Abstract
The association of A1513C (rs3751143) polymorphism of P2X7 gene with the risk of extrapulmonary tuberculosis (EPTB) has been extensively analyzed, but no consensus has been achieved. In this study, a meta-analysis was done to assess this precise association. Online web databases, like PubMed (MEDLINE) and EMBASE were searched for pertinent reports showing association of P2X7 A1513C polymorphism with EPTB risk. To assess the strength of this association, we calculated pooled odds ratios (ORs) and 95% confidence intervals (95% CIs). A total of eight reports involving 2237controls and 594 EPTB cases were included in this study. Four genetic models, viz. allele (C vs. A: p=0.011; OR= 1.677, 95% CI = 1.125–2.501), homozygous (CC vs. AA: p = 0.053; OR= 2.362, 95% CI = 0.991–5.632), heterozygous (AC vs. AA: p = 0.003; OR= 1.775, 95% CI = 1.209–2.607) and dominant (CC + AC vs. AA: p = 0.005; OR= 1.890, 95% CI = 1.207–2.962) showed significant associations compared with wild type genotypes. Subgroup analysis stratified by ethnicity was also performed and the results suggested that homozygous and heterozygous genotypes were associated significantly with increased susceptibility of EPTB in Asian population. Similarly, heterozygous and dominant models showed increased EPTB risk in Caucasian population. The present meta-analysis suggests that P2X7 A1513C polymorphism may be an important risk factor for EPTB. Also, our sub-group analysis indicates that P2X7 A1513C polymorphism confers increased EPTB risk among Asians and Caucasians. However, future larger studies are needed to provide more precise conclusion and endorse the present results.
Keywords: Extrapulmonary tuberculosis, P2X7 gene, polymorphism, genetic models, meta-analysis
INTRODUCTION
Tuberculosis (TB), an infectious disease caused by Mycobacterium tuberculosis (M. tb), is a dreadful disease with high rate of morbidity and mortality across the globe [1]. M. tb infects nearly one-third of the world’s population, out of which only 5-10% develop actual TB during their life time [2]. Previous studies reported that among all types of TB cases, 20% cases belong to extrapulmonary tuberculosis (EPTB) [3]. A key role played by the host genetic factors in risk of developing TB has been suggested by earlier studies [4-7]. Recently, genome-wide association studies (GWAS) demonstrate that host genetics factors are strongly linked with TB development [8]. Earlier reports showed that P2X7 receptor plays a major role in initiation of activity against mycobacteria and ATP works significantly in triggering of this activity along with human macrophages [9]. The gene for P2X7 encoding the P2X7 receptor is located on chromosome 12q24 of the human genome [10]. The P2X7 receptor is highly expressed by the cells of haemopoietic lineage and can trigger cell death, kill infectious organisms, and regulate inflammatory responses [11]. The involvement of P2X7 receptor in above mentioned pathways suggests that it plays a role of a major regulator of inflammation. The ATP treatment of macrophages infected with mycobacteria induces apoptosis and death of both the host cell and the internalized bacilli. The process is mediated via the P2X7 pathway [12]. The use of P2X7 antagonist has been shown to inhibit the ATP induced apoptosis and bacterial killing in infected macrophages in another study [13].
Many single nucleotide polymorphisms (SNPs) of P2X7 gene are reported in the literature indicating high polymorphic nature of this gene in humans [14]. A common SNP altering the function of the P2X7 receptor is A1513C, located in the carboxy terminal tail encoding region of the P2X7 gene [15, 16], which could affect an individual susceptibility to EPTB.
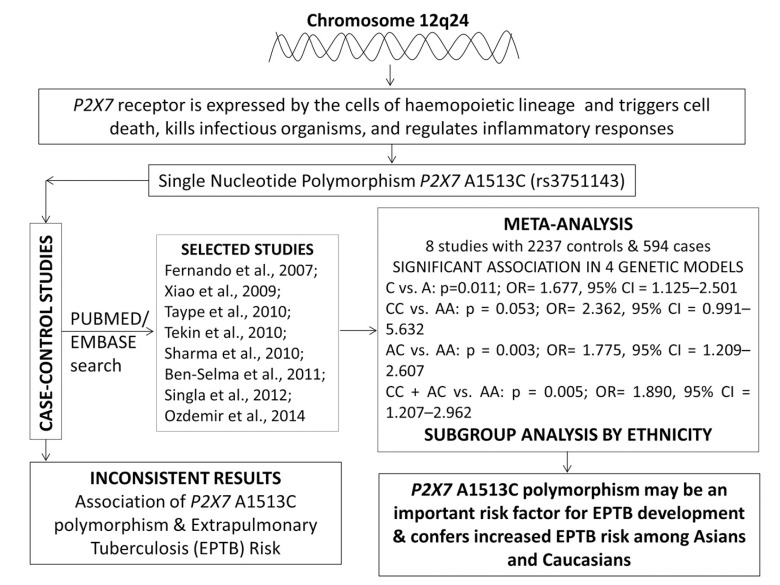
Keeping the biological significance of this genetic variant in view, the association of P2X7 A1513C polymorphism with the susceptibility of TB has been widely studied. Previously, it has been reported that the pathophysiology of EPTB is differ from other types of TB [17]. So, it is important to examine the genetic variants of P2X7 receptor gene specifically associated with EPTB. Till now, several research studies have been done to assess the possible association between P2X7 A1513C genetic polymorphism and the development of EPTB in different populations but their findings are either contradictory or inconclusive [18-25]. Therefore, data from various case-control studies were pooled and meta-analysis was performed to derive a more precise conclusion regarding the relationship between the overall effect of P2X7 1513 A>C genetic variant and the risk of developing EPTB. A meta-analysis is a potent method for evaluating cumulative data from different research studies to overcome the problem of small sample sizes and low statistical power [26], and has been successfully used for the pooling of the data for different case-control studies in relation with various polygenic diseases, e.g., cancer [27], tuberculosis [6, 7] etc. The schematic representation of the entire meta-analysis has been given as graphical abstract (Fig. 1).
Fig. (1).
Graphical abstract of the meta-analysis performed to evaluate the association of P2X7 A1513C (rs 3751143) polymorphism and EPTB risk.
MATERIALS AND METHODS
Identification and Eligibility of the Studies
Pertinent studies were cautiously identified by comprehensive search of PubMed (Medline) and EMBASE web databases using the following key words: “P2X7 gene AND (polymorphism OR variant OR mutation) AND (A1513C OR rs3751143) AND Extrapulmonary tuberculosis (EPTB)” to cover all the published research articles (last updated on June 2015). Research studies that express potential relevance for genetic association were weighed up by scrutinizing their titles and abstracts. All the studies matching with the above mentioned eligibility criteria were selected and included in the present meta-analysis.
Inclusion and Exclusion Criteria for the Studies
In order to reduce heterogeneity and to facilitate the accuracy of the findings in the present meta-analysis, preset criteria were followed for study inclusion/ exclusion. The study inclusion criteria were: (i) evaluation of P2X7 A1513C A>C polymorphism and EPTB risk, (ii) use of case-control design, (iii) enrollment of pathologically confirmed EPTB patients and TB-free controls, (iv) availability of genotype frequency in cases as well as in controls, and (v) published in the English language. In case, if the case-control study was reported by more than one article using the same case series, study with highest number of subjects was selected. Studies reporting overlapping data, case-only studies and review articles were the leading reasons for exclusion of the studies.
Data Extraction and Quality Assessment
The methodological quality assessment and the data extraction for all the selected publications were independently abstracted by two independent investigators in duplicate using the standard protocol. The data accuracy was ascertained by using a data-collection form according to the preset inclusion criteria as stated in the above section. The difference of opinion, if any, on the items collected from the retrieved studies was discussed in detail to achieve a common consensus. The abstracted characters from the selected studies assimilated the first author’s name, the publication year, the country of origin, sources of cases and controls and their numbers, genotype frequencies, and the study type.
Statistical Analysis
The pooled odds ratios (ORs) and their respective 95% confidence intervals (CIs) were estimated to assess the relation between -1513 A>C polymorphism and EPTB risk. Heterogeneity belief was investigated by the chi-square based Q-test [28]. The significance level (p-value) > 0.05 for the Q-test indicated a lack of heterogeneity among all the selected studies. Fixed/ random effects models were used to pool ORs [29, 30]. Additionally, I2 statistics was used to measure the inter-study variability ranging between 0 to 100%, wherein, 0% indicates no observed heterogeneity and larger values indicate an increasing degree of heterogeneity [31]. The Hardy-Weinberg equilibrium (HWE) was calculated using the chi-square test in the control group. The funnel plot asymmetry was calculated by the Egger’s linear regression test, which is a linear regression methodology to measure the funnel plot asymmetry on the natural logarithm scale of the OR. Student’s t-test (p-value <0.05 was maintained as a sign of statistically significant publication bias) was used to determine the significance of the intercept [32]. The entire statistical analysis for the current meta-analysis was executed with the help of comprehensive meta-analysis (CMA) Version 2 software program (Biostat, USA). The CMA V2 has many positive aspects in comparison to other meta-analysis software programs. A precise and comprehensive comparison of meta-analysis programs was performed for the selection of apt meta-analysis program using url. http://meta-analysis.com/pages/comparisons.html.
RESULTS
Characteristics of the Selected Published Studies
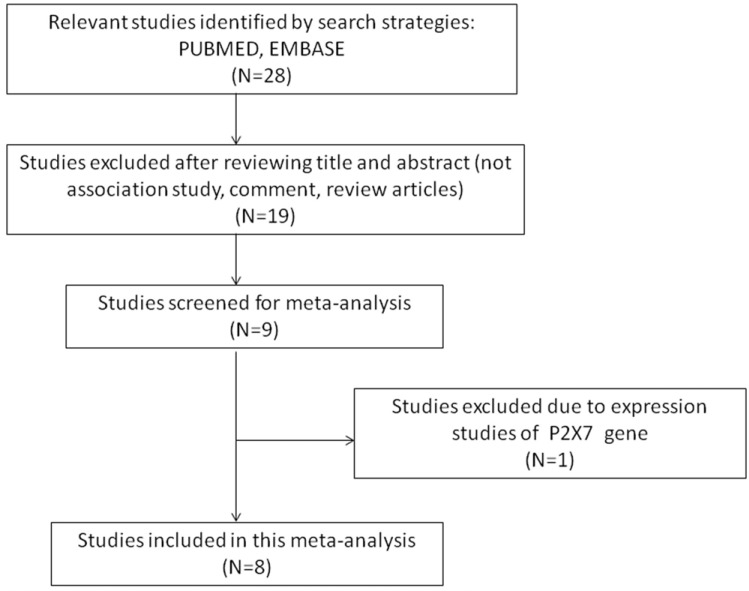
The number of hits obtained by doing a literature search on PubMed (Medline) and EMBASE database was twenty eight. All the retrieved hits (articles) were checked by reading their titles, and abstracts, and the full texts for the possibly relevant publications. In addition, the articles were further scrutinized for their aptness for the present meta-analysis (Fig. 2: PRISMA Flow-diagram). Similarly, the reference lists of the retrieved publications were also screened for other possible apposite articles. The studies of P2X7 polymorphism predicting survival and considering it as therapy response indicator were excluded straightaway. Research articles that report the levels of P2X7 mRNA or protein expression and also the review articles were disqualified. In order to derive a precise conclusion from this pooled analysis, a very stringent criterion was followed for searching the publication. Only case-control or cohort design studies with frequencies of all the three genotypes were included. After thorough analysis and following the stern inclusion and exclusion criteria, eight originally published studies representing the above mentioned possible association were found eligible and included in this study (Table 1). The selection process (inclusion/ exclusion) of the studies for this meta-analysis is shown in a PRISMA flow-diagram in (Fig. 2). Information regarding distribution of genotypes, HWE p-values in the controls, and susceptibility to EPTB is provided in (Table 2).
Fig. (2).
PRISMA flow-diagram showing the selection process (inclusion/exclusion) of the pertinent studies of P2X7 A1513C (rs 3751143) polymorphism and EPTB risk.
Table 1. Main characteristics of all the studies summarized for the present meta-analysis.
|
First Authors and
Reference No. |
Year | Country of Origin | Ethnicity | Genotyping Method | Cases | Controls |
Source of
Genotyping |
|---|---|---|---|---|---|---|---|
| Fernando et al. [18] | 2007 | Southeast Asia (Liverpool cohort) | Asian | ABI PRISM | 30 | 167 | Blood |
| Fernando et al. [18] | 2007 | Australia | Caucasian | ABI PRISM | 50 | 102 | Blood |
| Xiao et al. [19] | 2009 | China | Asian | PCR-RFLP | 55 | 384 | Blood |
| Taype et al. [20] | 2010 | Peru | Caucasian | PCR-RFLP | 121 | 513 | Blood |
| Tekin et al. [21] | 2010 | Turkey | Caucasian | PCR-RFLP | 100 | 192 | Blood |
| Sharma et al. [22] | 2010 | India | Asian | PCR-RFLP | 23 | 177 | Blood |
| Ben-Selma et al. [23] | 2011 | Tunisia | Caucasian | PCR-RFLP | 55 | 150 | Blood |
| Singla et al. [24] | 2012 | India | Asian | PCR-RFLP | 71 | 392 | Blood |
| Ozdemir et al. [25] | 2014 | Turkey | Caucasian | PCR-RFLP | 89 | 160 | Blood |
Table 2. Distribution of P2X7 A1513C gene polymorphism studies included in this meta-analysis.
| Authors, Year of Publication and Reference No. | Ethnicity | Controls | Cases | HWE | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Genotype | Minor Allele | Genotype | Minor Allele | |||||||
| AA | AC | CC | AA | AC | CC | |||||
| Fernando et al. 2007 [18] | Asian | 105 | 55 | 7 | 0.20 | 9 | 17 | 4 | 0.41 | 0.950 |
| Fernando et al. 2007 [18] | Caucasian | 64 | 34 | 4 | 0.20 | 18 | 28 | 4 | 0.36 | 0.840 |
| Xiao et al. 2009 [19] | Asian | 221 | 119 | 44 | 0.26 | 30 | 19 | 6 | 0.28 | 0.001 |
| Taype et al. 2010 [20] | Caucasian | 347 | 149 | 17 | 0.17 | 82 | 37 | 2 | 0.16 | 0.830 |
| Tekin et al. 2010 [21] | Caucasian | 141 | 46 | 5 | 0.14 | 39 | 28 | 7 | 0.28 | 0.590 |
| Sharma et al. 2010 [22] | Asian | 126 | 48 | 3 | 0.15 | 8 | 13 | 2 | 0.36 | 0.510 |
| Ben-Selma et al. 2011 [23] | Caucasian | 104 | 40 | 6 | 0.17 | 19 | 23 | 13 | 0.44 | 0.340 |
| Singla et al. 2012 [24] | Asian | 258 | 123 | 11 | 0.18 | 45 | 22 | 1 | 0.17 | 0.410 |
| Ozdemir et al. 2014 [25] | Caucasian | 76 | 63 | 21 | 0.32 | 47 | 34 | 8 | 0.28 | 0.170 |
HWE: Hardy-Weinberg equilibrium.
Evaluation of Publication Bias
Funnel plot and Egger’s test were employed to test publication bias among the selected studies (Table 3). The funnel plot remained symmetric for CC vs. AA, whereas CC vs. AA+AC showed absence of publication bias. Nevertheless, plots for C vs. A, AC vs. AA and CC+AC vs. AA were asymmetric (Table 3). To adjust the publication bias, Trim-and-fill technique was used for these three genetic models (C vs. A, AC vs. AA and CC+AC vs. AA) in order to re-compute the effect size [33].
Table 3. Statistics to test publication bias and heterogeneity for overall analysis.
| Comparisons | Egger’s Regression Analysis | Heterogeneity Analysis | Model Used for the Meta-analysis | ||||
|---|---|---|---|---|---|---|---|
| Intercept | 95% Confidence Interval | p-value | Q-value | Pheterogeneity | I2 (%) | ||
| C vs A | 10.47 | 1.26 to 19.69 | 0.031 | 50.83 | 0.001 | 84.26 | Random |
| CC vs AA | 1.76 | -4.49 to 8.02 | 0.520 | 32.63 | 0.001 | 75.48 | Random |
| AC vs AA | 6.46 | 2.73 to 10.19 | 0.001 | 26.36 | 0.001 | 69.65 | Random |
| CC+AC vs AA | 7.70 | 2.59 to 12.80 | 0.009 | 39.66 | 0.001 | 79.83 | Random |
| CC vs AA+AC | 0.88 | -4.40 to 6.18 | 0.700 | 23.06 | 0.001 | 65.31 | Random |
Test of Heterogeneity
Q-test and I2 statistics were used to evaluate the inter- and intra-study variations and based upon the significance value different models were selected for this meta-analysis (Table 3). We observed heterogeneity in all the genetic models, i.e., allele (C vs. A), heterozygous (AC vs. AA), homozygous (CC vs. AA), recessive (CC + AC vs. AA) and dominant (CC vs. AA+ AC), hence random effects model was applied.
Meta-analysis of P2X7 1513 A > C Polymorphism and EPTB Susceptibility
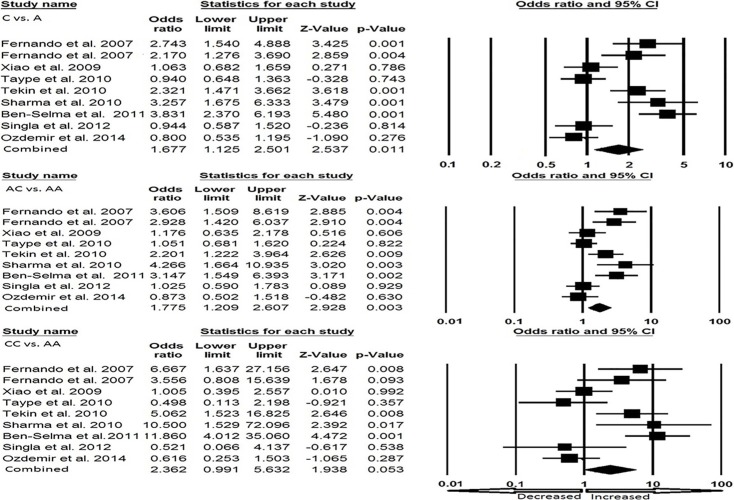
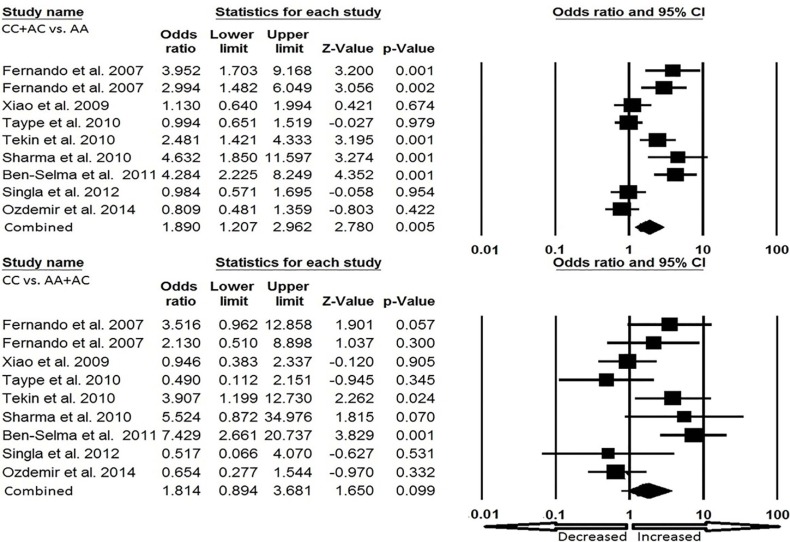
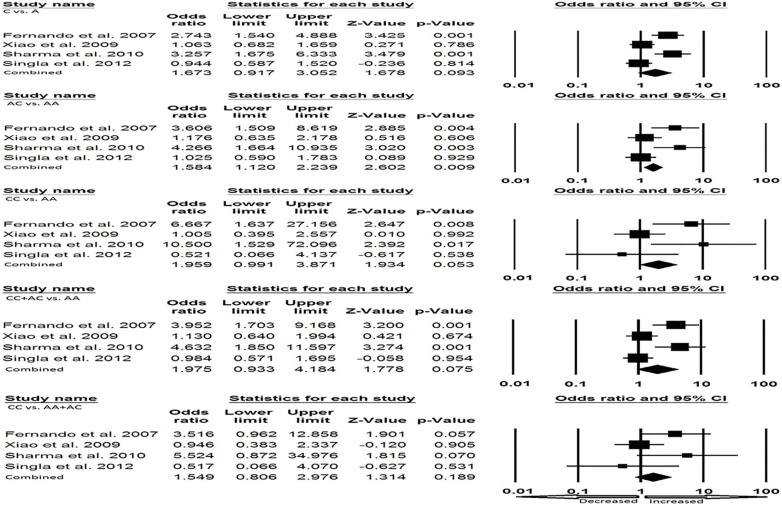
Cautious evaluation of all the data from eight selected studies was done and the data were pooled together which accumulated to 2237 controls and 594 EPTB cases. The random effects model was used to evaluate the overall association between the P2X7 1513 A>C polymorphism and the risk of EPTB (Fig. 3a and Fig. 3b). Overall, four genetic models, allele (C vs. A: p = 0.011; OR = 1.677, 95% CI = 1.125-2.501), homozygous (CC vs. AA: p = 0.053; OR= 2.362, 95% CI = 0.991-5.632), heterozygous (AC vs. AA: p = 0.003; OR = 1.775, 95% CI = 1.209–2.607), dominant (CC + AC vs. AA: p = 0.005; OR= 1.890, 95% CI = 1.207–2.962) were found to be significantly associated with an increased risk of EPTB as compared to wild type genotype. Whereas, recessive (CC vs. AA+ AC: p = 0.099; OR= 1.814, 95% CI = 0.894–3.681) genetic model did not show any risk of EPTB.
Fig. (3a).
a). Forest plot for overall analysis (C vs. A, AC vs. AA, CC vs. AA) showing OR with 95% CI to evaluate the association of the P2X7 A1513C (rs 3751143) gene polymorphism and EPTB risk.
Fig. (3b).
Forest plot for overall analysis (CC + AC vs. AA, CC vs. AA + AC) showing OR with 95% CI to evaluate the association of the P2X7 A1513C (rs 3751143) gene polymorphism and EPTB risk.
Sensitivity Analysis
To estimate the effect of each individual study on the pooled OR, one way sensitivity analysis was done where one single study was deleted serially each time. Omitting one
study at a time in the meta-analysis did not influence the pooled ORs that represents the robustness and credibility of the results obtained in the present meta-analysis (data not shown).
Subgroup Analysis
In this present pooled analysis, we have stratified the studies of P2X7 1513 A>C and EPTB risk for the subgroup analysis based upon the ethnicity to detect impact of this association among Asians and Caucasians. Four studies from Asian and five studies from Caucasian populations were included in this study.
Asian Population
In some of the genetic models, heterogeneity and publication bias were found in Asian ethnicity, so based on heterogeneity, random and fixed effect model were employed (Fig. 4). We found that homozygous genotype (CC vs. AA: p=0.053; OR=1.959, 95% CI=0.991-3.871) and heterozygous genotype (AC vs. AA: p = 0.009; OR= 1.584, 95% CI= 1.120-2.239) found significant increased risk of EPTB. Whereas, allele (C vs. A: p = 0.093; OR= 1.673, 95% CI= 0.917-3.052), dominant (CC + AC vs. AA: p = 0.075; OR= 1.975, 95% CI = 0.933–4.184) and recessive (CC vs. AA+ AC: p = 0.189; OR= 1.549, 95% CI = 0.806–2.976) genetic models did not show any risk of developing EPTB (Table 4; Fig. 4).
Fig. (4).
Forest plot for Asian population showing OR with 95% CI to evaluate the association of the P2X7 A1513C (rs 3751143) gene polymorphism and EPTB risk.
Table 4. Statistics to test publication bias and heterogeneity of asian population.
| Comparisons | Egger’s Regression Analysis | Heterogeneity Analysis | Model Used for the Meta-analysis | ||||
|---|---|---|---|---|---|---|---|
| Intercept | 95% Confidence Interval | p-value | Q-value | Pheterogeneity | I2 (%) | ||
| C vs A | 12.02 | -1.02 to 25.06 | 0.050 | 15.32 | 0.002 | 80.42 | Random |
| CC vs AA | 1.89 | -11.59 to 15.38 | 0.600 | 9.37 | 0.025 | 67.98 | Fixed |
| AC vs AA | 7.59 | 5.44 to 9.73 | 0.001 | 10.94 | 0.012 | 72.59 | Fixed |
| CC+AC vs AA | 8.46 | 6.52 to 10.39 | 0.002 | 14.05 | 0.003 | 78.69 | Random |
| CC vs AA+AC | 1.17 | -9.52 to 11.86 | 0.680 | 5.58 | 0.130 | 46.31 | Fixed |
Caucasian Population
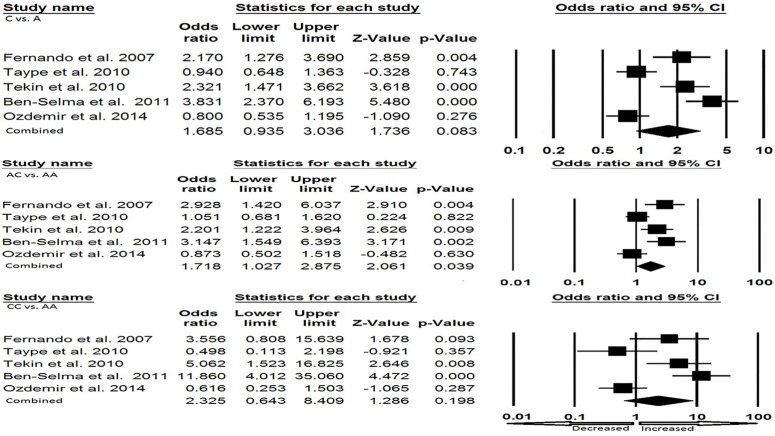
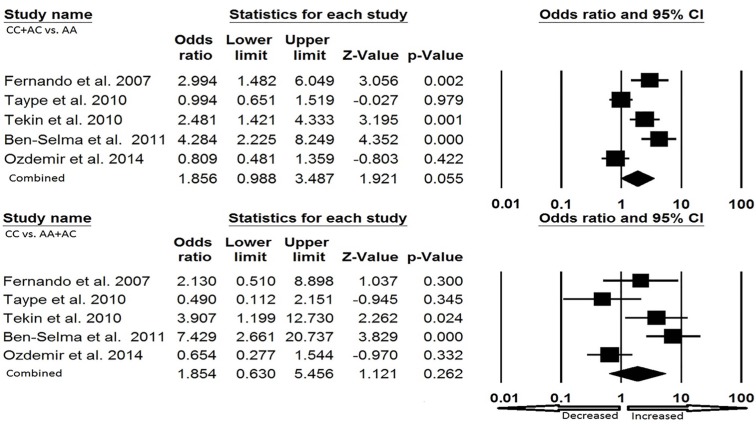
In Caucasian ethnicity, publication bias did not exist. We found heterogeneity in all the five genetic models, so random effect model was applied (Fig. 5a and Fig. 5b). Heterozygous genotype (AC vs. AA: p = 0.039; OR = 1.718, 95% CI = 1.027-2.875) and dominant (CC + AC vs. AA: p = 0.055; OR = 1.856, 95% CI = 0.988–3.487) showed increased risk of EPTB as compared to the wild type genotype. Other genotypic models, allele (C vs. A: p = 0.083; OR= 1.685, 95% CI = 0.935-3.036), homozygous (CC vs. AA: p = 0.198; OR= 2.325, 95% CI = 0.643–8.409) and recessive (CC vs. AA+ AC: p = 0.262; OR= 1.854, 95% CI = 0.630-5.456) failed to show any risk of developing EPTB (Table 5; Fig. 5a and Fig. 5b).
Fig. (5a).
a). Forest plot for Caucasian population (C vs. A, AC vs. AA, CC vs. AA) showing OR with 95% CI to evaluate the association of the P2X7 A1513C (rs 3751143) gene polymorphism and EPTB risk.
Fig. (5b).
Forest plot for Caucasian population (CC + AC vs. AA, CC vs. AA + AC) showing OR with 95% CI to evaluate the association of the P2X7 A1513C (rs 3751143) gene polymorphism and EPTB risk.
Table 5. Statistics to test publication bias and heterogeneity of caucasian population.
| Comparisons | Egger’s Regression Analysis | Heterogeneity Analysis | Model Used for the Meta-analysis | ||||
|---|---|---|---|---|---|---|---|
| Intercept | 95% Confidence Interval | p-value | Q value | Pheterogeneity | I2 (%) | ||
| C vs A | 17.23 | -5.29 to 39.76 | 0.090 | 35.49 | 0.001 | 88.73 | Random |
| CC vs AA | 2.00 | -17.96 to 21.99 | 0.770 | 23.20 | 0.001 | 82.76 | Random |
| AC vs AA | 7.60 | -1.73 to 16.95 | 0.080 | 15.37 | 0.004 | 73.99 | Random |
| CC+AC vs AA | 10.33 | -2.76 to 23.42 | 0.080 | 25.58 | 0.001 | 84.36 | Random |
| CC vs AA+AC | 0.74 | -16.01 to 17.50 | 0.890 | 17.34 | 0.002 | 76.93 | Random |
DISCUSSION
The presence of EPTB is generally determined by a combination of environmental and genetic factors. The precise mechanism conferring resistance against M. tb infection is still obscure, even though both innate and acquired immunity are known to play their part in eliminating this infection. However, involvement of various genes in M. tb susceptibility has been evident from the literature [34]. We also know that macrophages are the main cells allowing mycobacterial replication intracellularly, present the antigen at the site of infection during lymphocyte activation and are liable to eliminate the internalized bacilli [35]. The ligand-gated cation channel of the P2X7 receptor is highly expressed on macrophages in both humans and mice which allows the phenomenon of Ca2+ influx and K+ efflux involved in apoptosis initiation and phospholipase D activation that promotes the fusion of phagosome and lysosome and thus leads to death of the mycobacterium [36, 37].
Earlier studies revealed that P2X7 deficient animals have reduction in the incidence and severity of TB compared with wild-type animals [38]. M. tb infected P2X7 mice have also shown greater M. tb burden in their lung tissues as compared to the infected wild-type mice [39]. Keeping aforesaid facts about the key role of P2X7 receptor in view, a number of studies have been conducted to evaluate the relationship between P2X7 1513 A>C gene polymorphism and the development of EPTB. The results from these published studies are contradictory and, therefore, inconclusive which prompted us to do the meta-analysis to improve the statistical power of this association. This meta-analysis to assess the association between P2X7 1513 A>C polymorphism and EPTB risk was done from eight case-control studies, as combining of the data from different studies has an advantage of reducing the random error [40].
In this pooled study, we found that out of five types of genetic model comparisons, four genetic models of P2X7 1513 A>C polymorphism showed association with increased risk of EPTB (when compared to wild type alleles and genotypes). Furthermore, when stratified the different ethnicities (Asian and Caucasian), a significant association of homozygous variant and heterozygous genotype was found with EPTB in Asian population. In Caucasian stratification, heterozygous and dominant models showed significant increased risk of EPTB when compared with wild type genotype.
The homozygous subjects of P2X7 1513 A>C polymorphism found on exon 13 of P2X7 gene showed reduced ATP
induced apoptosis and ATP mediated killing of mycobacteria, whereas heterozygous subjects showed significantly impaired ATP responsiveness [41, 42]. Previous meta-analysis conducted by Wu et al. [43] also reported significant association of P2X7 1513 A>C polymorphism with EPTB risk, but they missed some of the pertinent studies due to limited database selection, as the research article existing in one database is not essentially available in another one. So, every meta-analysis has a major limitation of database selection and, additionally, some relevant studies might publish in other languages. Therefore, conducting this meta-analysis was necessary to derive a more robust conclusion. Comparatively, this study has major improvement(s) over previous one as in this meta-analysis we searched most famous and reliable databases i.e., Pubmed and EMBASE, and tried to increase the number of individuals as well as incorporate new reports in addition to the missing studies which were not taken into consideration in the previous meta-analysis.
As we know that the nature of genetic susceptibility to TB is polygenic [43]; hence, single genetic variants cannot be interpreted as adequate for the risk of this dreaded disease. The important aspect of this gene polymorphism is that its occurrence can vary sufficiently among different races or ethnic populations. Prior to reaching a conclusion, limitations of the present meta-analysis must be discussed. Firstly, we found heterogeneity in this study, which might be attributed to one or multiple reasons from the following, (a) the ethnicity of the patients, (b) recruiting control samples - ethnicity specific genetic variations may influence host immunity to TB, causing different TB susceptibilities [44]. Second, studies published only in English language were included. The third limitation is database selection, the studies indexed by the selected electronic databases (PubMed, EMBASE) were included for this analysis. There is a possibility that some relevant articles might have been published in other languages and indexed with other databases (not known to us) which we may have missed. The fourth limitation is that due to lack of sufficient information in the primary reports, the data was not stratified by other factors such as HIV status and TB severity.
Despite these limitations, there are some advantages as well associated with this meta-analysis. First, this meta-analysis incorporated more studies than the earlier published researches making this to be more balanced and robust. Second, the subgroup analysis was also done and found that P2X7 1513 A>C polymorphism have more likelihood to develop EPTB in Asian and Caucasian population.
CONCLUSION
In conclusion, the present meta-analysis shows significant association of P2X7 1513 A>C polymorphism with genetic susceptibility to overall EPTB risk. Separate subgroup analysis of both Asian and Caucasian populations also showed association of P2X7 1513 A>C polymorphism with risk of developing EPTB. However, future large well-designed studies which include more EPTB cases and healthy controls might be helpful to provide more precise conclusion and further validation of our results. Also, larger studies will help in understanding the role of P2X7 1513 A>C polymorphism in the pathophysiology of EPTB.
ACKNOWLEDGEMENTS
The authors are grateful to the Deanship of Scientific Research, Jazan University, Jazan, Saudi Arabia for providing the necessary dry-lab facility for the present study.
CONFLICT OF INTEREST
The authors confirm that this article content has no conflict of interest.
REFERENCES
- 1.Zumla A., George A., Sharma V., Herbert N., Baroness Masham of Ilton WHO’s 2013 global report on tuberculosis: successes, threats, and opportunities. Lancet. 2013;382(9907):1765–1767. doi: 10.1016/S0140-6736(13)62078-4. [DOI] [PubMed] [Google Scholar]
- 2.Horsburgh C.R., Jr Priorities for the treatment of latent tuberculosis infection in the United States. N. Engl. J. Med. 2004;350(20):2060–2067. doi: 10.1056/NEJMsa031667. [DOI] [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention (CDC) Decrease in reported tuberculosis cases - United States, 2009. MMWR Morb. Mortal. Wkly. Rep. 2010;59(10):289–294. [PubMed] [Google Scholar]
- 4.Mitsos L.M., Cardon L.R., Fortin A., Ryan L., LaCourse R., North R.J., Gros P. Genetic control of susceptibility to infection with Mycobacterium tuberculosis in mice. Genes Immun. 2000;1(8):467–477. doi: 10.1038/sj.gene.6363712. [DOI] [PubMed] [Google Scholar]
- 5.Kramnik I., Dietrich W.F., Demant P., Bloom B.R. Genetic control of resistance to experimental infection with virulent Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA. 2000;97(15):8560–8565. doi: 10.1073/pnas.150227197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Areeshi M.Y., Mandal R.K., Panda A.K., Bisht S.C., Haque S. CD14 -159 C>T gene polymorphism with increased risk of tuberculosis: evidence from a meta-analysis. PLoS One. 2013;8(5):e64747. doi: 10.1371/journal.pone.0064747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Areeshi M.Y., Mandal R.K., Panda A.K., Haque S. A meta-analysis of the association between the CC chemokine ligand 5 (CCL5) -403 G>A gene polymorphism and tuberculosis susceptibility. PLoS One. 2013;8(8):e72139. doi: 10.1371/journal.pone.0072139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mahasirimongkol S., Yanai H., Nishida N., Ridruechai C., Matsushita I., Ohashi J., Summanapan S., Yamada N., Moolphate S., Chuchotaworn C., Chaiprasert A., Manosuthi W., Kantipong P., Kanitwittaya S., Sura T., Khusmith S., Tokunaga K., Sawanpanyalert P., Keicho N. Genome-wide SNP-based linkage analysis of tuberculosis in Thais. Genes Immun. 2009;10(1):77–83. doi: 10.1038/gene.2008.81. [DOI] [PubMed] [Google Scholar]
- 9.Sikora A., Liu J., Brosnan C., Buell G., Chessel I., Bloom B.R. Cutting edge: purinergic signaling regulates radical-mediated bacterial killing mechanisms in macrophages through a P2X7-independent mechanism. J. Immunol. 1999;163(2):558–561. [PubMed] [Google Scholar]
- 10.Buell G.N., Talabot F., Gos A., Lorenz J., Lai E., Morris M.A., Antonarakis S.E. Gene structure and chromosomal localization of the human P2X7 receptor. Receptors Channels. 1998;5(6):347–354. [PubMed] [Google Scholar]
- 11.Bulanova E., Budagian V., Orinska Z., Koch-Nolte F., Haag F., Bulfone-Paus S. ATP induces P2X7 receptor-independent cytokine and chemokine expression through P2X1 and P2X3 receptors in murine mast cells. J. Leukoc. Biol. 2009;85(4):692–702. doi: 10.1189/jlb.0808470. [DOI] [PubMed] [Google Scholar]
- 12.Kusner D.J., Barton J.A. ATP stimulates human macrophages to kill intracellular virulent Mycobacterium tuberculosis via calcium-dependent phagosome-lysosome fusion. J. Immunol. 2001;167(6):3308–3315. doi: 10.4049/jimmunol.167.6.3308. [DOI] [PubMed] [Google Scholar]
- 13.Gargett C.E., Cornish E.J., Wiley J.S. Phospholipase D activation by P2Z-purinoceptor agonists in human lymphocytes is dependent on bivalent cation influx. Biochem. J. 1996;313(Pt 2):529–535. doi: 10.1042/bj3130529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Canaday D.H., Beigi R., Silver R.F., Harding C.V., Boom W.H., Dubyak G.R. ATP and control of intracellular growth of mycobacteria by T cells. Infect. Immun. 2002;70(11):6456–6459. doi: 10.1128/IAI.70.11.6456-6459.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gu B.J., Zhang W., Worthington R.A., Sluyter R., Dao-Ung P., Petrou S., Barden J.A., Wiley J.S. A Glu-496 to Ala polymorphism leads to loss of function of the human P2X7 receptor. J. Biol. Chem. 2001;276(14):11135–11142. doi: 10.1074/jbc.M010353200. [DOI] [PubMed] [Google Scholar]
- 16.Sluyter R., Shemon A.N., Wiley J.S. Glu496 to Ala polymorphism in the P2X7 receptor impairs ATP-induced IL-1 beta release from human monocytes. J. Immunol. 2004;172(6):3399–3405. doi: 10.4049/jimmunol.172.6.3399. [DOI] [PubMed] [Google Scholar]
- 17.Yang Z., Kong Y., Wilson F., Foxman B., Fowler A.H., Marrs C.F., Cave M.D., Bates J.H. Identification of risk factors for extrapulmonary tuberculosis. Clin. Infect. Dis. 2004;38(2):199–205. doi: 10.1086/380644. [DOI] [PubMed] [Google Scholar]
- 18.Fernando S.L., Saunders B.M., Sluyter R., Skarratt K.K., Goldberg H., Marks G.B., Wiley J.S., Britton W.J. A polymorphism in the P2X7 gene increases susceptibility to extrapulmonary tuberculosis. Am. J. Respir. Crit. Care Med. 2007;175(4):360–366. doi: 10.1164/rccm.200607-970OC. [DOI] [PubMed] [Google Scholar]
- 19.Xiao J., Sun L., Jiao W., Li Z., Zhao S., Li H., Jin J., Jiao A., Guo Y., Jiang Z., Mokrousov I., Shen A. Lack of association between polymorphisms in the P2X7 gene and tuberculosis in a Chinese Han population. FEMS Immunol. Med. Microbiol. 2009;55(1):107–111. doi: 10.1111/j.1574-695X.2008.00508.x. [DOI] [PubMed] [Google Scholar]
- 20.Taype C.A., Shamsuzzaman S., Accinelli R.A., Espinoza J.R., Shaw M.A. Genetic susceptibility to different clinical forms of tuberculosis in the Peruvian population. Infect. Genet. Evol. 2010;10(4):495–504. doi: 10.1016/j.meegid.2010.02.011. [DOI] [PubMed] [Google Scholar]
- 21.Tekin D., Kayaalti Z., Dalgic N., Cakir E., Soylemezoglu T., Isin Kutlubay B., Aydin Kilic B. Polymorphism in the p2x7 gene increases susceptibility to extrapulmonary tuberculosis in Turkish children. Pediatr. Infect. Dis. J. 2010;29(8):779–782. doi: 10.1097/INF.0b013e3181d9932e. [DOI] [PubMed] [Google Scholar]
- 22.Sharma S., Kumar V., Khosla R., Kajal N., Sarin B., Sehajpal P. Association of P2X7 receptor +1513 (A-->C) polymorphism with tuberculosis in a Punjabi population. Int. J. Tuberc. Lung Dis. 2010;14(9):1159–1163. [PubMed] [Google Scholar]
- 23.Ben-Selma W., Ben-Kahla I., Boukadida J., Harizi H. Contribution of the P2X7 1513A/C loss-of-function polymorphism to extrapulmonary tuberculosis susceptibility in Tunisian populations. FEMS Immunol. Med. Microbiol. 2011;63(1):65–72. doi: 10.1111/j.1574-695X.2011.00824.x. [DOI] [PubMed] [Google Scholar]
- 24.Singla N., Gupta D., Joshi A., Batra N., Singh J. Genetic polymorphisms in the P2X7 gene and its association with susceptibility to tuberculosis. Int. J. Tuberc. Lung Dis. 2012;16(2):224–229. doi: 10.5588/ijtld.11.0076. [DOI] [PubMed] [Google Scholar]
- 25.Ozdemir F.A., Erol D., Konar V., Yüce H., Kara Şenli E., Bulut F., Deveci F. Lack of association of 1513 A/C polymorphism in P2X7 gene with susceptibility to pulmonary and extrapulmonary tuberculosis. Tuberk. Toraks. 2014;62(1):7–11. doi: 10.5578/tt.4740. [DOI] [PubMed] [Google Scholar]
- 26.Cohn L.D., Becker B.J. How meta-analysis increases statistical power. Psychol. Methods. 2003;8(3):243–253. doi: 10.1037/1082-989X.8.3.243. [DOI] [PubMed] [Google Scholar]
- 27.Mandal R.K., Yadav S.S., Panda A.K. Meta-analysis on the association of nucleotide excision repair gene XPD A751C variant and cancer susceptibility among Indian population. Mol. Biol. Rep. 2014;41(2):713–719. doi: 10.1007/s11033-013-2910-y. [DOI] [PubMed] [Google Scholar]
- 28.Wu R., Li B. A multiplicative-epistatic model for analyzing interspecific differences in outcrossing species. Biometrics. 1999;55(2):355–365. doi: 10.1111/j.0006-341x.1999.00355.x. [DOI] [PubMed] [Google Scholar]
- 29.Mantel N., Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J. Natl. Cancer Inst. 1959;22(4):719–748. [PubMed] [Google Scholar]
- 30.DerSimonian R., Laird N. Meta-analysis in clinical trials. Control. Clin. Trials. 1986;7(3):177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 31.Higgins J.P., Thompson S.G., Deeks J.J., Altman D.G. Measuring inconsistency in meta-analyses. BMJ. 2003;327(7414):557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Egger M., Davey Smith G., Schneider M., Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315(7109):629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Duval S., Tweedie R. Trim and fill: A simple funnel-plot-based method of testing and adjusting for publication bias in meta-analysis. Biometrics. 2000;56(2):455–463. doi: 10.1111/j.0006-341x.2000.00455.x. [DOI] [PubMed] [Google Scholar]
- 34.Kramnik I., Dietrich W.F., Demant P., Bloom B.R. Genetic control of resistance to experimental infection with virulent Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA. 2000;97(15):8560–8565. doi: 10.1073/pnas.150227197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schaible U.E., Collins H.L., Kaufmann S.H. Confrontation between intracellular bacteria and the immune system. Adv. Immunol. 1999;71:267–377. doi: 10.1016/s0065-2776(08)60405-8. [DOI] [PubMed] [Google Scholar]
- 36.Coutinho-Silva R., Stahl L., Raymond M.N., Jungas T., Verbeke P., Burnstock G., Darville T., Ojcius D.M. Inhibition of chlamydial infectious activity due to P2X7R-dependent phospholipase D activation. Immunity. 2003;19(3):403–412. doi: 10.1016/s1074-7613(03)00235-8. [DOI] [PubMed] [Google Scholar]
- 37.Kusner D.J., Barton J.A. ATP stimulates human macrophages to kill intracellular virulent Mycobacterium tuberculosis via calcium-dependent phagosome-lysosome fusion. J. Immunol. 2001;167(6):3308–3315. doi: 10.4049/jimmunol.167.6.3308. [DOI] [PubMed] [Google Scholar]
- 38.Labasi J.M., Petrushova N., Donovan C., McCurdy S., Lira P., Payette M.M., Brissette W., Wicks J.R., Audoly L., Gabel C.A. Absence of the P2X7 receptor alters leukocyte function and attenuates an inflammatory response. J. Immunol. 2002;168(12):6436–6445. doi: 10.4049/jimmunol.168.12.6436. [DOI] [PubMed] [Google Scholar]
- 39.Santos A.A., Jr, Rodrigues-Junior V., Zanin R.F., Borges T.J., Bonorino C., Coutinho-Silva R., Takyia C.M., Santos D.S., Campos M.M., Morrone F.B. Implication of purinergic P2X7 receptor in M. tuberculosis infection and host interaction mechanisms: a mouse model study. Immunobiology. 2013;218(8):1104–1112. doi: 10.1016/j.imbio.2013.03.003. [DOI] [PubMed] [Google Scholar]
- 40.Ioannidis J.P., Boffetta P., Little J., O’Brien T.R., Uitterlinden A.G., Vineis P., Balding D.J., Chokkalingam A., Dolan S.M., Flanders W.D., Higgins J.P., McCarthy M.I., McDermott D.H., Page G.P., Rebbeck T.R., Seminara D., Khoury M.J. Assessment of cumulative evidence on genetic associations: interim guidelines. Int. J. Epidemiol. 2008;37(1):120–132. doi: 10.1093/ije/dym159. [DOI] [PubMed] [Google Scholar]
- 41.Fernando S.L., Saunders B.M., Sluyter R., Skarratt K.K., Wiley J.S., Britton W.J. Gene dosage determines the negative effects of polymorphic alleles of the P2X7 receptor on adenosine triphosphate-mediated killing of mycobacteria by human macrophages. J. Infect. Dis. 2005;192(1):149–155. doi: 10.1086/430622. [DOI] [PubMed] [Google Scholar]
- 42.Saunders B.M., Fernando S.L., Sluyter R., Britton W.J., Wiley J.S. A loss-of-function polymorphism in the human P2X7 receptor abolishes ATP-mediated killing of mycobacteria. J. Immunol. 2003;171(10):5442–5446. doi: 10.4049/jimmunol.171.10.5442. [DOI] [PubMed] [Google Scholar]
- 43.Wu G., Zhao M., Gu X., Yao Y., Liu H., Song Y. The effect of P2X7 receptor 1513 polymorphism on susceptibility to tuberculosis: A meta-analysis. Infect. Genet. Evol. 2014;24:82–91. doi: 10.1016/j.meegid.2014.03.006. [DOI] [PubMed] [Google Scholar]
- 44.Möller M., Hoal E.G. Current findings, challenges and novel approaches in human genetic susceptibility to tuberculosis. Tuberculosis (Edinb.) 2010;90(2):71–83. doi: 10.1016/j.tube.2010.02.002. [DOI] [PubMed] [Google Scholar]