Abstract
Background
Long non-coding RNA SPRY4 intronic transcript 1 (lncRNA SPRY4-IT1) has been reported to be associated with the progression of several cancers, but its expression level in colorectal cancer (CRC) has rarely been reported. The purpose of this study was to estimate the clinical significance of SPRY4-IT1 in CRC.
Material/Methods
The relative expression levels of SPRY4-IT1 were detected by quantitative real-time polymerase chain reaction (qRT-PCR) in diseased tissues and the adjacent normal tissues of 106 CRC patients. Chi-square method was used to evaluate the association between SPRY4-IT1 expression and the clinical features. Additionally, we assessed the overall survival at different expression levels of SPRY4-IT1 using Kaplan-Meier method. The prognostic significance of SPRY4-IT1 was estimated by Cox regression analysis.
Results
Up-regulated level of SPRY4-IT1 was detected in pathologic tissues of CRC patients compared with adjacent normal tissues (P=0.000). The relative expression of SPRY4-IT1 was associated with the tumor size, the depth of invasion, lymph node invasion, distant invasion, and tumor stage (P<0.05). Patients with high expression of SPRY4-IT1 had poor overall survival compared with those with high level (39.3 vs. 49.3 months, log-rank test, P=0.016). Cox regression analysis showed that SPRY4-IT1 could act as an independent prognostic factor in CRC (HR=2.341, 95% CI=1.136–4.826, P=0.021).
Conclusions
SPRY4-IT1 might be associated with tumorigenesis and progression of CRC, and it may be a promising biomarker for prognosis in patients with CRC.
MeSH Keywords: Colorectal Neoplasms, Lynch Syndrome II, Prognosis
Background
Colorectal cancer (CRC) is one of the most commonly diagnosed cancers in the world, with over 600,000 deaths per year [1,2]. Unfortunately, the morbidity of CRC has been rapidly rising in Asian countries in recent years [3]. However, the multiple known carcinogenic factors and complex genetic backgrounds make it difficult to estimate the key factor in CRC progression [4]. At the present time, the basic prognostic biomarker in CRC is the clinicopathologic tumor staging, based on the tumor-node-metastasis (TNM) system. Nevertheless, the TNM stage is not an ideal biomarker for CRC outcomes. Patients at the same TNM stage may have different progressions and clinical outcomes due to their various genetic and epigenetic backgrounds [5,6]. Therefore, it is necessary to identify sensitive and specific molecular biomarkers for CRC clinical outcomes.
Long non-coding RNAs (LncRNAs) transcribed by RNA polymerase II lack open reading frames (ORF) longer than 200 nucleotides [7]. Although the functions of most lncRNAs are unknown, more and more lncRNAs are characterized and many of them are reported to regulate gene expression in the development and differentiation of diseases [8–11]. LncRNAs also have been reported to influence the development of human cancers. For example, lncRNA CCHE1 promotes cervical cancer cell proliferation [12], HNF1A-AS1 regulates proliferation and metastasis in lung adenocarcinoma [13], and MALAT1 is associated with poor prognosis of glioma [14]. SPRY4-IT1 is significantly increased in plasma samples of NSCLC patients and can act as a biomarker in NSCLC [15]. TRPM2-AS can act as a novel biomarker and therapeutic target in prostate cancer [16]. In this study, we focused on lncRNA SPRY4 intronic transcript 1 (SPRY4-IT1), which is located within an intron of the SPRY4 gene. SPRY4-IT1 was previously reported to be up-regulated in melanoma, gastric cancer, breast cancer, and esophageal squamous cell carcinoma [17–20]. However, the effect of SPRY4-IT1 in CRC prognosis is unknown.
In the present study, we applied different methods in analyzing the association between SPRY4-IT1 expression and clinical features, aiming to determine the clinical influences of SPRY4-IT1 in CRC patients and to discover a reliable predictor for CRC.
Material and Methods
Patients and clinical features collection
In this study, 106 CRC patients confirmed by pathological and clinical diagnoses at the PLA General Hospital were enrolled from October 2008 to January 2014. This study was approved by the Ethics Committee of PLA General Hospital, and written consent was obtained from all the patients. Tumor and adjacent normal tissues were obtained from the CRC patients before they received any chemotherapy or radiotherapy. All the tissue samples were stored in liquid nitrogen until they were utilized.
In order to observe the results of the surgery, follow-up was performed every 3 months in the first 2 years and then every 6 months until the end of the study. All the patients were enrolled in the surgery. Overall survival was used to estimate the influence of SPRY4-IT1 on CRC patient prognosis.
RNA extraction
Total RNA was extracted from all the tissues using TRIzol reagent according to the manufacturer’s instructions. The extracted RNA was dissolved in diethyl pyrocarbonate (DEPC)-water and then treated by DNase to remove DNA. The concentration of the total RNA was detected by UV absorbance at 260 nm and 280 nm (A260/A280). We used 1% agarose gel electrophoresis to check the quality of the total RNA.
Fluorescence quantitative real-time PCR
Fluorescence quantitative real-time PCR (qRT-PCR) was used to assess the relative expression levels of SPRY4-IT1 in pathologic and adjacent normal tissues of CRC patients. The complementary DNA (cDNA) temples enrolling in the qRT-PCR were from the PrimeScript RT reagent kit (Takara, China). The qRT-PCR was performed with SYBR Green assay (Takara, China). The expression of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used for normalized control. The data were analyzed by 2−ΔΔCt method. The primers sequences are shown in Table 1.
Table 1.
The sequences of primers used in this study.
| Name | Sequences | |
|---|---|---|
| SPRY4-IT1 | Forward | 5′-ATCCGAAGCGCAGACACAATTCA-3′ |
| Reverse | 5′-CCTCGATGTAGTCTATGTCATAGGA-3′ | |
| GAPDH | Forward | 5′-AGACTCGCTGATGATCCATGC-3′ |
| Reverse | 5′-AGGTGACCACAGTGTTCTG-3′ | |
Statistical analysis
Statistical analysis was completed in SPSS 18.0 software. Student’s t-test was used to estimate the different expression levels of SPRY4-IT1 and the data are shown as mean ± standard deviation (SD). The association between the clinical features and SPRY4-IT expression was evaluated by chi-square method. Kaplan-Meier method with log-rank test was applied to analyze the overall survival of the CRC patients, and univariate and multivariate Cox regression analysis were used to evaluate the prognostic value of SPRY4-IT1. P<0.05 was considered statistical significance.
Results
Different expression of SPRY4-IT1 in CRC tissues and normal tissues
The 106 CRC patients enrolled in this study included 52 men and 54 women with an average age of 55.02 years old. The clinical data of the participators are summarized in Table 2. QRT-PCR was used to evaluate the relative expression of SPRY4-IT1 in CRC tissues and normal tissues. The results indicated that the relative expression of SPRY4-IT1 in pathologic tissues was significantly higher than that in the adjacent normal tissues (P=0.000, Figure 1).
Table 2.
The clinical features of the CRC patients in this study.
| Characteristics | Total number (n) | SPRY4-IT1 expression | χ2 | P | |
|---|---|---|---|---|---|
| High (n) | Low (n) | ||||
| Gender | 0.503 | 0.478 | |||
| Men | 57 | 33 | 24 | ||
| Women | 59 | 25 | 24 | ||
| Age | 0.022 | 0.882 | |||
| ≥55 | 61 | 33 | 28 | ||
| <55 | 45 | 25 | 20 | ||
| Tumor size | 6.177 | 0013 | |||
| ≥5 cm | 66 | 37 | 19 | ||
| <5 cm | 50 | 21 | 29 | ||
| Location | 0.412 | 0.521 | |||
| Colon | 60 | 29 | 21 | ||
| Rectum | 56 | 29 | 27 | ||
| Histological differentiation | 1.211 | 0.271 | |||
| Well | 67 | 34 | 23 | ||
| Poor | 49 | 24 | 25 | ||
| The depth of invasion | 8.183 | 0.004 | |||
| T1+T2 | |||||
| T3+T4 | |||||
| Lymph node metastasis | 5.665 | 0.017 | |||
| Absent | 55 | 24 | 31 | ||
| Present | 51 | 34 | 17 | ||
| Venous invasion | 0.101 | 0.751 | |||
| Absent | 57 | 32 | 25 | ||
| Present | 49 | 26 | 23 | ||
| Nervous invasion | 0.212 | 0.645 | |||
| Absent | 60 | 34 | 26 | ||
| Present | 56 | 24 | 22 | ||
| Distant invasion | 6.344 | 0.012 | |||
| Absent | 52 | 22 | 30 | ||
| Present | 54 | 36 | 18 | ||
| Tumor stage | 6.177 | 0.015 | |||
| I+II | 57 | 25 | 32 | ||
| III+IV | 49 | 33 | 16 | ||
Figure 1.
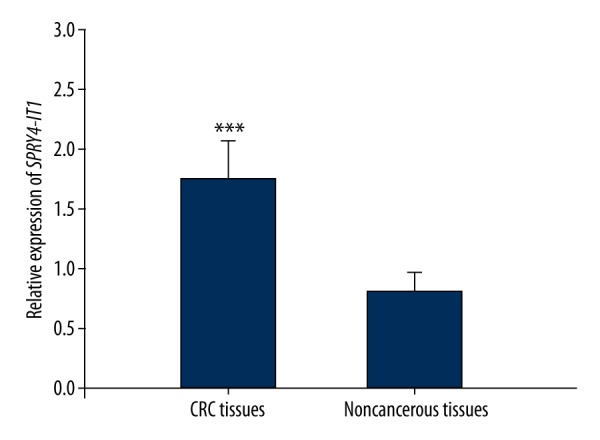
Relative expression of SPRY4-IT1 in CRC patients. Up-regulated level of SPRY4-IT1 was detected in CRC tissues compared with adjacent normal tissues (GAPDH as normalized control).* Indicated P<0.001.
Relationship between SPRY4-IT1 expression and clinical characteristics
To evaluate the association between SPRY4-IT1 expression and clinical features, the CRC patients were divided into high and low expression groups on the basis of their average expression of SPRY4-IT1. The chi-square results are shown in Table 2, which shows that the expression levels of SPRY4-IT1 were associated with the tumor size (P=0.013), the depth of invasion (P=0.004), lymph node metastasis (P=0.017), distant invasion (P=0.012), and tumor stage (P=0.015). However, there was not significant relationship between the expression levels and sex, age, tumor location, histological differentiation, venous invasion, or nerve invasion (P>0.05). The results suggest that the expression level of SPRY4-IT1 might be associated with the development of the CRC.
Overall survival analysis
Overall survival analysis was conducted by Kaplan-Meier method, showing that the CRC patients with high expression of SPRY4-IT1 had low overall survival (the average overall survival was 39.3 months), while the low expression patients had an average overall survival of 49.3 months (Figure 2). In other words, differences between the 2 groups were significant (log-rank test, P=0.016).
Figure 2.
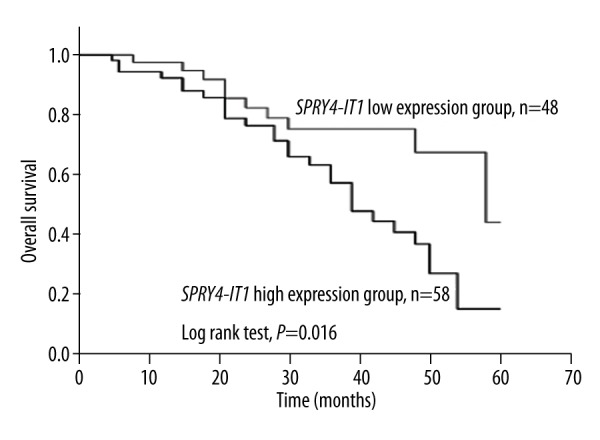
Overall survival analysis for patients with CRC. Patients with low level of SPRY4-IT1 had better outcomes compared with those with high level (log-rank test, P=0.016).
Univariate and multivariate Cox regression analysis for CRC prognosis
In this study, we used Cox regression analysis to estimate the prognostic value of SPRY4-IT1. The results of univariate analysis show that the levels of SPRY4-IT1 were significantly associated with the poor prognosis in CRC patients (P=0.021). Multivariate analysis suggests that SPRY4-IT1 is an independent factor for CRC prognosis (HR=2.341, 95% CI=1.136–4.826, P=0.021). The results of univariate and multivariate Cox regression analysis are summarized in Table 3.
Table 3.
Cox regression analysis for prognosis in CRC patients.
| Characteristics | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P | HR | 95% CI | P | |
| SPRY4-IT1 | 2.341 | 1.136–4.826 | 0.021 | 2.341 | 1.136–4.826 | 0.021 |
| Gender | 1.297 | 0.677–2.487 | 0.433 | – | – | – |
| Age | 0.897 | 0.470–1.713 | 0.742 | – | – | – |
| Tumor size | 1.299 | 0.684–2.467 | 0.423 | – | – | – |
| Location | 0.986 | 0.521–1.866 | 0.966 | – | – | – |
| Histological differentiation | 1.214 | 0.637–2.314 | 0.556 | – | – | – |
| The depth of invasion | 0.816 | 0.425–1.566 | 0.540 | – | – | – |
| Lymph node invasion | 0.949 | 0.500–1.801 | 0.872 | – | – | – |
| Venous invasion | 0.941 | 0.493–1.795 | 0.853 | – | – | – |
| Nervous invasion | 0.826 | 0.435–1.571 | 0.561 | – | – | – |
| Distant invasion | 1.236 | 0.648–2.357 | 0.521 | – | – | – |
| Tumor stage | 0.825 | 0.433–1.571 | 0.557 | – | – | – |
‘–’ – Indicated no available data.
Discussion
Despite great progress in early diagnosis, surgical techniques, and chemotherapy, the prognosis of patients with CRC is still unsatisfactory [21]. Prognostic factors are helpful in determining the therapeutic regimen in cancers. Due to the complex genetic background and epigenetic factors, there is still no highly sensitive and specific biomarker for CRC clinical outcomes, so the prognosis of CRC patients is poor [4]. Therefore, finding reliable biomarkers may improve the treatment of CRC.
Recently, many lncRNAs have been reported to play significant regulatory roles in human diseases [22]. Cancer-specific lncRNAs have been proven to contribute in tumor progression and serve as prognostic factors in many types of cancer. Wu et al. reported that lncRNA UCA1 could be a promising biomarker for early detection and prognosis in gastric cancer [23]. LncRNA BANCR was proved to regulate the growth and metastasis of the retinoblastoma cells and act as a prognostic target [24]. Chen et al. demonstrated that lncRNA HOTTIP promoted pancreatic cancer cell proliferation, survival, and migration [25]. As a result, the association between CRC and lncRNAs may also provide significant information about the development of cancer and clinical outcomes.
LncRNA SPRY4-IT1 is a novel lncRNA, which was first reported to be associated with molecular etiology in human melanoma [26]. It is a 687nt unspliced, polyadenylated transcript, localized at chromosome 5q31.3. In this study, we detected the relative expression levels of SPRY4-IT1 in CRC patients using qRT-PCR (GAPDH as normalized control), aiming to estimate the clinical significance of SPRY4-IT1 in CRC. The results showed that the relative expression of SPRY4-IT1 was much higher in diseased tissues than in the corresponding adjacent normal tissues and there were significant differences between them. The abnormal expression of SPRY4-IT1 might be associated with CRC progression. We also analyzed the relationship between the SPRY4-IT1 expression and the clinical characteristics by chi-square method. The results indicated that the relative expression levels of SPRY4-IT1 were related to the tumor size, the depth of invasion, lymph node invasion, distant invasion, and tumor stage. SPRY4-IT1 expression was not relevant for sex, age, tumor location, histological differentiation, venous invasion, or nervous invasion. These data suggest that SPRY4-IT1 may play a role in the tumorigenesis and progression in CRC.
The prognostic value of SPRY4-IT1 in CRC was also analyzed in this study. Over-expression of SPRY4-IT1 was correlated with low overall survival and the Cox regression analysis showed that SPRY4-IT1 could be an independent prognostic biomarker for CRC. Similar results were also found in other types of cancer, and higher expression of SPRY4-IT1 predicted poor prognosis in many cancers, such as gastric cancer [18], esophageal squamous cell carcinoma [20], clear cell renal cell carcinoma [27], and non-small cell lung cancer [28]. These studies indicated that SPRY4-IT1 might be useful for providing insights into mechanisms of cancers development.
Conclusions
This study proves that SPRY4-IT1 expresses aberrantly in diseased tissues of CRC patients compared with the adjacent normal tissues. Moreover, the expression level is associated with tumor size, the depth of invasion, lymph node invasion, distant invasion, and tumor stage. Additional, SPRY4-IT1 can act as an independent biomarker for CRC prognosis and over-expression of SPRY4-IT1 predicts poor prognosis in CRC. These results suggest that SPRY4-IT1 may be a promising target for CRC therapy.
Footnotes
Source of support: This work was supported by a grant from the National Natural Science Foundation of China (81502682)
References
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 3.Sung JJ, Ng SC, Chan FK, et al. An updated Asia Pacific Consensus Recommendations on colorectal cancer screening. Gut. 2015;64:121–32. doi: 10.1136/gutjnl-2013-306503. [DOI] [PubMed] [Google Scholar]
- 4.Han Y, Yang YN, Yuan HH, et al. UCA1, a long non-coding RNA up-regulated in colorectal cancer influences cell proliferation, apoptosis and cell cycle distribution. Pathology. 2014;46:396–401. doi: 10.1097/PAT.0000000000000125. [DOI] [PubMed] [Google Scholar]
- 5.Dong Y, Yu J, Ng SS. MicroRNA dysregulation as a prognostic biomarker in colorectal cancer. Cancer Manag Res. 2014;6:405–22. doi: 10.2147/CMAR.S35164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reimers MS, Zeestraten EC, Kuppen PJ, et al. Biomarkers in precision therapy in colorectal cancer. Gastroenterol Rep (Oxf) 2013;1:166–83. doi: 10.1093/gastro/got022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kornienko AE, Guenzl PM, Barlow DP, Pauler FM. Gene regulation by the act of long non-coding RNA transcription. BMC Biol. 2013;11:59. doi: 10.1186/1741-7007-11-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–41. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 9.Xu XF, Li J, Cao YX, et al. Differential expression of long noncoding RNAs in human cumulus cells related to embryo developmental potential: A microarray analysis. Reprod Sci. 2015;22:672–78. doi: 10.1177/1933719114561562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chalei V, Sansom SN, Kong L, et al. The long non-coding RNA Dali is an epigenetic regulator of neural differentiation. Elife. 2014;3:e04530. doi: 10.7554/eLife.04530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taft RJ, Pang KC, Mercer TR, et al. Non-coding RNAs: regulators of disease. J Pathol. 2010;220:126–39. doi: 10.1002/path.2638. [DOI] [PubMed] [Google Scholar]
- 12.Yang M, Zhai X, Xia B, et al. Long noncoding RNA CCHE1 promotes cervical cancer cell proliferation via upregulating PCNA. Tumour Biol. 2015;36:7615–22. doi: 10.1007/s13277-015-3465-4. [DOI] [PubMed] [Google Scholar]
- 13.Wu Y, Liu H, Shi X, et al. The long non-coding RNA HNF1A-AS1 regulates proliferation and metastasis in lung adenocarcinoma. Oncotarget. 2015;6:9160–72. doi: 10.18632/oncotarget.3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma KX, Wang HJ, Li XR, et al. Long noncoding RNA MALAT1 associates with the malignant status and poor prognosis in glioma. Tumour Biol. 2015;36:3355–59. doi: 10.1007/s13277-014-2969-7. [DOI] [PubMed] [Google Scholar]
- 15.Hu X, Bao J, Wang Z, et al. The plasma lncRNA acting as fingerprint in non-small-cell lung cancer. Tumour Biol. 2016;37:3497–504. doi: 10.1007/s13277-015-4023-9. [DOI] [PubMed] [Google Scholar]
- 16.Mouraviev V, Lee B, Patel V, et al. Clinical prospects of long noncoding RNAs as novel biomarkers and therapeutic targets in prostate cancer. Prostate Cancer Prostatic Dis. 2016;19:14–20. doi: 10.1038/pcan.2015.48. [DOI] [PubMed] [Google Scholar]
- 17.Mazar J, Zhao W, Khalil AM, et al. The functional characterization of long noncoding RNA SPRY4-IT1 in human melanoma cells. Oncotarget. 2014;5:8959–69. doi: 10.18632/oncotarget.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peng W, Wu G, Fan H, et al. Long noncoding RNA SPRY4-IT1 predicts poor patient prognosis and promotes tumorigenesis in gastric cancer. Tumour Biol. 2015;36:6751–58. doi: 10.1007/s13277-015-3376-4. [DOI] [PubMed] [Google Scholar]
- 19.Shi Y, Li J, Liu Y, et al. The long noncoding RNA SPRY4-IT1 increases the proliferation of human breast cancer cells by upregulating ZNF703 expression. Mol Cancer. 2015;14:51. doi: 10.1186/s12943-015-0318-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xie HW, Wu QQ, Zhu B, et al. Long noncoding RNA SPRY4-IT1 is upregulated in esophageal squamous cell carcinoma and associated with poor prognosis. Tumour Biol. 2014;35:7743–54. doi: 10.1007/s13277-014-2013-y. [DOI] [PubMed] [Google Scholar]
- 21.Sung JJ, Lau JY, Goh KL, Leung WK. Increasing incidence of colorectal cancer in Asia: Implications for screening. Lancet Oncol. 2005;6:871–76. doi: 10.1016/S1470-2045(05)70422-8. [DOI] [PubMed] [Google Scholar]
- 22.Bhan A, Mandal SS. Long noncoding RNAs: Emerging stars in gene regulation, epigenetics and human disease. Chem Med Chem. 2014;9:1932–56. doi: 10.1002/cmdc.201300534. [DOI] [PubMed] [Google Scholar]
- 23.Zheng Q, Wu F, Dai WY, et al. Aberrant expression of UCA1 in gastric cancer and its clinical significance. Clin Transl Oncol. 2015;17:640–46. doi: 10.1007/s12094-015-1290-2. [DOI] [PubMed] [Google Scholar]
- 24.Su S, Gao J, Wang T, et al. Long non-coding RNA BANCR regulates growth and metastasis and is associated with poor prognosis in retinoblastoma. Tumour Biol. 2015;36:7205–11. doi: 10.1007/s13277-015-3413-3. [DOI] [PubMed] [Google Scholar]
- 25.Cheng Y, Jutooru I, Chadalapaka G, et al. The long non-coding RNA HOTTIP enhances pancreatic cancer cell proliferation, survival and migration. Oncotarget. 2015;6:10840–52. doi: 10.18632/oncotarget.3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khaitan D, Dinger ME, Mazar J, et al. The melanoma-upregulated long noncoding RNA SPRY4-IT1 modulates apoptosis and invasion. Cancer Res. 2011;71:3852–62. doi: 10.1158/0008-5472.CAN-10-4460. [DOI] [PubMed] [Google Scholar]
- 27.Zhang HM, Yang FQ, Yan Y, et al. High expression of long non-coding RNA SPRY4-IT1 predicts poor prognosis of clear cell renal cell carcinoma. Int J Clin Exp Pathol. 2014;7:5801–9. [PMC free article] [PubMed] [Google Scholar]
- 28.Sun M, Liu XH, Lu KH, et al. EZH2-mediated epigenetic suppression of long noncoding RNA SPRY4-IT1 promotes NSCLC cell proliferation and metastasis by affecting the epithelial-mesenchymal transition. Cell Death Dis. 2014;5:e1298. doi: 10.1038/cddis.2014.256. [DOI] [PMC free article] [PubMed] [Google Scholar]


