SUMMARY
The physical organization of DNA enzymes at a replication fork enables efficient copying of two antiparallel DNA strands, yet dynamic protein interactions within the replication complex complicate replisome structural studies. We employed a combination of crystallographic, native mass spectrometry, and small-angle x-ray scattering experiments to capture alternative structures of a model replication system encoded by bacteriophage T7. Two molecules of DNA polymerase bind the ring-shaped primase-helicase in a conserved orientation and provide structural insight into how the acidic C-terminal tail of the primase-helicase contacts the DNA polymerase to facilitate loading of the polymerase onto DNA. A third DNA polymerase binds the ring in an offset manner that may enable polymerase exchange during replication. Alternative polymerase binding modes are also detected by small-angle x-ray scattering with DNA substrates present. Our collective results unveil complex motions within T7 replisome higher-order structures that are underpinned by multivalent protein-protein interactions with functional implications.
Keywords: DNA replication, x-ray crystallography, native mass spectrometry, small-angle x-ray scattering
eTOC
Wallen et al. describe the first crystal structure of the T7 primase-helicase bound to three copies of DNA polymerase and show that the shape of the replisome changes in the presence of DNA. The structure provides insight into how a replisome assembles to copy both strands of DNA.
INTRODUCTION
Replisomes are multi-protein complexes that catalyze the efficient duplication of cellular DNA through the coupled activities of enzymes that unwind duplex DNA, prime its synthesis, and copy two antiparallel strands of DNA (Crampton et al., 2011; Langston et al., 2009; Lee and Richardson, 2011; Pandey et al., 2010). These activities are well-characterized biochemically for several model replication systems, but few experimental data describe the higher order structures of the corresponding replisomes. The bacteriophage T7 replisome provides a prototypic core complex, containing all essential replication activities in just four proteins (Lee and Richardson, 2011). The T7 DNA polymerase catalytic subunit (gp5) forms a stable 1:1 complex with its processivity factor Escherichia coli thioredoxin (trx) that functions as both the leading and lagging strand DNA polymerase. The T7 primase-helicase protein (gp4) is a dual-function enzyme with an N-terminal primase domain that synthesizes short, tetra-ribonucleotides that prime Okazaki fragment synthesis on the lagging strand, whereas the C-terminal helicase domain catalyzes nucleotide-dependent DNA unwinding that advances the replication fork. The T7 single-stranded DNA (ssDNA) binding protein (gp2.5) covers the ssDNA generated by the gp4 helicase and directly contacts the gp4 and gp5/trx proteins during coupled DNA synthesis. The fully constituted T7 replisome copies double-stranded DNA at a rate of ~150 nucleotides s−1 (Hamdan et al., 2007; Pandey et al., 2010) with a processivity of ~17,000 nucleotides incorporated per DNA encounter (Hamdan et al., 2007).
Biochemical studies of several model replisomes have demonstrated the central importance of physical interactions between the ring-shaped helicase and multiple copies of DNA polymerase (Crampton et al., 2011; Langston et al., 2009; Lee et al., 1998; Lee and Richardson, 2011). Bulk solution and single-molecule studies of the T7 replisome have identified alternative modes of interaction between the gp4 primase-helicase and the gp5/trx DNA polymerase at different stages of the replication cycle. For example, gp5/trx binds to the acidic C-terminal tail of gp4 via two clusters of basic residues located in the DNA binding groove (front basic patch) and in the thioredoxin-binding loop of the polymerase (Huidong Zhang et al., 2011). Interactions between the gp4 tail and front basic patch may assist in loading the polymerase onto DNA, whereas the interaction with the thioredoxin-binding loop may promote exchange of DNA polymerases during ongoing replication and/or tether DNA polymerase to the moving replisome (Hamdan et al., 2007; Huidong Zhang et al., 2011). When gp5/trx is engaged on DNA, its binding affinity for gp4 increases independently of its interaction with the C-terminal tail of gp4 (Hamdan et al., 2007; Kulczyk et al., 2012; Huidong Zhang et al., 2011). A third distinct interaction occurs when an Okazaki fragment is synthesized on the lagging strand of the replication fork. DNA synthesis is primed by the tetraribonucleotide products of the gp4 primase, which operates at specific sequences termed primase recognition sites. The primed DNA template is then handed off from the gp4 primase to gp5/trx polymerase through a physical interaction between gp5/trx and the N-terminal zinc-binding domain (ZBD) of gp4. This primer handoff is an obligate mechanism for efficient utilization of short oligoribonucleotide primers synthesized by T7 primase (Kato et al., 2001; 2004; Kulczyk and Richardson, 2012; Wallen et al., 2013).
Although crystal structures of individual T7 replication proteins were determined (Doublié et al., 1998; Hollis et al., 2001; Toth et al., 2003), an understanding of the assembled T7 replisome has been hampered by alternative modes of protein interactions that generate heterogeneous protein complexes. To overcome these difficulties, we integrated crystallographic and biophysical methods to analyze the core architecture of the T7 replisome. Here we elucidate a crystal structure of a gp4 heptamer bound to three copies of gp5/trx complemented by electrospray ionization native mass spectrometry (native MS) and small-angle x-ray scattering (SAXS) studies. The crystal structure shows two copies of polymerase bound to gp4 in a conserved orientation while a third polymerase binds the gp4 ring in an alternate conformation. Native MS reveals that in the absence of DNA both hexamers and heptamers of gp4 bind DNA polymerase. The SAXS data illustrate the dynamic structure of the replisome with alternative binding modes of gp5/trx on the ring-shaped gp4 helicase in the presence of DNA. This combination of crystallographic and solution-based structure determination methods highlights the multivalent interactions underpinning stable replication complexes with dynamic conformations that accommodate the coupled synthesis of antiparallel DNA strands.
RESULTS
T7 Replisome Crystal Structure
Crystals were obtained in the absence of DNA of a T7 replisome core subassembly consisting of three of the four essential replication proteins: the gp4 primase-helicase, the gp5 DNA polymerase, and its processivity factor, thioredoxin. Despite its intrinsic flexibility, X-ray diffraction data was collected to 4.8 Å resolution for the complex, and the structure was determined by molecular replacement (Table 1; see Materials and Methods). The crystallized replisome consists of a heptamer of the gp4 primase-helicase bound to three copies of T7 DNA polymerase located around the outer rim of the helicase ring in a ~670 kDa protein complex (Figure 1). The ring-shaped assembly of helicase domains is an interaction hub for all three DNA polymerases that bind at the interfaces between adjacent helicase subunits (Figure 1). The gp4 primase domains do not contact the polymerases but adopt different conformations within the gp4 heptamer. Additionally, the fingers subdomain of all three polymerases is rotated away from the thumb in an open conformation that exposes the polymerase active site for binding to DNA and nucleotide substrates (Doublié et al., 1998; Doublié and Ellenberger, 1998).
Table 1.
X-Ray Diffraction Data Collection and Refinement Statistics for the T7 Replisome Structure.
| T7 Replisome* | |
|---|---|
| Data collection | |
| Space group | P212121 |
| Cell dimensions | |
| a, b, c (Å) | 174.87, 238.09, 243.47 |
| a, b, g (°) | 90, 90, 90 |
| Resolution (Å) | 4.8 |
| Complexes per ASU | 1 |
| Reflections | 180,737 |
| Unique Reflections | 41,183 |
| CC1/2 | (0.556) |
| Rmerge | 12.6 (86.6) |
| I / sI | 10.1 (1.3) |
| Completeness (%) | 97.9 (97.6) |
| Redundancy | 3.7 (3.3) |
| Refinement | |
| Resolution (Å) | 29.98-4.80 |
| No. reflections | 41,183 |
| Rwork / Rfree | 27.5 (35.3)/31.8 (40.8) |
| No. atoms | 43,846 |
| Protein | 43,846 |
| Ligand/ion | 0 |
| Water | 0 |
| B-factors | |
| Protein | 240.11 |
| R.M.S. deviations | |
| Bond lengths (Å) | 0.004 |
| Bond angles (°) | 0.736 |
Values in parentheses are for highest-resolution shell.
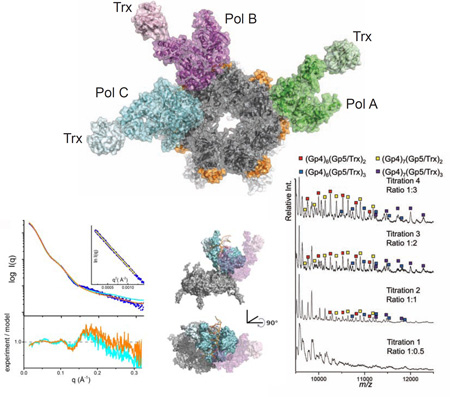
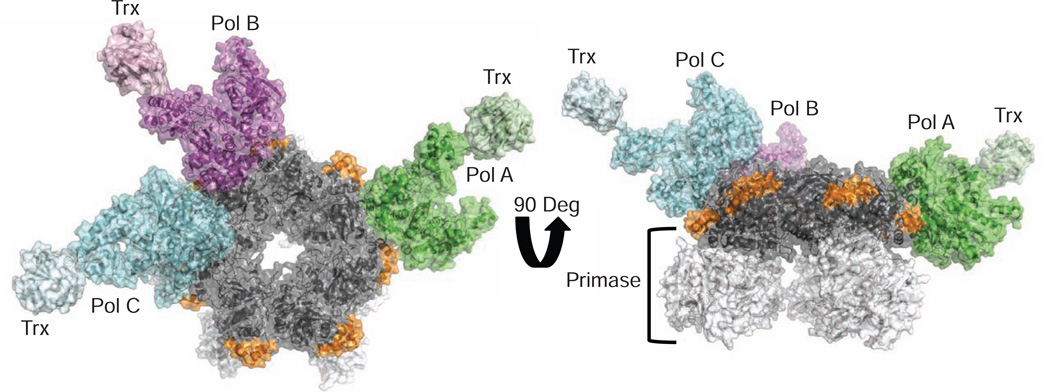
Figure 1.
A Three-Polymerase Replisome
View looking at the C-terminal face of the gp4 heptameric ring (colored gray; left). A helix-turn-helix motif (residues 366–387, colored orange) of gp4 lies on the outside surface of the ring and contacts all three polymerases. Pols A, B, and C are colored green, magenta, and cyan, respectively. A 90-degree rotation of the structure (right) illustrates that the primase domains of gp4 (colored white) do not contact the polymerases.
The DNA polymerases labelled Pol A and Pol B in Figure 1 are bound to gp4 in the same orientation, whereas a third molecule of gp5/trx (Pol C in Figure 1) binds to gp4 in an alternative conformation at a site adjacent to Pol B, where it makes limited interactions with the Pol B fingers subdomain. Gp4 chains F, G, and H constitute the binding interface for Pols B and C and lie in a coplanar configuration within the heptamer ring. The other four gp4 subunits tilt in and out of the plane of the ring to various extents, preventing other polymerases from binding to the heptamer in a similar manner (a polymerase binding next to Pol A, for example). Compared to the bound orientation of Pol A and Pol B on gp4, Pol C is rotated ~90 degrees around an axis that is tipped slightly out of the plane of the gp4 ring (Figure S1). The function of this “extra” copy of DNA polymerase in the T7 replisome is uncertain, yet it is noteworthy that more than two DNA polymerases are present in several prokaryotic, archaeal, and bacteriophage replisomes, including the bacteriophage T7 system (Geertsema et al., 2014; McInerney et al., 2007; Nossal et al., 2007; Lin et al., 2012).
The Front Basic Patch of Pol A and Pol B Engages the Acidic C-Terminal Tail of Gp4
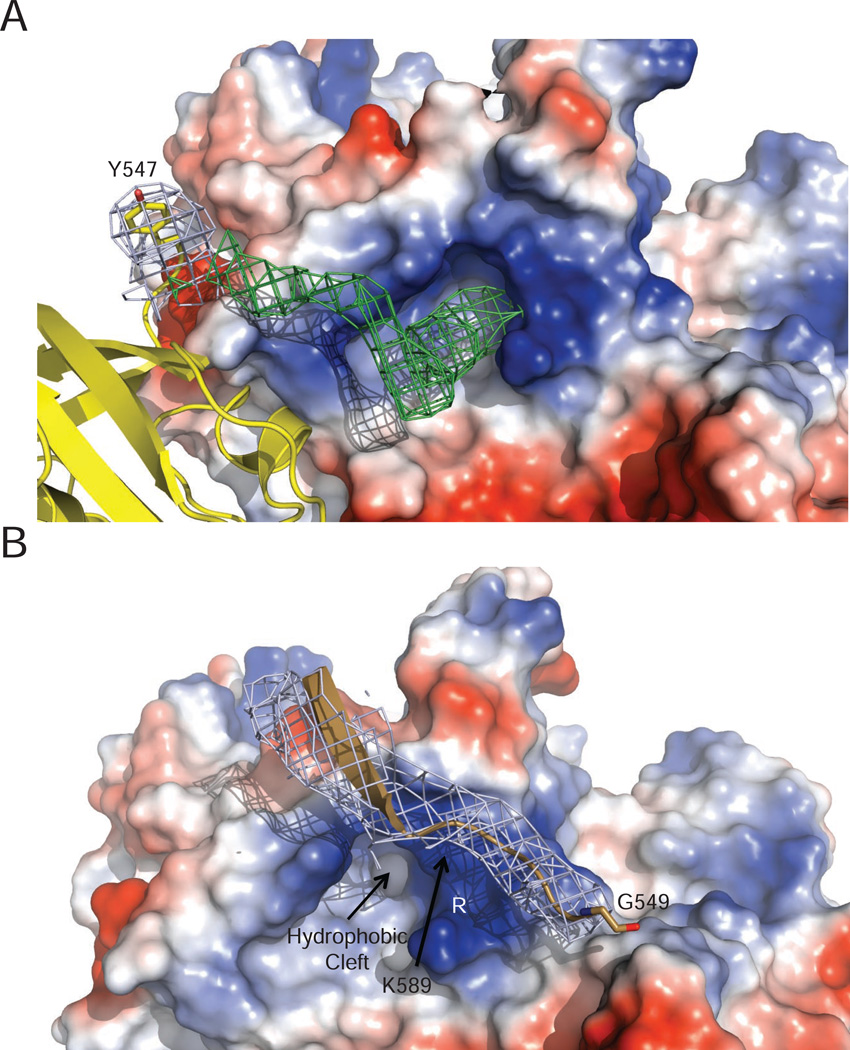
Biochemical studies show the importance of the C-terminal acidic tail of gp4 and its interaction with a cluster of basic residues (K587, K589, R590, and R591) on the gp5/trx polymerase, known as the “front basic patch” (Huidong Zhang et al., 2011). We find that all three DNA polymerases bind to gp4 with their front basic patch adjacent to the C-terminus of a gp4 subunit. Electron density is present next to R590 of Pols A and B, leading from the last residue modelled in the C-terminus of gp4 (Y547 of gp4 chain G) to the front basic patch of the polymerase (Figure 2A). The crystallographic data reveal the location of the binding site for the gp4 tail to be the base of the fingers subdomain in a hydrophobic pocket flanked by a-helices L, P, and Q (Figure 3A). This hydrophobic pocket on the surface of DNA polymerase is suitable for interaction with the side chain of gp4 F566, the C-terminal residue that is essential for the tail’s interaction with DNA polymerase (Lee et al., 2006). Positively charged residues of the front basic patch lie on one side of this hydrophobic binding pocket where they can interact with acidic residues of the gp4 tail (Figure 2A). The low contour of electron density observed in this region is consistent with flexibility or disorder in the gp4 tail, precluding accurate modelling of these residues and their inclusion in the final crystallographic model.
Figure 2.
The Gp4 C-terminal Tail Inserts into a Pocket near the Front Basic Patch of Gp5/trx
(A) A 2Fo-Fc map (blue), contoured at 1 sigma, is shown for the C-terminal amino acid (Y547) of gp4 included in the crystallographic model. A Fo-Fc map contoured at 2.5 sigma (green) reveals additional electron density leading from the C-terminal tail of gp4 (colored yellow) to a hydrophobic pocket adjacent to the front basic patch of Pol B. The electrostatic surface potential of Pol B is shown with residues K589 and R590 from the front basic patch labeled. An analogous patch of density is observed near the front basic patch of Pol A (data not shown).
(B) The C-terminal tail of gp4 (light brown) bypasses Pol C (colored by electrostatic surface potential). The front basic patch of Pol C is partially occluded by its interaction with gp4 so that the tail of gp4 does not engage in the binding interaction observed for Pol B and Pol A. 2Fo-Fc electron density (blue) contoured at 1 sigma is shown for the gp4 tail.
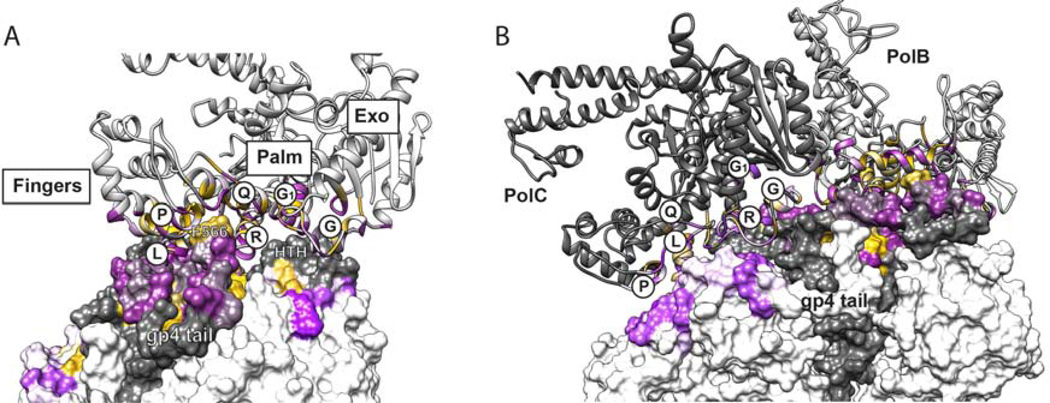
Figure 3.
Pol B Makes Extensive Interactions with Gp4 while Pol C Binds in a Skewed Orientation that Precludes Interactions with the Gp4 Tail
(A) Pol B contacts the gp4 ring on a convoluted interface that spans the fingers, palm, and exonuclease subdomains of Pol B and two subunits of gp4. A ribbon model of Pol B is colored for residues within 8 Å of the gp4 surface according to the Kyte Doolittle Hydropathy values (Kyte et al., 1982) ranging from polar residues (purple) to hydrophobic residues (yellow). The solvent accessible surface of gp4 is similarly colored according to hydrophobicity on a grey or white background depicting adjacent gp4 subunits. Pol B helices L, P, and Q circumscribe a hydrophobic pocket that accepts F566 from the tail of gp4. Residues from the front basic patch of Pol B, located in the connecting loop between helices P and Q, contact acidic residues (colored purple) in the gp4 tail. Another prominent interaction is made by the Helix-Turn-Helix (HTH) motif of gp4, which inserts in a groove located between Pol B helices R, G, and G1. Gp4 tail residues 550–566 are disordered in the crystal structure and were modeled here then subjected to MD simulation, which revealed a very stable interaction of F566 with Pol B (Table SI).
(B) Pol C (dark grey) is shown in the foreground bound next to Pol B on the surface of gp4. Many of the same residues of Pol C and Pol B make contact with gp4, but the rotated orientation of Pol C creates different sets of interactions with less buried surface area featuring predominately polar residues (purple) and fewer hydrophobic residues (yellow). Residues within 8 Å of the binding sites for Pol B and Pol C are colored according to the Kyte Doolittle hydropathy values on a scale ranging from polar residues (purple) to hydrophobic residues (yellow), as for panel A. Another important consequence of the alternative binding orientation of Pol C is that the gp4 tail-binding pocket comprising helices L, P, and Q of Pol C is occluded, preventing interactions with the tail, which are prominent for Pol A and Pol B (cf. Figure 2B). Absent these tail interactions, the Pol C interface supports fewer hydrogen bonding interactions with gp4 (Table S2) in comparison to the Pol B interface (Table SI).
The C-terminal tail of gp4 chain H lies near the front basic patch of Pol C (Figure 2B), but absent electron density beyond gp4 residue G549 suggests a disordered tail of gp4 chain H. Figure 2B also illustrates that the hydrophobic pocket and the front basic patch of Pol C are misaligned relative to the C-terminal residue of chain H (G549) such that the tail of gp4 cannot bind to Pol C in the mode seen for Pols A and B. In fact, the front basic patch of Pol C is partially occluded with residues K589 and R590 contacting another site on gp4, suggesting why the tail of gp4 chain H is disordered.
The Assembled Gp4 Helicase Makes Extensive Contacts with DNA Polymerase
The gp5/trx fingers, palm and proofreading exonuclease subdomains form an extensive contact surface with the gp4 helicase in the crystal structure. In Pol A and Pol B, helices L, P, Q, R, G, and G1 pack against the convoluted surface of two adjacent helicase domains, a binding site that is constituted only in the assembled gp4 ring (Figure 3A). A helix-turn-helix motif of gp4 (residues 366–387; colored orange in Figure 1) is a prominent feature that inserts into a groove between helices G and R of the DNA polymerase. A slight difference in the bound orientations of Pol A and Pol B suggests that movement is possible (Figure S1). The same aspect of Pol C contacts gp4, although the alternative mode of Pol C binding provides a less intimate fit with the peaks and valleys of gp4 (Figure 3B). In Pol C, a groove located at the base of helix R in the palm contacts the gp4 HTH motif, creating a less extensive region of interaction with gp4 that suggests weaker binding (Figure 3B). Polar residues predominate in the buried interaction surfaces of Pol A (1218 Å2), Pol B (1393 Å2), and Pol C (1104 Å2) that contact gp4.
The limited resolution of the x-ray diffraction data (4.8 Å; Table I) and relatively high atomic temperature factors are consistent with protein flexibility and resulting disorder. We therefore sought additional validations for the observed interactions between gp4 and gp5/trx. Molecular dynamics (MD) simulations of Pol B and Pol C in complex with gp4 are able to identify favorable protein-protein interactions. For MD simulations, the gp4 tail contacting Pol B was added to the crystallographic model and the gp4 tail adjacent to Pol C was modelled on the surface of the helicase ring. Polar interactions with gp4 are tallied for Pol B and Pol C in Tables SI and S2, respectively. Electrostatic interactions involving the tail of gp4 (highlighted in orange, Table SI) are prominent among the highest occupancy interactions with Pol B, with the main chain carboxyl of gp4 F566 topping the list. The remaining electrostatic interactions with Pol B are distributed across the interface. In comparison to Pol B, Pol C participates in fewer high occupancy polar interactions with gp4 (Table S2), reflective of its non-ideal binding geometry and resulting decrease in buried surface area interacting with gp4. Although Pol B and Pol C use an overlapping set of residues to interact with gp4, their significantly different binding orientations result in the selection of different hydrogen bonding partners in every instance.
The MD data also reveal side chain interactions that could underlie selective binding of Pol B versus Pol C to gp4. In Pol B, the side chains of R219 and E635 interact with gp4 residues E374 and S281, respectively (Table S1), and none of these residues participate in the binding of Pol C. Conversely, the side chains of K189 and K628 of Pol C are positioned to interact with E551 and E538 of gp4, respectively (Table S2), and these interactions are absent in Pol B. Additional interactions (highlighted in Tables SI and S2) are observed that are unique for either Pol B or Pol C binding to gp4, and these residues could promote the specific binding orientations observed in the crystal structure.
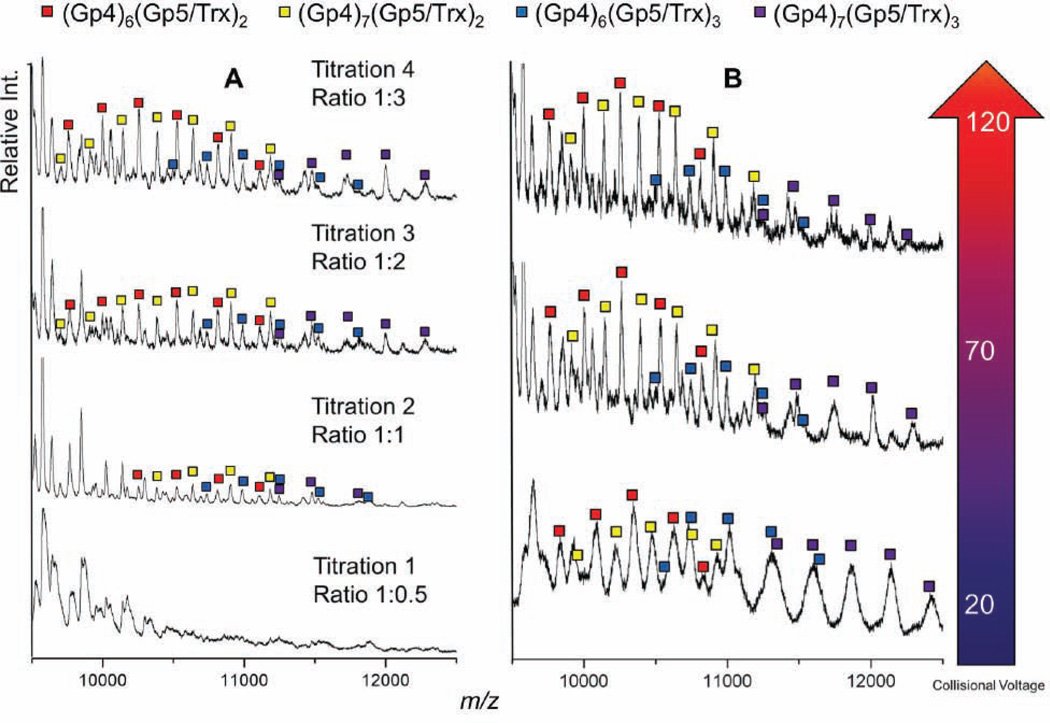
Native Mass Spectrometry Reveals a Three-Polymerase Replisome
To evaluate T7 replisome constituents in solution, we used native MS to identify the assembly states of these replication proteins in the absence of DNA (Hao Zhang et al., 2011). The solubility of protein complexes reconstituted in ammonium acetate buffer for electrospray ionization (EI) was limited, so we improved the solubility of replication complexes during EI by using a truncated gp4 construct lacking the N-terminal primase domain (gp4Δprimase, residues 241–566). It is noteworthy that the primase domains of gp4 do not interact with gp5/trx in the crystal structure (Figure 1) and that the truncated gp4Δprimase protein retains DNA unwinding activity and binds to T7 DNA polymerase (Bird et al., 1997; Guo et al., 1999). In native MS, the gp4Δprimase sprays as a distribution of oligomers ranging from monomers to heptamers (Figure S2), in agreement with biochemical studies (Crampton et al., 2006b; Toth et al., 2003). The gp5/trx polymerase binds to gp4 hexamers as well as heptamers with increasing polymerase concentration (Figure 4A). Although the hexameric form of gp4 is presumed to function in DNA replication (Picha and Patel, 1998), our native MS data provide the first evidence that both hexamers and heptamers of gp4 efficiently interact with gp5/trx polymerase. There was no detectable interaction of gp5/trx with smaller oligomers of the gp4Δprimase (monomers-tetramers; data not shown), suggesting that a fully assembled gp4 ring is required for a stable interaction.
Figure 4.
Native MS Reveals Different T7 Replisome Assembly States.
(A) A titration of gp5/trx against a fixed concentration of gp4 shows the assembled, ring-shaped gp4 helicase binds to one, two, and three copies of the polymerase.
(B) At low collision voltage, complexes containing two copies of gp5/trx are of comparable abundance to assemblies containing three copies of gp5/trx. The three-copy gp5/Trx complexes become less abundant upon collisional activation at higher voltages, indicating they are less stable than those with two copies of Gp5/Trx. The peak shapes are broadened at lower collision voltages, strongly suggesting incomplete desolvation and making it difficult to deduce quantitatively the relative binding affinity of the third gp5/Trx with the gp4 hexamer and heptamer.
Titrating the concentration of gp5/trx against a fixed concentration of gp4Δprimase reveals complexes with one, two or three polymerases bound to gp4 hexamers and heptamers (Figure 4A). Multiple copies of gp5/trx associate with gp4 hexamers and heptamers, even with limiting amounts of gp5/trx (see the 1:1 titration in Figure 4A), suggestive of a preference for assembling higher order complexes. Increasing the collision cell energy in a tandem MS reveals a selective loss of the 3:1 (gp5/trx:gp4 ring) complexes relative to the 2:1 complexes (Figure 4B), supporting the conclusion that the third polymerase is bound less strongly than the other two polymerases in the replisome complex. This interpretation proved to be consistent with the crystal structure, which subsequently revealed that fewer hydrogen-bonding interactions with gp4 are possible for Pol C in comparison to Pols A and B (Figure 3; Tables SI and S2). These complementary methods corroborate that 3:1 complexes of gp5/trx bound to gp4 hexamers and heptamers are formed, with the three-polymerase replisome being less stable than a 2:1 complex.
Dynamic Structures of the T7 Replisome Shown by SAXS
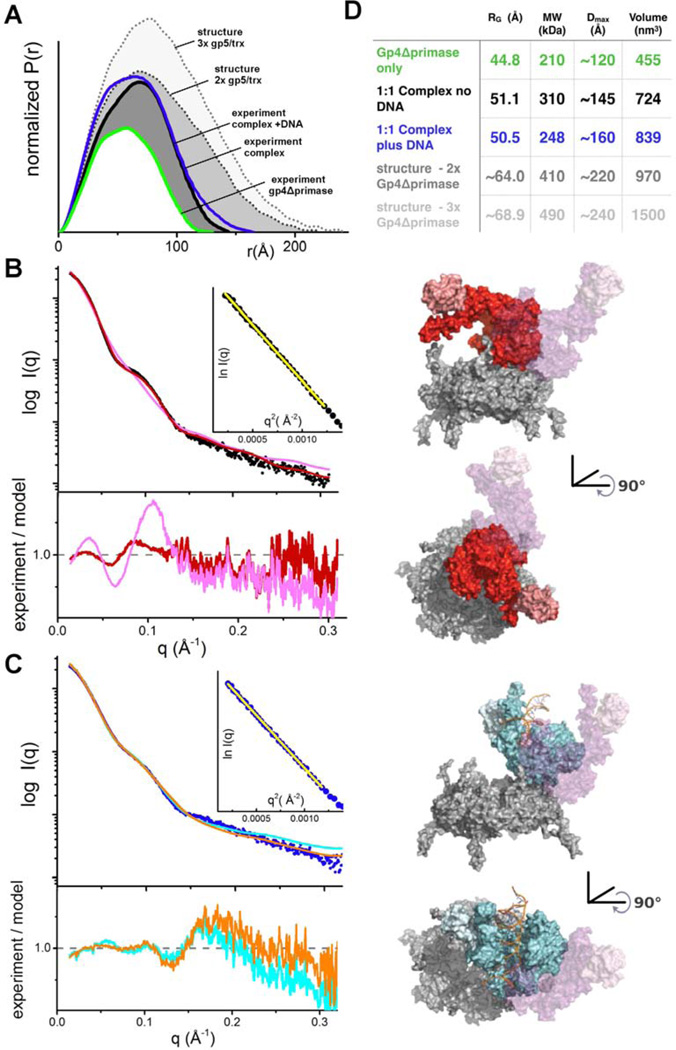
The shape and subunit composition of the T7 replisome were also examined in solution by SAXS, which in combination with crystal structures of components can accurately define multiprotein conformation and assembly in solution under near physiological conditions as SAXS data encode all interatomic distance information even from low-populated flexible conformations (Putnam et al., 2007). Different mixtures of gp5/trx and the gp4Δprimase were analyzed by SAXS, with a dT15 oligo and dTTP added to all samples to promote the formation of gp4 hexamers in preference to heptamers (Crampton et al., 2006b; Picha and Patel, 1998). These substrate ligands decreased sample heterogeneity (Figure S2), and under these conditions the gp4Δprimase is predominately hexameric with a radius of gyration (Rg) = 44.8 Å and a mass of 210 kDa calculated from the SAXS data (Rambo and Tainer, 2013) (Figure S3). Experimental conditions that shift gp4 to predominantly heptamers were not identified, preventing SAXS studies of gp5/trx bound to heptamers in solution. Addition of gp5/trx in an equimolar ratio to the gp4Δprimase hexamer causes a significant increase in Rg (51.1 Å), with Dmax increasing from ~120 Å to ~145 Å for the polymerase:gp4 complex. The calculated mass of 310 kDa based on the SAXS data is consistent with a 1:1 complex of gp5/trx:gp4Δprimase hexamer (Figure 5).
Figure 5.
SAXS Reveals a Dynamic Structure of the T7 Replisome
(A) Normalized P(r) functions for gp4Δprimase alone (green) and a 1:1 complex of gp4Δprimase hexamer and gp5/trx either in the absence (black) or presence (blue) of a primer/template DNA. Calculated P(r) functions are shown for structures of the gp4Δprimase hexamer bound to two copies of DNA polymerase oriented as observed for Pol A and Pol B in the structure (gray dots) and bound to three polymerase molecules (Pols A, B, and C) as observed in the crystal structure (light gray dots). The P(r) functions are normalized based on the molecular volume of the complexes determined by SAXS, as indicated in panel D.
(B) Raw scattering profiles of the 1:1 complex in the absence of a primer/template DNA (black) and a theoretical scattering profile obtained from our crystal structure (magenta) reveals that the structure in solution differs from a model based on the interaction of Pol B with gp4 in the crystallized replisome. An ab initio docking model generated by FoXSDock (red) shows a better fit (χ2 2.7) to the experimental data than the Pol B-gp4Δprimase complex model (χ2 7.2). Inset: Guinier plot for experimental data with a limit of (qxRg) < 1.6. In panels (B) and (C) the orientation of Pol B in the crystal structure is shown as a transparent surface (magenta), and the residuals between the experimental SAXS data and the models are shown below the raw scattering profiles.
(C) The scattering profile of the 1:1 complex with a primer/template DNA (blue) reveals a unique structure in solution as compared to the no DNA sample and the crystal structure. A FoXSDock model (cyan, (χ2 3.5)) shows the polymerase on the outside of the ring, which could be generated by a rotation of the polymerase as observed in our crystal structure. An ensemble model calculated by FoXS (orange trace) predicts that our experimental data is best described by a mixture of the FoXSDock model (57%) and the Pol B-gp4Δprimase model (43%). This ensemble of two models improves the fit (χ2 2.6) to the high q region of the experimental scattering data. Inset: Guinier plot for experimental data with a limit of (qxRg) < 1.6.
(D) SAXS-based structural parameters calculated from the experimental data shown in panels A-C.
To evaluate the shape of the polymerase:gp4 complex, we compared the experimental SAXS data with simulated scattering data for models of gp5/trx docked in a 1:1 complex with the gp4Δprimase hexamer (adapted from PDB 1E0K) (Singleton et al., 2000). Surprisingly, the scattering curve calculated from a model based on the orientation of Pol B in the T7 replisome does not fit the experimental SAXS data (Figure 5B). The Rg (56.0 Å) and Dmax (~210 Å) values for this model greatly exceed those calculated from the solution scattering data (Figure 5D). Alternative models were interrogated for compatibility with the SAXS data. Diverse docking models were generated for gp5/trx bound to the gp4Δprimase hexamer using FoXSDock (Schneidman-Duhovny et al., 2011). A model with the DNA polymerase lying across the C-terminal face of the gp4 helicase ring fits well with the experimental SAXS data (Figure 5B). This model represents a substantial rotation and repositioning of the polymerase on the C-terminal face of the gp4 helicase ring in comparison to the binding orientations of Pols A, B, and C, and this new orientation exposes the front basic patch of the polymerase for stabilizing interactions with a gp4 C-terminal tail. At higher ratios of gp5/trx:gp4Δprimase, no single model adequately matches the experimental SAXS data, consistent with aggregation or heterogeneity of assemblies that limited further SAXS analysis.
The addition of a double-stranded primer/template DNA to the samples did not significantly change the Rg (50.5 Å) and Dmax (~160 Å) values calculated from the SAXS data, yet the orientation of the bound DNA polymerase shifted, based on comparisons with the scattering curves of modelled complexes (Figure 5C). We found an excellent fit to the low resolution experimental SAXS data with a model that places the polymerase on the outside of the helicase ring but rotated away from the position of Pol B in the crystal structure (Figure 5C). This model fits the low q region of the SAXS data well, but a combination of two models is required to obtain a reasonable agreement with the higher q data. A minimal ensemble search (MES) (Pelikan et al., 2009) identifies a composite model fitting both low-q and high-q regions of the scattering curve, consisting of the model described above (~60% of the total population) and one superimposing Pol B from the crystal structure on the gp4Δprimase hexamer (~40% of the population). These SAXS studies support the dynamic structure for the T7 replisome, with movement of DNA polymerase on the surface of gp4 coincident with binding to DNA.
DISCUSSION
The T7 replisome is a dynamic, four-protein machine that takes on multiple shapes to coordinate DNA synthesis at the leading and lagging strands of a replication fork (Lee and Richardson, 2011). Here, we present structures of a T7 replisome subassembly consisting of three of the four essential replication proteins: the gp4 primase-helicase, the gp5 DNA polymerase, and its processivity factor, thioredoxin. Our structure reveals two distinct sites of interaction: the well characterized interaction of the gp4 tail to the front basic patch of the polymerase, and another large contact patch located along the equator of the gp4 helicase ring that engages the base of the polymerase. We also find that two subunits of a gp4 heptamer bind to a single gp5/trx (Figure 1), and use native MS data to demonstrate conclusively that only the ring-shaped hexamers and heptamers of gp4Δprimase bind to T7 DNA polymerase (Figure 4), indicating that gp5/trx preferentially binds to a fully assembled helicase.
Prevailing models of the T7 replisome feature gp4 hexamers, whereas the role of gp4 heptamers in replication is ambiguous (Lee and Richardson, 2011). Yet, heptamers may facilitate the loading of gp4 on DNA (Picha et al., 2000; Crampton et al., 2006b). In this model, an encounter with DNA causes the release of one subunit from the heptamer to open the ring and thereby allow gp4 to encircle the DNA. Previous studies have shown that gp4 and gp5/trx physically interact in the absence of DNA via electrostatic interactions between the gp4 acidic C-terminal tail and two basic patches of the polymerase (Huidong Zhang et al., 2011; Kulczyk et al., 2012; Crampton et al., 2006a). Our crystal structure (Figure 1) and native MS data (Figure 4) provide the first direct evidence that gp4 heptamers bind to gp5/trx, and we have used hybrid methods of structure determination to reveal the physical makeup of these interactions. We propose that the stable complexes of gp4-DNA polymerase we have characterized are structural intermediates that are competent to load onto DNA, and some of these interactions are likely to be retained during replication. However, the structure of the T7 replisome is highly dynamic. Our SAXS data (Figure 5) reveal a replisome conformational change that occurs when either gp4 or gp5/trx load onto DNA, an illustration of the dynamic shape of the replisome that is required to accommodate different stages of replication.
Our SAXS results support and extend the low resolution SAXS envelop analysis (scattering angle q=0.12) from Kulczyk et al., 2012, which also indicated a one-to-one complex in a sample prepared by mixing gp5/trx-primer/template-gp4Δprimase-ssDNA. We analyzed the complex in the absence of primer/template DNA (Figure 5B), which was not previously examined, and with the plus primer template DNA (Figure 5C) to a higher scattering angle (q=0.3). We find a ~180 degree different placement of the polymerase and a substantially lower radius of gyration (Rg= 50.5Å versus 60.5Å). These differences may reflect our separation of the complex, higher angle data, and more detailed structural analyses. Our samples were separated and buffer exchanged by gel filtration rather than mixing, our chromatographic separation removed mixed and aggregated states compared to the somewhat heterogeneous sample in the prior analysis, and we include flexibility by employing MES to find the minimum set of structures that together fit the data.
In model replisomes encoded by bacteriophage T4 (Salinas and Benkovic, 2000; Noble et al., 2015) and E. coli (Langston et al., 2009) the helicase and polymerase proteins communicate indirectly through additional replication factors. In the T7 replication system, gp4 and gp5/trx directly contact one another to coordinate replication activities. Here we report the first detailed biophysical characterization of these interactions whereby multiple copies of gp5/trx contact the C-terminal helicase domain of the gp4 ring through interactions involving the C-terminal tail of gp4 and a larger contact patch on the outer circumference of the ring-shaped helicase. The dominant side chain interactions observed in MD simulations (Tables SI and S2) reveal alternative sets of contacts made by Pol B and Pol C, which could impart selectivity for binding of each polymerase molecule to the helicase. Mutagenesis of these candidate selectivity determinants for Pol B and Pol C could shed additional light regarding the functions of the asymmetrically bound polymerases.
The architecture of a eukaryotic replisome was recently revealed by a single-particle electron microscopy study of the CMG helicase bound to multiple copies of DNA polymerase (Sun et al., 2015). As is reminiscent of the T7 replication proteins, the eukaryotic replisome shows a direct contact between the C-terminus of the CMG helicase and the leading strand polymerase ε. However, in the eukaryotic structure, polymerase ε and polymerase α bind to opposite faces of the helicase ring, with polymerase α indirectly associated with the CMG helicase through its interaction with the Ctf4 protein. Notably, the eukaryotic replisome was imaged in the presence of a fork-shaped DNA whereas our structure lacks a DNA substrate, and both structures likely represent one structural intermediate of a dynamic replication complex. Additional studies building upon these defined replisome states will shed more light on alternative conformations and conserved structural features that allow for coupled replication as well as for unique features that support specialized functions in higher organisms.
We previously reported that two gp5/trx molecules are stably bound to a gp4 hexamer loaded onto DNA in the T7 priming complex that initiates Okazaki fragment synthesis (Wallen et al., 2013). Native MS studies in this report show up to three copies of gp5/trx associated with gp4 hexamers and heptamers in the absence of DNA. For samples containing gp5/trx in excess of gp4 hexamer/heptamers at ratios higher than 3:1, the native mass spectra were dominated by peaks from free gp5/trx, and we could not accurately assign peaks corresponding to protein complexes. Single-molecule studies of the T7 replisome show that two or three copies of gp5/trx are bound to gp4 during DNA replication; however, under some conditions the gp4 ring can bind up to six copies of gp5/trx (Geertsema et al., 2014). With this in mind, we ask why only three copies of DNA polymerase bind the gp4 heptamer in the crystal structure? The tilting of gp4 subunits in and out of the plane of the ring suggests a mechanism for the translocation of the gp4 helicase on DNA (Singleton et al., 2000), and these conformational changes could also restrict the number of polymerases bound by changing the shape of the binding site, which spans two adjacent subunits. The active sites of the ring-shaped gp4 helicase are located at the subunit interfaces where changes in subunit packing interactions control enzymatic activity and drive gp4 translocation along DNA (Crampton et al., 2004; 2006a; Singleton et al., 2000). The binding sites of Pol B and Pol C in the crystal structure are a planar arrangement of the three contiguous gp4 subunits. A simple rearrangement of these three gp4 subunits may compromise interactions with the binding site and restrict the number of DNA polymerases bound to gp4. Our crystal structure thus reasonably represents one of many structural intermediates.
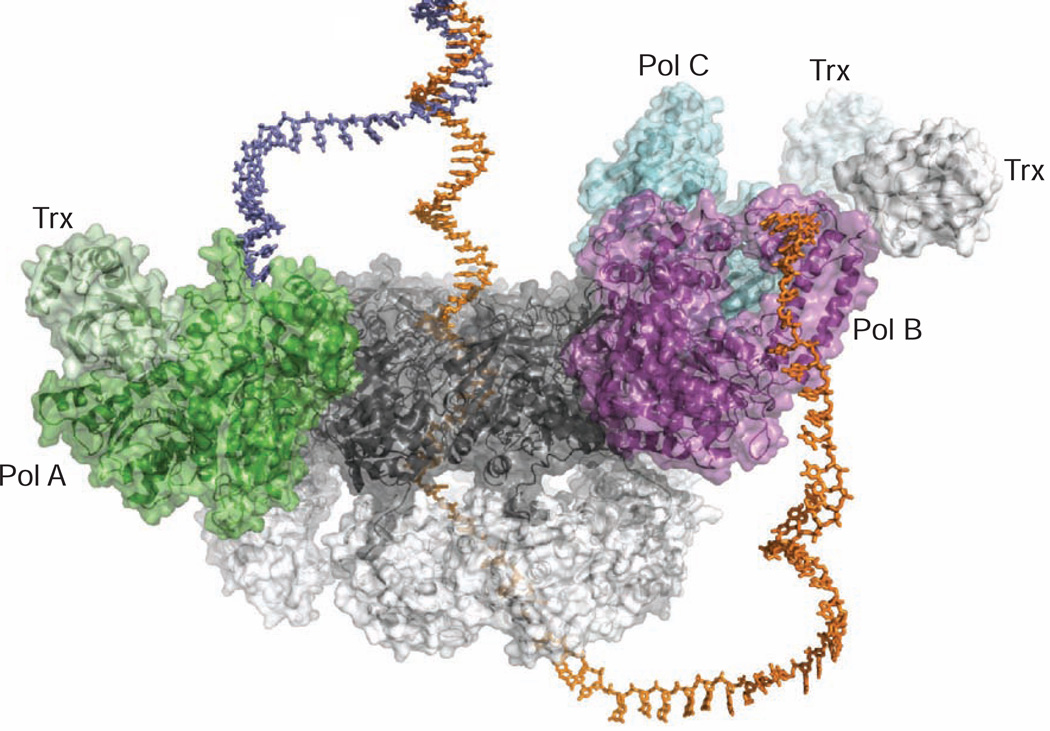
The crystal structure of a three-polymerase T7 replisome shows two polymerase molecules bound to gp4 in the same orientation with a third polymerase weakly associated in an alternative binding orientation (Figure 3), which may serve as a spare that facilitates the exchange of polymerases during replication (Geertsema et al., 2014). Based on the crystal structure of the T7 replisome, we generated a docking model using a branched DNA to mimic a replication fork (Figure 6). In the model, either Pol A or Pol B could serve as the leading or lagging strand polymerase. A replication loop reorients the lagging strand, establishing a conserved mode of interaction with Pol A and Pol B on both strands of the replication fork. This model implies that the leading and lagging strand DNA polymerases are structurally equivalent and could exchange roles during replication, while remaining bound to gp4. The model in Figure 6 could represent the initial configuration for loading DNA polymerases onto a replication fork via the interaction between the front basic patch of the polymerases and the acidic tails of gp4. Alternative conformations of the T7 replisome in solution are revealed by the SAXS data for complexes with a ssDNA template or a primer/template pair (Figure 5). Thus, the model in Figure 6 likely represents one of the multiple intermediate states during DNA replication.
Figure 6.
Model of DNA binding in the T7 Replisome
Docking model of the T7 replisome complex bound to a replication fork DNA. The leading and lagging strands are colored slate blue and orange, respectively. As single-stranded DNA passes through the central channel of the helicase, the displaced leading strand is diverted to Pol A for leading strand synthesis. The modeled lagging strand DNA is redirected by a replication loop to interact with Pol B. In this way, the crystal structure of the T7 replisome is suggestive of a simple mechanism for coupled replication of two antiparallel DNA strands.
The shapes of replication complexes determined by SAXS are in fact suggestive of alternative binding modes for gp5/trx, which can dock on different aspects of the gp4 helicase. These alternative-binding modes may be facilitated by the interaction of the front basic patch with the gp4 tail, which serves as a flexible tether between gp5/trx and the gp4 primase-helicase. The crystallized replisome complex positions the front basic patch of each polymerase near the tail of a gp4 subunit, consistent with a previously characterized replication pre-initiation complex (Huidong Zhang et al., 2011). The combined biophysical methods used here point to the dynamic nature of the T7 replisome and show that DNA binding alters the structure of the T7 replisome in solution. Crystal structures of the replisome in complex with DNA, including a model replication fork DNA as well as an Okazaki priming complex, will reveal more details about how the replisome changes shape to coordinate replication of both DNA strands.
EXPERIMENTAL PROCEDURES
Crystallization, X-ray Data Collection, and Refinement
A mixture of full-length gp4 with gp5/trx produced showers of small crystals that could not be optimized. Large, single crystals were formed when full-length gp4 was substituted with a truncated gp4 lacking the zinc-binding domain (ZBD). The best diffracting crystals appeared when gp4ΔZBD was mixed with excess gp5/trx without DNA. Additionally, the inclusion of the non-hydrolyzable ATP analog adenosine 5′-(β,γ-imido)triphosphate (AMPPNP) enhanced crystal growth, although there was no electron density present for the AMPPNP nucleotide at this resolution. Addition of either a ssDNA substrate for gp4 or a primer/template DNA for gp5/trx prevented crystal growth. Diffracting crystals of the T7 replisome complex were obtained from a mixture of 50 µM gp4ΔZBD (monomer concentration) with 75 µM gp5/trx in buffer containing 25 mM Tris pH 7.5, 200 mM potassium glutamate, 10 mM MgCl2, 2 mM DTT, and 5 mM AMPPNP. Crystals were grown by hanging drop vapor diffusion at 22 °C, with a mixture of 1 µL protein complex and 0.5 µL reservoir placed over a 500 µL reservoir containing 1.47–1.62 M sodium formate and 0.1M Tris pH 7.5. Plate-like crystals appeared within one day and grew to full size (~300–500 µm) within one week. To prepare for data collection, crystals were transferred to 3.4M sodium malonate pH 7.0 by a stepwise exchange of buffer then allowed to equilibrate overnight before crystals were flash-cooled in a gaseous nitrogen stream at 100 K. X-ray data were collected at a wavelength of 0.97872 at beamline 21-ID-F of the Life Sciences Collaborative Access Team (LS-CAT) at the Advanced Photon Source using a MarMosaic 225 detector. Data were processed using the HKL2000 suite (Otwinowski and Minor, 1997). X-ray diffraction data were corrected for anisotropy using the UCLA diffraction anisotropy server (Strong et al., 2006).
The structure of the T7 replisome was determined by molecular replacement. Full details of the structure solution and model refinement are provided in the Supplemental Experimental Procedures. The final refined model shows good Ramachandran statistics, with ϕ,ψ angles favored = 87.3%, ϕ,ψ angles allowed = 9.8%, and ϕ,ψ angles outliers = 2.9%. Structure factors and coordinates for the T7 replisome complex have been deposited in the Protein Data Bank under accession code 5IKN.
Native Mass Spectrometry
Gp4Δprimase and gp5/trx were buffer exchanged into 0.2 M ammonium acetate by gel filtration on a Waters Acquity UPLC BEH200 1.7 µm column and a Dionex ICS-5000 HPLC instrument. The column was run at 50 µL/min with 50 µL fractions collected. Gp4Δprimase and gp5/trx were either sprayed separately or were mixed at different molar ratios. Samples were loaded into an off-line, custom-made electrospray capillary and injected onto a hybrid ion mobility quadrupole time-of flight mass spectrometer (Q-IM-TOF, SYNAPT G2 HDMS, Waters Inc., Milford, MA). The instrument was operated in sensitive mode under gentle ESI conditions (capillary voltage 1.5–2.3 kV, source temperature 30 °C). The source parameters for sampling cone (100 V) and extraction cone (2 V) voltages were adjusted to optimize the signal for each protein complex. The collision voltage at the trap was adjusted from 10 to 120 V for different experiments whereas the collision voltage at the transfer region was maintained at 10 V for all experiments. The pressure of the vacuum/backing region was 5.23 mbar. Each spectrum was acquired from m/z 100–20000 every 1 second for between 10 minutes-1 hour. The instrument was externally calibrated with a NaI solution producing cluster ions. Native mass spectra were exported from Masslynx (Waters, Milford, MA,) as xy files that were directly loaded into the MASSIGN software for peak assignment. Peaks representing multiply charged protein ions were grouped (at least three charge states for each protein oligomer state or complex state) based on their deconvoluted mass and (near Gaussian) distribution. The MASSIGN software was described (Morgner and Robinson, 2012) along with a data analysis protocol that is available online (http://massign.chem.ox.ac.uk/).
Small-Angle X-ray Scattering
Gp4Δprimase E343Q,H465Y and gp5/trx were buffer-exchanged into 50 mM potassium phosphate pH 7.0, 0.5mM EDTA, 5% glycerol, and 1mM DTT by gel filtration using a 10/300 Superose 6 column (GE Healthcare). Fractions containing protein were pooled and concentrated, then dialyzed overnight in 50 mM potassium phosphate pH 7.0, 0.5mM EDTA, 2mM MgCl2, 0.2mM dTTP, 5% glycerol, and 1mM DTT. Samples containing gp4Δprimase E343Q,H465Y alone, gp5/trx alone, or mixtures of gp4Δprimase E343Q,H465Y and gp5/trx were prepared for SAXS analysis. A dT15 ssDNA oligo was present in all samples containing gp4Δprimase E343Q,H465Y to reduce hexamer/heptamer heterogeneity and drive specific hexamer formation (Crampton et al., 2006b). SAXS data were collected at beamline 12.3.1 (SIBYLS, structurally integrated biology for life sciences) at the Advanced Light Source, Lawrence Berkeley National Laboratory (Classen et al., 2013; Hura et al., 2009). The wavelength was set to λ=1.0 Å and sample-to-detector distance to 1.5 m for the collection of data corresponding to scattering vectors (q) ranging from 0.01 Å−1 to 0.32 Å−1. The scattering vector is defined as q = 4π sinθ/λ, where 2θ is the scattering angle. All experiments were performed at 20 °C and data were processed as previously described (Hura et al., 2009). A series of scattering curves were collected at 1.25 mg/mL, 2.5 mg/mL, and 5 mg/mL sample concentrations. Initial data processing, determination of radius of gyration (Rg), and pair distribution function P(r) were done with the program SCÅTTER. Aggregation-free states of samples were evaluated using data from the linear regions of the Guinier plots (Guinier and Fournet, 1955). The distance r where P(r) functions approach zero intensity identified the maximal dimension (Dmax) of the macromolecule. The reported Dmax values were determined from the P(r) functions calculated by SCÅTTER. The displayed P(r) functions were normalized to the Porod volumes of the multiprotein assemblies as calculated by SCÅTTER (Rambo and Tainer, 2011). Molecular weights were calculated based on the Volume of Correlation (Vc) (Rambo and Tainer, 2013). To evaluate discrete replisome structures within a mixture of states, the docking algorithm in FoXSDock (Schneidman-Duhovny et al., 2011) was applied using FoXS (Schneidman-Duhovny et al., 2013) to calculate the theoretical scattering profiles of atomic models. A Minimal Ensemble Search (MES) (Pelikan et al., 2009) was used to evaluate further ensembles of various replisome conformers in the samples.
Computational Modeling of Protein Interactions
Protein-protein interactions predicted by the low-resolution crystal structure of the T7 replisome were further examined by molecular dynamics simulations. All simulations were run with GROMACS 4.6.7 (Pronk et al., 2013; van der Spoel et al., 2005). Details of the molecular dynamics simulations are provided in the Supplemental Experimental Procedures. Electrostatic interactions between subunits of the replisome were quantified based on occupancy by using the hydrogen-bond analysis module of the VMD molecular visualization program (Humphrey et al., 1996).
Supplementary Material
Highlights.
Structure of the primase-helicase bound to three copies of DNA polymerase.
Hexamers and heptamers of primase-helicase bind multiple copies of DNA polymerase.
Presence of DNA alters the binding mode of DNA polymerase on the primase-helicase.
Acknowledgments
This work was supported by grants from the National Institutes of Health (GM055390 to T.E., GM105404 to M.H. and J.A.T., GM103422 to M.L.G., and CA92584 to J.A.T. and T.E.) and the U.S. Department of Energy (DE-SC0001035 to M.L.G). Native MS studies were conducted at the Mass Spectrometry Facility with partial support of the Photosynthetic Antenna Research Center, an Energy Frontier Research Center funded by the U.S. Department of Energy. J.A.T. acknowledges added support of a Robert A. Welch Chemistry Chair, plus startup funds from the Cancer Prevention and Research Institute of Texas, and the University of Texas STARs program. Efforts to combine SAXS and crystallography at the SIBYLS beamline of the Advanced Light Source (Lawrence Berkeley National Laboratory) were supported in part by United States Department of Energy program IDAT. We thank our many colleagues for creative ideas and advice.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
J.R.W., H.Z., W.C., and C.M.W.H. designed and performed experiments and analyzed the data. M.H., C.W., and B.M.F. analyzed SAXS data and performed modeling studies. J.A.T., M.L.G., and T.E. provided advice and oversight of experiments and data analysis. J.R.W. and T.E. wrote the manuscript and the other authors contributed their data, figures, and an editorial review.
REFERENCES
- Bird LE, Hâkansson K, Pan H, Wigley DB. Characterization and crystallization of the helicase domain of bacteriophage T7 gene 4 protein. Nucleic Acids Res. 1997;25:2620–2626. doi: 10.1093/nar/25.13.2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Classen S, Hura GL, Holton JM, Rambo RP, Rodic I, McGuire PJ, Dyer K, Hammel M, Meigs G, Frankel KA, Tainer JA. Implementation and performance of SIBYLS: a dual endstation small-angle X-ray scattering and macromolecular crystallography beamline at the Advanced Light Source. J. Appl. Crystallogr. 2013;46:1–13. doi: 10.1107/S0021889812048698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crampton DJ, Guo S, Johnson DE, Richardson CC. The arginine finger of bacteriophage T7 gene 4 helicase: role in energy coupling. Proc. Natl. Acad. Sci. U.S.A. 2004;101:4373–4378. doi: 10.1073/pnas.0400968101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crampton DJ, Mukherjee S, Richardson CC. DNA-induced switch from independent to sequential dTTP hydrolysis in the bacteriophage T7 DNA helicase. Mol. Cell. 2006a;21:165–174. doi: 10.1016/j.molcel.2005.11.027. [DOI] [PubMed] [Google Scholar]
- Crampton DJ, Ohi M, Qimron U, Walz T, Richardson CC. Oligomeric states of bacteriophage T7 gene 4 primase/helicase. J. Mol. Biol. 2006b;360:667–677. doi: 10.1016/j.jmb.2006.05.037. [DOI] [PubMed] [Google Scholar]
- Crampton DJ, Patel SS, Guo S, Pandey M, Johnson DE, Nandakumar D, Richardson CC. Dynamic coupling between the motors of DNA replication: hexameric helicase, DNA polymerase, and primase. Curr. Opin. Chem. Biol. 2011;15:595–605. doi: 10.1016/j.cbpa.2011.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doublié S, Ellenberger T. The mechanism of action of T7 DNA polymerase. Curr. Opin. Struct. Biol. 1998;8:704–712. doi: 10.1016/s0959-440x(98)80089-4. [DOI] [PubMed] [Google Scholar]
- Doublié S, Tabor S, Long AM, Richardson CC, Ellenberger T. Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 A resolution. Nature. 1998;391:251–258. doi: 10.1038/34593. [DOI] [PubMed] [Google Scholar]
- Geertsema HJ, Kulczyk AW, Richardson CC, van Oijen AM. Single-molecule studies of polymerase dynamics and stoichiometry at the bacteriophage T7 replication machinery. Proc. Natl. Acad. Sci U.S.A. 2014;111:4073–4078. doi: 10.1073/pnas.1402010111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guinier A, Fournet G. Small-angle scattering of X-rays. New York: John Wiley & Sons; 1955. [Google Scholar]
- Guo S, Tabor S, Richardson CC. The linker region between the helicase and primase domains of the bacteriophage T7 gene 4 protein is critical for hexamer formation. J. Biol. Chem. 1999;274:30303–30309. doi: 10.1074/jbc.274.42.30303. [DOI] [PubMed] [Google Scholar]
- Hamdan SM, Johnson DE, Tanner NA, Lee J-B, Qimron U, Tabor S, van Oijen AM, Richardson CC. Dynamic DNA helicase-DNA polymerase interactions assure processive replication fork movement. Mol. Cell. 2007;27:539–549. doi: 10.1016/j.molcel.2007.06.020. [DOI] [PubMed] [Google Scholar]
- Hollis T, Stattel JM, Walther DS, Richardson CC, Ellenberger T. Structure of the gene 2.5 protein, a single-stranded DNA binding protein encoded by bacteriophage T7. Proc. Natl. Acad. Sci. U.S.A. 2001;98:9557–9562. doi: 10.1073/pnas.171317698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Hura GL, Menon AL, Hammel M, Rambo RP, Poole FL, Tsutakawa SE, Jenney FE, Classen S, Frankel KA, Hopkins RC, et al. Robust, high-throughput solution structural analyses by small angle X-ray scattering (SAXS) Nature methods. 2009;6:606–612. doi: 10.1038/nmeth.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Frick DN, Lee J, Tabor S, Richardson CC, Ellenberger T. A complex of the bacteriophage T7 primase-helicase and DNA polymerase directs primer utilization. J. Biol. Chem. 2001;276:21809–21820. doi: 10.1074/jbc.M101470200. [DOI] [PubMed] [Google Scholar]
- Kato M, Ito T, Wagner G, Ellenberger T. A molecular handoff between bacteriophage T7 DNA primase and T7 DNA polymerase initiates DNA synthesis. J. Biol. Chem. 2004;279:30554–30562. doi: 10.1074/jbc.M403485200. [DOI] [PubMed] [Google Scholar]
- Kato M, Ito T, Wagner G, Richardson CC, Ellenberger T. Modular architecture of the bacteriophage T7 primase couples RNA primer synthesis to DNA synthesis. Mol. Cell. 2003;11:1349–1360. doi: 10.1016/s1097-2765(03)00195-3. [DOI] [PubMed] [Google Scholar]
- Kulczyk AW, Akabayov B, Lee S-J, Bostina M, Berkowitz SA, Richardson CC. An interaction between DNA polymerase and helicase is essential for the high processivity of the bacteriophage T7 replisome. J. Biol. Chem. 2012;287:39050–39060. doi: 10.1074/jbc.M112.410647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulczyk AW, Richardson CC. Molecular interactions in the priming complex of bacteriophage T7. Proc. Natl. Acad. Sci. U.S. A. 2012;109:9408–9413. doi: 10.1073/pnas.1207033109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyte J, Doolittle R. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Langston LD, Indiani C, O'Donnell M. Whither the replisome: emerging perspectives on the dynamic nature of the DNA replication machinery. Cell Cycle. 2009;8:2686–2691. doi: 10.4161/cc.8.17.9390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Chastain PD, Kusakabe T, Griffith JD, Richardson CC. Coordinated leading and lagging strand DNA synthesis on a minicircular template. Mol. Cell. 1998;1:1001–1010. doi: 10.1016/s1097-2765(00)80100-8. [DOI] [PubMed] [Google Scholar]
- Lee SJ, Marintcheva B, Hamdan SM, Richardson CC. The C-terminal residues of bacteriophage T7 gene 4 helicase-primase coordinate helicase and DNA polymerase activities. J. Biol. Chem. 2006;281:25841–25849. doi: 10.1074/jbc.M604602200. [DOI] [PubMed] [Google Scholar]
- Lee SJ, Richardson CC. Choreography of bacteriophage T7 DNA replication. Curr. Opin. Chem. Biol. 2011;15:580–586. doi: 10.1016/j.cbpa.2011.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin HK, Chase SF, Laue TM, Jen-Jacobson L, Trakselis MA. Differential temperature-dependent multimeric assemblies of replication and repair polymerases on DNA increase processivity. Biochemistry. 2012;51:7367–7382. doi: 10.1021/bi300956t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInerney P, Johnson A, Katz F, O'Donnell M. Characterization of a triple DNA polymerase replisome. Mol. Cell. 2007;27:527–538. doi: 10.1016/j.molcel.2007.06.019. [DOI] [PubMed] [Google Scholar]
- Morgner N, Robinson CV. Massign: an assignment strategy for maximizing information from the mass spectra of heterogeneous protein assemblies. Anal. Chem. 2012;84:2939–2948. doi: 10.1021/ac300056a. [DOI] [PubMed] [Google Scholar]
- Noble E, Spiering MM, Benkovic SJ. Coordinated DNA Replication by the Bacteriophage T4 Replisome. Viruses. 2015;6:3186–3200. doi: 10.3390/v7062766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nossal NG, Makhov AM, Chastain PD, Jones CE, Griffith JD. Architecture of the bacteriophage T4 replication complex revealed with nanoscale biopointers. J. Biol. Chem. 2007;282:1098–1108. doi: 10.1074/jbc.M606772200. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods in Enzymology. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Pandey M, Syed S, Donmez I, Patel G, Ha T, Patel SS. Coordinating DNA replication by means of priming loop and differential synthesis rate. Nature. 2010;462:940–943. doi: 10.1038/nature08611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelikan M, Hura GL, Hammel M. Structure and flexibility within proteins as identified through small angle X-ray scattering. Gen. Physiol. and Biophys. 2009;28:174–189. doi: 10.4149/gpb_2009_02_174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picha KM, Patel SS. Bacteriophage T7 DNA helicase binds dTTP, forms hexamers, and binds DNA in the absence of Mg2+. The presence of dTTP is sufficient for hexamer formation and DNA binding. J. Biol. Chem. 1998;273:27315–27319. doi: 10.1074/jbc.273.42.27315. [DOI] [PubMed] [Google Scholar]
- Picha KM, Ahnert P, Patel SS. DNA binding in the central channel of bacteriophage T7 helicase-primase is a multistep process. Nucleotide hydrolysis is not required. Biochemistry. 2000;39:6401–6409. doi: 10.1021/bi992857i. [DOI] [PubMed] [Google Scholar]
- Pronk S, Páll S, Schulz R, Larsson P, Bjelkmar P, Apostolov R, Shirts MR, Smith JC, Kasson PM, van der Spoel D, et al. GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics. 2013;29:845–854. doi: 10.1093/bioinformatics/btt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam CD, Hammel M, Hura GL, Tainer JA. X-ray solution scattering (SAXS) combined with crystallography and computation: defining accurate macromolecular structures, conformations and assemblies in solution. Q. Rev. Biophys. 2007;40:191–285. doi: 10.1017/S0033583507004635. [DOI] [PubMed] [Google Scholar]
- Rambo RP, Tainer JA. Characterizing flexible and intrinsically unstructured biological macromolecules by SAS using the Porod-Debye law. Biopolymers. 2011;95:559–571. doi: 10.1002/bip.21638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambo RP, Tainer JA. Accurate assessment of mass, models and resolution by small5 angle scattering. Nature. 2013;496:477–481. doi: 10.1038/nature12070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salinas F, Benkovic SJ. Characterization of bacteriophage T4-coordinated leading- and lagging-strand synthesis on a minicircle substrate. Proc. Natl. Acad. Sci. U.S.A. 2000;97:7196–7201. doi: 10.1073/pnas.97.13.7196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawaya MR, Guo S, Tabor S, Richardson CC, Ellenberger T. Crystal structure of the helicase domain from the replicative helicase-primase of bacteriophage T7. Cell. 1999;99:167–177. doi: 10.1016/s0092-8674(00)81648-7. [DOI] [PubMed] [Google Scholar]
- Schneidman-Duhovny D, Hammel M, Sali A. Macromolecular docking restrained by a small angle X-ray scattering profile. J. Struc. Biol. 2011;173:461–471. doi: 10.1016/j.jsb.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneidman-Duhovny D, Hammel M, Tainer JA, Sali A. Accurate SAXS profile computation and its assessment by contrast variation experiments. Biophys. J. 2013;105:962–974. doi: 10.1016/j.bpj.2013.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton MR, Sawaya MR, Ellenberger T, Wigley DB. Crystal structure of T7 gene 4 ring helicase indicates a mechanism for sequential hydrolysis of nucleotides. Cell. 2000;101:589–600. doi: 10.1016/s0092-8674(00)80871-5. [DOI] [PubMed] [Google Scholar]
- Strong M, Sawaya MR, Wang S, Phillips M, Cascio D, Eisenberg D. Toward the structural genomics of complexes: crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 2006;103:8060–8065. doi: 10.1073/pnas.0602606103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Shi Y, Georgescu RE, Yuan Z, Chait BT, Li H, O'Donnell ME. The architecture of a eukaryotic replisome. Nat. Struc. Mol. Biol. 2015;12:976–982. doi: 10.1038/nsmb.3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth EA, Li Y, Sawaya MR, Cheng Y, Ellenberger T. The crystal structure of the bifunctional primase-helicase of bacteriophage T7. Mol. Cell. 2003;12:1113–1123. doi: 10.1016/s1097-2765(03)00442-8. [DOI] [PubMed] [Google Scholar]
- van der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJC. GROMACS: fast, flexible, and free. J. of Comput. Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- Wallen JR, Majka J, Ellenberger T. Discrete interactions between bacteriophage T7 primase-helicase and DNA polymerase drive the formation of a priming complex containing two copies of DNA polymerase. Biochemistry. 2013;52:4026–4036. doi: 10.1021/bi400284j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Hao, Cui W, Wen J, Blankenship RE, Gross ML. Native electrospray and electron-capture dissociation FTICR mass spectrometry for top-down studies of protein assemblies. Anal. Chem. 2011;83:5598–5606. doi: 10.1021/ac200695d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Huidong, Lee SJ, Zhu B, Tran NQ, Tabor S, Richardson CC. Helicase-DNA polymerase interaction is critical to initiate leading-strand DNA synthesis. Proc. Natl. Acad. Sci. U.S.A. 2011;108:9372–93771. doi: 10.1073/pnas.1106678108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.