cholesterol, as reported by the Global Lipids Genetics Consortium (GLGC);14 low pairwise linkage disequilibrium (r2≤0·30) with SNPs within the same and adjacent genes (1000 Genomes CEU data); high prior probability of being a functional variant based on the combined annotation dependent depletion (CADD) score, or the SNP being non-synonymous, or both;15 or previous reported associations with CHD.16 On the basis of these criteria, we selected the SNPs rs11583680 (minor allele frequency 0·14), rs11591147 (0·01), rs2479409 (0·36), and rs11206510 (0·17; appendix).
Individual participant-level and summary-level data
Data were analysed from two sources. Participating studies executed a common analysis script on their own data, submitting summary estimates to a central analysis centre at University College London, London, UK. Main effect estimates from the participating studies were then meta-analysed with pooled summary estimates from the public domain data repositories of relevant genetic (genome-wide association study [GWAS]) consortia, but only if the study-level estimates had not previously contributed to consortia results, to prevent double counting. All studies contributing data to these analyses were approved by their local ethics committees.
Data were collected for LDL cholesterol, insulin (fasting and non-fasting), glucose (fasting and non-fasting), HbA1c, insulin resistance and secretion via basal homeostatic model assessments (HOMA-IR and HOMA-B), bodyweight, height, BMI, waist-to-hip ratio, and history or incidence of type 2 diabetes.
Publicly available summary-level data were available on blood lipids from the GLGC;14 type 2 diabetes-related biomarkers (plasma insulin, glucose, HbA1c, HOMA-IR, and HOMA-B) from the Meta-Analyses of Glucose and Insulin-related traits Consortium (MAGIC);17, 18, 19 bodyweight, height, BMI, and waist-to-hip ratio from the Genetic Investigation of Anthropometric Traits consortium (GIANT);20, 21 and type 2 diabetes from the Diabetes Genetics Replication and Meta-analysis consortium (DIAGRAM)22 and Exome chip 80K.23 Additionally, cross-sectional data were obtained for adiposity traits and the prevalence of type 2 diabetes from UK Biobank.24
Statistical analyses
In all analyses we assumed an additive allele effect with genotypes coded as 0, 1, and 2, representing the number of minor alleles. We analysed continuous biomarkers using linear regression models; the composite endpoint of prevalent or incident type 2 diabetes was analysed with logistic regression. Study-specific associations were pooled for each SNP by use of the inverse-variance weighted method for fixed-effect and random-effects meta-analysis. We assessed between-study heterogeneity using the Q-test and the I2 statistic25 with a one-sided upper 97·5% CI. Study-specific associations were excluded if the SNP was not in Hardy-Weinberg equilibrium (appendix).
Our approach to SNP selection was designed to prune the number of SNPs at PCSK9 used in the analysis, without loss of information. We decided a priori to combine the four approximately independent SNPs in a weighted gene-centric score (GS) using the inverse-variance weighted method for fixed and random effects.26 The GS provides a more precise estimate of the downstream effects of variation at PCSK9 by incorporating maximum biological variation. Furthermore, if the four SNP effects are homogeneous (assessed by the heterogeneity measures Q-test and I2), the GS estimates will be more powerful and precise compared with individual SNPs in isolation. If, however, the SNP effects are heterogeneous (meaning that the PCSK9 effects are different according to which part of the gene is assessed), the GS method will be less powerful than the individual SNP tests (depending on the degree of heterogeneity). Our aim was to estimate the effect of the PCSK9 locus as a whole, but SNP-specific estimates are also reported. Other important assumptions of the GS approach are (approximate) independence of the included SNPs (assessed by pairwise linkage disequilibrium (r2) and use of multivariable regression models) and the additivity of allele effects. We also investigated whether the association of individual SNPs with diabetes risk was in proportion to the association with LDL cholesterol lowering.
Estimates are presented as mean differences or odds ratios (ORs) with 95% CIs, presented either per LDL cholesterol-decreasing allele or, in the case of GS, per 1 mmol/L (38·67 mg/dL) lower LDL cholesterol. The per 1 mmol/L GS effect estimates were derived by multiplying point estimates and their variances by the multiplicative inverse of the estimated SNP-LDL cholesterol effects. Similar to most genetic studies, missing data were excluded in an available case manner, assuming a missing-completely-at-random mechanism.27, 28 To avoid potential bias due to population stratification and non-modelled ancestry interactions, analyses excluded individuals of non-European ancestry. Differences in ancestry can be a potential source of confounding bias (ie, population stratification bias) when environment is related to both the genes and the outcome of interest. Analyses were done with the statistical programme R (version 3.3.0).
Sensitivity analyses
We assumed that the allele effects were additive, which we assessed in available individual participant data by comparing an additive model to a non-additive model (allowing for dominance or recessiveness) using a likelihood ratio test (meta-analysed by Fisher's method).29 Because measurement error might be larger in prevalent cases (ascertained, for example, from hospital records) we did a further sensitivity analysis in which we separately analysed incident and prevalent type 2 diabetes. This sensitivity analysis was done not because we expect the true associations of PCSK9 to be different with respect to prevalent and incident case status, but merely reflected a quality-control check. Although SNPs were selected to be independent, there was some degree of residual dependency (appendix; maximum r2 0·26). To explore the effect of this residual correlation between the four study SNPs (appendix), we compared results from a multivariable analysis (including the four SNPs in the same model) in studies with individual participant data (correcting for this correlation) to pairwise results (ignoring any between-SNP correlation) based on the same data.
Role of the funding source
The funder of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the report. The corresponding author (AFS) had full access to all the data in the study and shared final responsibility for the decision to submit for publication with all authors.
Results
50 studies shared participant-level data from up to 245 942 individuals, which was supplemented by summary effect estimates from data repositories, resulting in a maximum available sample size of 568 448 individuals, including 51 623 cases of incident or prevalent type 2 diabetes. Individual studies were similar with respect to the distribution of biochemical measures (assessed by the median of study-specific means): LDL cholesterol 3·41 mmol/L (IQR 0·39), fasting glucose 5·38 mmol/L (0·58), and HbA1c 5·50% (appendix). Pooled pairwise linkage disequilibrium estimates for the four PCSK9 SNPs all had r2 values less than 0·30 (appendix), confirming that the selected SNPs were in low correlation in the collected data.
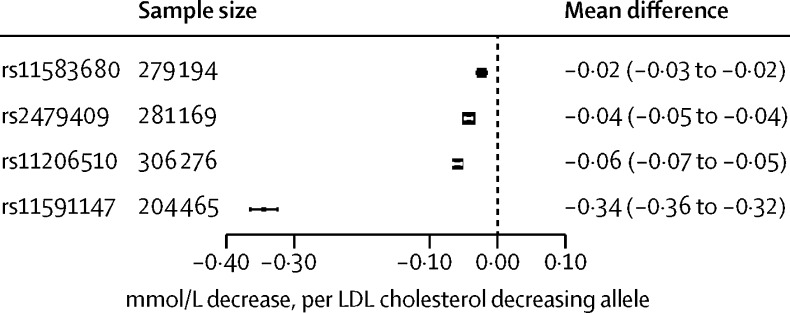
The four PCSK9 SNPs were associated with reductions in LDL cholesterol ranging from −0·02 mmol/L (95% CI −0·03 to −0·02) for rs11583680 to −0·34 mmol/L (−0·36 to −0·32) for rs11591147 per LDL cholesterol-decreasing allele (figure 1).
Figure 1.
Association of genetic variants in PCSK9 with circulating LDL cholesterol concentration
Effect estimates are presented as mean difference in LDL cholesterol (mmol/L) per LDL cholesterol-lowering allele, with 95% CIs. Results are pooled by use of a fixed-effect model. The size of the black dots representing the point estimates is proportional to the inverse of the variance. Note that results from individual participant data are supplemented by repository data from the Global Lipids Genetics Consortium.
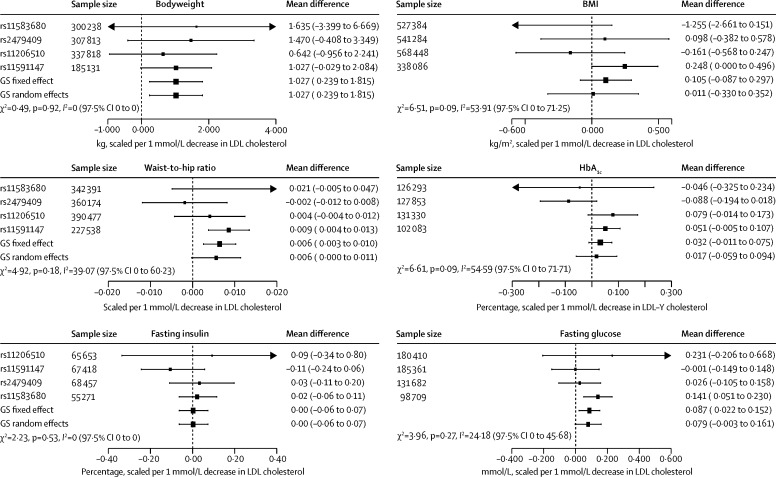
Figure 2 depicts the associations of the four PCSK9 SNPs after scaling the SNP effect to 1 mmol/L lower LDL cholesterol. Results of the PCSK9 GS analysis show that a 1 mmol/L lower LDL cholesterol was associated with an increase in bodyweight of 1·03 kg (95% CI 0·24 to 1·82; and an increase of 0·006 (0·003 to 0·010) in waist-to-hip ratio, but we observed a potentially neutral association with BMI (0·11 kg/m2, −0·09 to 0·30). Associations of the PCSK9 GS with glycaemia measures were 0·09 mmol/L (0·02 to 0·15) higher fasting plasma glucose, HbA1c of 0·03% (−0·01 to 0·08; and for fasting insulin 0·00%, −0·06 to 0·07). SNP-specific forest plots are presented in the appendix. The estimates were similar when corrected for linkage disequilibrium (appendix), and no systematic deviations from an additive model were identified (appendix). Finally, we noted an unanticipated effect on height (mean difference 0·008 m, 0·0008 to 0·015; appendix).
Figure 2.
Association of genetic variants in PCSK9 with glycaemic and anthropometric biomarkers
Effect estimates are presented as mean difference with 95% CIs. Associations were scaled to a 1 mmol/L reduction in LDL cholesterol. SNP-specific results are pooled by use of a fixed-effect model; weighted gene-centric score (GS) models combining all four SNP-specific estimates are presented as fixed-effect and random-effects estimates. The size of the black dots representing the point estimates is proportional to the inverse of the variance. Between-SNP heterogeneity was measured as a two-sided Q-test (χ2) and an I2 with one-sided 97·5% CI. Note that results from individual participant data are supplemented by repository data from the Global Lipids Genetics Consortium, the Meta-Analyses of Glucose and Insulin-related traits Consortium, and the Genetic Investigation of Anthropometric Traits consortium.
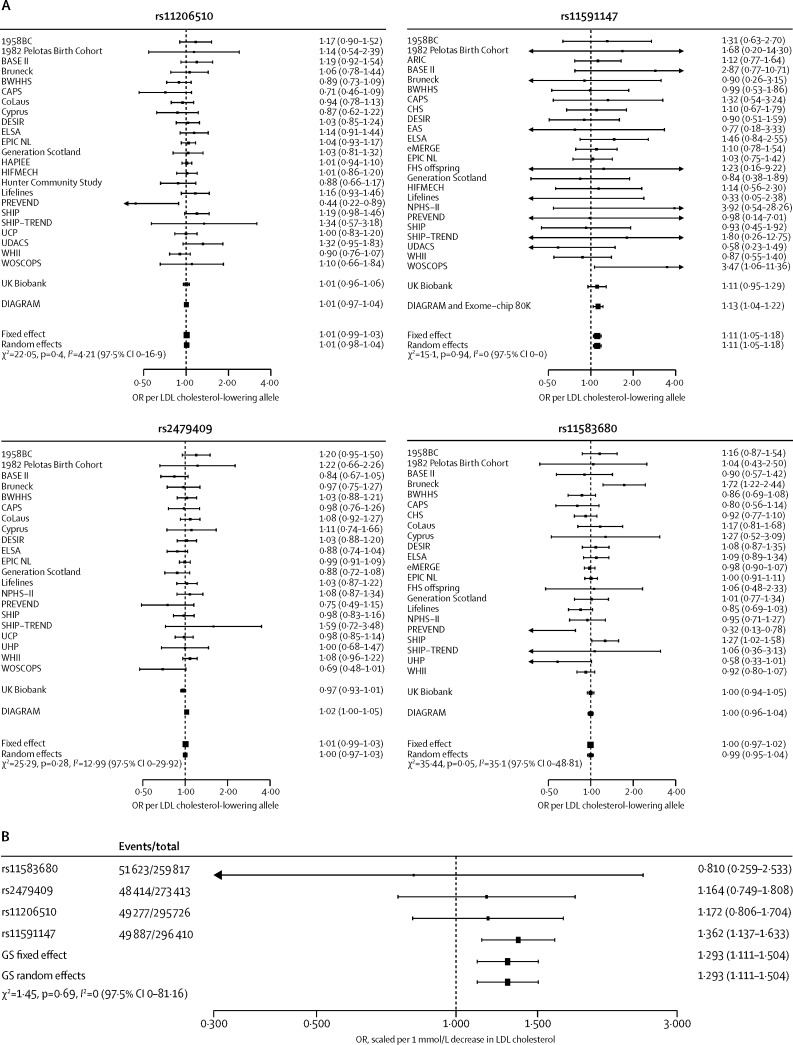
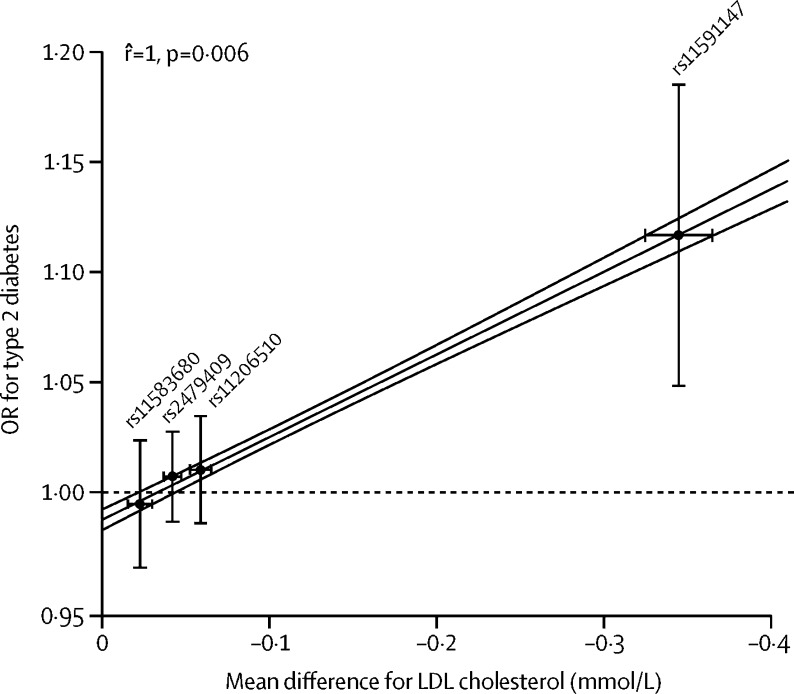
Figure 3 shows the associations of individual PCSK9 variants and the GS with risk of type 2 diabetes. Using the PCSK9 GS, 1 mmol/L lower LDL cholesterol was associated with an increased risk of type 2 diabetes (OR 1·29, 95% CI 1·11 to 1·50). Exploring the PCSK9 associations with incident (appendix) or prevalent (appendix) type 2 diabetes separately showed directional concordance of this effect (incident type 2 diabetes OR 1·15, 0·76 to 1·72; prevalent type 2 diabetes OR 1·26, 0·88 to 1·80). Associations of individual SNPs with LDL cholesterol and risk of type 2 diabetes showed a dose-response relation (figure 4).
Figure 3.
Association of genetic variants in PCSK9 with risk of type 2 diabetes, individually (A) and as weighted gene-centric score (B)
Effect estimates are presented as odds ratios (ORs) for the incidence or prevalence of type 2 diabetes, with 95% CIs. Associations were scaled to a 1 mmol/L reduction in LDL cholesterol. SNP-specific results are pooled by use of a fixed-effect model; weighted gene-centric score (GS) models combining all four SNP-specific estimates are presented as fixed-effect and random-effects estimates. The size of the black dots representing the point estimates is proportional to the inverse of the variance. Between-SNP heterogeneity was measured as a two-sided Q-test (χ2) and an I2 with one-sided 97·5% CI. Results from individual participant data are supplemented by repository data from the Diabetes Genetics Replication and Meta-analysis consortium.
Figure 4.
Correlation between PCSK9 associations with LDL cholesterol concentration and type 2 diabetes
Effect estimates are presented as mean difference in LDL cholesterol concentration (mmol/L) and odds ratios (ORs) for the incidence or prevalence of type 2 diabetes, with 95% CIs. Associations are presented per LDL cholesterol-decreasing allele. The Pearson correlation coefficient, regression line (grey), and its 95% CI (red) were calculated by weighting the SNPs for the inverse of the variance in the type 2 diabetes association. Excluding the SNP with the largest effect on LDL cholesterol (rs11591147) resulted in a correlation coefficient of 0·993 and a p value of 0·437.
Discussion
In this mendelian randomisation study, genetic variants in PCSK9, used as a proxy for pharmacological inhibition of PCSK9, were associated with lower LDL cholesterol concentration and increased risk of type 2 diabetes. The same variants were also associated with higher fasting glucose, bodyweight, and waist-to-hip ratio, and with directionally concordant but non-significant associations for BMI and HbA1c and a seemingly neutral association for fasting insulin. These results are in agreement with previous findings for variants in the HMGCR gene encoding the target of statin drugs, with statins modestly increasing bodyweight and the risk of type 2 diabetes.3
When scaled to 1 mmol/L lower LDL cholesterol, the risk for type 2 diabetes based on HMGCR variants13 was an OR of 1·39 (95% CI 1·12 to 1·73), similar to the corresponding scaled estimate for this PCSK9 GS (1·29, 1·11 to 1·50), and similar to an estimate based on SNPs affecting LDL cholesterol selected from throughout the genome (1·27, 1·14 to 1·41).5 However, effect estimates obtained from mendelian randomisation studies proxy lifetime exposure to natural genetic variation, and might therefore not directly translate to the size of effect of any corresponding pharmacological treatment introduced much later in life and thus for a shorter duration of time.30 For example, in a meta-analysis of randomised controlled trials of statin treatment,31 the OR for type 2 diabetes was 1·12 (95% CI 1·06 to 1·18).
In the case of statins, the treatment benefit in terms of CHD risk reduction greatly outweighs any potential adverse effect on risk of type 2 diabetes, partly because the size of the risk reduction in CHD is greater than the risk increase in type 2 diabetes, and partly because the absolute risk of CHD in primary prevention populations eligible for statin treatment is greater than the absolute risk of type 2 diabetes.32 A similarly precise risk assessment for PCSK9 inhibitors awaits results from larger and longer-term randomised trials. In a recent pooled analysis,33 researchers reported that treatment with alirocumab was associated with an OR for type 2 diabetes of 0·89 (95% CI 0·62 to 1·28) compared with placebo, based on 133 type 2 diabetes events.
Variants that affect circulating LDL cholesterol have been reported previously to affect the probability of being prescribed a lipid-lowering drug.34 We were unable to account for this effect in the analysis because prescription data for these treatments were often not available, and when they were recorded they were only available for a single follow-up point. For lipid-lowering treatments, one record of treatment does not properly reflect the time-varying therapy received, and adjusting for only a single record when in fact treatment varies over follow-up might increase bias.35 Typically, diabetes drug treatments are much less variable over time and correction for this treatment might seem advisable; however, because of the strong correlation between history of type 2 diabetes and use of type 2 diabetes-related drugs, any correction for the latter would essentially correct for prevalent type 2 diabetes as well. Importantly, any effect of lipid-lowering drug therapy would attenuate rather than inflate any associations.
We have previously reported examples of common variants in genes encoding a protein drug target mimicking the on-target effects of pharmacological interventions on biomarkers and disease outcomes in type, direction, and relative size.3, 36, 37 However, such analyses cannot predict off-target effects of treatments. We refer to on-target effects as those that are due to a drug effect on the intended target (in this case PCSK9) and off-target effects as those that might occur because of the drug also binding to an unintended target (in this case, any target other than PCSK9). Although monoclonal antibody therapeutics are often highly specific, perhaps more so than small molecule therapeutics, they retain the potential for off-target effects. Hence, in the presence of off-target effects, results from ongoing randomised controlled trials could differ from the genetic associations reported here.
Our main findings are based on four PCSK9 SNPs in combination and scaled to 1 mmol/L lower LDL cholesterol. This approach assumes additive effects across the SNPs, an assumption that held well in sensitivity analyses. A potentially unobserved non-additive effect might explain why we identified a genetic association with fasting glucose and a concordant (although non-significant) association with HbA1c, whereas fasting insulin seemed unaffected. Conflicting evidence exists about a possible role of PCSK9 and PCSK9 monoclonal antibodies in disruption of pancreatic islet function.38, 39 Although concordant with fasting glucose, the HbA1c association was non-significant in the collected data, which might be related to the large amount of heterogeneity between the four SNPs (upper-bound I2 72%). Interestingly, the association of the PCSK9 GS with BMI was smaller than that with bodyweight, which might be (partially) explained by a slightly greater average height among individuals with PCSK9 variants associated with lower LDL cholesterol concentrations. A further potential reason for the slight discrepancy between the BMI and bodyweight associations could be the greater heterogeneity in the associations of PCSK9 SNPs with BMI than with weight. Notably, the GS effect estimates were often driven by a large effect of SNP rs11591147; as our dose-response analysis shows (figure 4), the larger influence of this SNP appropriately reflects the proportionally larger LDL cholesterol effect of this SNP. Finally, we did not have access to measures of PCSK9 concentration in this analysis, but others40 have shown associations between common and rare PCSK9 alleles (including some of the same SNPs used here) and circulating PCSK9 concentrations.
Setting aside associations with glycaemia and weight, risk of type 2 diabetes could also be increased because lifelong exposure to genetic variation in PCSK9 might reduce mortality, making it conceivable that individuals with these variants survive longer and hence have more time to develop type 2 diabetes. However, whether PCSK9 genotype reduces mortality has not be conclusively shown.8, 41 Irrespective of the nature of the PCSK9 association with type 2 diabetes, large randomised trials should determine whether this relation also holds for PCSK9 monoclonal antibodies.
In a recent study,13 investigators used a single SNP in PCSK9 and also reported evidence of an association with type 2 diabetes (OR 1·19, 95% CI 1·02 to 1·38; per 1 mmol/L reduction in LDL cholesterol). In the present study, we incorporated data from four SNPs, instead of a single SNP, in a PCSK9 gene score with participant data from 50 studies supplemented by large genetic consortia and are able to confirm their results, and also show this increase in type 2 diabetes risk is likely to be related to PCSK9-related increases in bodyweight and glucose. Previous studies of LDL cholesterol lowering HMGCR3 and NPC1L113 variants (encoding pharmacological targets of statins and ezetimibe, respectively) and more widely on LDL cholesterol-lowering variants from multiple GWAS-associated loci,5 as well as analyses of patients with monogenic hypercholesterolaemia,6 have provided evidence of a link between LDL cholesterol and type 2 diabetes, compatible with the findings from the present study. However, it is far from certain that all LDL cholesterol-lowering interventions will increase risk of type 2 diabetes, as not all share the same mechanism of action. The major site of both statins and PCSK9 inhibitors is thought to be the liver, through increased cellular membrane expression of the LDL receptor. The liver is also the site of action of the investigational apolipoprotein B antisense oligonucleotide mipomersen, whereas ezetimibe, the other licensed LDL cholesterol lowering drug, acts in the intestine to limit LDL cholesterol absorption. A potential unifying mechanism might be pancreatic β cell LDL receptor upregulation, increased lipid accumulation, and β cell dysfunction,6 but this suggestion will need to be tested experimentally.
In conclusion, genetic variants in PCSK9 that associate with lower concentrations of LDL cholesterol are also associated with a modestly higher risk of type 2 diabetes and with associated differences in measures of glycaemia and bodyweight. Investigators of ongoing and future randomised controlled trials of PCSK9 inhibitors should carefully monitor changes in metabolic markers, including bodyweight and glycaemia, and the incidence of type 2 diabetes in study participants. Genetic studies of the type used here could be more widely used to interrogate the safety and efficacy of novel drug targets.
Acknowledgments
Acknowledgments
This work was supported by a British Heart Foundation Programme Grant (RG/10/12/28456). AFS is funded by University College London Hospitals NHS Foundation Trust (UCLH) National Institute for Health Research (NIHR) Biomedical Research Centre (BRC10200) and by a UCL springboard population science fellowship. FWA is supported by a Dekker scholarship-Junior Staff Member 2014T001–Netherlands Heart Foundation and UCL Hospitals NIHR Biomedical Research Centre. ADH is an NIHR Senior Investigator. Funding information and acknowledgments for studies contributing data are reported in the appendix.
Contributors
AFS, DIS, MVH, RSP, FWA, J-PC, BJK, ADH, DP, and NS contributed to the conception and design of the study. AFS, DIS, and MVH designed the analysis scripts shared with individual centres. AFS did the meta-analysis and had access to all the data. AFS, DIS, and MVH drafted the report. RSP, ZF-H, DML, FPH, BLH, EHy, CP, MM, EvI, GKH, ID, KN, ES-T, JD, LB, TL, SC, JWi, SK, KW, DM, JWr, RM, GW, PW, YB-S, SMc, JFP, MKi, CW, AS-G, PM-V, AN, AGP, NCO-M, YTvdS, GM, GF, SGua, CS, NJW, CL, RS, JL, MBo, SMa, AP, RK, ATa, HP, LLNH, NG, OP, TH, AL, KSS, JC, SEH, MBr, TK, HH, DSC, CAM, HLK, EBL, DRC, MdA, DMR, JCD, CC, SH, JA, EHo, MO'D, SY, MC, GP, PvdH, MAS, RNE, NV, HS (for the LifeLines Cohort study group), TC, DOM-K, SGus, LL, EI, RP, OF, AH, AU, AD, ATe, SB, MD, MML, UV, HV, JWa, JPP, DJS, TM, AHM-vdZ, EVB, RY, IF, AC, SP, MLB, DEG, PF, DT, BB, AB, BC, MS, YB, MKu, AM, PMR, DIC, APR, LAL, MDR, FWA, BJK, J-PC, ADH, DP, and NS were responsible for study-specific analyses and critically revised the report.
Declaration of interests
KH or his institution have received honoraria for consultancy, advisory board participation, or conduct of clinical trials from AMGEN, Aegerion, Pfizer, AstraZeneca, Sanofi, Regeneron, KOWA, Ionis pharmaceuticals, and Cerenis. GKH has received research support from Aegerion, AMGEN, Sanofi, AstraZeneca, and Synageva. BC has received research funding from Pfizer and Sanofi, has received personal fees for lectures from AstraZeneca, Pierre Fabre, Janssen, Eli Lilly, MSD Merck & Co, Novo Nordisk, Sanofi, and Takeda, and has acted as a consultant or advisory panel member for Amgen, Eli Lilly, Novo Nordisk, Sanofi, and Regeneron. DP has consulted for Sanofi on two occasions in previous employment (related to PCSK9 inhibitors) and was an investigator on clinical trials of PCSK9 inhibition funded by Amgen. NS has consulted for Amgen and Sanofi (related to PCSK9 inhibitors) and was an investigator on clinical trials of PCSK9 inhibition funded by Amgen. He has also consulted for MSD, Boehringer Ingelheim, Janssen, and Novo Nordisk. DIS has consulted for Pfizer for work unrelated to this report. AP has been a consultant and a member of an advisory panel for Amgen. EI is a scientific advisor and consultant for Precision Wellness Inc and a scientific advisor for Cellink, for work unrelated to this paper. All other authors declare no competing interests.
Contributor Information
Amand F Schmidt, Email: amand.schmidt@ucl.ac.uk.
Naveed Sattar, Email: naveed.sattar@glasgow.ac.uk.
Supplementary Material
References
- 1.Sattar N, Preiss D, Murray HM. Statins and risk of incident diabetes: a collaborative meta-analysis of randomised statin trials. Lancet. 2010;375:735–742. doi: 10.1016/S0140-6736(09)61965-6. [DOI] [PubMed] [Google Scholar]
- 2.Preiss D, Seshasai SR, Welsh P. Risk of incident diabetes with intensive-dose compared with moderate-dose statin therapy: a meta-analysis. JAMA. 2011;305:2556–2564. doi: 10.1001/jama.2011.860. [DOI] [PubMed] [Google Scholar]
- 3.Swerdlow DI, Preiss D, Kuchenbaecker KB. HMG-coenzyme A reductase inhibition, type 2 diabetes, and bodyweight: evidence from genetic analysis and randomised trials. Lancet. 2015;385:351–361. doi: 10.1016/S0140-6736(14)61183-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fall T, Xie W, Poon W. Using genetic variants to assess the relationship between circulating lipids and type 2 diabetes. Diabetes. 2015;64:2676–2684. doi: 10.2337/db14-1710. [DOI] [PubMed] [Google Scholar]
- 5.White J, Swerdlow DI, Preiss D. Association of lipid fractions with risks for coronary artery disease and diabetes. JAMA Cardiol. 2016;1:692–699. doi: 10.1001/jamacardio.2016.1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Besseling J, Kastelein JJ, Defesche JC, Hutten BA, Hovingh GK. Association between familial hypercholesterolemia and prevalence of type 2 diabetes mellitus. JAMA. 2015;313:1029–1036. doi: 10.1001/jama.2015.1206. [DOI] [PubMed] [Google Scholar]
- 7.Abifadel M, Varret M, Rabes JP. Mutations in PCSK9 cause autosomal dominant hypercholesterolemia. Nat Genet. 2003;34:154–156. doi: 10.1038/ng1161. [DOI] [PubMed] [Google Scholar]
- 8.Cohen JC, Boerwinkle E, Mosley TH, Jr, Hobbs HH. Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N Engl J Med. 2006;354:1264–1272. doi: 10.1056/NEJMoa054013. [DOI] [PubMed] [Google Scholar]
- 9.Navarese EP, Kolodziejczak M, Schulze V. Effects of proprotein convertase subtilisin/kexin type 9 antibodies in adults with hypercholesterolemia: a systematic review and meta-analysis. Ann Intern Med. 2015;163:40–51. doi: 10.7326/M14-2957. [DOI] [PubMed] [Google Scholar]
- 10.Stein EA, Mellis S, Yancopoulos GD. Effect of a monoclonal antibody to PCSK9 on LDL cholesterol. N Engl J Med. 2012;366:1108–1118. doi: 10.1056/NEJMoa1105803. [DOI] [PubMed] [Google Scholar]
- 11.Hingorani A, Humphries S. Nature's randomised trials. Lancet. 2005;366:1906–1908. doi: 10.1016/S0140-6736(05)67767-7. [DOI] [PubMed] [Google Scholar]
- 12.Sofat R, Hingorani AD, Smeeth L. Separating the mechanism-based and off-target actions of cholesteryl ester transfer protein inhibitors with CETP gene polymorphisms. Circulation. 2010;121:52–62. doi: 10.1161/CIRCULATIONAHA.109.865444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lotta LA, Sharp SJ, Burgess S. Association between low-density lipoprotein cholesterol-lowering genetic variants and risk of type 2 diabetes: a meta-analysis. JAMA. 2016;316:1383–1391. doi: 10.1001/jama.2016.14568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Global Lipids Genetics Consortium Discovery and refinement of loci associated with lipid levels. Nat Genet. 2013;45:1274–1283. doi: 10.1038/ng.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kircher M, Witten DM, Jain P, O'Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.CARDIoGRAMplusC4D Consortium. Deloukas P, Kanoni S. Large-scale association analysis identifies new risk loci for coronary artery disease. Nat Genet. 2013;45:25–33. doi: 10.1038/ng.2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scott RA, Lagou V, Welch RP. Large-scale association analyses identify new loci influencing glycemic traits and provide insight into the underlying biological pathways. Nat Genet. 2012;44:991–1005. doi: 10.1038/ng.2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Soranzo N, Sanna S, Wheeler E. Common variants at 10 genomic loci influence hemoglobin A1c levels via glycemic and nonglycemic pathways. Diabetes. 2010;59:3229–3239. doi: 10.2337/db10-0502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dupuis J, Langenberg C, Prokopenko I. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet. 2010;42:105–116. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Randall JC, Winkler TW, Kutalik Z. Sex-stratified genome-wide association studies including 270,000 individuals show sexual dimorphism in genetic loci for anthropometric traits. PLoS Genet. 2013;9:e1003500. doi: 10.1371/journal.pgen.1003500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Locke AE, Kahali B, Berndt SI. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518:197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morris AP, Voight BF, Teslovich TM. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet. 2012;44:981–990. doi: 10.1038/ng.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fuchsberger C, Flannick J, Teslovich TM. The genetic architecture of type 2 diabetes. Nature. 2016;536:41–47. doi: 10.1038/nature18642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sudlow C, Gallacher J, Allen N. UK Biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12:e1001779. doi: 10.1371/journal.pmed.1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson T. Efficient calculation for multi-SNP genetic risk scores. American Society of Human Genetics Annual Meeting; San Francisco, CA; Nov 6–10, 2012.
- 27.Groenwold RH, Donders AR, Roes KC, Harrell FE, Jr., Moons KG. Dealing with missing outcome data in randomized trials and observational studies. Am J Epidemiol. 2012;175:210–217. doi: 10.1093/aje/kwr302. [DOI] [PubMed] [Google Scholar]
- 28.Little RJA, Rubin DB. Statistical analysis with missing data. 2nd edn. John Wiley & Sons; Hoboken: 2002. [Google Scholar]
- 29.Fisher R. Statistical methods for research workers. Oliver and Boyd; Edinburgh: 1934. [Google Scholar]
- 30.Burgess S, Butterworth A, Malarstig A, Thompson SG. Use of Mendelian randomisation to assess potential benefit of clinical intervention. BMJ. 2012;345:e7325. doi: 10.1136/bmj.e7325. [DOI] [PubMed] [Google Scholar]
- 31.Taylor F, Huffman MD, Macedo AF. Statins for the primary prevention of cardiovascular disease. Cochrane Database Syst Rev. 2013;1 doi: 10.1002/14651858.CD004816.pub5. CD004816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ridker PM, Pradhan A, MacFadyen JG, Libby P, Glynn RJ. Cardiovascular benefits and diabetes risks of statin therapy in primary prevention: an analysis from the JUPITER trial. Lancet. 2012;380:565–571. doi: 10.1016/S0140-6736(12)61190-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Colhoun HM, Ginsberg HN, Robinson JG. No effect of PCSK9 inhibitor alirocumab on the incidence of diabetes in a pooled analysis from 10 ODYSSEY phase 3 studies. Eur Heart J. 2016;37:2981–2989. doi: 10.1093/eurheartj/ehw292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shah S, Casas JP, Gaunt TR. Influence of common genetic variation on blood lipid levels, cardiovascular risk, and coronary events in two British prospective cohort studies. Eur Heart J. 2013;34:972–981. doi: 10.1093/eurheartj/ehs243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robins JM, Hernan MA, Brumback B. Marginal structural models and causal inference in epidemiology. Epidemiology. 2000;11:550–560. doi: 10.1097/00001648-200009000-00011. [DOI] [PubMed] [Google Scholar]
- 36.Holmes MV, Simon T, Exeter HJ. Secretory phospholipase A2-IIA and cardiovascular disease: a mendelian randomization study. J Am Coll Cardiol. 2013;62:1966–1976. doi: 10.1016/j.jacc.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Interleukin-6 Receptor Mendelian Randomisation Analysis Consortium The interleukin-6 receptor as a target for prevention of coronary heart disease: a mendelian randomisation analysis. Lancet. 2012;379:1214–1224. doi: 10.1016/S0140-6736(12)60110-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mbikay M, Sirois F, Mayne J. PCSK9-deficient mice exhibit impaired glucose tolerance and pancreatic islet abnormalities. FEBS Lett. 2010;584:701–706. doi: 10.1016/j.febslet.2009.12.018. [DOI] [PubMed] [Google Scholar]
- 39.Langhi C, Le May C, Gmyr V. PCSK9 is expressed in pancreatic delta-cells and does not alter insulin secretion. Biochem Biophys Res Commun. 2009;390:1288–1293. doi: 10.1016/j.bbrc.2009.10.138. [DOI] [PubMed] [Google Scholar]
- 40.Chernogubova E, Strawbridge R, Mahdessian H. Common and low-frequency genetic variants in the PCSK9 locus influence circulating PCSK9 levels. Arterioscler Thromb Vasc Biol. 2012;32:1526–1534. doi: 10.1161/ATVBAHA.111.240549. [DOI] [PubMed] [Google Scholar]
- 41.Benn M, Nordestgaard BG, Grande P, Schnohr P, Tybjaerg-Hansen A. PCSK9 R46L, low-density lipoprotein cholesterol levels, and risk of ischemic heart disease: 3 independent studies and meta-analyses. J Am Coll Cardiol. 2010;55:2833–2842. doi: 10.1016/j.jacc.2010.02.044. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.