Abstract
Cannabinoid receptor interacting protein 1a (CRIP1a) is a CB1 receptor (CB1R) distal C-terminal-associated protein that alters CB1R interactions with G-proteins. We tested the hypothesis that CRIP1a is capable of also altering CB1R interactions with β-arrestin proteins that interact with the CB1R at the C-terminus. Coimmunoprecipitation studies indicated that CB1R associates in complexes with either CRIP1a or β-arrestin, but CRIP1a and β-arrestin fail to coimmunoprecipitate with each other. This suggests a competition for CRIP1a and β-arrestin binding to the CB1R, which we hypothesized could attenuate the action of β-arrestin to mediate CB1R internalization. We determined that agonist-mediated reduction of the density of cell surface endogenously expressed CB1Rs was clathrin and dynamin dependent and could be modeled as agonist-induced aggregation of transiently expressed GFP-CB1R. CRIP1a overexpression attenuated CP55940-mediated GFP-CB1R as well as endogenous β-arrestin redistribution to punctae, and conversely, CRIP1a knockdown augmented β-arrestin redistribution to punctae. Peptides mimicking the CB1R C-terminus could bind to both CRIP1a in cell extracts as well as purified recombinant CRIP1a. Affinity pull-down studies revealed that phosphorylation at threonine-468 of a CB1R distal C-terminus 14-mer peptide reduced CB1R-CRIP1a association. Coimmunoprecipitation of CB1R protein complexes demonstrated that central or distal C-terminal peptides competed for the CB1R association with CRIP1a, but that a phosphorylated central C-terminal peptide competed for association with β-arrestin 1, and phosphorylated central or distal C-terminal peptides competed for association with β-arrestin 2. Thus, CRIP1a can compete with β-arrestins for interaction with C-terminal CB1R domains that could affect agonist-driven, β-arrestin-mediated internalization of the CB1R.
Introduction
The CB1 cannabinoid receptor (CB1R) is among the most abundantly expressed 7-transmembrane receptor in the brain. In the central nervous system, CB1Rs are predominately expressed on presynaptic neurons and respond to the endocannabinoids 2-arachidonoylglycerol (2-AG) and anandamide to suppress neurotransmitter release (Pertwee, 2006; Turu and Hunyady, 2010). CB1R activation leads to coupling of pertussis toxin-sensitive Gi/o proteins to inhibit voltage-sensitive Ca2+ channels and adenylyl cyclase and activate inwardly rectifying K+ channels and mitogen-activated protein kinases including ERK1/2 (Turu and Hunyady, 2010). Clinically targeted pharmacotherapies at CB1R have been of considerable interest in the treatment of nausea, obesity, cancer, substance abuse, and neurodegenerative disorders (Pertwee, 2006). However, the clinical success of agonist compounds has been limited by untoward psychoactive side effect profiles, abuse liability, and the development of tolerance. Thus, a better understanding of the underlying mechanisms and proteins involved in regulating CB1R activity is needed.
The majority of experiments that center on G-protein-coupled receptor (GPCR) functioning have used agonist-mediated activity as the primary means to study receptor signal transduction. However, the emergence of GPCR interacting proteins as important modulators of GPCR ligand specificity, signaling, cell surface expression, and trafficking has opened up a new avenue of investigation in GPCR regulation. As such, insights into CB1R accessory proteins may help to uncover novel mechanisms involved in the regulation of CB1R signaling. To date, numerous C-terminal associated proteins for CB1R exist, including G-protein receptor kinases (GRKs), β-arrestins, adaptor protein complex-3, Src homology 3-domain growth factor receptor-bound 2-like (endophilin) interacting protein 1 (Hajkova et al., 2016), and GPCR-associated sorting protein, which appear to be involved in the regulation of CB1R protein expression, trafficking, cellular localization, and signaling (for review, see Howlett et al., 2010; Smith et al., 2010).
Cannabinoid receptor interacting protein 1a (CRIP1a) is another CB1R-associated protein, shown to selectively bind to the last nine C-terminal residues of the CB1R but not CB2R (Niehaus et al., 2007). Efforts to define a physiologic relevance for CRIP1a have been slow in developing. In superior cervical ganglion neurons, CRIP1a overexpression was capable of suppressing CB1R-mediated tonic inhibition of N-type voltage-gated Ca2+ channels, and as such was suggested to function as an inhibitor of CB1R constitutive activity (Niehaus et al., 2007). Gene expression and immunocytochemical analyses of hippocampal tissue revealed that CRIP1a and CB1R were coexpressed in glutamatergic pyramidal neurons, and data suggest a role for CRIP1a in G-proteins associated with extended suppression of excitatory currents that may play a role in curtailing seizure activity (Ludanyi et al., 2008; Guggenhuber et al., 2016). We reported that CRIP1a overexpression in striatal GABAergic neurons reduced CB1R-stimulated ERK1/2 phosphorylation and opioid peptide gene expression (Blume et al., 2013).
Recent studies have sought to identify the cellular mechanisms for regulation of CB1R by CRIP1a. In primary neuronal cortical cultures, it was found that CRIP1a overexpression switched CB1R-mediated neuroprotection from an agonist to an antagonist-mediated mechanism (Stauffer et al., 2011). We determined that CRIP1a overexpression in cultured N18TG2 or HEK293 cells was associated with reductions in CB1R G-protein-mediated signal transduction (Blume et al., 2015; Smith et al., 2015). CRIP1a overexpression decreased CB1R-stimulated [35S]GTPγS binding to Gi3 (Blume et al., 2015), an interaction believed to involve the C-terminus of the CB1R (Mukhopadhyay et al., 2000; Mukhopadhyay and Howlett, 2001). Moreover, we found that CRIP1a overexpression in CB1R-expressing HEK293 cells inhibited CB1R downregulation in response to prolonged agonist occupancy (Smith et al., 2015), whereas genetic deletion of β-arrestin 2 inhibited CB1R downregulation in mice (Nguyen et al., 2012). These findings led us to hypothesize that CRIP1a could also influence the interaction of the CB1R with β-arrestins, known to function in internalization via the CB1R C-terminus (Hsieh et al., 1999; Daigle et al., 2008). In the present study, we demonstrate that CRIP1a and β-arrestins both are coimmunoprecipitated in a complex with CB1Rs, but not in a complex with each other, suggesting competition for interaction with the CB1R. We provide fluorescence microscopy evidence that CB1R agonist-induced redistribution of both β-arrestin and GFP-CB1R are significantly attenuated by overexpression of CRIP1a, suggesting that CRIP1a may functionally interfere with the mechanisms of clathrin- and dynamin-dependent internalization. Peptides mimicking the CB1R C-terminus could bind to both CRIP1a in cell extracts as well as purified recombinant CRIP1a. These peptides competed for the CB1R-CRIP1a complex in coimmunoprecipitation studies. Phosphorylation of these peptides reduced binding to CRIP1a but increased the competition for the CB1R association with β-arrestin in coimmunoprecipitation complexes. Taken together, these studies provide evidence that the CB1R accessory protein CRIP1a competes with β-arrestin binding to critical sites in the CB1R and serves to functionally attenuate agonist-mediated β-arrestin redistribution. These novel findings suggest a functional influence of CRIP1a on agonist-driven CB1R internalization.
Materials and Methods
Materials.
The National Institute of Drug Abuse drug supply program kindly provided CP55940 [(−)-cis-3R-[2-hydroxy-4-(1,1-dimethylheptyl)phenyl]-trans-4R-3(3-hydroxypropyl)-1R-cyclohexanol]. Tetrahydrolipstatin (THL, orlistat) was purchased from Cayman Chemical (Ann Arbor, MI). Chlorpromazine, carbachol, and d-ala2-d-leu5-enkephalin were purchased from Sigma-Aldrich (St. Louis, MO), dynasore [3-hydroxynaphthalene-2-carboxylic acid (3,4-dihydroxybenzylidene)hydrazide] was purchased from Tocris Biosciences (Minneapolis, MN), and the GRK2 inhibitor [methyl 5-[2-(5-nitro-2-furyl)vinyl]-2-furoate] was purchased from EMD Millipore (Billerica, MA). The following antibodies were obtained from Santa Cruz Biotechnology (Santa Cruz, CA): anti-β-arrestin 1/2 (A-1), anti-β-arrestin 1 (N-19), anti-β-arrestin 2 (B-4), anti-CB1R (N-15), anti-CB1R (H-150), and anti-CRIP1a (K-12). We previously reported the generation and specificity of a rabbit CRIP1a polyclonal antibody (D20) developed against amino acids D20-F32 of human CRIP1a (Blume et al., 2013). Nitrocellulose membranes, Odyssey blocking buffer, and secondary IR dye-conjugated antibodies (800CW goat anti-rabbit, 800CW goat anti-mouse, 800CW donkey anti-goat, 680CW donkey anti-rabbit, 680CW donkey anti-goat, and 680CW donkey anti-mouse) were from LI-COR Biosciences (Lincoln, NE). Alexa Fluor 350 goat anti-mouse IgG (A-11045) and Prolong Gold Anti-fade reagent were from Life Technologies (Grand Island, NY).
Cell Culture and Immunocytochemistry Determination of CB1R Internalization.
We previously reported the development and characterization of stable CRIP1a overexpression (XS-1 and XS-5) and siRNA-knockdown (KD-2C and KD-2F) N18TG2 neuroblastoma clones (passage numbers 30–38) (Blume et al., 2015; Smith et al., 2015). Wild-type, empty-vector control, and transgenic clones were maintained at 37°C under a 5% CO2 atmosphere in Dulbecco's modified Eagle's medium:Ham's F12 (1:1) complete with GlutaMax, sodium bicarbonate, and pyridoxine-HCl, supplemented with penicillin (100 units/ml) and streptomycin (100 μg/ml) (Gibco Life Technologies, Gaithersburg, MD) and 10% heat-inactivated bovine serum (Gemini Bioproducts, West Sacramento, CA).
CB1R cell surface density and internalization were quantified using a 96-well format “On-cell-Western” immunocytochemistry assay, as previously reported (Miller, 2004; Blume et al., 2015, 2016; Smith et al., 2015). N18TG2 cells have been reported to synthesize the full agonist endocannabinoid 2-AG (Bisogno et al., 1997). To eliminate any possible effect of 2-AG generation on CB1R-mediated signaling, cells at 90% confluency were serum-starved (18 hours) and pretreated with the diacylglycerol lipase inhibitor THL (1 μM, 2 hours) before stimulation with cannabinoid compounds. For drug treatment assays, an aliquot of cannabinoid drug stock (stored at −20°C as 10 mM solutions in ethanol) or ethanol (control) was air-dried under sterile conditions in trimethylsilyl-coated glass test tubes, taken up in 100 volumes of 5 mg/ml fatty acid-free bovine serum albumin, and serially diluted before being added to cells. Cells were treated with vehicle or 10 nM CP55940 at 37°C for the indicated times, media were removed, and plates were placed on ice and fixed with ice-cold 1.2% phosphate-buffered formalin (15 minutes at 21–23°C), washed three times, blocked in LI-COR blocking buffer, and incubated with a primary antibody against the amino-terminus of CB1R (1:800). Plates were then washed in phosphate buffered saline (PBS) containing 0.1% Tween-20 (PBST), and incubated simultaneously for 1 hour with a secondary IRDye 800CW donkey anti-goat (1:1,500) and the nuclear stain DRAQ5 (1:5,000) (Cell Signaling) to normalize for well-to-well variations in cell density, and washed four times with PBST. Immunofluorescence was imaged using the LI-COR Odyssey, and CB1R fluorescence intensity was normalized to DRAQ5. Receptor internalization was quantified as percent of cell surface immunoreactive CB1Rs relative to wild type (WT) at time 0 minute expressed as 100%.
Imaging of GFP-CB1R and β-Arrestin 1/2.
For imaging of transiently expressed GFP-CB1R, cells were seeded at a density of 20,000 cells per well onto 4-well Nunc Laboratory-Tek II chamber slides coated with poly-d-lysine. Cells were grown to ∼30% confluence, the medium was replaced with serum-free and antibiotic-free Dulbecco's modified Eagle's medium:F12 media (400 μl) before Lipofectamine 2000-facilitated transfection with 1 μg of a pcDNA1 vector carrying the full-length CB1R N-terminally tagged with GFP preceded by a signal sequence from human growth hormone (SS-GFP-CB1R, provided by A.J. Irving) (McDonald et al., 2007). Cells undergoing transient transfections were incubated at 37°C in a humidified atmosphere containing 95% air and 5% CO2 for 6 hours, then washed twice and grown with serum-containing, antibiotic-free media for 32 hours. SS-GFP-CB1R-transfected cells were then serum-starved overnight (16 hours) and pretreated with THL (1 μM, 2 hours) before challenge with the CB1R agonist CP55940 (10 nM) for 15 minutes. Immediately after drug exposure, cells were fixed with 4% phosphate-buffered formalin for 15 minutes at 21–23°C, washed four times with PBS, once with ultra-pure water, and then mounted with Prolong Gold Anti-fade. GFP-CB1R images were acquired with a Zeiss LSM510 (64× oil immersion objective; excitation at 405 nm) using Zen software. Neuronal cells from all experimental conditions were acquired with all imaging parameters held constant to permit intensity comparisons. These included excitation intensity, gain settings, pinhole, and scan parameters. Initial image capture settings were carefully established such that fluorescence emissions in all images were below saturation limits.
For immunofluorescence of β-arrestin, cells were seeded at a density of 30,000 cells/well onto poly-d-lysine coated 4-well Nunc Laboratory-Tek II glass chamber slides or 22 mm glass coverslips maintained in 6-well dishes. Cells were grown to ∼40% confluency, serum-starved overnight (16 hours), and pretreated with THL (1 μM, 2 hours) before challenge with either vehicle or agonist for either 5 or 15 minutes as indicated. Cells were fixed with 4% phosphate-buffered formalin for 15 minutes at 21–23°C, washed twice with PBS, and once for 5 minutes with PBS containing 0.3% glycine, and then permeabilized for 10 minutes with 0.3% Triton X-100. Slides or coverslips were then incubated in blocking buffer (PBS, 1% bovine serum albumin, 5% normal goat serum, 0.1% Triton X-100) for 30 minutes at 21–23°C, followed by anti-β-arrestin 1/2 (1:200) overnight at 4°C. Slides were washed four times with PBS, incubated with Alexa Fluor 405 rabbit anti-mouse IgG (1:500) for 45 minutes at 21–23°C, washed five times with PBS, one time with ultra-pure water, and then mounted with Prolong Gold Anti-fade. β-Arrestin 1/2 images were acquired with an Olympus IX71 (40×/0.6Ph2, 0.55 NA) equipped with a Hamamatsu Digital CCD C8484-03G02 camera and digital image CellSens software. Images were pseudocolored green using Adobe Photoshop.
Quantitation of the agonist-mediated redistribution of GFP-CB1R or β-arrestin 1/2 is based upon aggregation referred to as punctae (such as in clathrin-coated pits/vesicles), which creates enhanced pixel intensity relative to diffuse staining. This is conceptually comparable to fluorescent βarr2-GFP aggregate analyses in which the primary assay read-out was fluorescent spots representing pits or vesicles per cell (Kotsikorou et al., 2011). To quantify changes in fluorescence intensity, analyses of the unmodified images were performed using NIH Image J for GFP-CB1R and by CellSens software for β-arrestin 1/2 images. The average pixel intensity after subtraction of background was obtained for each cell “region of interest” in a field and averaged to yield the results for the groups within each individual experiment. By a second method used for β-arrestin 1/2 analysis, the number of pixels was obtained for each cell “region of interest” in a field using the Olympus Cell-Sens image acquisition software. To determine total pixels per cell, the software sliding scale was used to set the minimum fluorescence intensity value as that calculated to be the background for each field, and the maximum fluorescence intensity was allowed to be determined by the imaging software. Then the sliding scale was moved to set the minimum value at the fluorescence intensity that was calculated to be one standard deviation above the mean for the vehicle-treated WT cells, and values above that were considered to be in the “high-intensity fluorescence” range. This provided the number of pixels within each cell “region of interest” that exhibited fluorescence in the high-intensity range. The high-intensity pixels are reported as a percent of the total pixels, and the values were averaged to yield the results for the groups within individual experiments.
Expression, Purification, and Characterization of Recombinant CRIP1a.
The gene from rat CRIP1a was codon-optimized for Escherichia coli by GenScript and subcloned into the NdeI-BamHI restriction sites of pET15b. C41(DE3) E. coli cells containing the expression plasmid were grown at 37°C in LB broth containing ampicillin to an OD600 of ∼0.8 within a 10-l fermentor. The temperature was then lowered to 16°C, and protein expression induced by the addition of 0.1 mM isopropyl-β-d-thiogalactopyranoside overnight. The cells were harvested by centrifugation and stored at −80°C until use. The cells were thawed and resuspended in 150 ml buffer A (20 mM HEPES, pH 7.9, 500 mM KCl, 5 mM imidazole, 0.1% Triton X-100, 10% glycerol) containing 0.1 mM of the protease inhibitors phenylmethyl-sulfonyl fluoride and benzamidine, 1 mM MgCl2, 40 µg/ml DNAase I. The cells were lysed using an Avestin EmulsiFlex-C3 cell homogenizer, and the supernatant was loaded onto a 10-ml HisPur Cobalt Resin (Thermo Scientific) column. The column was washed with several column volumes of buffer B (20 mM HEPES, pH 7.9, 500 mM KCl, 5 mM imidazole) before applying a linear 5 to 500 mM imidazole gradient in buffer B. The fractions containing CRIP1a, as visualized by SDS-PAGE, were pooled and dialyzed overnight against 4 l of 20 mM MES pH 6.5, 10% glycerol. The protein was then loaded onto a Sepharose HP column equilibrated with the same buffer. The protein was eluted using a linear gradient of the same buffer containing 1 M NaCl. The fractions containing CRIP1a were pooled, and 0.2 U/mg of biotinylated thrombin was added. After overnight incubation at 4°C, removal of the His-tag was confirmed by MALDI MS. The protein was then loaded onto a Superdex 75 HiLoad column equilibrated with 20 MES pH 6.5, 100 mM NaCl, 10% glycerol. The fractions containing CRIP1a were pooled, concentrated to 9 mg/ml, aliquoted, and flash frozen with liquid N2 for storage at −80°C. For some experiments, CRIP1a was dialyzed into the same buffer without the presence of glycerol. The secondary structure of recombinant CRIP1a was assessed by circular dichroism using a Jasco 720 spectrometer and cuvettes with a 0.05 cm path length. Spectra from 180 to 250 nm were scanned in triplicate and averaged. The melting point of CRIP1a was determined by monitoring the molar ellipticity at 220 nm as the temperature was increased from 20 to 85°C.
Determine Binding Parameters of Recombinant CRIP1a and CB1R C-terminal Peptides in Physiologic Solution.
Peptides containing the last 9 and 12 amino acids of the CB1R were synthesized by GenScript to contain a fluorescein molecule attached to the N-terminus. These 9-mer and 12-mer peptides were used as reporter probes for peptide-protein interactions with CRIP1a in fluorescence polarization anisotropy assays in a 96-well format, following procedures used successfully to characterize other peptides (Cushing et al., 2008; Jonsson et al., 2008; Moerke, 2009). Surface plasmon resonance spectroscopy used a Biacore T100 instrument. An FITC-CB1R-12-mer peptide was immune-captured by an anti-fluorescein monoclonal antibody covalently bound to a CM5 chip to achieve a sparse monolayer. CRIP1a binding was detected by delivering increasing CRIP1a concentrations (up to 10,000 nM) to both sample (immune-captured peptide) and reference (antibody only) channels, with the resonance unit differences (sample-reference signals) reporting specific binding (Hantgan and Stahle, 2009).
CB1R and CRIP1a Coimmunoprecipitation, Peptide Affinity Pull-down, and Western Immunoblotting Procedures.
N18TG2 WT cells were grown to 80% confluence, harvested in PBS-EDTA, and cell pellets were resuspended in Lysis buffer (25 mM Tris-HCl pH 7.4, 150 mM NaCl, 1 mM EDTA, 0.25 mM NP40, and 5% glycerol with a protease inhibitor cocktail) on ice for 30 minutes. Whole cell detergent lysates were collected after centrifugation at 2,500 g at 4°C for 5 minutes, and protein concentrations were determined using the BCA assay (BioRad). Each protein sample (500 μg protein) was diluted into IP buffer (100 mM NaCl, 2.7 mM KCl, 1.5 mM KH2PO4, 8 mM Na2HPO4, 0.1 mM EDTA, 0.01 mM NP40, pH 7.0) and immunoprecipitated with antibodies (1–2 μg) specific for CB1R, CRIP1a, or β-arrestin 1/2 at 4°C for 18 hours. Immuno-bound proteins were collected with protein A/G PLUS-agarose beads (Santa Cruz), sedimented at 10,000 g for 5 minutes at 4°C, washed three times with ice-cold IP buffer, and heated in Laemmli's sample buffer (62.5 mM Tris-HCl, pH 6.8, 2% SDS, 10% glycerol, 0.002% bromophenol blue, 710 mM β-mercaptoethanol; 65°C for 10 minutes). Eluted proteins were separated from agarose beads by centrifugation at 10,000 g for 5 minutes at 4°C, and supernatants were analyzed by Western blotting.
For pull-down assays, peptides representing the last 16 residues (149–164) of the C-terminus of CRIP1a, the last 14 residues (460–473) of the C-terminus of CB1R, or the CB1R C-terminal 14-mer peptide phosphorylated at threonine 468 were each synthesized with an N-terminal cysteine residue (purity ≥98%) (GenScript) and immobilized onto agarose beads through irreversible thioether bonds (Pierce SulfoLink Immobilization kit). Immobilized peptide beads were incubated overnight with whole cell NP40 detergent lysates (500 μg protein) from N18TG2 WT cells. The agarose-peptide complex was then centrifuged at 1,000 g for 5 minutes at 4°C, and was washed four times with PBS. Proteins were eluted with 0.1 M glycine-HCl (pH 2.8), immediately neutralized on ice with 1 M Tris-HCl (pH 9), and centrifuged at 10,000 g for 5 minutes at 4°C. Eluted proteins (supernatant) were then subjected to Western blotting. β-Arrestin proteins were detected with antibodies specific for the N-terminal 19 residues of human β-arrestin 1 or an antibody against the C-terminal residues 385–408 residues of human β-arrestin 2 (Santa Cruz Biotechnology product datasheets).
For CB1R C-terminal peptide competition coimmunoprecipitation, peptides were synthesized (GenScript) with or without phosphorylation on the underlined amino acids: central, TAQPLDNSMGDSDCLHKH; distal, VTMSVSTDTSAEAL. N18TG2 whole cell detergent lysates (300 μg protein) were incubated with a final concentration of 3 μM peptide in IP buffer for 1.5 hours at 4°C. After reaching equilibrium, the mixture was incubated overnight with agarose beads conjugated to CB1R antibodies (rabbit, H-150, Santa Cruz). Immuno-bound proteins on agarose beads were sedimented at 10,000 g for 5 minutes at 4°C, washed three times with ice-cold IP buffer, and boiled in Laemmli's sample buffer and resolved by SDS-PAGE.
For Western blotting, proteins denatured and reduced in Laemmli sample buffer were resolved on gradient SDS-PAGE gels (4–20%; Bio-Rad) at 21–23°C for 1 hour at 150 V. Proteins were transferred to nitrocellulose membranes in Towbin's buffer at 4°C for 8 hours at 20 V. Blots were rinsed for 5 minutes with PBS, blocked for 60 minutes with LI-COR blocking buffer, and then incubated with primary antibodies at 4°C for 18 hours: CB1R (1:750), CRIP1a D20 (1:500), CRIP1a K-12 (1:500), β-arrestin 1/2 (1:500), β-arrestin1 (1:500), or β-arrestin 2 (1:500). Blots were washed four times with PBST, incubated with an appropriate IR dye-conjugated secondary antibody (1:10,000) for 1 hour at 21–23°C [800CW goat anti-rabbit, 800CW donkey anti-goat, 680CW donkey anti-rabbit, 680CW donkey anti-goat (LI-COR)], and washed three times with PBST followed by one wash with PBS. Some studies used a Quick Western Kit (LI-COR) using a 1:500 primary antibody in LI-COR blocking buffer with 0.2% Tween and LI-COR IR Dye 680RD. Bands on immunoblots were imaged and quantified by densitometry using Odyssey Infrared Imaging System software (LI-COR Biosciences).
Statistical Analyses.
Statistical differences were determined using Student’s t test for comparison of two independent variables or one-way or two-way analysis of variance and either Bonferroni or Dunnett’s post hoc test for experiments comparing two or more sets of independent variables. Statistical analyses were conducted using GraphPad Prism VI (GraphPad Software). Data shown are mean ± S.E.M. values and were considered significant when the P value ≤ 0.05.
Results
The cytoplasmic tail of CB1R is a key regulator of G-protein binding, desensitization, and cellular trafficking (see reviews Howlett, 2009; Stadel et al., 2011). Studies of exogenously expressed CB1R indicate that the CB1R distal C-terminus is required for agonist-mediated CB1R internalization (Hsieh et al., 1999; Daigle et al., 2008). In HEK293 cells, phosphorylation of a cluster of 4–6 serine and threonine residues at the CB1R C-terminus, presumed to be by GRKs or cell signaling kinases such as protein kinase C, is an important prerequisite step in the rapid (1–5 minutes) recruitment of β-arrestin 2 to activated CB1Rs and CB1R internalization (Daigle et al., 2008; Gyombolai et al., 2013). Inasmuch as CRIP1a also binds to the CB1R distal C-terminus (Niehaus et al., 2007), we investigated the interactions between CRIP1a, β-arrestin, and the CB1R.
CRIP1a Competes for Interaction of β-Arrestin1/2 with CB1R.
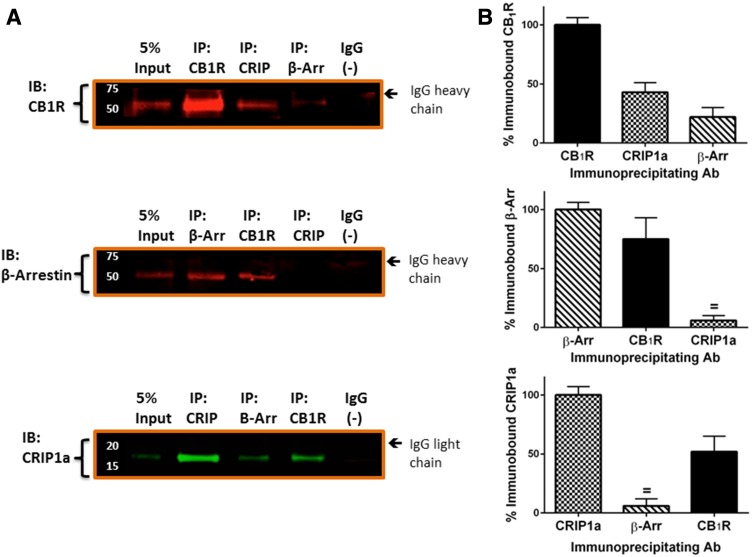
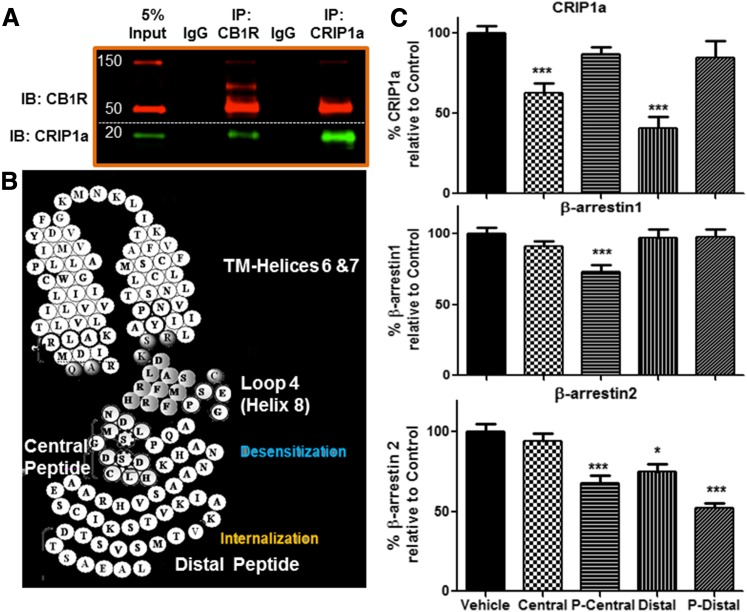
We performed immunoprecipitations to determine protein association between CB1R and CRIP1a or β-arrestin 1/2. CB1R, β-arrestin 1/2, and CRIP1a were immunoprecipitated from N18TG2 whole cell NP40 lysates, and the immunoprecipitated protein plus those proteins associated in a complex with it were identified by Western blotting. The antibodies for CB1R and CRIP1a target epitopes that are distant from the predicted protein binding domains, and these were previously characterized for their capacity for immunoprecipitation and Western blotting (Howlett et al., 1998; Mukhopadhyay and Howlett, 2005; Blume et al., 2013,2015). The antibody used for β-arrestin immunoprecipitation was a mouse monoclonal antibody generated to the N-terminal human β-arrestin 1 (residues R6 to D290) and recognizes both β-arrestin 1 and β-arrestin 2 from multiple species (Santa Cruz, Biotechnology Product Data Sheet). This antibody has previously been characterized for Western blotting and coimmunoprecipitation of 7-transmembrane and other receptors (Por et al., 2012; Burns et al., 2014; Daniele et al., 2014; Zappelli et al., 2014). We found that CB1R could be observed in an immune complex precipitated by either CRIP1a or β-arrestin 1/2 antibodies (Fig. 1, A and B, upper panel). β-Arrestin 1/2 (Fig. 1, A and B, middle panel) and CRIP1a (Fig. 1, A and B, lower panel) were found in an immune complex precipitated by CB1R antibodies. However, immunoprecipitation using a CRIP1a antibody failed to show immune-staining for β-arrestin 1/2 (Fig. 1, middle panel). Furthermore, immunoprecipitation using a β-arrestin 1/2 antibody failed to show immune-staining for CRIP1a (Fig. 1, lower panel). The lack of β-arrestin 1/2 and CRIP1a coimmunoprecipitation suggests that these two proteins do not exist simultaneously in a complex with each other or with CB1R in N18TG2 NP40 lysates. However, an alternative interpretation is that CRIP1a might bind to a CB1R-β-arrestin 1/2 complex at the same site as the antibody, thereby creating a steric interference in the ability of the antibody to precipitate the protein complex. Control immunoprecipitations with nonimmune IgG failed to yield an interaction with either of the proteins examined. These results led us to propose a role for CRIP1a in modulating the ability of CB1R to interact with β-arrestin.
Fig. 1.
CB1R associates in complexes with β-arrestin 1/2 and CRIP1a, but β-arrestin 1/2 and CRIP1a do not associate in complexes with each other. (A) Representative Western blots of immunoprecipitation protein complexes (IP) from N18TG2 WT whole cell NP40 lysates to show the abundance of 5% input as a reference to determine the relative amount of each protein in the complex and to confirm the mobility of the bands detected by the immunoblotting antibodies (lanes 1 of each panel). Lysates were immunoprecipitated with antibodies against the CB1R N-terminus, β-arrestin 1/2, an internal region of CRIP1a, or a preimmune IgG as negative control (−), as indicated above each lane. Immunocomplex proteins were resolved on SDS-PAGE as described in Materials and Methods and immunoblotted (IB) with antibodies against the CB1R (top); β-arrestin 1/2 (middle), or CRIP1a (bottom). (B) Quantification of immunoprecipitation complexes resolved on Western blot. For each immunoblotted protein panel, the band density of immunoprecipitated protein is represented as 100%, and the densities of bands that were coimmunopreciptated in a complex with other proteins are presented relative to 100%. Data were calculated as the mean ± S.E.M. from three independent experiments; = indicates not significantly different from 0 (background) by Student t test.
Functional Ramifications of CRIP1a Competition for β-Arrestin Binding.
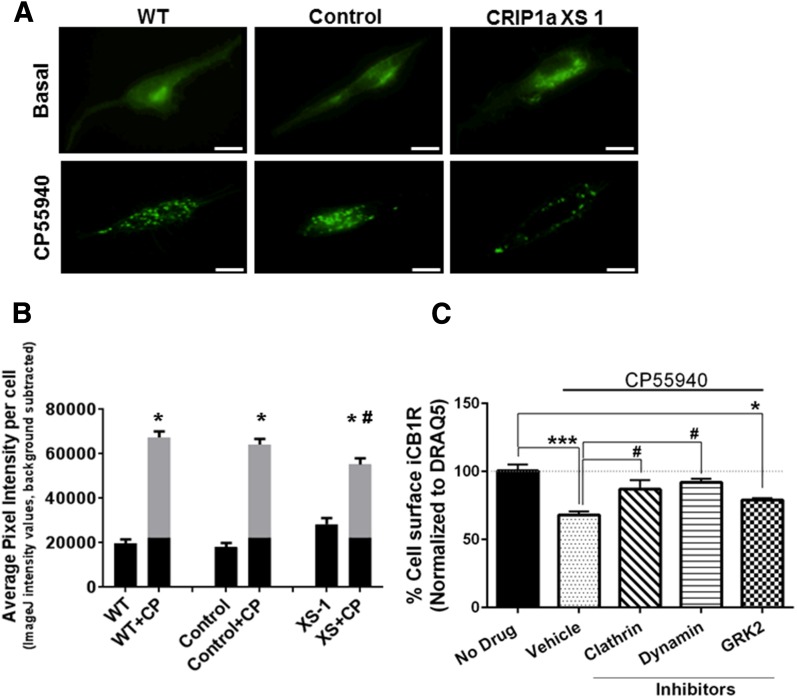
To determine the influence of CRIP1a on agonist-mediated CB1R redistribution, we examined WT, empty-vector control and CRIP1a XS cells that were transiently transfected with N-terminally tagged green fluorescent protein-CB1R fusion protein (GFP-CB1R) (Fig. 2A). Confocal microscope images taken along the membrane near the point of contact with the surface illustrate relatively diffuse fluorescence distribution of GFP-CB1R under basal conditions 48 hours after transfection. At this posttransfection stage, the GFP-CB1R fluorescence has migrated away from the source at the perinuclear Golgi-ER complex. Interestingly, CRIP1a XS cells appeared to exhibit some punctae fluorescence along the soma (not located at the perinuclear Golgi-ER complex) (Fig. 2A, top panel); however, this increase was not statistically different (P = 0.06) from WT (Fig. 2B). The clustering of GFP-CB1R in the CRIP1a XS cells before agonist stimulation is a phenomenon that will require greater investigation, because data from our studies of exposure to agonist ligands suggest that CRIP1a could play an inhibitory role in trafficking of the CB1R to the plasma membrane (Blume et al., 2016). Exposure to CP55940 (10 nM, 15 minutes) resulted in a pronounced redistribution of GFP-CB1R fluorescence into discrete punctate aggregates along the membrane surface in WT and empty-vector control cells. Figure 2B shows quantification of average pixel intensity per cell, which increased 3- to 4-fold in response to CP55940 in WT and empty-vector control cells. The average pixel intensity for GFP-CB1R fluorescence evoked by CP55940 in CRIP1a XS cells was significantly lower than WT. We interpret this to mean that CRIP1a overexpression significantly reduced agonist-mediated GFP-CB1R redistribution into high-intensity fluorescence aggregates.
Fig. 2.
CRIP1a overexpression attenuates agonist-induced GFP-CB1R redistribution in N18TG2 cells. (A) Visualization of the redistribution of GFP-CB1R by confocal microscopy. N18TG2 WT, empty vector Control, and CRIP1a XS cells were transiently transfected with N-terminal GFP-CB1R, as described in Materials and Methods, and treated with vehicle or CP55940 (10 nM) for 15 minutes; scale bar, 10 μm. (B) Quantification of GFP-CB1R aggregates after 15-minute treatment with vehicle or CP55940 (10 nM). The average pixel intensity per cell (11 to 15 cells) was averaged for each experimental group. Data are reported as the mean ± S.E.M. from three independent experiments and statistical comparisons were made using two-way ANOVA and Student’s t test. *P < 0.05 indicates a significant difference between vehicle and CP55940-treated; #P < 0.05 indicates a significant difference from CP55940-treated WT. (C) Agonist-driven internalization of endogenously expressed surface CB1R in N18TG2 cells is dependent upon clathrin and dynamin and partially dependent on GRK2. N18TG2 WT cells were pretreated for 15 minutes with vehicle (DMSO) or inhibitors of clathrin (chlorpromazine, 25 μM), dynamin (dynasore, 80 μM), or GRK2 (1 μM), and then challenged with the CB1R agonist CP55940 (10 nM) for 5 minutes. Quantification of CB1R cell surface expression was determined using an On-cell-Western immunohistochemical method as described in Materials and Methods. CB1R surface expression was determined as the ratio of immunoreactive CB1R to DRAQ5 (nuclear stain), which was expressed as a 100% for vehicle. Data are expressed as mean ± S.E.M. from three independent experiments performed in duplicate, and statistical comparisons were made using one-way ANOVA with Dunnett’s post hoc test. ***P < 0.005; *P < 0.05 indicates significant difference between no drug versus CP55940-treated groups; #P < 0.05 indicates significant difference between CP55940 vehicle versus CP55940 plus a clathrin inhibitor or a dynamin inhibitor.
The distal C-terminus of the CB1R has been implicated as a region critical for regulating agonist-mediated receptor internalization (Hsieh et al., 1999; Daigle et al., 2008; Singh et al., 2011). For GPCRs, the canonical pathway for internalization includes a GRK-mediated phosphorylation, β-arrestin mobilization to the site of phosphorylated GPCRs, accumulation via clathrin-coated pits, and cleavage to clathrin-coated vesicles by dynamin (Drake et al., 2006; Hanyaloglu and von Zastrow M., 2008). GRK2 holds a prominent role in phosphorylation of some GPCRs by interaction with the βγ dimer after agonist-dependent dissociation of the G-protein heterotrimer (Penela et al., 2003). As a control experiment to demonstrate that the CB1R can use this pathway in N18TG2 cells, we quantitated endogenously expressed CB1R on the cell surface before and after treatment of cells with the CB1 agonist CP55940 for 5 minutes. In Fig. 2C, we demonstrate that agonist treatment led to a statistically significant 33% diminution of extracellular surface CB1Rs. We confirmed the importance of clathrin in the process by application of chlorpromazine at a concentration shown to effectively inhibit clathrin internalization (Wang et al., 1993; Rejman et al., 2005). We confirmed the importance of dynamin using dynasore, a dynamin inhibitor that selectively blocks dynamin-mediated internalization at two steps in the process (Macia et al., 2006; Kirchhausen et al., 2008). In the presence of these internalization process inhibitors, there was no significant diminution in cell surface CB1R after treatment with CP55940. The GRK2 inhibitor resulted in only a minor decrement (not statistically significant) in agonist-mediated cell surface CB1R diminution in N18TG2 cells. However, we determined that the N18TG2 neuronal cell line expresses multiple GRKs (2 through 6) using a Western blot screen (Howlett and Kabler, data not shown). This suggests that alternate GRKs [e.g., GRK3 (Jin et al., 1999)] or other cell signaling kinases (Garcia et al., 1998; Lee et al., 2003; Asimaki and Mangoura, 2011) that have been implicated in agonist-mediated modifications of the CB1R may play a determinant role in the phosphorylation of CB1R.
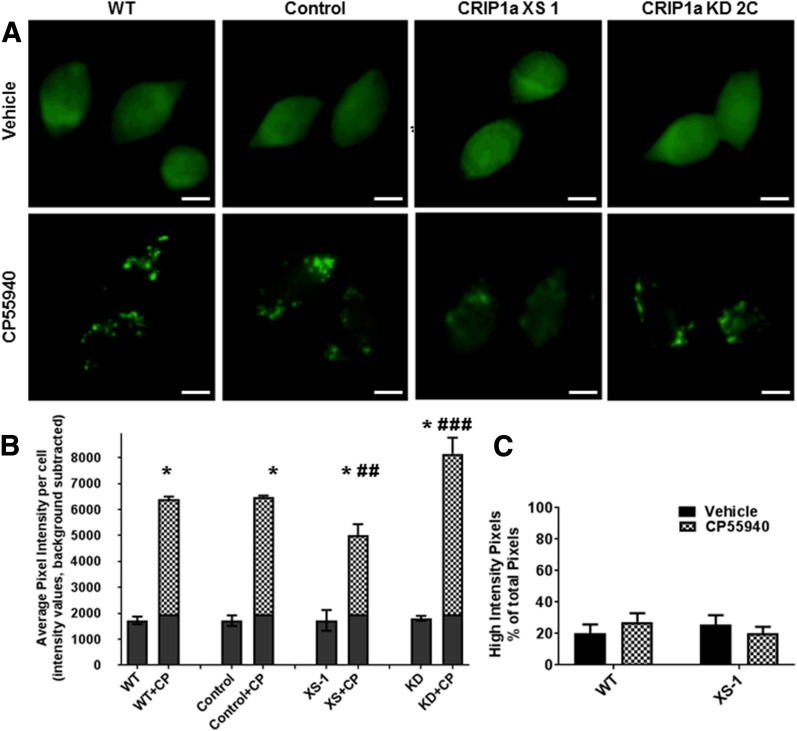
To determine whether CRIP1a could affect agonist-promoted redistribution of β-arrestins, we used the β-arrestin 1/2 antibody that we had characterized for its ability to detect a CB1R-β-arrestin immune complex (Fig. 1). Under basal conditions (16-hour serum-starvation and blockade of 2-AG biosynthesis, vehicle treated), we observed a diffuse immunofluorescence staining pattern in all cell lines (Fig. 3A, top panel). The average pixel intensity per cell of β-arrestin 1/2 was not different between cells expressing endogenous levels (WT or empty-vector control), overexpression, or knockdown of CRIP1a (Fig. 3B). A 5-minute exposure to CP55940 (10 nM) promoted a redistribution of β-arrestin 1/2, resulting in discrete punctate staining in WT and empty-vector control N18TG2 clones (Fig. 3A, bottom panel). In contrast, CRIP1a XS clones displayed a reduced agonist-dependent redistribution of β-arrestin 1/2 compared with WT or control cells. CRIP1a KD clones exhibited a pronounced increase in β-arrestin 1/2 punctate staining relative to WT cells (Fig. 3A, bottom panel). Quantification of average pixel intensity per cell of β-arrestin 1/2 immunofluorescence confirmed that CRIP1a overexpression significantly reduced, whereas CRIP1a depletion significantly augmented the CP55940-stimulated redistribution of β-arrestin 1/2 relative to WT (Fig. 3, B and C). This redistribution appears to be due to greater intensity per pixel rather than to an increased number of pixels contributing to the high intensity (i.e., greater than basal) fluorescence (Fig. 3C). As a negative control, WT cells transiently transfected with enhanced GFP, a soluble protein, showed diffuse cellular staining and the enhanced GFP was not responsive to agonist treatment by forming the characteristic aggregates (data not shown). In addition, we treated N18TG2 cells with agonists for other Gi/o-coupled receptors (15 minutes): carbachol, M4 muscarinic; d-ala2,d-leu5-enkephalin, δ-opioid), without observing a difference between agonist-stimulated pcDNA control and CRIP1a-XS cells in β-arrestin 1/2 average fluorescence intensity per cell except for the CP55940-treated cells (Supplemental Fig. S1). These studies are consistent with dose-related CRIP1a effects on agonist-mediated β-arrestin redistribution that parallel agonist-mediated, clathrin- and dynamin-dependent CB1R redistribution. This finding supports the contention that β-arrestin translocation is a key step in agonist-mediated CB1R internalization (Daigle et al., 2008; Nguyen et al., 2012; Gyombolai et al., 2013) and provides novel data suggesting that CRIP1a could be a physiologic modulator of this response.
Fig. 3.
CB1R agonist-mediated redistribution of β-arrestin 1/2 is influenced by CRIP1a. A, Visualization of the redistribution of β-arrestin 1/2 by immunofluorescence microscopy. N18TG2 WT, empty vector control, CRIP1a XS, and CRIP1a KD cells were treated for 5 minutes with vehicle or CP55940 (10 nM); scale bar, 10 μm. (B) Quantification of β-arrestin 1/2 staining after 5-minute treatment with vehicle or CP55940 (10 nM). (B) Quantification of β-arrestin 1/2 staining after 5-minute treatment with vehicle or CP55940 (10 nM). Data are shown as average pixel intensity per cell after subtracting the background, from a total of 29 to 44 cells evaluated for each treatment group. The values were averaged for each treatment group and reported as the mean ± S.E.M. from n = three independent experiments. Statistical analysis was performed using 2-way ANOVA and Bonferroni multiple comparisons test. No significant differences in vehicle-treated samples were observed between WT, control, CRIP1a XS, and CRIP1a KD clones. *P < 0.05 indicates a significant difference between vehicle and CP55940-treatment. ##P < 0.01, ###P < 0.001 indicate significant difference from CP55940-treated WT. (C) Quantification of pixels in the high-intensity range as a fraction of total pixels. Using CellSens software, the total pixel number emitting fluorescence was determined after the sliding scale bar had been set to a minimum that was equivalent to the background. To determine the number of pixels emitting fluorescence in the high intensity range, we arbitrarily defined high intensity as “above the population of basal.” We calculated this nonbasal average intensity level to be one standard deviation above the mean of the average pixel intensity of all the vehicle-treated control cells (see gray area at the bottom of the bars in Fig. 3B). The sliding scale bar was moved to bring the minimum up to the nonbasal level, and the number of pixels emitting fluorescence in this high-intensity range was determined. The ratio of high intensity (i.e., above basal) pixels to total (i.e., above background) pixels was calculated as a percent.
CB1R C-terminal Phosphorylation Influences Association with β-Arrestin versus CRIP1a.
Peptides that represent domains of the CB1R have been used in the Howlett laboratory to discern interactions with G-proteins, either by their ability to compete for receptor-G-protein interactions (Mukhopadhyay et al., 2000; Mukhopadhyay and Howlett, 2001,2005) or their ability to promote cellular signaling (Howlett et al., 1998; Mukhopadhyay et al., 1999; Mukhopadhyay and Howlett, 2001). Deletion mapping of the C-terminus of CB1R identified the distal nine residues as the minimal domain needed to bind CRIP1a (Niehaus et al., 2007). To determine the affinity of CRIP1a to 9-mer and 12-mer distal CB1R peptides, CRIP1a was recombinantly expressed and purified from Escherichia coli. Circular dichroism analysis of the protein supports that the protein contains ∼14% α-helical content (Supplemental, Fig. S2A), but was quite thermostable with a melting temperature of ∼56°C (Supplemental Fig. S2B).
The binding of CRIP1a to the peptide was determined by two orthogonal approaches. Fluorescence polarization anisotropy determines the ability of the small fluorescent peptide to emit light that is unpolarized as the peptide exhibits a high rate of molecular rotation in solution. When the reporter peptide engages the much larger target protein (CRIP1a), its rate of rotation is limited and the emitted light is now more polarized (Moerke, 2009). In an alternative procedure, the surface plasmon resonance determined the refractive index of a light source reported as a change in angle upon binding of the two biomolecules (Wilson, 2002; Hantgan et al., 2008; Tanious et al., 2008; Piliarik et al., 2009). Supplemental Fig. S3 shows the equilibrium binding of CRIP1a at increasing concentrations to FITC-12-mer peptide (Kd = 3.4 ± 3.6 µM) and the surface plasmon resonance equilibrium binding of CRIP1a when passed over a surface of an immunocaptured fluorescein-labeled CB1R distal C-terminal 12-mer peptide (Kd = 6.8 ± 4.1 µM). Relative affinities from both procedures are in agreement that CRIP1a binds to the peptide in the µM range in aqueous solution.
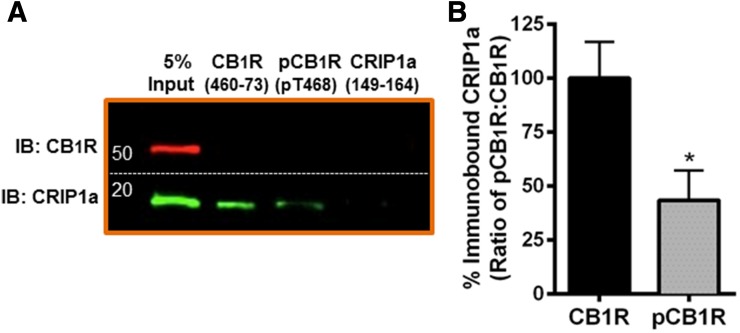
We hypothesized that phosphorylation of the distal CB1R C-terminus could alter the association of CRIP1a with CB1R. Affinity pull-down assays indicate that CRIP1a from whole cell lysates of N18TG2 WT cells binds to a CB1R distal C-terminal (residues 460–473) peptide (Fig. 4A, lane 2). We also demonstrated that recombinant CRIP1a (see Supplemental Figs. S2 and S3) binds to this peptide in affinity pull-down assays (Blume, Lowther, Kabler, and Howlett, unpublished observations). The C-terminus of CRIP1a (149–164) was not able to bind to CB1R or CRIP1a (Fig. 4B, lane 3), indicating that the distal C-terminus of CRIP1a is not required for its interaction with CB1R, nor capable of interacting in complexes containing endogenously expressed CRIP1a proteins. It is interesting to note that neither the CB1R distal C-terminal peptide nor the pT468 phosphopeptide were able to pull down the CB1R, suggesting that this is not a binding domain for CB1R multimerization.
Fig. 4.
Pulldown of CRIP1a by a CB1R distal carboxy terminus peptide is attenuated by phosphorylation. (A) Pull-down of proteins using agarose beads coupled to peptides corresponding to the CB1R C-terminus, T468-phosphorylated CB1R C-terminus, or CRIP1a C-terminus. Bound proteins were washed, eluted, and subjected to immunoblot analysis as described in Materials and Methods. (B) CB1R C-terminus phosphorylation disrupts peptide association with CRIP1a. Quantification of CRIP1a immunoblot band densities is presented as binding to the nonphosphorylated CB1R expressed as 100%. Data are calculated from three independent experiments and are expressed as the mean ± S.E.M. *P < 0.001 indicates significant difference from non-phosphorylated peptide using Student’s t test.
Consensus sequence and protein structural analyses (NetPhosK1.0, NetPhos2.0) predict that the distal C-terminus has a 94.8% probability of being phosphorylated at T468 by protein kinase C (Blom et al., 1999, 2004). Because T468 lies within the CB1R region required for association with CRIP1a, peptide affinity pull-down assays were employed to establish whether phosphorylation of the CB1R at T468 could impact the association with CRIP1a. We found that the pull-down of CRIP1a by the pT468 phosphopeptide was reduced by 66 ± 14% compared with the nonphosphorylated peptide (Fig. 4B). It can be concluded that the CB1R-CRIP1a interaction can be dynamically regulated by phosphorylation.
To further explore the putative locations and the role of phosphorylation on CRIP1a and β-arrestin binding, we used peptides and phosphopeptides mimicking the C-terminus to compete for the ability of CB1R to associate with CRIP1a or the β-arrestins in immune complexes. We coimmunoprecipitated endogenously expressed CB1R and CRIP1a from whole cell lysates of N18TG2 WT cells with an antibody recognizing the N-terminus of CB1R (Fig. 5A, lane 3) as well as with a CRIP1a antibody recognizing an epitope proximal to a region proposed to interact with CB1R (Fig. 5A, lane 5). This demonstrates that these antibodies are mutually effective in maintaining an immune complex in NP40 detergent solution. Peptides representing a central C-terminal 18-mer or its homolog phosphorylated at S426 and S430, or the distal C-terminal 14-mer or its homolog phosphorylated at T468, were incubated with the N18TG2 whole cell lysates. After equilibrium had been established, we performed immunoprecipitations with the CB1R antibody and immunoblotted to detect CRIP1a, β-arrestin 1, or β-arrestin 2 as shown in Fig. 5. The interaction of CB1R with CRIP1a was significantly reduced by competition with both the unphosphorylated central as well as the distal C-terminal 14-mer. This suggests that in addition to the distal C-terminus, CRIP1a contacts other CB1R sites. The only peptide used in this study that was able to significantly compete for the CB1R-β-arrestin 1 interaction was the phosphorylated central domain peptide. The interaction of CB1R with β-arrestin 2 was significantly competed by the phosphorylated pT468 distal C-terminal peptide, the nonphosphorylated distal C-terminal 14-mer, and the phosphorylated central C-terminal peptide. Considered together, these data suggest that CRIP1a can interact with the CB1R at multiple C-terminal loci in the absence of phosphorylation, whereas after phosphorylation, β-arrestin 1 may interact with the central domain and β-arrestin 2 may interact with both domains of CB1R.
Fig. 5.
Competition by peptides representing CB1R C-terminal central or distal domains for immunoprecipitated CB1R-CRIP1a protein complexes. (A) Representative Western blot of an immunoprecipitation (IP) experiment performed on whole cell NP40 lysates from N18TG2 cells, using antibodies targeting the N-terminus of CB1R, an internal region of CRIP1a (D20-F32), or a preimmune IgG as negative control. Immunocomplexes were resolved and immunoblotting (IB) was performed as described in Materials and Methods. The input shows an aliquot of 5% of whole cell NP40 lysate used for each experiment and serves as a reference to determine the relative amount of each immunoprecipitated complex and the relative mobility of the proteins in the native lysate. (B) Helical amino acid depiction of CB1R’s C-terminal domain representing regions corresponding to central and distal CB1R peptides and putative regions for internalization and desensitization. (C) Peptides representing the central or distal C-terminal domains, either nonphosphorylated or phosphorylated (see Materials and Methods), were incubated with NP40 whole cell extracts as described in Materials and Methods. Immunopreciptations were performed using an antibody to the CB1R N-terminal, washed, and eluted proteins from the immunoprecipitated complex were subjected to SDS-PAGE. Immunoblotting was performed with antibodies specific for CRIP1a, β-arrestin 1, or β-arrestin 2, and band densities were determined as relative to the band density of the vehicle control in the absence of peptides as 100%. The data are mean ± S.E.M. (n = four to seven independent experiments); *P < 0.05; ***P < 0.001 indicates significant difference from vehicle using one-way ANOVA with Dunnett’s post hoc test.
Discussion
The mechanisms responsible for agonist-dependent CB1R internalization appear to involve a sequence of events leading to the activation of GRKs and the subsequent phosphorylation and recruitment of β-arrestin 2 to the C-terminus (Hsieh et al., 1999; Daigle et al., 2008; Stadel et al., 2011). β-Arrestins play a prominent role in CB1R desensitization and internalization (Jin et al., 1999; Daigle et al., 2008; Nguyen et al., 2012; Gyombolai et al., 2013; Delgado-Peraza et al., 2016). Our observation of discrete β-arrestin aggregate staining after a short (5 minute) exposure to CP55940 confirms that CB1R activation results in cellular redistribution of β-arrestin (van der Lee et al., 2009). Furthermore, our coimmunoprecipitation data demonstrate that CB1Rs interact directly with β-arrestin 1 and β-arrestin 2 in a native expression system. Our results demonstrate that overexpression of CRIP1a coincided with a reduction in β-arrestin 1/2 translocation, suggesting that a disruption occurs in the coupling between β-arrestins and CB1R. Our data that CB1R can associate with either CRIP1a or β-arrestin, but with limited binding to both simultaneously, support a model of competition of these two proteins for CB1R binding. We demonstrated that binding competition by the distal C-terminal peptide phosphorylated at T468 or the phosphorylated central peptide disrupts CB1R-β-arrestin 2 association; however, only the phosphorylated central peptide disrupts the CB1R-β-arrestin 1 association. The finding that nonphosphorylated peptides from both the central and distal CB1R C-terminus compete for the CRIP1a-CB1R association suggests that the interaction surface is greater than the distal terminus as was first proposed (Niehaus et al., 2007). These findings not only identify the receptor locus of interaction, but also demonstrate that the phosphorylation state is critical to directing protein interaction with the CB1R.
In previous studies (Blume et al., 2015), we found by using an antibody-targeted scintillation proximity [35S]GTPγS binding assay, that CRIP1a overexpression elucidated a switch in Gi/o subtype preference. Activation of Gi3 and Go subtypes was reduced by CRIP1a overexpression, whereas activation of Gi1 and Gi2 subtypes was increased. If Gi3 and Go are more efficacious regulators of cellular signal transduction than are Gi1 and Gi2, then CRIP1a would have a functional impact on cellular signaling. A cycle of activity initiated by agonist occupancy of the CB1R-Gi protein complex is expected to result in Gi dissociation, GRK-phosphorylation, and β-arrestin-mediated internalization. Thus, we cannot rule out the possibility that if CRIP1a bound to the CB1R precludes a CB1R-Gi3 or Go complex, then a subsequent phosphorylation and interactions with β-arrestin might not be initiated. In studies of the agonist-mediated depletion of cell surface CB1R, we found that CRIP1a overexpression prevented CB1R internalization (Blume et al., 2016). This finding is completely consistent with a competition by CRIP1a for a binding site for β-arrestin on the CB1R C-terminal and the amelioration of the β-arrestin-mediated internalization processes.
Reports are unclear regarding β-arrestin subtype binding to CB1R C-terminal sites to initiate the internalization process. In our previous study (Smith et al., 2015), CRIP1a overexpression attenuated CB1R downregulation but not desensitization of G-protein activation induced by prolonged agonist exposure. Evidence suggests that the distal C-terminus mediates CB1R internalization required for CB1R downregulation (Daigle et al., 2008; Jin et al., 1999; Martini et al., 2007), but that central CB1R C-terminal sites are required for desensitization (Jin et al., 1999; Daigle et al., 2008; Morgan et al., 2014). Gyombolai and colleagues (2013) recently reported that β-arrestin 2 is required for agonist-induced, but not the constitutive, internalization of CB1R in Neuro2A and HeLa cells. Bakshi and colleagues (2007) reported a conformational change in a phosphorylated, central C-terminal CB1R peptide after β-arrestin 1 binding, but those studies did not investigate the internalization function. Studies of Delgado-Peraza and colleagues (2016) used an S426A/S430A mutated CB1R expressed in HEK cells to investigate cellular signaling via β-arrestin pathways. Their studies suggested that internalization is dependent upon β-arrestin 2 (Ahn et al., 2013; Flores-Otero et al., 2014), whereas sustained ERK phosphorylation is dependent upon β-arrestin 1(Delgado-Peraza et al., 2016). These authors demonstrated that the nonphosphorylated central domain S426A/S430A mutation decreases recruitment of β-arrestin 2 as well as internalization compared with WT CB1R. They noted that, although there was no difference from WT in the recruitment of β-arrestin 1 by the S426A/S430A mutant receptor, there was an increase in the prolonged ERK phosphorylation by β-arrestin 1 (Flores-Otero et al., 2014; Delgado-Peraza et al., 2016). These data contrast with our demonstration of displacement of β-arrestin 1 as well as β-arrestin 2 from the CB1R by the competing central domain phosphopeptide. Our data indicate that, when not phosphorylated, this domain can be occupied by CRIP1a.
A question that the present data elicits is whether β-arrestin 1 or β-arrestin 2 would be silenced functionally by the presence of CRIP1a and how that functional silencing would affect the β-arrestin signalosome. In addition to the GRK family proteins that mediate receptor phosphorylation patterns that regulate specificity in β-arrestin subtype binding (Dewire et al., 2007; Liggett, 2011), it is plausible that CRIP1a binding would regulate not only CB1R internalization but also the cellular signaling functions of the β-arrestins. The crystal structure of the visual arrestin-rhodopsin complex indicates that arrestin interacts at multiple sites on the receptor including intracellular loops 2 and 3 and transmembrane helices 5,6, 7, and helix 8 in addition to the C-terminus (Kang et al., 2015,2016). These studies of β-arrestin functioning suggest that the protein can bind to GPCRs either as a “hanging” conformation or a “core binding” conformation. One very interesting recent finding is that the “hanging” conformation can accommodate the concurrent GPCR interactions with G-protein subunits, thereby allowing continued G-protein signaling due to “super-complexes” of signaling proteins localized on internalized endosomes (Thomsen et al., 2016). This finding begs the question of whether CRIP1a binding might be compatible with β-arrestin 1 or β-arrestin 2 binding if it occurs at an alternative site.
Overall these findings identify a novel function for CRIP1a in regulating agonist-promoted β-arrestin interaction with CB1Rs. We propose that CRIP1a interferes with β-arrestin recruitment to agonist-activated CB1Rs and subsequent β-arrestin-mediated internalization. Contemporary drug development strategies are seeking compounds that exhibit biased agonism by which a ligand can promote selective interaction of a 7-transmembrane receptor with either a G-protein or β-arrestin to promote specific cellular signaling pathways. The ability of CRIP1a to selectively modulate CB1R interaction with defined subtypes of Gi/o proteins (Blume et al., 2015), as well as β-arrestin subtypes (demonstrated here), offers an alternative and potentially promising approach to the development of novel, selective CB1R pharmacotherapies. For example, using peptides to block a protein functional site is a strategy that could preclude a protein-protein interaction. Peptides binding to CRIP1a at its CB1R-binding locus would thereby increase the probability of functional interactions between β-arrestin and the CB1R C-terminal domains. Computational modeling has predicted a structure based upon the primary sequence (Ahmed et al., 2014; Singh, Howlett, and Cowsik, unpublished data), but the complete structure has yet to be identified. Subsequent structural determinations of the protein-protein surface interactions will need to be accomplished to rationally design high-affinity competitive peptides to functionally silence the endogenous CRIP1a for pharmacotherapeutic purposes.
Acknowledgments
The authors thank Dr. Ken Grant in the Department of Pathology at Wake Forest School of Medicine for providing critical support for preliminary confocal microscopy experiments. Dr. Andrew J. Irving generously provided us with the SS-GFP-CB1R plasmid construct. The authors are also grateful for biophysical chemistry equipment support from Molecular Partners Core Laboratory Services. The Crystallography and Computational Bioscience Shared Resource is supported by the Wake Forest School of Medicine and the Wake Forest Baptist Comprehensive Cancer Center.
Abbreviations
- CB1R
CB1 cannabinoid receptor
- CP55940
(−)-cis-3-[2-hydroxy-4-(1,1-dimethylheptyl)phenyl]-trans-4-(3-hydroxypropyl)cyclohexanol
- CRIP1a
cannabinoid receptor interacting protein 1a
- GFP
green fluorescent protein
- GPCR
G-protein-coupled receptor
- GRK
G-protein-coupled receptor kinase
- PBS
phosphate-buffered saline
- SS
signal sequence
- SS-GFP-CB1R
human CB1R with an N-terminal artificial SS insert and enhanced GFP
- THL
tetrahydrolipstatin
- 2-AG
2-arachidonoylglycerol
- WT
wild type
Authorship Contributions:
Participated in research design: Blume, Lowther, Selley, and Howlett.
Conducted experiments: Blume, Eldeeb, Ilyasov, Hantgan, Keegan, Leone-Kabler, Lowther, O’Neal, and Patten.
Contributed new reagents or analytic tools: Blume, Bass.
Performed data analysis: Blume, Eldeeb, Leone-Kabler, Hantgan, Lowther, Selley, and Howlett.
Wrote or contributed to the writing of the manuscript: Blume, Patten, Lowther, and Howlett.
Footnotes
This work was supported by National Institutes of Health National Institute on Drug Abuse [Grants R01-DA03690, R21-DA025321, K01-DA024763, T32-DA007246, and F31-DA032215, R03-DA035424, P50-DA006634]; National Institute on Alcoholism and Alcohol Abuse [Grant T32-AA007565]; National Cancer Institute [Grant P30-CA012197]; and National Institute of General Medical Sciences [Grants R25-GM064249, K12-GM102773]. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Current affiliation: Department of Pharmacology, University of Pennsylvania, Philadelphia, PA 19104.
Current affiliation: Department of Pharmacology, Jerry M. Wallace School of Osteopathic Medicine, Campbell University, Buies Creek, NC 27506.
Current affiliation: Department of Natural Resources and Environmental Design, North Carolina A&T State University, Greensboro, NC 27411.
Current affiliation: Department of Pharmacology and Toxicology, School of Medicine and Biomedical Sciences, University at Buffalo, Buffalo, NY 14260.
 This article has supplemental material available at molpharm.aspetjournals.org.
This article has supplemental material available at molpharm.aspetjournals.org.
References
- Ahmed MH, Kellogg GE, Selley DE, Safo MK, Zhang Y. (2014) Predicting the molecular interactions of CRIP1a-cannabinoid 1 receptor with integrated molecular modeling approaches. Bioorg Med Chem Lett 24:1158–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn KH, Mahmoud MM, Shim JY, Kendall DA. (2013) Distinct roles of β-arrestin 1 and β-arrestin 2 in ORG27569-induced biased signaling and internalization of the cannabinoid receptor 1 (CB1). J Biol Chem 288:9790–9800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asimaki O, Mangoura D. (2011) Cannabinoid receptor 1 induces a biphasic ERK activation via multiprotein signaling complex formation of proximal kinases PKCε, Src, and Fyn in primary neurons. Neurochem Int 58:135–144. [DOI] [PubMed] [Google Scholar]
- Bakshi K, Mercier RW, Pavlopoulos S. (2007) Interaction of a fragment of the cannabinoid CB1 receptor C-terminus with arrestin-2. FEBS Lett 581:5009–5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisogno T, Sepe N, Melck D, Maurelli S, De Petrocellis L, Di Marzo V. (1997) Biosynthesis, release and degradation of the novel endogenous cannabimimetic metabolite 2-arachidonoylglycerol in mouse neuroblastoma cells. Biochem J 322:671–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blom N, Gammeltoft S, Brunak S. (1999) Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J Mol Biol 294:1351–1362. [DOI] [PubMed] [Google Scholar]
- Blom N, Sicheritz-Pontén T, Gupta R, Gammeltoft S, Brunak S. (2004) Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics 4:1633–1649. [DOI] [PubMed] [Google Scholar]
- Blume LC, Bass CE, Childers SR, Dalton GD, Roberts DC, Richardson JM, Xiao R, Selley DE, Howlett AC. (2013) Striatal CB1 and D2 receptors regulate expression of each other, CRIP1A and δ opioid systems. J Neurochem 124:808–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blume LC, Eldeeb K, Bass CE, Selley DE, Howlett AC. (2015) Cannabinoid receptor interacting protein (CRIP1a) attenuates CB1R signaling in neuronal cells. Cell Signal 27:716–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blume LC, Leone-Kabler S, Luessen DJ, Marrs GS, Lyons E, Bass CE, Chen R, Selley DE, Howlett AC. (2016) Cannabinoid receptor interacting protein suppresses agonist-driven CB1 receptor internalization and regulates receptor replenishment in an agonist-biased manner. J Neurochem 139:396–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns RN, Singh M, Senatorov IS, Moniri NH. (2014) Mechanisms of homologous and heterologous phosphorylation of FFA receptor 4 (GPR120): GRK6 and PKC mediate phosphorylation of Thr347, Ser350, and Ser357 in the C-terminal tail. Biochem Pharmacol 87:650–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cushing PR, Fellows A, Villone D, Boisguérin P, Madden DR. (2008) The relative binding affinities of PDZ partners for CFTR: a biochemical basis for efficient endocytic recycling. Biochemistry 47:10084–10098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daigle TL, Kwok ML, Mackie K. (2008) Regulation of CB1 cannabinoid receptor internalization by a promiscuous phosphorylation-dependent mechanism. J Neurochem 106:70–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniele S, Trincavelli ML, Fumagalli M, Zappelli E, Lecca D, Bonfanti E, Campiglia P, Abbracchio MP, Martini C. (2014) Does GRK-β arrestin machinery work as a “switch on” for GPR17-mediated activation of intracellular signaling pathways? Cell Signal 26:1310–1325. [DOI] [PubMed] [Google Scholar]
- Delgado-Peraza F, Ahn KH, Nogueras-Ortiz C, Mungrue IN, Mackie K, Kendall DA, Yudowski GA. (2016) Mechanisms of biased β-arrestin-mediated signaling downstream from the cannabinoid 1 receptor. Mol Pharmacol 89:618–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeWire SM, Ahn S, Lefkowitz RJ, Shenoy SK. (2007) Beta-arrestins and cell signaling. Annu Rev Physiol 69:483–510. [DOI] [PubMed] [Google Scholar]
- Drake MT, Shenoy SK, Lefkowitz RJ. (2006) Trafficking of G protein-coupled receptors. Circ Res 99:570–582. [DOI] [PubMed] [Google Scholar]
- Flores-Otero J, Ahn KH, Delgado-Peraza F, Mackie K, Kendall DA, Yudowski GA. (2014) Ligand-specific endocytic dwell times control functional selectivity of the cannabinoid receptor 1. Nat Commun 5:4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia DE, Brown S, Hille B, Mackie K. (1998) Protein kinase C disrupts cannabinoid actions by phosphorylation of the CB1 cannabinoid receptor. J Neurosci 18:2834–2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guggenhuber S, Alpar A, Chen R, Schmitz N, Wickert M, Mattheus T, Harasta AE, Purrio M, Kaiser N, Elphick MR, et al. (2016) Cannabinoid receptor-interacting protein Crip1a modulates CB1 receptor signaling in mouse hippocampus. Brain Struct Funct 221:2061–2074. [DOI] [PubMed] [Google Scholar]
- Gyombolai P, Boros E, Hunyady L, Turu G. (2013) Differential β-arrestin2 requirements for constitutive and agonist-induced internalization of the CB1 cannabinoid receptor. Mol Cell Endocrinol 372:116–127. [DOI] [PubMed] [Google Scholar]
- Hájková A, Techlovská Š, Dvořáková M, Chambers JN, Kumpošt J, Hubálková P, Prezeau L, Blahos J. (2016) SGIP1 alters internalization and modulates signaling of activated cannabinoid receptor 1 in a biased manner. Neuropharmacology 107:201–214. [DOI] [PubMed] [Google Scholar]
- Hantgan RR, Stahle MC. (2009) Integrin priming dynamics: mechanisms of integrin antagonist-promoted alphaIIbbeta3:PAC-1 molecular recognition. Biochemistry 48:8355–8365. [DOI] [PubMed] [Google Scholar]
- Hantgan RR, Stahle MC, Horita DA. (2008) Entropy drives integrin alphaIIbbeta3:echistatin binding--evidence from surface plasmon resonance spectroscopy. Biochemistry 47:2884–2892. [DOI] [PubMed] [Google Scholar]
- Hanyaloglu AC, von Zastrow M. (2008) Regulation of GPCRs by endocytic membrane trafficking and its potential implications. Annu Rev Pharmacol Toxicol 48:537–568. [DOI] [PubMed] [Google Scholar]
- Howlett AC. (2009) Functional selectivity at receptors for cannabinoids and other lipids, in Functional Selectivity in G Protein Coupled Receptor Ligands: New Opportunities in Drug Discovery (Neve KA, ed) pp 211–242, Humana Press, Totowa, NJ. [Google Scholar]
- Howlett AC, Blume LC, Dalton GD. (2010) CB(1) cannabinoid receptors and their associated proteins. Curr Med Chem 17:1382–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howlett AC, Song C, Berglund BA, Wilken GH, Pigg JJ. (1998) Characterization of CB1 cannabinoid receptors using receptor peptide fragments and site-directed antibodies. Mol Pharmacol 53:504–510. [DOI] [PubMed] [Google Scholar]
- Hsieh C, Brown S, Derleth C, Mackie K. (1999) Internalization and recycling of the CB1 cannabinoid receptor. J Neurochem 73:493–501. [DOI] [PubMed] [Google Scholar]
- Jin W, Brown S, Roche JP, Hsieh C, Celver JP, Kovoor A, Chavkin C, Mackie K. (1999) Distinct domains of the CB1 cannabinoid receptor mediate desensitization and internalization. J Neurosci 19:3773–3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jönsson TJ, Johnson LC, Lowther WT. (2008) Structure of the sulphiredoxin-peroxiredoxin complex reveals an essential repair embrace. Nature 451:98–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y, Gao X, Zhou XE, He Y, Melcher K, Xu HE. (2016) A structural snapshot of the rhodopsin-arrestin complex. FEBS J 283:816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y, Zhou XE, Gao X, He Y, Liu W, Ishchenko A, Barty A, White TA, Yefanov O, Han GW, et al. (2015) Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser. Nature 523:561–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchhausen T, Macia E, Pelish HE. (2008) Use of dynasore, the small molecule inhibitor of dynamin, in the regulation of endocytosis. Methods Enzymol 438:77–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotsikorou E, Madrigal KE, Hurst DP, Sharir H, Lynch DL, Heynen-Genel S, Milan LB, Chung TD, Seltzman HH, Bai Y, et al. (2011) Identification of the GPR55 agonist binding site using a novel set of high-potency GPR55 selective ligands. Biochemistry 50:5633–5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MC, Smith FL, Stevens DL, Welch SP. (2003) The role of several kinases in mice tolerant to delta 9-tetrahydrocannabinol. J Pharmacol Exp Ther 305:593–599. [DOI] [PubMed] [Google Scholar]
- Liggett SB. (2011) Phosphorylation barcoding as a mechanism of directing GPCR signaling. Sci Signal 4:pe36. [DOI] [PubMed] [Google Scholar]
- Ludányi A, Eross L, Czirják S, Vajda J, Halász P, Watanabe M, Palkovits M, Maglóczky Z, Freund TF, Katona I. (2008) Downregulation of the CB1 cannabinoid receptor and related molecular elements of the endocannabinoid system in epileptic human hippocampus. J Neurosci 28:2976–2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macia E, Ehrlich M, Massol R, Boucrot E, Brunner C, Kirchhausen T. (2006) Dynasore, a cell-permeable inhibitor of dynamin. Dev Cell 10:839–850. [DOI] [PubMed] [Google Scholar]
- Martini L, Waldhoer M, Pusch M, Kharazia V, Fong J, Lee JH, Freissmuth C, Whistler JL. (2007) Ligand-induced down-regulation of the cannabinoid 1 receptor is mediated by the G-protein-coupled receptor-associated sorting protein GASP1. FASEB J 21:802–811. [DOI] [PubMed] [Google Scholar]
- McDonald NA, Henstridge CM, Connolly CN, Irving AJ. (2007) Generation and functional characterization of fluorescent, N-terminally tagged CB1 receptor chimeras for live-cell imaging. Mol Cell Neurosci 35:237–248. [DOI] [PubMed] [Google Scholar]
- Miller JW. Tracking G protein-coupled receptor trafficking using Odyssey (R) imaging. LI-COR Biosciences Application Notes 2004. 2004.
- Moerke NJ. (2009) Fluorescence polarization (FP) assays for monitoring peptide-protein or nucleic acid-protein binding. Curr Protoc Chem Biol 1:1–15. [DOI] [PubMed] [Google Scholar]
- Morgan DJ, Davis BJ, Kearn CS, Marcus D, Cook AJ, Wager-Miller J, Straiker A, Myoga MH, Karduck J, Leishman E, et al. (2014) Mutation of putative GRK phosphorylation sites in the cannabinoid receptor 1 (CB1R) confers resistance to cannabinoid tolerance and hypersensitivity to cannabinoids in mice. J Neurosci 34:5152–5163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay S, Cowsik SM, Lynn AM, Welsh WJ, Howlett AC. (1999) Regulation of Gi by the CB1 cannabinoid receptor C-terminal juxtamembrane region: structural requirements determined by peptide analysis. Biochemistry 38:3447–3455. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay S, Howlett AC. (2001) CB1 receptor-G protein association. Subtype selectivity is determined by distinct intracellular domains. Eur J Biochem 268:499–505. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay S, Howlett AC. (2005) Chemically distinct ligands promote differential CB1 cannabinoid receptor-Gi protein interactions. Mol Pharmacol 67:2016–2024. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay S, McIntosh HH, Houston DB, Howlett AC. (2000) The CB(1) cannabinoid receptor juxtamembrane C-terminal peptide confers activation to specific G proteins in brain. Mol Pharmacol 57:162–170. [PubMed] [Google Scholar]
- Nguyen PT, Schmid CL, Raehal KM, Selley DE, Bohn LM, Sim-Selley LJ. (2012) β-arrestin2 regulates cannabinoid CB1 receptor signaling and adaptation in a central nervous system region-dependent manner. Biol Psychiatry 71:714–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niehaus JL, Liu Y, Wallis KT, Egertová M, Bhartur SG, Mukhopadhyay S, Shi S, He H, Selley DE, Howlett AC, et al. (2007) CB1 cannabinoid receptor activity is modulated by the cannabinoid receptor interacting protein CRIP 1a. Mol Pharmacol 72:1557–1566. [DOI] [PubMed] [Google Scholar]
- Penela P, Ribas C, Mayor F., Jr (2003) Mechanisms of regulation of the expression and function of G protein-coupled receptor kinases. Cell Signal 15:973–981. [DOI] [PubMed] [Google Scholar]
- Pertwee RG. (2006) Cannabinoid pharmacology: the first 66 years. Br J Pharmacol 147 (Suppl 1):S163–S171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piliarik M, Vaisocherová H, Homola J. (2009) Surface plasmon resonance biosensing. Methods Mol Biol 503:65–88. [DOI] [PubMed] [Google Scholar]
- Por ED, Bierbower SM, Berg KA, Gomez R, Akopian AN, Wetsel WC, Jeske NA. (2012) β-Arrestin-2 desensitizes the transient receptor potential vanilloid 1 (TRPV1) channel. J Biol Chem 287:37552–37563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rejman J, Bragonzi A, Conese M. (2005) Role of clathrin- and caveolae-mediated endocytosis in gene transfer mediated by lipo- and polyplexes. Mol Ther 12:468–474. [DOI] [PubMed] [Google Scholar]
- Singh SN, Bakshi K, Mercier RW, Makriyannis A, Pavlopoulos S. (2011) Binding between a distal C-terminus fragment of cannabinoid receptor 1 and arrestin-2. Biochemistry 50:2223–2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith TH, Blume LC, Straiker A, Cox JO, David BG, McVoy JR, Sayers KW, Poklis JL, Abdullah RA, Egertová M, et al. (2015) Cannabinoid receptor-interacting protein 1a modulates CB1 receptor signaling and regulation. Mol Pharmacol 87:747–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith TH, Sim-Selley LJ, Selley DE. (2010) Cannabinoid CB1 receptor-interacting proteins: novel targets for central nervous system drug discovery? Br J Pharmacol 160:454–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadel R, Ahn KH, Kendall DA. (2011) The cannabinoid type-1 receptor carboxyl-terminus, more than just a tail. J Neurochem 117:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stauffer B, Wallis KT, Wilson SP, Egertová M, Elphick MR, Lewis DL, Hardy LR. (2011) CRIP1a switches cannabinoid receptor agonist/antagonist-mediated protection from glutamate excitotoxicity. Neurosci Lett 503:224–228. [DOI] [PubMed] [Google Scholar]
- Tanious FA, Nguyen B, Wilson WD. (2008) Biosensor-surface plasmon resonance methods for quantitative analysis of biomolecular interactions. Methods Cell Biol 84:53–77. [DOI] [PubMed] [Google Scholar]
- Thomsen AR, Plouffe B, Cahill TJ, 3rd, Shukla AK, Tarrasch JT, Dosey AM, Kahsai AW, Strachan RT, Pani B, Mahoney JP, et al. (2016) GPCR-G Protein-β-Arrestin Super-Complex Mediates Sustained G Protein Signaling. Cell 166:907–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turu G, Hunyady L. (2010) Signal transduction of the CB1 cannabinoid receptor. J Mol Endocrinol 44:75–85. [DOI] [PubMed] [Google Scholar]
- van der Lee MM, Blomenröhr M, van der Doelen AA, Wat JW, Smits N, Hanson BJ, van Koppen CJ, Zaman GJ. (2009) Pharmacological characterization of receptor redistribution and beta-arrestin recruitment assays for the cannabinoid receptor 1. J Biomol Screen 14:811–823. [DOI] [PubMed] [Google Scholar]
- Wang LH, Rothberg KG, Anderson RG. (1993) Mis-assembly of clathrin lattices on endosomes reveals a regulatory switch for coated pit formation. J Cell Biol 123:1107–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson WD. (2002) Tech.Sight. Analyzing biomolecular interactions. Science 295:2103–2105. [DOI] [PubMed] [Google Scholar]
- Zappelli E, Daniele S, Abbracchio MP, Martini C, Trincavelli ML. (2014) A rapid and efficient immunoenzymatic assay to detect receptor protein interactions: G protein-coupled receptors. Int J Mol Sci 15:6252–6264. [DOI] [PMC free article] [PubMed] [Google Scholar]