Abstract
The sleep electroencephalogram is highly heritable in humans and yet little is known about the genetic basis of inter-individual differences in sleep architecture. The aim of this study was to identify associations between candidate circadian gene variants and the polysomnogram, recorded under highly controlled laboratory conditions during a baseline, overnight, 8-h sleep opportunity. A candidate gene approach was employed to analyze single nucleotide polymorphisms from five circadian-related genes in a two-phase analysis of 84 healthy young adults (28 F; 23.21 ± 2.97 years) of European ancestry.
A common variant in Period2 (PER2) was associated with 20 minutes less slow wave sleep (SWS) in carriers of the minor allele than in non-carriers, representing a 22% difference in SWS duration. Moreover, spectral analysis in a subset of samples (n=37), showed the same PER2 polymorphism was associated with reduced EEG power density in the low delta range (0.25–1.0 Hz) during non-REM sleep and lower slow-wave activity (0.75–4.5 Hz) in the early part of the sleep episode. These results indicate the involvement of PER2 in the homeostatic process of sleep. Additionally, a rare variant in Melatonin Receptor 1B was associated with longer REM sleep latency, with minor allele carriers exhibiting an average of 65 minutes (87%) longer latency from sleep onset to REM sleep, compared to non-carriers. These findings suggest that circadian-related genes may modulate sleep architecture and the sleep EEG, including specific parameters previously implicated in the homeostatic regulation of sleep.
Keywords: circadian genes, sleep EEG, slow wave sleep, slow-wave activity
Introduction
Sleep is regulated by two main physiological processes: 1) the circadian timing system (Process C), regulating daily rhythms of sleep and wakefulness, and 2) the sleep homeostatic process (Process S) which tracks accumulating sleep pressure with time awake and which decreases with time asleep (Borbély, 1982). The prevalence and amplitude of slow waves in the sleep electroencephalogram (EEG), typically quantified by slow-wave activity (SWA; 0.75–4.5 Hz power density) or slow wave sleep (SWS), serve as markers of homeostatic sleep pressure (Achermann et al., 1993). The EEG is highly heritable, both during wakefulness (h2=76–89%) (Lennox et al., 1945; van Beijsterveldt & van Baal, 2002) and during sleep (h2=50–96%) (Ambrosius et al., 2008; Landolt & Dijk, 2010; Linkowski, 1999). Surprisingly, despite the high heritability of EEG traits, little is known about the genetic basis of these inter-individual differences in the sleep EEG.
Several sleep-associated genetic variants have been identified in human candidate-gene studies but associations have not been consistently reproduced; namely in PER3, DQB1*0602, ADA, ADORA2A, BDNF, and COMT (Bachmann et al., 2012; Bodenmann et al., 2012; Bodenmann & Landolt, 2010; Guindalini et al., 2014; Mignot et al., 1999; Retey et al., 2005; Viola et al., 2007). Of these genes, however, only PER3 is considered a genetic component of the circadian molecular timing system shown to alter the sleep EEG in humans (Viola et al., 2007). Gene variants in BDNF, a circadian-related gene, have also been implicated in changes to EEG during sleep (Bollen & Curran, 2005; Guindalini et al., 2014).
In the current study, we used a candidate gene approach to identify associations between single nucleotide polymorphisms (SNPs) in circadian-related genes and polysomnographic (PSG) measures collected under controlled laboratory conditions, an ideal setting for reducing environmental and behavioral variation to identify genetic variants with a strong effect. Common single nucleotide polymorphisms (SNPs) in 5 circadian-related genes were genotyped in our laboratory sample: Circadian Locomotor Output Cycles Kaput (CLOCK), Cryptochrome2 (CRY2), basic helix-loop-helix family, member e41 (BHLHE41; or DEC2), Period2 (PER2), and Melatonin Receptor1B (MTNR1B). As an initial exploration of the role of circadian genes in sleep physiology, we selected these genes as candidates based on a) previously reported associations with sleep phenotypes in the case of CLOCK, DEC2, and PER2 (Allebrandt et al., 2010; He et al., 2009; Toh et al., 2001) and b) association with type 2 diabetes (T2D) risk (Dupuis et al., 2010; Lyssenko et al., 2008; Prokopenko et al., 2008), in the case of CRY2 and MTNR1B. Furthermore, MTNR1B gene variants were included in this analysis because of previously documented association with metabolic measures (Dupuis et al., 2010; Lyssenko et al., 2008; Prokopenko et al., 2008) and its role as an output of the clock, not because of a role in the core molecular clock. Because sleep disturbances and short sleep duration lead to increased risk of T2D (Nedeltcheva & Scheer, 2014; Spiegel et al., 2005), we hypothesized that changes in sleep quality and/or duration may provide a mechanistic link between known variants in this gene and T2D risk (Hanlon & Van Cauter, 2011).
Methods
Overview
This genetic study includes data from individuals who previously participated in one of several inpatient physiological protocols in which their sleep EEG was recorded in the Intensive Physiological Monitoring Unit of the Brigham and Women’s Hospital. Written informed consents, both for the original in-laboratory studies and for the genetic studies, were obtained from each study participant. The Partners Human Research Committee approved the protocol and study procedures conformed to the Declaration of Helsinki.
Study Participants and Screening Procedures
A total of 101 healthy adults aged 18–35 years (34 F; mean age ± SD 23.10 ± 3.28 y) were enrolled in this study. A PSG recording collected during an 8-hour baseline overnight sleep episode (1st or 3rd night [see below]; Fig. 1) and genetic samples obtained from each participant were used for association analyses.
Figure 1.
Raster plots of laboratory protocols show PSG recorded during 8-h sleep episodes for a representative participant with the habitual sleep timing of 00:00–08:00. For all study protocols the ambient room light was dim (gray) for at least 6h before the sleep episodes (black) during which PSGs were recorded. A) In the Phase 1 cohort PSG was recorded (red box) during the 3rd baseline night. B) The first baseline night was recorded in the Phase 2 cohort.
Prior to admission, study participants completed extensive screening procedures to ensure eligibility. Inclusion criteria required that participants were 18–35 years of age and in good health, free from medical or psychological conditions or disorders. While the protocols in which the volunteers participated varied slightly from each other with respect to specific screening procedures, all protocols included a physical examination, laboratory tests of blood and urine samples, a 12-lead electrocardiogram (ECG), and an interview with a clinical psychologist to confirm good medical and psychological health. Exclusion criteria included night work in the prior 3 years and travel across >1 time zone in the prior 3 months. During the 3-week pre-admission screening interval, participants were asked to refrain from taking medications, alcohol, caffeine, and nicotine; and they maintained a self-selected, stable, 8-hour sleep schedule each night, which was verified with daily sleep diaries, telephone calls at bedtime and wake time, and wrist actigraphy for a minimum of 1 week prior to admission.
Genotyping
DNA was extracted from whole blood using Qiagen Autopure LS (Qiagen, Limburg, The Netherlands). A total of 116 SNPs tagging common variation and rare missense variants from the 5 candidate genes (CLOCK, CRY2, BHLHE41, PER2, and MTNR1B; +/− 20kb) were selected, providing full coverage for each gene (see Supplementary Table 1 for full list). SNPs with a minor allele frequency (MAF) > 0.05 were selected using a pairwise tagging threshold (r2 > 0.8) in 4 populations: Northern and Western European (CEU), Yoruba African (YRI), Han Chinese (CHB), and Japanese (JPT) in HapMap release 21 in Tagger on Haploview (Barrett et al., 2005). Additionally, 58 African-American and Hispanic ancestry informative markers were genotyped to test and correct for population stratification. Genotyping was performed using the Sequenom platform (Broad Institute, Cambridge, Massachusetts).
Standard quality control criteria included SNP genotyping rate > 90%, MAF > 1%, and Hardy Weinberg Equilibrium (HWE) p-value > 10−6, and were assessed using PLINK (Purcell et al., 2007). Eleven SNPs were excluded due to low genotyping rate, 20 SNPs were excluded due to low minor allele frequency, and 3 SNPs were excluded on the basis of departure from HWE; several of the SNPs were excluded by more than one criterion leaving 87 SNPs that passed all quality control filters. For primary association analysis, 84/101 participants that were genetically categorized as being of European ancestry (within 4 SD of the HapMap release 3 CEU panel based on principal components analysis) were selected (see Supplementary Fig 1).
PSG Recordings and Sleep Measures
PSG recordings were continuously monitored and recorded throughout an 8-h baseline sleep episode using a Vitaport-3 recording system (TEMEC Instruments, B.V. Kerkrade, The Netherlands). Recordings included the EEG (derivations C3, C4, O1, and O2, referenced to contralateral mastoids), right and left electrooculogram (EOG), electromyogram (EMG), and the electrocardiogram (ECG). Electrode impedances were <10 kΩ prior to the beginning of each recording. EEG signals were filtered (high-pass EEG filter 0.23 Hz; low-pass EEG filter 70.1 Hz; 24 dB/octave, sampling rate 256 Hz). PSG records were scored (blind to genotype) visually in 30-second epochs according to conventional criteria of Rechtschaffen and Kales (Rechtschaffen & Kales, 1968). Sleep measures included: total sleep time (TST); sleep efficiency (TST/time in bed as a %); sleep latency (interval between lights off and first stage of sleep); duration for stages 1, 2, slow wave sleep (SWS; S3 + S4 combined), REM sleep, and wakefulness; wake after sleep onset (WASO); wake after final awakening (WAFA); number of awakenings longer than 30 seconds. Duration of sleep stages and wakefulness was calculated in minutes. For sleep stage 2, SWS, and REM sleep, latencies were computed from sleep onset. PSG measures that were not normally distributed were transformed to achieve a normal distribution, including SWS duration and number of awakenings (log transformation) and REM sleep latency (inverse normal transformation).
Spectral analysis of C3/A2 EEG recordings was also completed in the subset of male participants whose PSG was recorded on the 3rd baseline night (n=37; see below). Signals were first visually inspected, and 4-second epochs containing artifacts arising from body movements, eye blinks, or eye movements were removed. The remaining 4-second epochs were subjected to fast-Fourier transformation, using a 10%-cosine-tapered window (Vitascore, TEMEC, The Netherlands). A maximum of ten, consecutive, overlapping 4-second epochs were averaged into 30-second epochs and matched with sleep scores. Data were reduced further by averaging power densities into 0.5-Hz bins. For a specific SNP that was associated with SWS, power spectra were compared between genotype groups (risk-allele carriers vs. non-carriers) by ANOVA (SAS 9.2).
Genetic Analysis and Protocol Groups
Genetic analysis was initiated in a cohort in whom PSG was recorded during the 8-h sleep episode on the 3rd baseline night (Phase 1; n=59; Fig. 1A). Potential associations with significance (p < 0.001) were then tested in a separate cohort of different individuals in whom PSG recordings were collected during the 8-h sleep episode on the 1st baseline night in the laboratory (Phase 2; n=25; Fig. 1B). Procedures for the collection of PSG, sleep schedule, room conditions, and ambient light levels prior to the sleep episode were consistent between Phase 1 and Phase 2 cohorts. Both groups were independent with no overlap of individuals, and the cohorts were selected a priori. The Phase 2 group in which PSG was collected during the 1st baseline night was selected to provide a more stringent test of replication for any potential associations that were identified in the Phase 1 cohort, assuming greater variability in measures due to first night effects.
An additive genetic model was used to test for association of SNPs with sleep measures in linear regression analyses adjusting for covariates age, sex and ancestry (5 principal components). Permutation testing was used to evaluate significance after correction for multiple comparisons. As power for single variant association testing was low and individual effects may be subtle, an additional gene-based analysis was conducted using sequence kernel association testing (SKAT) to detect combined effects of variants (Ionita-Laza et al., 2013). This analysis provides a more comprehensive view of potential associations between candidate genes and phenotypes by including all variants from one gene, rather than each individual SNP, in the analysis. Furthermore, this allowed for a reduced number of comparisons (the number of genes as opposed to the number of SNPs).
Results
Summary sleep measures for both cohorts are shown in Table 1. The cohorts differed significantly only in REM sleep duration, with a shorter REM sleep duration in the Phase 2 cohort, which may have been due to a first night effect in that study (Agnew et al., 1966). Results for SNPs most strongly associated with sleep phenotype measures in the Phase 1 cohort, in the Phase 2 cohort, and from meta-analysis of the entire sample are listed in Table 2. In Phase 1, a SNP in the PER2 gene (rs6753456) showed nominal association with SWS duration; a second PER2 SNP (rs3739064) was associated with the number of awakenings; and a MTNR1B SNP (rs7942988) was associated with REM sleep latency. None of these associations remained significant in the smaller Phase 2 cohort in which only first night sleep data were assessed. Nonetheless, 2 associations remained significant in the meta-analysis of both phases of analysis: the PER2 SNP with SWS duration, and the MTNR1B SNP with REM sleep latency. For the 2 phenotypes that showed significant association in the meta-analysis the association with all tested SNPs are shown in Supplementary Figure 2; and association plots for remaining sleep phenotypes with all SNPs are shown in Supplementary Figure 3. Results from association testing of the two sleep-associated SNPs with circadian phenotypes are shown in Supplementary Table 2.
Table 1.
Summary of sleep measures for Phase 1 (n=59) and Phase 2 (n=25) cohorts.
| PSG measure | Phase 1 mean (SD) | Phase 2 mean (SD) | T-test p |
|---|---|---|---|
| TST (min) | 440.98 (21.56) | 431.76 (38.79) | 0.272 |
| Sleep efficiency (%) | 91.85 (4.49) | 89.92 (8.07) | 0.269 |
| Sleep latency (min) | 10.59 (7.02) | 12.26 (8.89) | 0.410 |
| Stage 1 duration (min) | 26.17 (10.80) | 21.38 (11.19) | 0.077 |
| Stage 2 duration (min) | 209.90 (35.16) | 219.18 (32.62) | 0.250 |
| Stage 2 latency (min) | 3.54 (6.86) | 4.46 (3.22) | 0.407 |
| SWS duration (min) | 87.53 (34.74) | 92.08 (28.99) | 0.539 |
| REM duration (min) | 118.08 (23.52) | 99.40 (20.96) | 7.31E-04 |
| REM latency (min) | 73.28 (36.75) | 80.02 (37.85) | 0.456 |
| WASO (min) | 22.05 (20.44) | 32.88 (34.49) | 0.153 |
| Awakenings >30 sec | 10.46 (6.92) | 11.44 (8.43) | 0.610 |
| WAFA (min) | 3.76 (14.02) | 3.40 (7.68) | 0.880 |
Significant p-values (p<0.05) are shown in bold.
Table 2.
Genetic association results with PSG phenotypes.
| Gene | PER2 | PER2 | MTNR1B |
|---|---|---|---|
| SNP | rs6753456 | rs3739064 | rs7942988 |
| Minor allele | G | G | T |
| Frequency | 37.5% | 21.3% | 3.7% |
| PSG measure | SWS duration (min) | # awakenings/night | REM latency (min) |
|
| |||
| Phase 1 cohort | |||
| n | 59 | 57 | 57 |
| Beta (SE) | −24.37 (7.00) | −9.64 (2.42) | 81.19 (17.50) |
| Beta (SE)* | −0.29 (0.08) | −0.93 (0.25) | 3.90 (1.00) |
| p* | 9.2E-04 | 6.0E-04 | 3.1E-04 |
| Minor hom mean (SE) | 78.63 (13.27) | 2.50 (0.50) | na |
| Het mean (SE) | 80.13 (5.63) | 11.06 (1.88) | 163.00 (38.00) |
| Major hom mean (SE) | 98.83 (7.70) | 11.03 (1.11) | 78.85 (3.16) |
|
| |||
| Phase 2 cohort | |||
| n | 25 | 25 | 25 |
| Beta (SE) | −19.38 (10.41) | −2.22 (4.51) | 27.27 (27.66) |
| Beta (SE)* | −0.19 (0.11) | −0.02 (0.39) | 1.47 (1.43) |
| p* | 0.103 | 0.959 | 0.32 |
| Minor hom mean (SE) | 97.00 (3.54) | 5.50 (1.50) | na |
| Het mean (SE) | 84.71 (8.06) | 12.57 (3.69) | 115.17 (27.53) |
| Major hom mean (SE) | 99.72 (11.88) | 11.69 (2.09) | 90.52 (8.92) |
|
| |||
| Meta-analysis | |||
| n | 84 | 82 | 82 |
| Beta (SE) | −20.46 (5.58) | −6.30 (2.23) | 64.55 (14.46) |
| Beta (SE)* | −0.24 (0.06) | −0.54 (0.21) | 0.61 (0.17) |
| p* | 4.4E-04 | 1.3E-02 | 7.4E-04 |
| p-adj* | 4.6E-02 | 0.505 | 4.6E-02 |
| Minor hom mean (SE) | 84.75 (9.09) | 3.50 (0.81) | na |
| Het mean (SE) | 81.54 (4.57) | 11.52 (1.68) | 139.08 (23.55) |
| Major hom mean (SE) | 99.08 (6.37) | 11.23 (0.99) | 82.23 (3.44) |
Three SNPs (rs6753456, rs3739064, and rs7942988) showed association (p<0.001) with PSG sleep phenotypes in the Phase 1 cohort. The beta and p-value of transformed data are denoted by the asterisk. Adjusted p-values (p-adj) using covariates for age, sex and X principal components were corrected for multiple comparisons. Mean (SE) values for the associated phenotypes are listed by genotype for each SNP for Phase 1, Phase 2, and meta-analysis of both cohorts; note these values are unadjusted. Genotype groups included 1) homozygous individuals for the minor allele (minor hom), 2) heterozygous individuals (het) and, 3) homozygous individuals for the major allele (major hom).
PER2
A common polymorphism in the PER2 gene (rs6753456; MAF 38%) was nominally significant in the Phase 1 sample for SWS duration with 24 minutes less in SWS in risk allele carriers compared to non-carriers, a difference of 27% (p=9.2×10−4; Table 2). While the magnitude of the effect of this PER2 SNP was similar in the Phase 2 cohort (19 minutes less of SWS), this association was not statistically significant (p=0.103). Meta-analysis of both phases showed association with 20 minutes of less SWS duration, equivalent to a 22% difference compared to non-carriers (p=4.43×10−4). This difference was significant after correction for multiple-comparisons (padj=0.046). Consistent results of a study-wide significant association (p=3.39×10−4) with no heterogeneity of genetic effect between the more controlled Phase 1 cohort and the noisier Phase 2 cohort (I2=0%, phet=0.49; (Han & Eskin, 2011)) were seen using a modified random effect meta-analysis approach, indicating that the genetic signal was robust to differences between the datasets. Model-estimated mean differences ± SE for SWS duration for the Phase 1, Phase 2, and meta-analysis groups are shown in Figure 2A. Results from meta-analysis of both phase groups revealed lower SWS in homozygous and heterozygous groups compared to the non-risk allele carriers (Fig. 2A).
Figure 2.
Model-estimated mean differences by genotype for (A) PER2 SNP rs6753456 and SWS duration and (B) MTNR1B SNP rs7942988 and REM sleep latency. Model-estimated means are set to 0 minutes for each cohort and mean differences were adjusted for covariates. Mean difference, standard error (SE), and corresponding n for each genotype group are shown for homozygous (black bars) and heterozygous (grey bars) carriers for the risk allele and non-carriers (white bars).
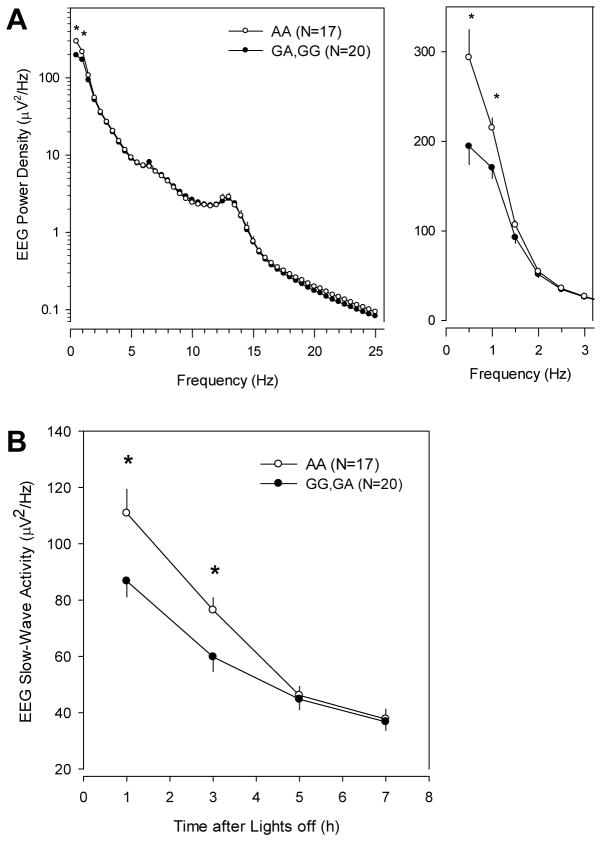
EEG spectral analysis was carried out in the 37 male participants from the Phase 1 sample in order to eliminate heterogeneity by sex, given that slow wave sleep power density differs between men and women (Armitage, 1995; Dijk et al., 1989; Ehlers & Kupfer, 1997), and because there was a very unequal ratio of men to women in the genotype groups. Genotype affected the power spectrum during non-REM sleep in a frequency-specific manner (Genotype x EEG Frequency bin, p=0.049; Fig 3A). Post-hoc analysis showed that this effect arose mainly from lower power density in the low delta range (0.25–1.0 Hz; p<0.044, F-test) in individuals with one or two copies of the risk allele compared to non-carriers. Moreover, genotype affected the time course of the typical SWA decline across the night that is a measure of the decrease in homeostatic sleep pressure across the night. The biggest effects of genotype was observed in the first half of the night, with lower EEG SWA in the risk allele carriers and a shallower decline of SWA across the sleep episode in the carriers (Fig 3B). EEG power spectra during REM sleep were not affected by genotype (data not shown).
Figure 3.
Effect of PER2 rs6753456 genotype on EEG power density during NREM sleep in the male subset (n=37) of the Phase 1 cohort. A) Mean power density spectra (left: 0.25–25.0Hz; right (enlarged): 0.25–3.0Hz) are shown for risk allele carriers (filled symbols) and non-carriers (open symbols). Risk allele carriers have significantly reduced spectral power density in the low delta-frequency range (0.25–1.0Hz). B) Mean slow-wave activity (SWA; 0.75–4.5 Hz) is plotted for consecutive 2-hour bins after lights off. Significant differences between genotypes (p<0.05, F-tests) are denoted by the asterisks and error bars show SE.
Another common PER2 SNP (rs3739064; MAF 21.3%) showed notable association with the number of awakenings in the Phase 1 cohort (p=6.0×10−4; Table 2). Minor allele homozygotes (G/G) had an average of 8 fewer awakenings per night than either those with a single copy of the minor allele (G/A) or non-carriers (A/A). This association was not significant in the Phase 2 sample and meta-analysis did not show significant association after correction of multiple comparisons (p=0.013, padj=0.505).
MTNR1B
A SNP in the MTNR1B gene (rs7942988; MAF 3.7%) was significantly associated with REM sleep latency from sleep onset (p=3.10×10−4; Table 2) in the Phase 1 cohort. Individuals carrying the risk allele showed much longer latencies to REM sleep (by 81 minutes) than non-carriers, corresponding to a doubling of REM sleep latency (111% increase). This association was not significant in the Phase 2 sample alone (p>0.1), but significant (p=4.14×10−4, padj=0.046; Table 2) in the overall sample, with the latency from sleep onset to REM sleep 65 minutes (87%) longer in carriers than in non-carriers. In a secondary meta-analysis assuming random effects and testing for heterogeneity, this association demonstrated a consistent effect between the Phase 1 and Phase 2 cohorts (p=6.5×10−3, I2=8.7%, phet=0.30). The risk allele of rs7942988 (T) is relatively rare with a minor allele frequency (MAF) of 3.7% in our population (6 T/C and 0 T/T individuals). The distribution of REM sleep latency values from both the Phase 1 and Phase 2 cohorts are plotted in Supplementary Figure 4 and show that half of the risk allele carriers had REM sleep latencies that were more than twice as long as the group average (>120 minutes). Post-hoc analysis found that genotype had a significant effect on the chance of having a very long sleep latency using both Chi-squared analysis (OR=8.79; CI=1.48–52.07; p=0.008) and Fisher’s exact test (p=0.015). Figure 2B displays the model-estimated mean differences for REM sleep latency in the discovery and replication cohorts by genotype.
This same MTNR1B gene variant rs7942988 that associated with REM sleep latency had a significant association (p = 0.013; padj = 0.039) with the duration of melatonin secretion, calculated as the difference between the dim light melatonin onset (DLMO) and offset (DLMOff). Because melatonin was collected only in the Phase 1 sample, there were only 3 individuals with this rare allele (T), but they had significantly reduced duration of nocturnal melatonin secretion by 1.33 hours compared to non-carriers, corresponding to a 13.1% reduction (Supplementary Table 2). This association with the melatonin secretion duration remained significant after correcting for multiple testing of the 3 SNPs. This SNP was also associated with an earlier diurnal preference (p = 0.041), although this association was no longer significant after correction for multiple testing of 3 SNPs.
Gene-based Analysis
Results of SKAT gene-based analysis showed associations for BHLHE41 (DEC2) with the duration of REM sleep (p = 0.005); PER2 with SWS (p = 0.027); and MTNR1B with REM sleep latency (p = 0.028) in stage 1. Only the BHLHE41 association remained significantly associated with REM sleep duration after correcting for multiple comparisons (5 genes; padj = 0.01).
Discussion
Associations between single variants in two candidate genes and sleep PSG measures were identified in our Phase 1 sample. Meta-analysis also showed study-wide significant associations for PER2 SNP rs6753456 with SWS duration and MTNR1B SNP rs7942988 with REM sleep latency that withstood correction for multiple tests. Neither association was observed in the smaller Phase 2 sample alone; however, similar effect estimates and concordant direction of the allelic effects were seen in both samples. While statistical power ranged from 83–89% (with alpha=0.05) in the Phase 2 sample to replicate the effect that was observed in the Phase 1 sample, lack of replication may be due to “winner’s curse” in the initial sample, the small size of the Phase 2 sample, and/or less favorable/more noisy experimental conditions during which the PSG was recorded (first night in Phase 2 cohort versus third night in Phase 1 cohort). As expected for a ‘first-night’ effect in the Phase 2 cohort, during which sleep is typically more disturbed due to sleeping in an unfamiliar laboratory environment and PSG recordings for the first time, there was significantly less REM sleep in the Phase 1 cohort (Agnew et al., 1966). First-night effects in the Phase 2 cohort therefore could have confounded associations with the SNPs. Indeed, we purposefully selected a ‘noisier’ Phase 2 cohort to perform a stringent validation of significant findings from the Phase 1 cohort, reasoning that replication despite the increased instability of first night measurements would favor true associations over false positives. Replication in additional large studies with EEG measurements will be important to validate these associations. Our study identified a common variant of PER2 (rs6753456) that was significantly associated with decreased SWS duration in the combined analysis of both cohorts after correction for multiple comparisons, and showed a large effect size (20-min reduction of SWS).
This SNP (rs6753456) is located in the upstream promoter region of PER2, a core circadian clock gene, and is in strong linkage disequilibrium (r2 = 0.8) in Europeans with rs11894491, a regulatory variant annotated with promoter and enhancer histone marks, DNAse hypersensitivity sites and Polymerase II binding in multiple cell lines (Ward & Kellis, 2012). However, these SNPs are uncorrelated to another PER2 SNP in the 5′ untranslated region that was reported to be associated with diurnal preference (Carpen et al., 2005). In the current study, PER2 gene variant rs6753456 was not associated with diurnal preference or any other circadian phenotype, such as timing of sleep, as was seen with a rare PER2 missense mutation in a family with Advanced Sleep Phase Disorder (Toh et al., 2001). A study in mice showed that the effects of an acute 6-hour sleep deprivation on increasing PER2 expression were dependent on the time of day (Curie et al., 2013). In that report, the authors suggest that PER2 acts as an integrator of both circadian and homeostatic signals. Our findings are consistent with a role of PER2 in homeostatic sleep regulation. First, the greatest genotype-dependent differences in power density were found in the low delta range of the non-REM sleep EEG, which is particularly sensitive to changes in homeostatic sleep pressure (Aeschbach et al., 1996). Second, the differences in SWA were specifically seen during the early part of sleep, i.e., a time when homeostatic sleep pressure and its manifestation in the EEG are typically highest. To further corroborate the potential role of circadian genes in homeostatic sleep regulation, it will be important to subject individuals with different genotypes to challenges of the homeostatic system (e.g., sleep deprivation).
The MTNR1B variant rs7942988 showed significant association with REM sleep latency. The low prevalence of this allele in our population may explain the lack of replication. Although only 6/84 individuals were carriers for the risk allele, the effect size was considerable (>1 hour longer REM sleep latency) and half of the carriers displayed REM sleep latencies of more than two hours (120 minutes) indicating a “skipped” first REM sleep episode. It is remarkable that 3 of the 6 minor allele carriers were in the small subgroup of 9 individuals who displayed a long REM sleep latency, consistent with skipping the first REM sleep episode (see Supplemental Figure 4). Post-hoc analysis to test whether the chance for skipping of the first REM sleep episode was significantly affected by the genotype found a significant association using both Chi-squared analysis and Fisher’s exact test. Since REM sleep is the sleep state most influenced by circadian phase (Czeisler et al., 1980), and MTNR1B is expressed in the central circadian pacemaker, the suprachiasmatic nucleus (SCN) (Pandi-Perumal et al., 2008), the association between an MTNR1B gene variant and REM sleep latency may indicate that MTNR1B function modulates the SCN control of REM sleep propensity. This would need to be determined in future studies. It is also notable that we observed a significant association with a circadian phenotype (1.33 h shorter duration of melatonin profile), and a nominally significant association with diurnal preference (see Supplementary Table 2).
Although it is not clear how longer REM sleep latency and shorter duration of melatonin secretion are related, these findings suggest a possible role for MTNR1B in the circadian regulation of sleep, specifically REM sleep. Interestingly, and consistent with such a potential relationship, REM sleep latency has been reported to be increased in tetraplegia patients with complete cervical spinal cord transection and abolished nighttime peak in circulating melatonin (Berlowitz et al., 2012; Scheer et al., 2006). More recently, in a separate analysis, we have found that the MTNR1B variant, rs10830963, previously associated with increased fasting glucose levels and increased risk of T2D is associated with longer duration of elevated melatonin levels and a delay in circadian phase of melatonin offset in a similar inpatient sample as the one in this analysis (Lane et al., 2015). The variant in the current study, rs7942988, is not itself associated with risk of T2D (http://diagram-consortium.org) or in strong linkage disequilibrium with common or rare MTNR1B variants previously associated with T2D (Dupuis et al., 2010; Lyssenko et al., 2008; Prokopenko et al., 2008).
Results of the gene-based analysis showed trends of these associations for the PER2 and MTNR1B variants with SWS duration and REM sleep latency, respectively. In addition, the gene-based analysis showed a significant association in a 3rd gene, BHLHE41, with REM sleep duration, which was not identified by the analysis of individual SNPs. A rare variant in this gene (also known as DEC2) was associated with shorter sleep duration in humans (hDEC2-P385R) and less sleep time in transgenic mice carrying this mutation as compared to controls (He et al., 2009). Furthermore, sleep-deprivation studies of both transgenic hDEC2-P385R mice and Dec2 knockout mice showed slower recovery following sleep loss indicating the importance of this gene in sleep regulation (He et al., 2009). We did not find this rare variant in our entire sample, not surprising given the very low frequency of this allele and thus, were not able to replicate this previously reported association. Taken together, however, our findings support the evidence that BHLHE41 plays a role in the genetic regulation of sleep.
The small sample size was a limitation of this study, and due to limited power in this sample, we examined relatively common variants. A small sample size in addition to an unequal male/female ratio in the genotype groups for PER2 SNP rs6753456 was a limitation for the EEG spectral analysis. It will be important to replicate these findings in large samples, especially for the MTNR1B rare variant. Given the few number of minor allele carriers for this variant (6) and the finding that 3 of these showed significantly longer REM sleep latencies, it is theoretically possible that skipping of the first REM sleep episode could have influenced assessment of SWS and SWA, measures of sleep homeostasis. We found no direct association between MTNR1B SNP rs7942988 and SWS, however, making it unlikely that carriers were under increased homeostatic sleep pressure. Additionally, we focused our primary analysis on a European subset of the sample in order to examine a homogeneous group. Another limitation was that we tested only a few candidate genes as opposed to a more comprehensive panel or GWAS, although this reduced our multiple-testing burden. Selection of candidate genes was partially based on previously reported associations with circadian rhythms (Toh et al., 2001), sleep phenotypes (Allebrandt et al., 2010; He et al., 2009), and/or diabetes risk (Dupuis et al., 2010; Lyssenko et al., 2008; Prokopenko et al., 2008), however our aim was full coverage of each candidate gene by tagged SNPs rather than only previously-associated SNPs. Notably, we found associations with SNPs in 2 of the 5 genes selected for this study. These positive results are likely due to the use of the phenotypic measures that were collected under exceedingly well-controlled conditions in carefully screened participants, and involved the application of consistent and stringent procedures for the collection and analysis of data across protocols. Further studies examining these and other circadian gene variants with sleep EEG collected under different conditions is needed for replication of our results, to determine functional pathways and tissue specificity for gene expression, and for identification of other genetic associations.
Supplementary Material
Acknowledgments
We thank the study participants, the staff of the Center for Clinical Investigation of the Brigham and Women’s Hospital (BWH) for collection of the genetic samples, and the staff of the Broad Institute, Cambridge, MA for genotyping assays. We also thank the BWH Sleep & EEG Core, in particular Brandon Lockyer, for PSG analysis and support; and Joseph Ronda for support with an automated sleep summary program. We thank Alana O’Malley, David Klements, Dayna Bradstreet, and the recruitment office of the Division of Sleep Medicine for recruitment and screening of study participants; and Shantha Rajaratnam Ph.D. and Melanie Rüger Ph.D. for their contributions to the study protocols.
Footnotes
Declaration of Interest Statement
This study was funded by National Institute of Health (NIH) grant R21-DK089378, a pilot grant from Harvard Catalyst, Harvard Clinical and Translational Science Center (UL1 RR025758) and financial contributions from Harvard University. Additional funding was provided by NIH grants HL080978, MH045130, and HL077453; National Space Biomedical Research Institute (NSBRI) grant HPF01601; the BWH General Clinical Research Center (NIH M01-02635); and a BWH Faculty Career Development Award to AMC. AMC was supported in part by NIH grant K01-HL115458. DA was supported in part by the German Aerospace Center. SAS was supported in part by NIH grant K24-HL076446. FAJLS and RS were supported by a Harvard Catalyst Pilot Grant, R21-DK089378, R01-DK102696, and R21-HL121728. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH, Harvard Catalyst, Harvard University and its affiliated academic healthcare centers, NSBRI or BWH.
None of the authors have conflicts of interest with the work presented but the following financial support is stated in the interest of full disclosure.
Dr. Buxton reports research grant support from the National Institutes of Health outside of the submitted work; consulting with Matsutani America (scientific advisory board) and Harvard T.H. Chan School of Public Health. Travel support and honoraria for speaking from American Academy of Craniofacial Pain; Chevron; New York University; Academy of Nutrition and Dietetics; and Harvard T.H. Chan School of Public Health. Dr. Buxton received two investigator-initiated grants from Sepracor Inc (now Sunovion; ESRC-0004 and ESRC-0977, ClinicalTrials.gov Identifiers NCT00555750, NCT00900159), and two investigator-initiated grants from Cephalon Inc (now Teva; ClinicalTrials.gov Identifier: NCT00895570). OMB received Speaker’s Bureau, CME and non-CME lecture honoraria and an unrestricted educational grant from Takeda Pharmaceuticals North America. OMB serves as a consultant and expert witness for Dinsmore LLC, consulting fees for serving on the Scientific Advisory Board of Matsutani America, and consulting fees from the Wake Forest University Medical Center (NC). OMB received speaking fees and/or travel support for speaking from American Academy of Craniofacial Pain, NHLBI, NIDDK, National Postdoctoral Association, Oklahoma State University, Oregon Health Sciences University, SUNY Downstate Medical Center, American Diabetes Association, and New York University.
Dr. Anderson has served as consultant to the Rail, Bus and Tram Union. National Transport Commission (Australia) and VicPolice. She has also received research support from VicRoads and Sanofi-Aventis, and has received lecturing fees from Brown Medical School/Rhode Island Hospital and Ausmed.
Dr. Czeisler has received consulting fees from or served as a paid member of scientific advisory boards for: Bombardier, Inc.; Boston Bruins; Boston Celtics; Boston Red Sox; Cephalon, Inc. (acquired by Teva Pharmaceutical Industries Ltd. October 2011); Koninklijke Philips Electronics, N.V. (acquired Respironics, Inc. March 2008); Michael Jackson’s mother and children; Novartis; Sleep Multimedia, Inc.; United Parcel Service (UPS); Vanda Pharmaceuticals, Inc.; and Zeo Inc. CAC owns an equity interest in Apple; Lifetrac, Inc.; Microsoft; Somnus Therapeutics, Inc.; Vanda Pharmaceuticals, Inc., and Zeo Inc., and received royalties from McGraw Hill, Penguin Press/Houghton Mifflin Harcourt, and Philips Respironics, Inc. He has also received research support from Cephalon, National Football League Charities, ResMed and Philips Respironics; and has received lecture fees from APSS (Associated Professional Sleep Societies); AWHONN (Association of Women’s Health, Obstetric and Neonatal Nurses); Harvard School of Public Health; Hokkaido University Graduate School of Medicine; Japan Society for Occupational Health; Society of Thoracic Surgeons; University of Washington; the World Federation of Sleep Research and Sleep Medicine Societies and WME Entertainment LLC. CAC has also received research prizes with monetary awards from the American Academy of Sleep Medicine. The Harvard Medical School Division of Sleep Medicine (HMS/DSM), which CAC directs, has received unrestricted research and educational gifts and endowment funds from: Boehringer Ingelheim Pharmaceuticals, Inc., Cephalon, Inc., George H. Kidder, Esq., Gerald McGinnis, GlaxoSmithKline, Herbert Lee, Hypnion, Jazz Pharmaceuticals, Jordan’s Furniture, Merck & Co., Inc., Peter C. Farrell, Ph.D., Pfizer, ResMed, Respironics, Inc., Sanofi-Aventis, Inc., Sealy, Inc., Sepracor, Inc., Simmons, Sleep Health Centers LLC, Spring Aire, Takeda Pharmaceuticals and Tempur-Pedic. The HMS/DSM has received gifts from many outside organizations and individuals including: Cephalon, Inc., Committee for Interns and Residents, Eisai, Inc., Farrell Family Foundation, Fisher & Paykel Healthcare Corporation, Gerald McGinnis, Jazz Pharmaceuticals, Jordan’s Furniture, Lilly USA, LLC, Neurocare Center for Sleep, NeuroScience, Novartis Consumer Health, Philips-Respironics, Inc., Praxair US Homecare, Purdue Pharma, ResMed Foundation, Safeway, Sanofi-Aventis, Inc., Select Comfort Corporation, Sleep HealthCenters LLC, Somaxon Pharmaceuticals, Transcept Pharmaceuticals, United Healthcare, Vanda Pharmaceuticals, Inc., Wake Up Narcolepsy, Inc., Watermark Medical, Weight Watchers International, YMCA of the USA and Zeo, Inc. The HMS/DSM Sleep and Health Education Program has received Educational Grant funding from Cephalon, Inc., Takeda Pharmaceuticals, Sanofi-Aventis, Inc. and Sepracor, Inc. CAC is the incumbent of an endowed professorship provided to Harvard University by Cephalon, Inc. and holds a number of process patents in the field of sleep/circadian rhythms (e.g., photic resetting of the human circadian pacemaker). Since 1985, CAC has also served as an expert witness on various legal cases related to sleep and/or circadian rhythms.
Dr. Lockley has received consulting fees from Naturebright, Sound Oasis, Thomas Jefferson University and Wyle Integrated Science and Engineering (NASA), Blackrock, Cowen & Co, Endurant Capital Management, Far West Capital Management, Fidelity, Frankel Group, Impax Laboratories, Kearney Venture Partners, Lazard Capital Markets, New Horizon Capital, Perceptive Advisors, Polar Capital, ResearchWorks Inc, and Wyvern Funds. He has also received unrestricted equipment gifts from Biological Illuminations LLC, Bionetics Corporation, and Philips Lighting; an unrestricted monetary gift from Swinburne University of Technology, Australia; a fellowship gift from Optalert, Pty, Melbourne, Australia; equity in iSLEEP, Pty, Melbourne, Australia; and advance author payment and royalties from Oxford University Press and from Elsevier. SWL has received honoraria and/or travel and accommodation support from 8th International Conference on Managing Fatigue; 14th Annual Tennessee Perfusion Conference; Brown University; Connecticut Business & Industry Association Health and Safety Conference; Emergency Services Steering Committee; Estee Lauder; Harvard University; Lighting Science Group Corp; Massachusetts General Hospital; MediCom Worldwide, Inc (CME); Midwest Lighting Institute; National Research Council Canada; New England College of Occupational and Environmental Medicine; Ontario Association of Fire Chiefs; Rio Tinto; UMass Memorial; Woolcock Institute of Medical Research; and Wyle Integrated Science and Engineering. He has received investigator-initiated research grants from Biological Illuminations LLC, Philips Lighting, and Vanda Pharmaceuticals and has served as a paid expert on behalf of six public bodies in arbitration hearings related to sleep, circadian rhythms and work hours in firefighters and police.
References
- Achermann P, Dijk DJ, Brunner DP, Borbély AA. A model of human sleep homeostasis based on EEG slow-wave activity: Quantitative comparison of data and simulations. Brain Research Bulletin. 1993;31:97–113. doi: 10.1016/0361-9230(93)90016-5. [DOI] [PubMed] [Google Scholar]
- Aeschbach D, Cajochen C, Landolt H-P, Borbély AA. Homeostatic sleep regulation in habitual short sleepers and long sleepers. American Journal of Physiology. 1996;270:R41–R53. doi: 10.1152/ajpregu.1996.270.1.R41. [DOI] [PubMed] [Google Scholar]
- Agnew HW, Jr, Webb WB, Williams RL. The first night effect: An EEG study of sleep. Psychophysiol. 1966;2:263–266. doi: 10.1111/j.1469-8986.1966.tb02650.x. [DOI] [PubMed] [Google Scholar]
- Allebrandt KV, Teder-Laving M, Akyol M, Pichler I, Muller-Myhsok B, Pramstaller P, Merrow M, Meitinger T, Metspalu A, Roenneberg T. CLOCK gene variants associate with sleep duration in two independent populations. Biological Psychiatry. 2010;67:1040–1047. doi: 10.1016/j.biopsych.2009.12.026. [DOI] [PubMed] [Google Scholar]
- Ambrosius U, Lietzenmaier S, Wehrle R, Wichniak A, Kalus S, Winkelmann J, Bettecken T, Holsboer F, Yassouridis A, Friess E. Heritability of sleep electroencephalogram. Biological Psychiatry. 2008;64:344–348. doi: 10.1016/j.biopsych.2008.03.002. [DOI] [PubMed] [Google Scholar]
- Armitage R. The distribution of EEG frequencies in REM and NREM sleep stages in healthy young adults. Sleep. 1995;18:334–341. doi: 10.1093/sleep/18.5.334. [DOI] [PubMed] [Google Scholar]
- Bachmann V, Klein C, Bodenmann S, Schafer N, Berger W, Brugger P, Landolt HP. The BDNF Val66Met polymorphism modulates sleep intensity: EEG frequency- and state-specificity. Sleep. 2012;35:335–344. doi: 10.5665/sleep.1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Berlowitz DJ, Spong J, Gordon I, Howard ME, Brown DJ. Relationships between objective sleep indices and symptoms in a community sample of people with tetraplegia. Archives of Physical Medicine and Rehabilitation. 2012;93:1246–1252. doi: 10.1016/j.apmr.2012.02.016. [DOI] [PubMed] [Google Scholar]
- Bodenmann S, Hohoff C, Freitag C, Deckert J, Retey JV, Bachmann V, Landolt HP. Polymorphisms of ADORA2A modulate psychomotor vigilance and the effects of caffeine on neurobehavioural performance and sleep EEG after sleep deprivation. British Journal of Pharmacology. 2012;165:1904–1913. doi: 10.1111/j.1476-5381.2011.01689.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodenmann S, Landolt HP. Effects of modafinil on the sleep EEG depend on Val158Met genotype of COMT. Sleep. 2010;33:1027–1035. doi: 10.1093/sleep/33.8.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollen KA, Curran PJ. Latent growth curve models: A structural equation perspective. Hoboken, NJ: Wiley; 2005. [Google Scholar]
- Borbély AA. A two process model of sleep regulation. Human Neurobiology. 1982;1:195–204. [PubMed] [Google Scholar]
- Carpen JD, Archer SN, Skene DJ, Smits M, von schantz M. A single-nucleotide polymorphism in the 5′-untranslated region of the hPER2 gene is associated with diurnal preference. J Sleep Res. 2005;14:293–297. doi: 10.1111/j.1365-2869.2005.00471.x. [DOI] [PubMed] [Google Scholar]
- Curie T, Mongrain V, Dorsaz S, Mang GM, Emmenegger Y, Franken P. Homeostatic and circadian contribution to EEG and molecular state variables of sleep regulation. Sleep. 2013;36:311–323. doi: 10.5665/sleep.2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czeisler CA, Weitzman ED, Moore-Ede MC, Zimmerman JC, Knauer RS. Human sleep: Its duration and organization depend on its circadian phase. Science. 1980;210:1264–1267. doi: 10.1126/science.7434029. [DOI] [PubMed] [Google Scholar]
- Dijk DJ, Beersma DGM, Bloem G. Sex differences in the sleep EEG of young adults: Visual scoring and spectral analysis. Sleep. 1989;12:500–507. doi: 10.1093/sleep/12.6.500. [DOI] [PubMed] [Google Scholar]
- Dupuis J, Langenberg C, Prokopenko I, Saxena R, Soranzo N, Jackson AU, Wheeler E, Glazer NL, Bouatia-Naji N, Gloyn AL, Lindgren CM, Magi R, Morris AP, Randall J, Johnson T, Elliott P, Rybin D, Thorleifsson G, Steinthorsdottir V, Henneman P, Grallert H, Dehghan A, Hottenga JJ, Franklin CS, Navarro P, Song K, Goel A, Perry JR, Egan JM, Lajunen T, Grarup N, Sparso T, Doney A, Voight BF, Stringham HM, Li M, Kanoni S, Shrader P, Cavalcanti-Proenca C, Kumari M, Qi L, Timpson NJ, Gieger C, Zabena C, Rocheleau G, Ingelsson E, An P, O’Connell J, Luan J, Elliott A, McCarroll SA, Payne F, Roccasecca RM, Pattou F, Sethupathy P, Ardlie K, Ariyurek Y, Balkau B, Barter P, Beilby JP, Ben Shlomo Y, Benediktsson R, Bennett AJ, Bergmann S, Bochud M, Boerwinkle E, Bonnefond A, Bonnycastle LL, Borch-Johnsen K, Bottcher Y, Brunner E, Bumpstead SJ, Charpentier G, Chen YD, Chines P, Clarke R, Coin LJ, Cooper MN, Cornelis M, Crawford G, Crisponi L, Day IN, de Geus EJ, Delplanque J, Dina C, Erdos MR, Fedson AC, Fischer-Rosinsky A, Forouhi NG, Fox CS, Frants R, Franzosi MG, Galan P, Goodarzi MO, Graessler J, Groves CJ, Grundy S, Gwilliam R, Gyllensten U, Hadjadj S, Hallmans G, Hammond N, Han X, Hartikainen AL, Hassanali N, Hayward C, Heath SC, Hercberg S, Herder C, Hicks AA, Hillman DR, Hingorani AD, Hofman A, Hui J, Hung J, Isomaa B, Johnson PR, Jorgensen T, Jula A, Kaakinen M, Kaprio J, Kesaniemi YA, Kivimaki M, Knight B, Koskinen S, Kovacs P, Kyvik KO, Lathrop GM, Lawlor DA, Le Bacquer O, Lecoeur C, Li Y, Lyssenko V, Mahley R, Mangino M, Manning AK, Martinez-Larrad MT, McAteer JB, McCulloch LJ, McPherson R, Meisinger C, Melzer D, Meyre D, Mitchell BD, Morken MA, Mukherjee S, Naitza S, Narisu N, Neville MJ, Oostra BA, Orru M, Pakyz R, Palmer CN, Paolisso G, Pattaro C, Pearson D, Peden JF, Pedersen NL, Perola M, Pfeiffer AF, Pichler I, Polasek O, Posthuma D, Potter SC, Pouta A, Province MA, Psaty BM, Rathmann W, Rayner NW, Rice K, Ripatti S, Rivadeneira F, Roden M, Rolandsson O, Sandbaek A, Sandhu M, Sanna S, Sayer AA, Scheet P, Scott LJ, Seedorf U, Sharp SJ, Shields B, Sigurethsson G, Sijbrands EJ, Silveira A, Simpson L, Singleton A, Smith NL, Sovio U, Swift A, Syddall H, Syvanen AC, Tanaka T, Thorand B, Tichet J, Tonjes A, Tuomi T, Uitterlinden AG, van Dijk KW, van Hoek M, Varma D, Visvikis-Siest S, Vitart V, Vogelzangs N, Waeber G, Wagner PJ, Walley A, Walters GB, Ward KL, Watkins H, Weedon MN, Wild SH, Willemsen G, Witteman JC, Yarnell JW, Zeggini E, Zelenika D, Zethelius B, Zhai G, Zhao JH, Zillikens MC, Borecki IB, Loos RJ, Meneton P, Magnusson PK, Nathan DM, Williams GH, Hattersley AT, Silander K, Salomaa V, Smith GD, Bornstein SR, Schwarz P, Spranger J, Karpe F, Shuldiner AR, Cooper C, Dedoussis GV, Serrano-Rios M, Morris AD, Lind L, Palmer LJ, Hu FB, Franks PW, Ebrahim S, Marmot M, Kao WH, Pankow JS, Sampson MJ, Kuusisto J, Laakso M, Hansen T, Pedersen O, Pramstaller PP. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nature Genetics. 2010;42:105–116. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers CL, Kupfer DJ. Slow-wave sleep: Do young adult men and women age differently? J Sleep Res. 1997;6:211–215. doi: 10.1046/j.1365-2869.1997.00041.x. [DOI] [PubMed] [Google Scholar]
- Guindalini C, Mazzotti DR, Castro LS, D’Aurea CV, Andersen ML, Poyares D, Bittencourt LR, Tufik S. Brain-derived neurotrophic factor gene polymorphism predicts interindividual variation in the sleep electroencephalogram. Journal of neuroscience research. 2014;92:1018–1023. doi: 10.1002/jnr.23380. [DOI] [PubMed] [Google Scholar]
- Han B, Eskin E. Random-effects model aimed at discovering associations in meta-analysis of genome-wide association studies. Am J Hum Genet. 2011;88:586–598. doi: 10.1016/j.ajhg.2011.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanlon EC, Van Cauter E. Quantification of sleep behavior and of its impact on the cross-talk between the brain and peripheral metabolism. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(Suppl 3):15609–15616. doi: 10.1073/pnas.1101338108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Jones CR, Fujiki N, Xu Y, Guo B, Holder JL, Jr, Rossner MJ, Nishino S, Fu YH. The transcriptional repressor DEC2 regulates sleep length in mammals. Science. 2009;325:866–870. doi: 10.1126/science.1174443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ionita-Laza I, Lee S, Makarov V, Buxbaum JD, Lin X. Sequence kernel association tests for the combined effect of rare and common variants. Am J Hum Genet. 2013;92:841–853. doi: 10.1016/j.ajhg.2013.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landolt HP, Dijk D. Genetic basis of sleep in healthy humans. In: Kryger MH, Roth T, Dement WC, editors. Principles and Practice of Sleep Medicine. St. Louis, MO: Elsevier Saunders; 2010. pp. 175–183. [Google Scholar]
- Lane JM, Chang AM, Consortium C, Aeschbach D, Cain SW, Czeisler CA, Klerman E, Lockley SW, St Hilaire M, Shea SA, Duffy JF, Buxton OM, Redline S, Scheer FA, Saxena R. Impact of common variation at diabetes trait loci MTNR1B and CRY2 on sleep, circadian and melatonin physiology. Sleep. 2015;38:A5–6. [Google Scholar]
- Lennox WG, Gibbs EL, Gibbs FA. The brain-wave pattern, an hereditary trait. The Journal of Heredity. 1945;36:233–243. [Google Scholar]
- Linkowski P. EEG sleep patterns in twins. J Sleep Res. 1999;8(Suppl 1):11–13. doi: 10.1046/j.1365-2869.1999.00002.x. [DOI] [PubMed] [Google Scholar]
- Lyssenko V, Nagorny CL, Erdos MR, Wierup N, Jonsson A, Spegel P, Bugliani M, Saxena R, Fex M, Pulizzi N, Isomaa B, Tuomi T, Nilsson P, Kuusisto J, Tuomilehto J, Boehnke M, Altshuler D, Sundler F, Eriksson JG, Jackson AU, Laakso M, Marchetti P, Watanabe RM, Mulder H, Groop L. Common variant in MTNR1B associated with increased risk of type 2 diabetes and impaired early insulin secretion. Nature Genetics. 2008;41:82–88. doi: 10.1038/ng.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mignot E, Young T, Lin L, Finn L. Nocturnal sleep and daytime sleepiness in normal subjects with HLA-DQB1*0602. Sleep. 1999;22:347–352. [PubMed] [Google Scholar]
- Nedeltcheva AV, Scheer FA. Metabolic effects of sleep disruption, links to obesity and diabetes. Current opinion in endocrinology, diabetes, and obesity. 2014;21:293–298. doi: 10.1097/MED.0000000000000082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandi-Perumal SR, Trakht I, Srinivasan V, Spence DW, Maestroni GJ, Zisapel N, Cardinali DP. Physiological effects of melatonin: role of melatonin receptors and signal transduction pathways. Progress in Neurobiology. 2008;85:335–353. doi: 10.1016/j.pneurobio.2008.04.001. [DOI] [PubMed] [Google Scholar]
- Prokopenko I, Langenberg C, Florez JC, Saxena R, Soranzo N, Thorleifsson G, Loos RJ, Manning AK, Jackson AU, Aulchenko Y, Potter SC, Erdos MR, Sanna S, Hottenga JJ, Wheeler E, Kaakinen M, Lyssenko V, Chen WM, Ahmadi K, Beckmann JS, Bergman RN, Bochud M, Bonnycastle LL, Buchanan TA, Cao A, Cervino A, Coin L, Collins FS, Crisponi L, de Geus EJ, Dehghan A, Deloukas P, Doney AS, Elliott P, Freimer N, Gateva V, Herder C, Hofman A, Hughes TE, Hunt S, Illig T, Inouye M, Isomaa B, Johnson T, Kong A, Krestyaninova M, Kuusisto J, Laakso M, Lim N, Lindblad U, Lindgren CM, McCann OT, Mohlke KL, Morris AD, Naitza S, Orru M, Palmer CN, Pouta A, Randall J, Rathmann W, Saramies J, Scheet P, Scott LJ, Scuteri A, Sharp S, Sijbrands E, Smit JH, Song K, Steinthorsdottir V, Stringham HM, Tuomi T, Tuomilehto J, Uitterlinden AG, Voight BF, Waterworth D, Wichmann HE, Willemsen G, Witteman JC, Yuan X, Zhao JH, Zeggini E, Schlessinger D, Sandhu M, Boomsma DI, Uda M, Spector TD, Penninx BW, Altshuler D, Vollenweider P, Jarvelin MR, Lakatta E, Waeber G, Fox CS, Peltonen L, Groop LC, Mooser V, Cupples LA, Thorsteinsdottir U, Boehnke M, Barroso I, Van Duijn C, Dupuis J, Watanabe RM, Stefansson K, McCarthy MI, Wareham NJ, Meigs JB, Abecasis GR. Variants in MTNR1B influence fasting glucose levels. Nature Genetics. 2008;41:77–81. doi: 10.1038/ng.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rechtschaffen A, Kales A. A manual of standardized terminology, techniques and scoring system for sleep stages of human Subjects. Washington, D.C: U.S. Government Printing Office; 1968. [DOI] [PubMed] [Google Scholar]
- Retey JV, Adam M, Honegger E, Khatami R, Luhmann UF, Jung HH, Berger W, Landolt HP. A functional genetic variation of adenosine deaminase affects the duration and intensity of deep sleep in humans. PNAS. 2005;102:15676–15681. doi: 10.1073/pnas.0505414102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer FA, Zeitzer JM, Ayas NT, Brown R, Czeisler CA, Shea SA. Reduced sleep efficiency in cervical spinal cord injury; association with abolished night time melatonin secretion. Spinal Cord. 2006;44:78–81. doi: 10.1038/sj.sc.3101784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegel K, Knutson K, Leproult R, Tasali E, Van Cauter E. Sleep loss: A novel risk factor for insulin resistance and Type 2 diabetes. Journal of Applied Physiology. 2005;99:2008–2019. doi: 10.1152/japplphysiol.00660.2005. [DOI] [PubMed] [Google Scholar]
- Toh KL, Jones CR, He Y, Eide EJ, Hinz WA, Virshup DM, Ptácek LJ, Fu Y-H. An h Per2 phosphorylation site mutation in familial advanced sleep-phase syndrome. Science. 2001;291:1040–1043. doi: 10.1126/science.1057499. [DOI] [PubMed] [Google Scholar]
- van Beijsterveldt CE, van Baal GC. Twin and family studies of the human electroencephalogram: a review and a meta-analysis. Biol Psychol. 2002;61:111–138. doi: 10.1016/s0301-0511(02)00055-8. [DOI] [PubMed] [Google Scholar]
- Viola AU, Archer SN, James LM, Groeger JA, Lo JCY, Skene DJ, von schantz M, Dijk DJ. PER3 Polymorphism Predicts Sleep Structure and Waking Performance. Current Biology. 2007;17:1–6. doi: 10.1016/j.cub.2007.01.073. [DOI] [PubMed] [Google Scholar]
- Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40:D930–934. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.