Abstract
Promising drugs to treat Ebola virus (EBOV) infection are currently being developed, but so far none has shown efficacy in clinical trials. Drugs that can stimulate host innate defense responses may retard the progression of EBOV disease. We report here the dramatic effect of hemin, the natural inducer of the heme catabolic enzyme heme oxygenase-1 (HO-1), in the reduction of EBOV replication. Treatment of primary monocyte-derived macrophages (MDM), Vero E6 cells, HeLa cells, and human foreskin fibroblasts (HFF1) with hemin reduced EBOV infection by >90%, and showed minimal toxicity to infected cells. Inhibition of HO-1 enzymatic activity and silencing HO-1 expression prevented the hemin-mediated suppression of EBOV infection, suggesting an important role for induction of this intracellular mediator in restricting EBOV replication. The inverse correlation between hemin-induced HO-1 and EBOV replication provides a potentially useful therapeutic modality based on the stimulation of an innate cellular response against Ebola infection.
Keywords: Ebola, monocytes/macrophages, hemin, heme oxygenase-1
INTRODUCTION
Ebola virus (EBOV) is an enveloped, pleomorphic, negative-sense RNA virus that belongs to the Filoviridae family [1, 2]. The recent 2013–2015 EBOV epidemic in West Africa caused >20,000 infections and >9,000 deaths (as of January 2016). No licensed vaccines or drugs are currently available to treat EBOV infection [3–5]. Therefore, development of medical countermeasures such as antiviral drugs and other preventive or therapeutic strategies are of the highest priority.
EBOV is a highly pathogenic virus that induces direct and indirect cell death, replicates very rapidly, and has a short incubation period, and a significant number of infected individuals die within a few days after the onset of symptoms [1]. Because EBOV blocks innate and adaptive immune functions [6–9], we hypothesized that activation of a safe and effective host innate response, with an understanding of its mechanisms, is a logical approach to treat EBOV patients. We and others have previously identified roles for heme oxygenase-1 (HO-1), an endogenous cellular protective enzyme, which upon stimulation promotes host resistance against multiple pathogens [10–21]. Blocking EBOV replication early in infection by inducing HO-1 could allow patients time to develop adaptive immunity.
In the present study, we show that treatment of cells with hemin, which activates HO-1, markedly reduces EBOV replication. Our study defines specific cellular interfering mechanistic pathways that control EBOV replication that may facilitate the development of potentially novel therapeutic interventions to retard pathogenesis by stimulating the cellular innate response against Ebola virus infection.
MATERIALS AND METHODS
The FDA-approved drug Panhematin®, containing hemin as the active component, was purchased from Lundbeck, Deerfield, IL (manufactured by APP Pharmaceuticals, Raleigh, NC). Monocytes were isolated from peripheral blood mononuclear cells of donors seronegative for HIV-1 and hepatitis B after leukophoresis and purification by countercurrent centrifugal elutriation [22]. Monocytes were cultured 5 days in Dulbecco’s Modified Eagle Medium (DMEM) with 10% fetal bovine serum (FBS), 20 µg/ml gentamicin, and 1000 U/ml macrophage-colony stimulating factor (M-CSF). After 5 days, monocyte-derived macrophages (MDM) were washed three times with PBS and transferred to DMEM containing 10% FBS and 20 µg/ml gentamicin. HeLa human adenocarcinoma cells (ATCC, CCL-2) were cultured in Minimal Essential Medium (MEM) supplemented with 10% FBS, 1% L-glutamine, 10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), 1% non-essential amino acids, and 1% penicillin/streptomycin. HFF1 human foreskin fibroblast (ATCC, SCRC-1041) cells were cultured in MEM supplemented with 10% FBS and 0.5 mM sodium pyruvate. Cells were cultured for 3 days passaging or for plating in assay plates 24 hours before viral infection.
MDM were infected with a replication-competent recombinant vesicular stomatitis virus (VSV), in which the VSV-G envelope gene was replaced by the EBOV glycoprotein (GP), followed by the enhanced green fluorescence protein (GFP) gene (rVSV-EBOVgp-GFP) (Konduru et al. 2016, submitted for publication) at different multiplicities of infection (MOI). It should be pointed out that GFP is not incorporated into the rVSV-EBOVgp-GFP particles but it is expressed in infected cells and can be detected by fluorescence microscopy 4–8 hours after infection. After 2 hours of infection, virus was removed and fresh medium added with or without hemin. Cells were examined using a Keyence fluorescence microscope and images were acquired using the BIOREVO BZ-9000 application system. Ebola virus infection was scored by counting the number of GFP-positive cells per microscopic field with assays performed in triplicate.
Uninfected and rVSV-ZEBOVgp-GFP-infected MDM cultured 48 hours without or with 100 µM hemin were washed twice with cold Hanks balanced salt solution (HBSS), gently detached from the dish with a cell scraper, and fixed in 4% paraformaldehyde (PFA) for 30 minutes on ice. Cells were then washed with cold HBSS containing 10 mM glycine, stained with 4',6-diamidino-2-phenylindole (DAPI), and examined by image flow cytometry (ImageStream X Mark II Imaging Flow Cytometer, Amnis Corporation, Seattle, WA). The GFP/DAPI-positive population was gated and analyzed to quantify rVSV-ZEBOVgp-GFP-infected cells.
Small interfering RNA (siRNA) targeting human HO-1-coding sequences (Hs_HMOX1_1 and Hs_HMOX_10) and AllStars Negative Control siRNA (Qiagen Inc.) were used in expression knockdown experiments. Vero E6 cell monolayers in 6-well culture plates were transfected with 50 nM control or HO-1 siRNA for 6 hours in serum-free OPTIMEM media, using Lipofectamine® RNAiMAX (Invitrogen, Carlsbad, CA). Cells were then treated with 100 µM hemin 24 h prior to infection with rVSV-ZEBOVgp-GFP at MOI 0.1. Efficiency of HO-1 knockdown was assessed by Western blot analysis using specific HO-1 monoclonal antibody (Enzo) and rabbit anti-actin polyclonal antibody as negative control. Western blots were performed by Kendrick Labs, Inc. (Madison, WI). Virus infection was determined by high content imaging microscopy.
For high content imaging assay HeLa and HFF-1 cells were seeded in 384-well plates 24 hours before treatment. Treatment with hemin solution was done 2 hours prior to infection and performed in quadruplicate of 10 doses starting from 100 µM final concentration and diluted down with 2 fold steps to 0.195 µM to generate dose response for better assessment of potency. Experiment was repeated on two independent days. HeLa and HFF1 cells were infected with an authentic Kikwit-95 Zaire strain of Ebola virus at MOI 0.5 and 2.5, respectively. Infection was stopped after 48 hours by fixing cells in 4% paraformaldehyde (PFA) solution. Immunostaining was performed using murine anti-GP antibody, followed by Alexa488-conjugated anti-mouse IgM as secondary antibody. Images were acquired using the PE Opera confocal platform with 10× air objective and analysis by Acapella software. The signal for GP-staining was quantified and expressed as percent inhibition. Nuclear stains DRAQ5 (Thermo Fisher) (for HeLa cells) and 33342Hoechst (Life Technologies) (for HFF1) were used to determine total cell number and expressed as percent cellular viability. CellMask staining of cytoplasm was performed in assay with HFF1 cells for better image analysis. Data acquired form Opera/Acapella were analysed and normalized on the plate level using GeneData Screener software to percent inhibition of infection and percent of cellular viability. Analysis of the dose response curve in order to determine EC50 was performed using GeneData Condoseo software by applying the Levenberg-Marquardt algorithm (LMA) for curve fitting strategy. The fitting strategy is considered acceptable if it produced a conversion with R2 > 0.8.
RESULTS AND DISCUSSION
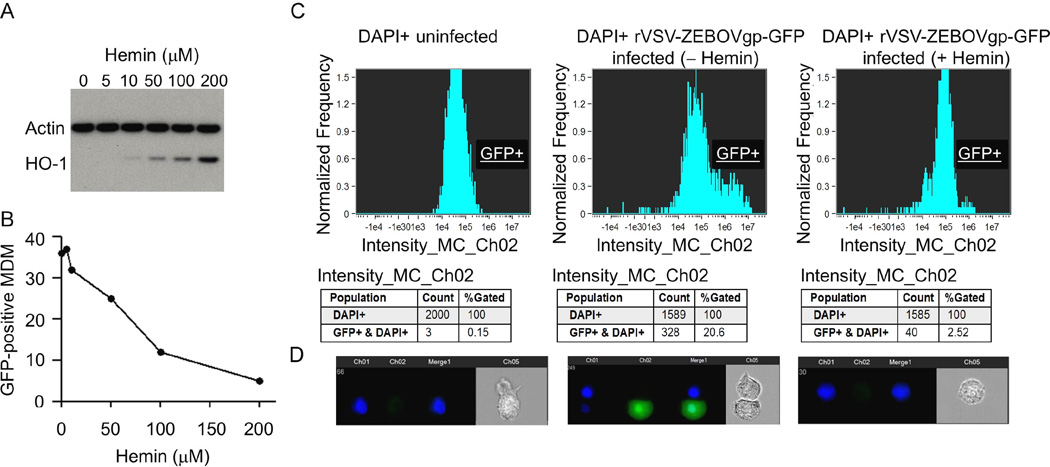
In our present work, we used an FDA-approved pharmaceutical formulation of hemin as an inducer for endogenous HO-1. First, we determined the effect of hemin on HO-1 expression and then examined effects on Ebola virus infection. We incubated monocyte-derived macrophages (MDM) in media containing increasing concentrations of hemin and examined for HO-1 expression by Western blot analysis after 48 hours in culture. As expected from its mode of action as a physiological activator of HO-1, treatment of MDM with hemin resulted in Hemin inhibits Ebola virus infection 121 increased HO-1 protein expression in a dose-dependent manner with a maximal effect at 100–200 µM concentration without altering the expression of the housekeeping protein actin (Fig. 1A). This HO-1 induction was accompanied by a >90% inhibition of the GFP fluorescence in MDM cells infected with rVSV-ZEBOVgp-GFP (Fig. 1B).
Fig. 1.
Hemin treatment inhibits EBOV infection of monocyte-derived macrophages (MDM). (A) Western blot analysis of hemin-treated MDM. Cells were incubated with the indicated concentrations of hemin, cellular proteins were separated on a sodium dodecyl sulfate (SDS)-polyacrylamide gel, transferred to polyvinylidene difluoride (PVDF) nitrocellulose membrane, and probed simultaneously with HO-1 and actin antibodies. (B) Data demonstrating reduction of rVSV-EBOVgp-GFP replication in MDM treated with increased concentrations of hemin, expressed as the proportion of cells positive for GFP. (C) Quantitative analysis of rVSV-EBOVgp-GFP-infected MDM in the absence or presence of 100 µM hemin by ImageStream. (D) Representative cell images of DAPI detection and rVSV-EBOVgp-GFP-positive cells. A total of 10,000 cell events were collected from each of the samples for data analysis, and fluorescence intensity was determined by IDEAS v5.0 software (Amnis Corporation, Seattle, WA).
Flow cytometry analysis (Fig. 1C) of uninfected (left) and rVSV-EBOVgp-GFP-infected cells cultured for 48 hours without (middle) or with 100 µM hemin (right panel) also showed a significant hemin-dependent inhibition of GFP in rVSV-EBOVgp-GFP-infected cells. The DAPI+ cell population was gated and quantified for GFP expression as a measure of virus replication. Approximately 21% of the cells were infected with rVSV-EBOVgp-GFP (middle panel), which was markedly reduced to ~2.5% by hemin treatment (right panel). Individual cell images showed nuclei (DAPI+, blue) in uninfected cells, GFP (green) co-localized with DAPI after VSV-EBOVgp-GFP infection, and substantially reduced GFP fluorescence after hemin treatment (Fig. 1D).
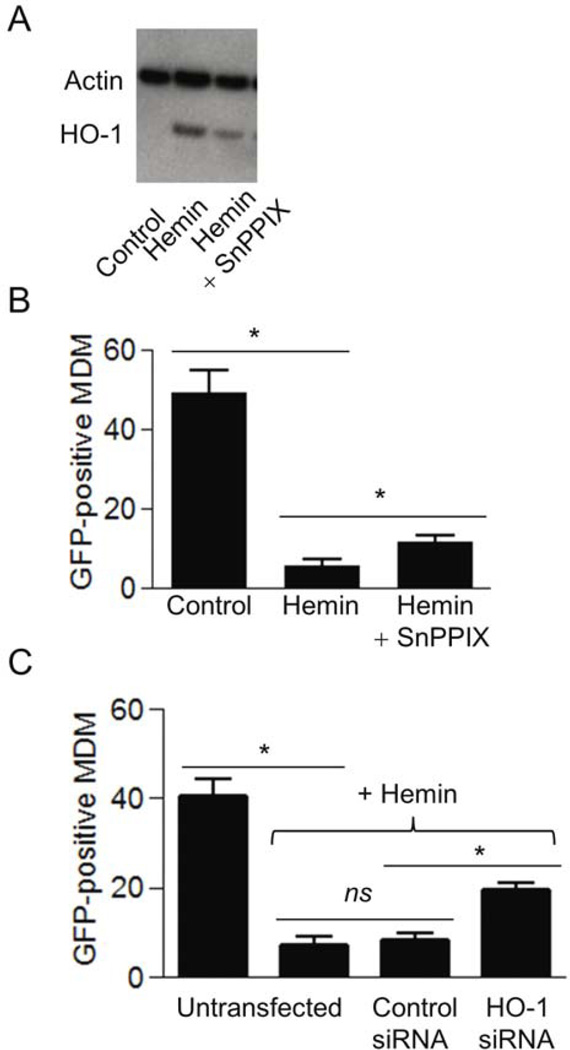
Addition of SnPPIX, a specific inhibitor of HO-1 enzymatic activity, also reduced hemin-induced HO-1 protein expression (Fig. 2A). Pretreatment with 10 µM SnPPIX attenuated the effects of hemin-induced down-regulation of rVSV-ZEBOVgp-GFP replication by two-fold, indicating that HO-1 activity was required for the inhibitory effect of hemin on viral replication (Fig. 2B). Silencing of the HO-1 gene by siRNA transfection had similar effects. This reduction in HO-1 diminished the protective effect of hemin against rVSV-ZEBOVgp-GFP infection (Fig. 2C). These results establish that: (a) HO-1 activity and expression are important for the inhibitory effect on Ebola virus replication, in agreement with a recently published report [18], and (b) reducing HO-1 correlates with increased rVSV-EBOVgp replication, suggesting HO-1 as a key mediator of hemin-induced host protective effects.
Fig. 2.
(A) Hemin-induced HO-1 protein expression in MDM remaining after treatment with the HO-1 enzyme inhibitor SnPPIX. Cells were cultured in the presence of 10 µM SnPPIX and 25 µM hemin for 24 hours and examined for the expression of HO-1 and actin by Western blot analysis using mouse-anti-human HO-1 monoclonal antibody and rabbit-anti-human actin polyclonal antibody. (B) Inhibition of HO-1 activity attenuates hemin-induced protection of MDM against rVSV-EBOVgp-GFP infection. MDM were pre-treated with 10 µM SnPPIX for 2 hours followed by treatment for 24 hours with 25 µM hemin. The cells were then infected with rVSV-EBOVgp-GFP, and after 24 hours, infected cells were quantified by fluorescence microscopy. (C) rVSV-EBOVgp-GFP replication in Vero E6 cells transfected with control siRNA and HO-1 siRNA. Untransfected and transfected cells were infected in the absence or presence of 100 µM hemin for 48 hours, and examined for infectivity by fluorescence microscopy as described in Materials and Methods. Data from two independent experiments are presented as mean ± SEM. *P<0.05; ns: not significant.
We further studied whether hemin induces HO-1 expression and provides cellular protection against EBOV infection. To do so, we infected HeLa and primary human foreskin fibroblasts (HFF1) cells with EBOV under BSL-4 conditions. Consistent with the data described above, treatment of both HeLa and HFF1 cells with 100 µM hemin efficiently induced HO-1 expression (Fig. 3A). Treatment with 100 µM hemin resulted in a substantial reduction of virus-infected cells, as evident by confocal microscopy images of these cells infected with EBOV (Fig. 3B).
Fig. 3.
(A) Western blot analysis of hemin-treated HeLa and HFF1 cells. HO-1 expression in total-protein lysates isolated from HeLa cells and HFF1 cells cultured for 24 hours in the absence or presence of 100 µM hemin. Cells were incubated with 100 µM hemin, cellular proteins were separated on a SDS-polyacrylamide gel, transferred to PVDF nitrocellulose membrane, and probed simultaneously with HO-1 and actin antibodies. (B) Confocal microscopy of HeLa and HFF1 cells infected with EBOV under BSL-4 conditions in the absence or presence of 100 µM hemin. Images were acquired using the PE Opera confocal platform with a 10× objective. The data were analyzed using Acapella software. Green: Ebola infection; blue: nuclei (DRAQ5 stain); magenta: cytoplasm (HFF1 cells).
To quantify the hemin concentration for maximal virus inhibition, HeLa cells and HFF1 cells were cultured for 2 h at 37 °C with hemin and subsequent infection with EBOV at MOI of 0.5 and 2.5 PFU/cell, respectively. Hemin treatment inhibited EBOV infection in a dose-dependent manner with a maximum effect at 100 µM hemin in both cell types (Fig. 4A and B). These results demonstrate >90% protection of hemin-treated HeLa and HFF1 cells against EBOV infection using levels of hemin within the range approved by the FDA for blood levels, further supporting our concept of cellular protection against EBOV by stimulating innate inducible host responses via HO-1.
Fig. 4.
Dose response to hemin treatment of EBOV-infected HeLa and HFF1 cells. HeLa cells (panel A) and HFF1 cells (panel B) were infected with Kikwit-95 Zaire strain of Ebola virus under BSL-4 conditions for 48 hours in the presence of the indicated concentrations of hemin. It gave activity with EC50 = 17–27 µM in HeLa cells and CC50 > 100 µM and EC50 = 26–30 µM in HFF1 and CC50 > 100 µM, n = 4 done in two independent experiments. Cellular viability of the infected cells at various hemin concentrations are illustrated in panels C and D.
CONCLUSION
There is currently no approved therapeutic or licensed vaccine against EBOV. The use of hemin to activate host innate responses is a logical therapeutic approach to treat EBOV-infected individuals.
Acknowledgments
We thank Dr. Paul Buehler and Dr. Nitin Verma for critical review of the manuscript. The findings and conclusions in this paper have not been formally disseminated by the Food and Drug Administration and should not be construed to represent any agency determination or policy. This work was supported by FDA and NIDCR Intramural Research Programs.
Footnotes
CONFLICT OF INTEREST STATEMENT
The authors declare no conflict of interest.
REFERENCES
- 1.Sanchez A, Geisbert TW, Feldmann H. Filoviridae: Marburg and Ebola Viruses. Philadelphia: Fields Virology Lippincott-Williams & Wilkins; 2007. [Google Scholar]
- 2.Ascenzi P, Bocedi A, Heptonstall J, Capobianchi MR, Caro AD, Mastrangelo E, Bolognesi M, Ippolito G. Mol. Aspects Med. 2008;29:151–185. doi: 10.1016/j.mam.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 3.Zhang Y, Feng S, Cowling BJ. Lancet. 2016;387:509. doi: 10.1016/S0140-6736(16)30127-1. [DOI] [PubMed] [Google Scholar]
- 4.Fang LQ, Yang Y, Jiang JF, Yao HW, Kargbo D, Li XL, Jiang BG, Kargbo B, Tong YG, Wang YW, Liu K, Kamara A, Dafae F, Kanu A, Jiang RR, Sun Y, Sun RX, Chen WJ, Ma MJ, Dean NE, Thomas H, Longini IM, Jr, Halloran ME, Cao WC. Proc. Natl. Acad. Sci. USA. 2016;113:4488–4493. doi: 10.1073/pnas.1518587113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moll R, Reece S, Cosford P, Kessel A. Br. Med. Bull. 2016;117:15–23. doi: 10.1093/bmb/ldw007. [DOI] [PubMed] [Google Scholar]
- 6.Basler CF. Virology. 2015;479–480:122–130. doi: 10.1016/j.virol.2015.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kumar A. Infect. Disord. Drug Targets. 2016;16:79–94. doi: 10.2174/1871526516666160108114644. [DOI] [PubMed] [Google Scholar]
- 8.Messaoudi I, Amarasinghe GK, Basler CF. Nat. Rev. Microbiol. 2015;13:663–676. doi: 10.1038/nrmicro3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoenen T, Groseth A, Feldmann H. Expert Opin. Biol. Ther. 2012;12:859–872. doi: 10.1517/14712598.2012.685152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Devadas K, Dhawan S. J. Immunol. 2006;176:4252–4257. doi: 10.4049/jimmunol.176.7.4252. [DOI] [PubMed] [Google Scholar]
- 11.Pamplona A, Ferreira A, Balla J, Jeney V, Balla G, Epiphanio S, Chora A, Rodrigues CD, Gregoire IP, Cunha-Rodrigues M, Portugal S, Soares MP, Mota MM. Nat. Med. 2007;13:703–710. doi: 10.1038/nm1586. [DOI] [PubMed] [Google Scholar]
- 12.Protzer U, Seyfried S, Quasdorff M, Sass G, Svorcova M, Webb D, Bohne F, Hosel M, Schirmacher P, Tiegs G. Gastroenterology. 2007;133:1156–1165. doi: 10.1053/j.gastro.2007.07.021. [DOI] [PubMed] [Google Scholar]
- 13.Seixas E, Gozzelino R, Chora A, Ferreira A, Silva G, Larsen R, Rebelo S, Penido C, Smith NR, Coutinho A, Soares MP. Proc. Natl. Acad. Sci. USA. 2009;106:15837–15842. doi: 10.1073/pnas.0903419106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hou WH, Rossi L, Shan Y, Zheng JY, Lambrecht RW, Bonkovsky HL. World J. Gastroenterol. 2009;15:4499–4510. doi: 10.3748/wjg.15.4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Devadas K, Hewlett IK, Dhawan S. J. Leukoc. Biol. 2010;87:915–924. doi: 10.1189/jlb.0307172. [DOI] [PubMed] [Google Scholar]
- 16.Schmidt WN, Mathahs MM, Zhu Z. Front. Pharmacol. 2012;3:129. doi: 10.3389/fphar.2012.00129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou ZH, Kumari N, Catalano J, Nekhai S, Wise J, Yamada KM, Dhawan S. Curr. Trends Immunol. 2013;14:53–56. [PMC free article] [PubMed] [Google Scholar]
- 18.Dhawan S, Degrabant A, Yamada KM. Curr. Trends Immunol. 2013;14:65–70. [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou ZH, Kumari N, Nekhai S, Clouse KA, Wahl LM, Yamada KM, Dhawan S. Biochem. Biophys. Res. Commun. 2013;435:373–377. doi: 10.1016/j.bbrc.2013.04.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Siegert SWK, Holt RJ. Adv. Ther. 2008;25:842–857. doi: 10.1007/s12325-008-0094-y. [DOI] [PubMed] [Google Scholar]
- 21.Hill-Batorski L, Halfmann P, Neumann G, Kawaoka Y. J. Virol. 2013;87:13795–13802. doi: 10.1128/JVI.02422-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wahl LM, Katona IM, Wilder RL, Winter CC, Haraoui B, Scher I, Wahl SM. Cell. Immunol. 1984;85:373–383. doi: 10.1016/0008-8749(84)90251-x. [DOI] [PubMed] [Google Scholar]