Abstract
BRCA1 is a well-known DNA repair pathway component and a tissue-specific tumor suppressor. However, its role in hematopoiesis is uncertain. Here we report that a cohort of patients heterozygous for BRCA1 mutations experienced more hematopoietic toxicity from chemotherapy than those with BRCA2 mutations. To test whether this reflects a requirement for BRCA1 in hematopoiesis, we generated mice with Brca1 mutations in hematopoietic cells. Mice homozygous for a null Brca1 mutation in the embryonic hematopoietic system (Vav1-iCre;Brca1F22–24/F22-24) developed hematopoietic defects in early adulthood that included reduced hematopoietic stem cells (HSCs). Although mice homozygous for a huBRCA1 knock-in allele (Brca1BRCA1/BRCA1) were normal, mice with a mutant huBRCA1/5382insC allele and a null allele (Mx1-Cre;Brca1F22-24/5382insC), had severe hematopoietic defects marked by a complete loss of hematopoietic stem and progenitor cells. Our data show that Brca1 is necessary for HSC maintenance and normal hematopoiesis, and that distinct mutations lead to different degrees of hematopoietic dysfunction.
E-TOC
Mgbemena et al. report that hematopoietic stem cells have an absolute requirement for Brca1 to survive. They also show that humanization of the mouse Brca1 gene with a knocked-in human BRCA1 cDNA, but not a mutant BRCA1/5382insC cDNA, fully substitutes for mouse Brca1 during both embryonic development and hematopoiesis.
INTRODUCTION
Hematopoietic stem cells (HSCs) depend upon DNA repair mechanisms to maintain their genomic integrity (Beerman et al., 2014; Mohrin et al., 2010; Rossi et al., 2007). Deficiencies in DNA repair proteins impair HSC function, although the nature and severity of the defects vary among DNA repair pathways. Deficiency for proteins involved in non-homologous end-joining does not affect HSC frequency or hematopoiesis in normal young adult mice but does reduce HSC function in response to stress (Rossi et al., 2007) and can lead to HSC depletion during aging (Nijnik et al., 2007). Deficiency for proteins involved in DNA mismatch repair does not appear to have major effects on hematopoiesis under normal conditions, but impairs the capacity of HSCs to reconstitute irradiated mice (Reese et al., 2003). Deficiency for homologous recombination-mediated double strand break repair proteins, however, can lead to hematopoietic failure in patients (Kottemann and Smogorzewska, 2013) and impair hematopoiesis in mice as well as impairing HSC function upon transplantation into irradiated mice (Bender et al., 2002; Carreau et al., 1999; Haneline et al., 1999; Ito et al., 2004; Navarro et al., 2006).
Fanconi anemia is caused by at least 18 different autosomal recessive mutants in the FA-BRCA repair pathway, including BRCA2 (Howlett et al., 2002; Xia et al., 2007), PALB2 (Reid et al., 2007), and BRIP1 (Seal et al., 2006). All three of these proteins physically interact with BRCA1 during DNA repair (Baer and Ludwig, 2002; Prakash et al., 2015; Xia et al., 2006; Zhang et al., 2009), raising the question of whether mutations in BRCA1 could also influence HSC function or hematopoiesis. Two individuals with developmental defects consistent with Fanconi anemia have been identified with genetic variants in both BRCA1 alleles (Domchek et al., 2013; Sawyer et al., 2015); however, it is not clear that these were all deleterious mutations and neither individual was reported to have hematopoietic defects. If loss-of-function mutations in BRCA1 impair DNA repair in hematopoietic cells, this would have broad implications for patients with BRCA1 mutations as these patients are at increased risk of certain cancers that are commonly treated with DNA-damaging chemotherapies.
Homozygosity for germline loss-of-function in Brca1 is embryonic lethal in mice (Drost and Jonkers, 2009). Conditional deletion of Brca1 from breast epithelium in mice leads to the development of breast cancer, but only when combined with p53 deficiency (Drost and Jonkers, 2009; McCarthy et al., 2007). Two recent studies conditionally deleted Brca1 from hematopoietic cells (Santos et al., 2014; Vasanthakumar et al., 2016). One showed that leukemia cells transformed by MLL-AF9 exhibited reduced proliferation and increased differentiation in the absence of Brca1 (Santos et al., 2014). The second study showed that conditional Brca1 deletion reduced blood cell counts and colony-forming progenitors. Transplantation of Brca1-deficient bone marrow cells into irradiated mice was associated with lower blood cell counts in recipient mice and a trend toward lower levels of donor cell reconstitution 10–15 days after transplantation. However, this study did not detect a significant reduction in HSC frequency and the consequences for the long-term reconstituting capacity of bone marrow cells was not assessed (Vasanthakumar et al., 2016). Therefore, it has not yet been tested whether Brca1 is required for HSC function or whether heterozygosity for BRCA1 mutations affects recovery after chemotherapy in humans or in mice.
We evaluated the hematologic effects of chemotherapy on cancer patients with germline BRCA1 or BRCA2 mutations and found that in our small cohort, BRCA1 mutations were associated with an increased risk of hematopoietic toxicity. Based on these clinical observations, we tested the effects of BRCA1 mutations on hematopoiesis in mice. To do this, we characterized the effects of two different mutant Brca1 alleles on mouse hematopoiesis. We show that Brca1 is necessary for HSC maintenance and normal hematopoiesis but that different alleles exhibit differences in the severity of the HSC phenotype that do not correlate with differences in the severity of their effects on embryonic development.
RESULTS
Association of BRCA1 mutations with hematopoietic toxicity from chemotherapy
Patients with germline BRCA1 mutations commonly develop cancers that are treated with DNA-damaging chemotherapies. To test whether those patients are at increased risk for hematopoietic complications, we analyzed hematopoietic parameters in patients heterozygous for deleterious mutations in BRCA1 at baseline (healthy patients) and after chemotherapy. We compared our patients to those who carry deleterious mutations in BRCA2 for two reasons. First, it allowed for a comparison cohort that was similar in gender and age (Table 1), and second because prior data suggested that BRCA2 mutation carriers may experience fewer episodes of neutropenia compared BRCA1 mutation carriers (Shanley et al., 2006). In the latter report it was not possible to match for type of chemotherapy regimen and most of the patients did not receive doxorubicin, a standard component of current breast cancer treatment. In our cohorts, most of the patients with BRCA1 or BRCA2 mutations that received chemotherapy had breast cancer and were treated with four cycles of dose dense doxorubicin plus cyclophosphamide (both drugs are DNA damaging) followed by four cycles of paclitaxel.
Table 1.
Characteristics of human BRCA1 and BRCA2 mutation carriers
| Population parameters | BRCA1 | BRCA2 | Total | |
|---|---|---|---|---|
| Patients (n) | Total | 104 | 96 | 200 |
| Female | 90 | 87 | 177 | |
| Male | 14 | 9 | 23 | |
| Age | Range | 20–79 | 20–77 | - |
| Median | 42 | 46 | - | |
| Race (n) | Black | 23 | 17 | 40 |
| Asian | 8 | 7 | 15 | |
| Caucasian | 33 | 44 | 77 | |
| Hispanic | 40 | 28 | 68 | |
| Cancer (n) | Total | 45 | 23 | 68 |
| Breast* | 33 | 17 | 50 | |
| Ovarian | 7 | 1 | 8 | |
| Other | 5 | 5 | 10 | |
| Chemo (n)** | Total | 29 | 13 | 42 |
| Breast | 24 | 11 | 35 | |
| Ovarian | 1 | 1 | 2 | |
| Other | 4 | 1 | 5 | |
Twenty-five of the BRCA1 mutation carriers had ER/PR/HER2 triple negative breast cancer (TNBC). None of the 23 BRCA2 mutation carriers had TNBC.
This represents the patients treated at our institution with chemotherapy: most breast cancer patients received four cycles of dose dense doxorubicin & cyclophosphamide followed by four cycles of paclitaxel; ovarian or peritoneal cancer patients received 4–8 cycles of carboplatin & paclitaxel (Table S1).
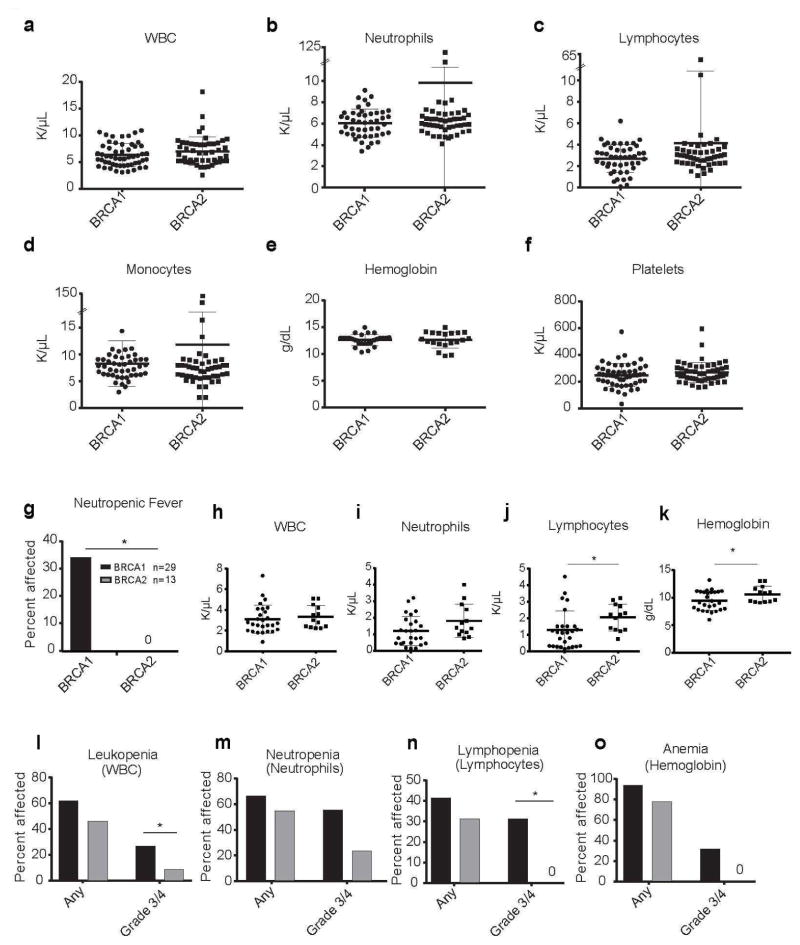
BRCA1 and BRCA2 mutation carriers at our institution had normal blood cell counts at steady state (Figure 1a–f). Both sets of patients experienced hematopoietic toxicity after chemotherapy, though BRCA1 mutation carriers tended to experience more frequent and severe hematopoietic toxicity from chemotherapy than BRCA2 mutation carriers (Figure 1g–k). This was evident only in a subset of the patients as shown by the wide variations in the post-chemotherapy blood counts (Figure 1h–k; blood counts from each patient at the time of maximum neutrophil toxicity). Severity of maximal toxicity for blood parameters was also quantified as recommended by the NCI’s criteria for adverse events (Figure 1l–o; Table 2). Since prophylactic G-CSF is standard for patients receiving dose dense doxorubicin plus cyclophosphamide and paclitaxel therapy, the overall rate of febrile neutropenia (neutropenia with fever) in breast cancer patients at our institution during the last three years has averaged ~5%. This is consistent with a recently reported 3.4% overall febrile neutropenia rate for this regimen in the United States (Caggiano et al., 2005). In contrast, 34% of BRCA1 mutation carriers experienced febrile neutropenia, a significantly higher frequency than observed among BRCA2 mutation carriers (0%; P<0.0001; Figure 1g). The BRCA1 and BRCA2 mutation carriers were relatively young patients (median age 41.5 and 46, respectively; Table 1) without co-morbidities, in whom febrile neutropenia would not be expected (Kouroukis et al., 2008). Consistent with the increased incidence of febrile neutropenia, we observed a trend toward an increased incidence of neutropenia among BRCA1 mutation carriers as compared to BRCA2 mutation carriers (Figure 1i; P<0.1).
Figure 1. Cancer patients with BRCA1 mutations have increased hematopoietic toxicity after chemotherapy.
(A–F) Complete blood cell counts from individuals with BRCA1/2 mutations who had not been exposed to chemotherapy. No significant differences were observed in (A) white blood cells (WBC), (B) Neutrophils, (C) Lymphocytes, (D) Monocytes, (E) Hemoglobin, or (F) Platelets between BRCA1 and BRCA2 mutation carriers (n= 63 BRCA1 and 50 BRCA2 patients). Bars in (A–F) represent means ± standard deviation. (G) Frequency of febrile neutropenia (FEN) in BRCA1 or BRCA2 cancer patients treated with chemotherapy. (H–K) Parameters from blood counts with the lowest absolute neutrophil counts after BRCA1 or BRCA2 patients were treated with chemotherapy. (L–O) Frequency and severity of hematopoietic toxicity in BRCA1 or BRCA2 cancer patients treated with chemotherapy. Black bar = BRCA1 mutation carriers. Grey bar = BRCA2 mutation carriers. Percent of patients affected with any grade and severe grade 3 or grade 4 (L) leukopenia (M) neutropenia, (N) lymphopenia or (O) anemia are shown. Grades of blood cell count toxicity were assigned as standardized by the National Cancer Institute. Common Terminology Criteria for Adverse Events v3.0 (Trotti et al., 2003). Neutropenic fever was defined as an ANC < 500 cells/mm3 and a sustained fever > 38°C (100.4°F). Statistical significance in (A–F, H–K) was assessed using a two-tailed Student’s t-test and in (G, L–O) using a Fisher’s Exact test (*P<0.05).
Table 2.
NCI CTCAE grading system for hematologic toxicity
| lineage | Grade 1 | Grade 2 | Grade 3 | Grade 4 |
|---|---|---|---|---|
| Neutrophils | <LLN to 1,500/mm3 | 1,000 to 1,500/mm3 | 500 to 1,000/mm3 | <500/ mm3 |
| Platelets | <LLN to 75,000/mm3 | 50,000 to 75,000/mm3 | 25,000 to 50,000/mm3 | <25,000/ mm3 |
| Hemoglobin | <LLN to 10 g/dL | 8.0 to 10.0 g/dL | <8.0 g/dL | Life-threatening consequences |
| Lymphocytes (total) | <LLN to 800/mm3 | 500 to 800/mm3 | 200 to 500/mm3 | <200/ mm3 |
Neutropenia, thrombocytopenia, anemia, and lymphopenia were determined from the complete blood count after chemotherapy and the lowest count was used for calculating grade of toxicity. All patients with sustained fever of > 100.4°F in the midst of chemotherapy-induced grade 4 neutropenia received a first course of IV antibiotics in-hospital. Taken from NCI CTCAE, version 3.0: National Cancer Institute Common Terminology Criteria for Adverse Events; LLN: lower limit of normal.
BRCA1 mutation carriers also had a significantly higher frequency of grade 3/4 leukopenia (28% for BRCA1 versus 8% for BRCA2; Figure 1l; P<0.05) and grade 3/4 lymphopenia (31% for BRCA1 versus 0% for BRCA2; Figure 1n; P<0.05) as compared to BRCA2 mutation carriers. There was also a trend toward an increased incidence of grade 3/4 anemia (31% for BRCA1 versus 0% for BRCA2; Figure 1o) after chemotherapy. There were no differences in platelet counts between the two cohorts (data not shown).
There were no correlations of specific BRCA1 mutations with febrile neutropenia. The small numbers of individuals with each mutation precluded our ability to determine whether or not specific mutations in the BRCA1 gene were associated with toxicity (Table S1). Nevertheless, these data suggest that collectively germline BRCA1 mutations are associated with a higher than expected risk for chemotherapy-associated hematopoietic complications.
Brca1 deficiency in mice causes pancytopenia
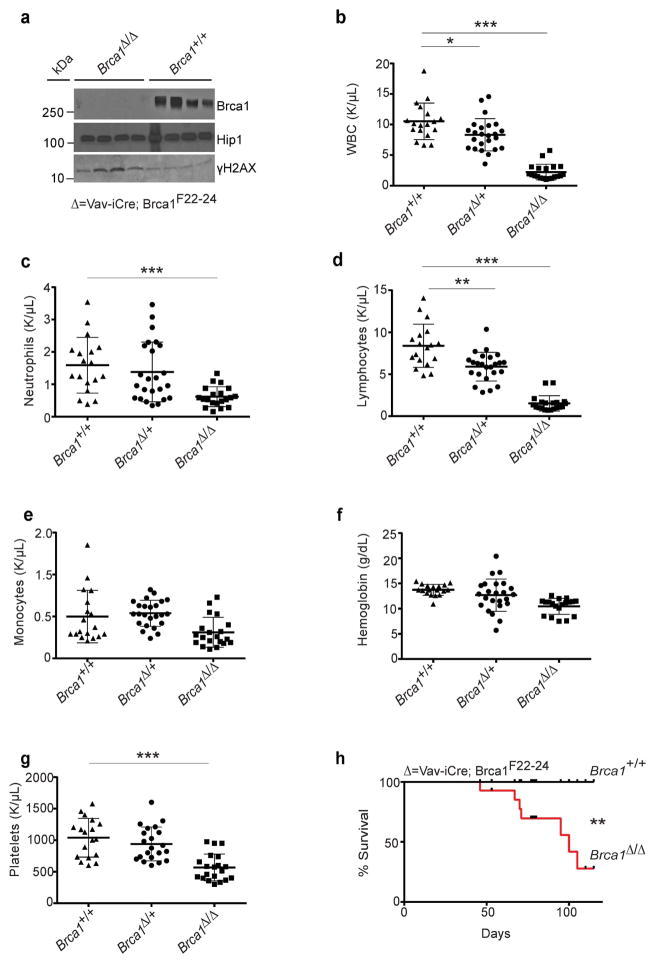
The high rate of hematopoietic toxicity in BRCA1 mutation carriers after chemotherapy raised the question of whether reduced BRCA1 function impairs the capacity to regenerate hematopoiesis after myeloablation. To test this, we generated Vav1-iCre;Brca1F22-24/F22-24 mutant mice (Figure S1) (McCarthy et al., 2007). When the conditional Brca1F22-24 allele is recombined, it is considered a null allele as mice that have homozygous deletion of this allele in the germline die before birth, and conditional deletion in the breast epithelium leads to breast cancer (McCarthy et al., 2007). Vav1-iCre deletes in embryonic and adult hematopoietic cells, including HSCs (Georgiades et al., 2002). Vav1-iCre;Brca1F22-24/F22-24 mice at weaning had normal body weight and appeared healthy. As reported previously (McCarthy et al., 2007), there was no detectable Brca1 protein in spleen cells isolated from Vav1-iCre;Brca1F22-24/F22-24 mice (Figure 2a; top panel). Phosphorylation of H2AX was increased in Vav1-iCre;Brca1F22-24/F22-24 splenocytes, as would be expected for a functionally null DNA repair gene (Figure 2a; bottom panel).
Figure 2. Brca1 deficiency leads to severe hematologic abnormalities.
(A) Western blot analysis for mouse Brca1, Hip1 (loading control) and γH2AX in spleen extracts (100 μg per lane) from Vav1-iCre;Brca1F22-24/F22-24 (Δ/ Δ; n=4) and control (+/+; n=4) mice. (B–G) Blood counts from 3–6 week old wild-type control mice (+/+; n=18), Vav1-iCre;Brca1F22-24/+ (Δ/+; n=24) and Vav1-iCre;Brca1F22-24/F22-24 (Δ/ Δ; n=20) mice. (B) White blood cells (WBC), (C) neutrophils, (D) lymphocytes, (E) monocytes, (F) hemoglobin, and (G) platelets are shown. (H) Kaplan-Meier survival curve of control (black line; +/+; n=14) and Vav1-iCre;Brca1F22-24/F22-24 (red line; Δ/ Δ; n=14) mice. All data represent means ± standard deviation. Statistical significance was assessed using a two-tailed Student’s t-test except in (H) where a log-rank test was used (*P<0.05, **P<0.01, ***P<0.001).
Vav1-iCre;Brca1F22-24/F22-24 mice developed severe pancytopenia. Three to six week old Vav1-iCre;Brca1F22-24/F22-24 mice had significantly decreased absolute numbers of white blood cells, including neutrophils and lymphocytes, as well as significantly reduced numbers of platelets compared to controls (Figure 2b–g). Deletion of a single allele of Brca1 was sufficient to slightly but significantly reduce WBC and lymphocyte levels (Figure 2b and 2d).
Vav1-iCre;Brca1F22-24/F22-24 mice had a shortened life-span and most died spontaneously without appearing ill, likely as a result of hematopoietic failure and its consequences (acute infection, bleeding etc.). Half of the Vav1-iCre;Brca1F22-24/F22-24 mice died by 75 days (Figure 2h). A fraction (27%; 7/26) of the Vav1-iCre;Brca1F22-24/F22-24 mice that survived beyond 3 months of age did become moribund (hunched, immobile, cold) prior to death and developed lymphocyte infiltrated splenomegaly (Figure S2a). Unlike the prior report that found p53 mutations in some spleens that were Brca1 null (Vasanthakumar et al., 2016), when we used RNA-seq to analyze for mutations in expressed genes, only wild type p53 was found in these enlarged spleens. However, consistent with the same report, T-cell infiltration was present based on a decreased B-cell specific gene expression and an increased T-cell gene expression pattern in the enlarged spleens (Figure S2b). These data suggest that deletion of Brca1 from mouse hematopoietic cells has the potential to promote the development of hematopoietic malignancies from surviving progenitors (Vasanthakumar et al., 2016).
Brca1 deficiency causes HSC depletion
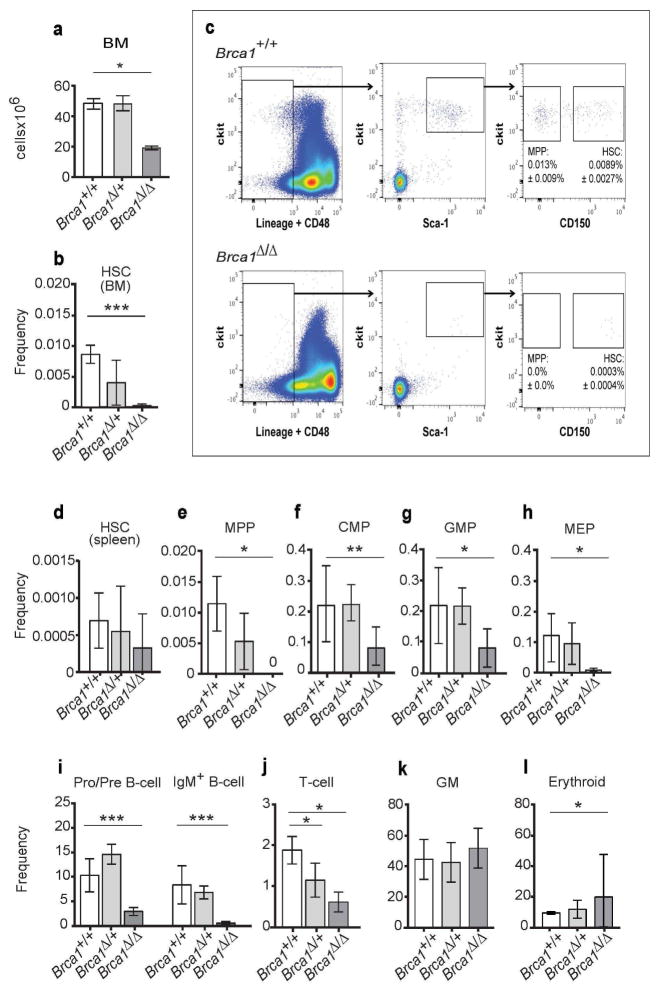
To identify the cause of the pancytopenia in Vav1-iCre;Brca1F22-24/F22-24 mice, we examined hematopoiesis in their bone marrow. Six-week-old Vav1-iCre;Brca1F22-24/F22-24 mice had a significant reduction in bone marrow cellularity compared to littermate controls (Figure 3a; P<0.05). Strikingly, the bone marrow of Vav1-iCre;Brca1F22-24/F22-24 mice had only 3% of the CD150+CD48−Lineage−Sca-1+ckit+ (CD150+CD48−LSK) (Kiel et al., 2005) HSCs observed in controls (Figure 3b and c). This decline in HSC frequency did not reflect HSC mobilization, as HSC frequency was not increased in the spleens of Vav1-iCre;Brca1F22-24/F22-24 mice (Figure 3d). Although there was a trend toward reduced HSC frequency in the heterozygotes (Vav1-iCre;Brca1F22-24/+), the difference was not statistically significant. There was also a severe reduction in the frequency and absolute number of hematopoietic progenitor cells in the bone marrow, including CD150−CD48−LSK multipotent progenitors (MPPs) (Oguro et al., 2013), CD34+CD16/32lowCD127−Sca-1−LK common myeloid progenitors (CMPs), CD34+CD16/32highCD127−Sca-1−LK granulocyte macrophage progenitors (GMPs), CD34−CD16/32lowCD127−Sca-1−LK megakaryocyte erythroid progenitors (MEPs) (Akashi et al., 2000) (Figure 3e-h, S3b-e), B220+IgM− B cell progenitors, B220+IgM+ B cells (Figure 3i, S3f), and CD3+ T cells (Figure 3j, S3g) in Vav1-iCre;Brca1F22-24/F22-24 bone marrow compared to controls. The frequency and absolute number of CD11b+Gr1+ myeloid cells (GM) was not significantly reduced in Vav1-iCre;Brca1F22-24/F22-24 bone marrow (Figure 3k, S3h). The frequency of CD71+Ter119+ erythroid progenitors was significantly increased in Vav1-iCre;Brca1F22-24/F22-24 bone marrow (Figure 3l), but absolute number were unchanged (Figure S3i). These data indicate that Brca1 deficiency in Vav1-iCre;Brca1F22-24/F22-24 mice depletes HSCs and hematopoietic progenitor cells in young adult mice.
Figure 3. Brca1 deficiency causes HSC depletion.
(A) Bone marrow (BM) cellularity (1 femur and 1 tibia) of control (+/+; n=4), Vav1-iCre;Brca1F22-24/+ (Δ/+; n=5), and Vav1-iCre;Brca1F22-24/F22-24 mice (Δ/Δ; n=4). (B) Frequency (number of HSCs/total BM cells) of HSCs in the BM of control (+/+; n=7; white bar), Vav1-iCre;Brca1F22-24/+(Δ/+; n=5; light grey bar), and Vav1-iCre;Brca1F22-24/F22-24 mice (Δ/Δ; n=4; dark grey bar). (C) Representative flow cytometry plot showing CD150+CD48−LSK HSCs in wild type mice (+/+; top panel) and Vav1-iCre;Brca1F22-24/F22-24 mice (Δ/Δ; bottom panel). Frequency of (D) HSCs in the spleen of control (n=3), Vav1-iCre;Brca1F22-24/+ (n=3), and Vav1-iCre;Brca1F22-24/F22-24 mice (n=3). Frequency of (E) MPPs, (F) CMPs, (G) GMPs, (H) MEPs, (I) B lineage cells, (J) T lineage cells, (K) myeloid cells, and (L) erythroid cells in the bone marrow of control (+/+; n=7; white bar), Vav1-iCre;Brca1F22-24/+(Δ/+; n=5; light grey bar), and Vav1-iCre;Brca1F22-24/F22-24 mice (Δ/Δ; n=4; dark grey bar). Statistical significance was assessed using a two-tailed Student’s t-test (*P<0.05, **P<0.01, ***P<0.001).
Competitive bone marrow transplantation assays were performed to functionally analyze HSCs from Vav1-iCre;Brca1F22-24/F22-24 mice (n=3 donors into 5 recipients each). Bone marrow cells from six week old Vav1-iCre;Brca1F22-24/F22-24 (CD45.2+) mice and Brca1F22-24/F22-24 (CD45.2+) controls were each transplanted with equal numbers of wild-type congenic bone marrow cells (CD45.1+) into lethally irradiated mice (CD45.1+). Four weeks after transplantation, few donor derived Vav1-iCre;Brca1F22-24/F22-24 (CD45.2+) cells were detected in the peripheral blood of recipient mice (Figure S3j). By 8 weeks after transplantation, there were no donor-derived B-, T- or myeloid cells in the peripheral blood of these recipients, some of which were followed out to 20 weeks without any detectable donor cell reconstitution (Figure S3k–m). Consistent with the decline in the frequency of HSCs based on surface marker phenotype (Figure 3b), Vav1-iCre;Brca1F22-24/F22-24 mice had no functional HSCs capable of reconstituting irradiated mice.
Heterozygosity for Brca1 reduces HSC reconstituting capacity
To begin to determine if the bone marrow of Vav1-iCre;Brca1F22-24/+ mice has increased sensitivity to DNA stress, we treated a cohort of Vav1-iCre;Brca1F22-24/+ mice with two cycles of cyclophosphamide and monitored their blood counts for recovery abnormalities. Under these specific conditions, we did not observe consistent differences between heterozygous and wild type mice (Figure S3r–w). To observe more subtle differences, it may be necessary to either give different doses of cyclophosphamide, treat with several more drug cycles or treat with other drugs used in our patients such as cisplatin, doxorubicin and paclitaxel.
We have also tested heterozygous bone marrow sensitivity to proliferative DNA stress by evaluating whether haploinsufficiency for Brca1 influences the bone marrow reconstituting capacity of HSCs. To do this bone marrow cells from six week old Vav1-iCre;Brca1F22-24/+ (CD45.2+) mice and Brca1+/+ (CD45.2+) controls were each transplanted with equal numbers of wild-type congenic bone marrow cells (CD45.1+) into lethally irradiated mice (CD45.1+). We observed no significant differences between Vav1-iCre;Brca1F22-24/+ and control cells in the reconstitution of primary recipient mice (Fig. S3j–m). These results from challenges with chemotherapy and transplantation suggest that haploinsufficiency of Brca1 in mice does not exhibit exceptionally high bone marrow sensitivity.
To further test if heterozygosity impaired HSC self-renewal potential, we serially transplanted bone marrow cells from the primary recipient mice into secondary recipient mice. Sixteen weeks after primary transplantation, we transplanted bone marrow cells from primary recipient mice with levels of donor cell reconstitution (CD45.2+) nearest the median values in each treatment. Vav1-iCre;Brca1F22-24/+ cells gave significantly lower levels of donor cell reconstitution in all lineages as compared to Brca1+/+ control cells in secondary recipients (Figure S3n–q). These results suggest that Vav1-iCre;Brca1F22-24/+ HSCs do in fact exhibit a reduced self-renewal capacity as compared to wild-type HSCs. Although single Vav1-iCre transgenic mice were not used as controls in this cohort, CBC abnormalities or defects in primary or secondary reconstitution by bone marrow cells from Vav1-iCre mice in the same pure C57/BL6 background have not been observed previously (Foley et al., 2013). These observations in mice are consistent with greater chemotoxicity in humans with BRCA1 mutations and suggest that heterozygosity for a loss-of-function mutation in Brca1 can impair the ability to regenerate hematopoiesis after one or more cycles of chemotherapy (Figure 1g).
Because serial bone marrow transplantation was required to observe a deleterious effect of proliferative stress on Brca1 haploinsufficient HSCs, it remains possible that the heterozygous genotype does not impose as much chemotherapeutic toxicity as our human data would suggest. Since our patient data are from a retrospective analysis of a small cohort (Figure 1g), further work to prospectively observe more humans with inherited cancer predisposition mutations who are treated with chemotherapy is necessary. Also, generation and serial treatment of more Brca1 haploinsufficient mice with various bone marrow stresses that include repeated cycles of cyclophosphamide, doxorubicin, cisplatin and paclitaxel will need to be completed.
Generation of wild-type and mutant knock-in alleles of human BRCA1
We and others have found that frameshift or stop-gain mutations in the last few exons of the BRCA1 gene encode non-functional mutant proteins (Scully et al., 1999) expressed from messages that do not experience RNA decay (Perrin-Vidoz et al., 2002; Soyombo et al., 2013). These C-terminal mutations frequently lead to cancer phenotypes distinct from those caused by mutations elsewhere in the gene (Rebbeck et al., 2015). We have found that the common Ashkenazi Jewish BRCA1 5382insC founder mutation results in expression of a mutant transcript in the same amounts as BRCA1 wild-type message in fibroblasts, induced pluripotent stem cells and teratomas (Soyombo et al., 2013). The 5382insC mutation leads to a C-terminal frame-shift mutation. We wondered whether this hypomorphic allele would have a less severe hematopoietic phenotype as compared to the null mutation.
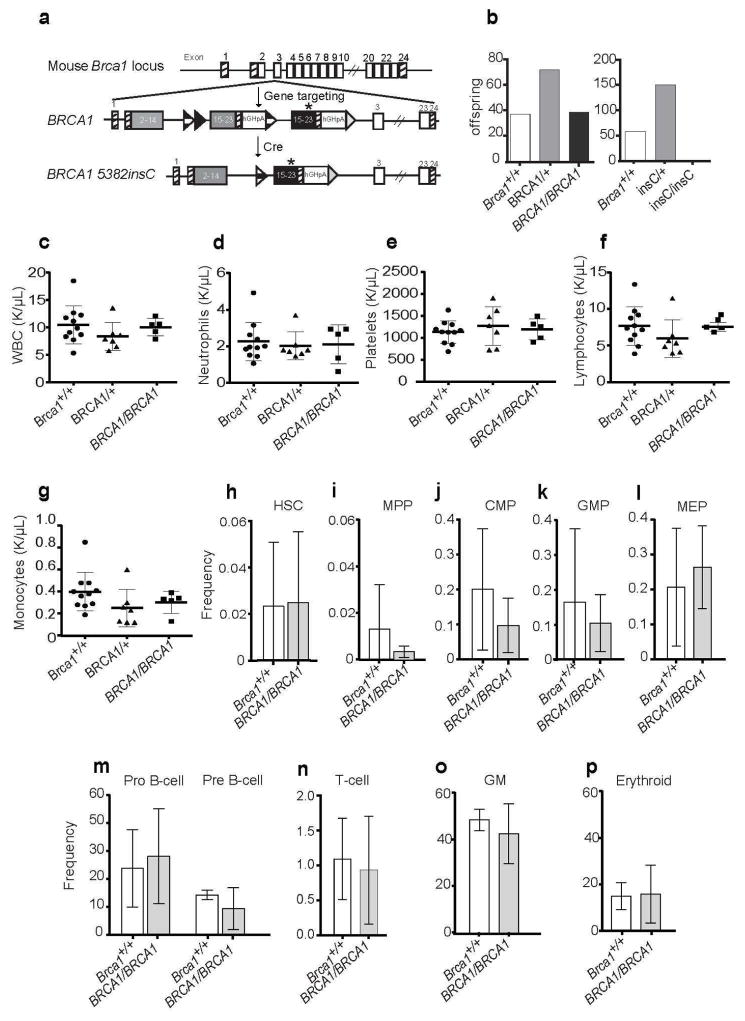
To assess the 5382insC mutation’s effects on embryogenesis and hematopoiesis, and to ensure that any abnormalities observed with humanization of the Brca1 locus with the BRCA1 5382insC allele were due to the abnormal BRCA1, we generated mice that were humanized with wild-type human BRCA1 in place of mouse Brca1. To do this, we designed a targeting vector that allowed for expression of both a wild-type BRCA1 allele and, upon Cre mediated recombination, the BRCA1 5382insC mutation (Figure 4a; Figure S4a). C57BL/6 embryonic stem cells (ESCs) were electroporated with the targeting construct and screened for correctly targeted knock-in alleles (Figure S4b). Two lines that were correctly targeted and expressed human BRCA1 (Figure S4c, lanes 3–6) were also electroporated with CMV-Cre to generate ESC lines that expressed the recombined Brca15382insC mutant allele (Figure S4c, lanes 7, 8). The Brca1BRCA1 allele substituted for wild-type mouse Brca1 function as evidenced by the fact that fully humanized homozygotes (Brca1BRCA1/BRCA1) were born at Mendelian frequencies (Figure 4b, left) and remained alive and well with no hematopoietic abnormalities (Figure 4d–p) for up to 1.5 years of age (Figure S4d).
Figure 4. Humanization of the Brca1 allele with wild type BRCA1 or BRCA1 5382insC knocked-in cDNA sequences.
(A) Targeting vector used to knock BRCA1 into the Brca1 locus. This allowed for humanization of the Brca1 gene with a wild type BRCA1 or the BRCA1 5382insC point mutation. (B) BRCA1 5382insC knock-in is embryonic lethal. (Left) Number of offspring and the genotypes produced from 15 heterozygous Brca1BRCA1/+ mating pairs. (Right) Number of offspring and the genotypes produced from 30 heterozygous Brca15382insC/+ (InsC) mating pairs. (C–G) Blood counts from wild-type control mice (+/+; n=11), homozygous Brca1huBRCA1/huBRCA1 (BRCA1/BRCA1; n=5) and heterozygous Brca1huBRCA1/+ (BRCA1/+; n=7). (C) White blood cells (WBC), (D) neutrophils, (E) platelets, (F) lymphocytes and (G) monocytes are shown.(H) HSCs, (I) MPPs, (J) CMPs, (K) GMPs (L) MEPs (M) B lineage cells, (N) T lineage cells, (O) myeloid cells, and (P) erythroid cells in the bone marrow of control (+/+; n=3; white bar) and Brca1huBRCA1/huBRCA1 (BRCA1/BRCA1; n=3; light grey bar). Statistical significance was assessed using a two-tailed Student’s t-test. There were no significant differences between the genotypes.
To generate mice with the BRCA1 5382inC allele (Brca15382insC) in the germline, Brca1BRCA1/BRCA1 mice were mated with CMV-Cre deletor mice (Dupe et al., 1997). The progeny with recombination in the germline were then used for further analysis of mice who carried this human mutation. In contrast to the Brca1BRCA1 allele, homozygosity for the Brca15382insC allele was embryonic lethal, as 208 progeny from the heterozygous Brca15382insC/+ parents included no Brca15382insC/5382insC homozygotes and an expected frequency of heterozygotes and wild type mice (Figure 4b, right, p<0.0001). This lethality confirms that the 5382insC allele encodes a severe loss-of-function mutation. These data thus indicate that the wild-type human BRCA1 cDNA rescues mouse embryonic lethality (suggesting that alternative splicing is not necessary for this gene to function properly in mice), and that the BRCA1 5382insC mutant does not.
Hematopoiesis in mice with the Brca15382insC mutation
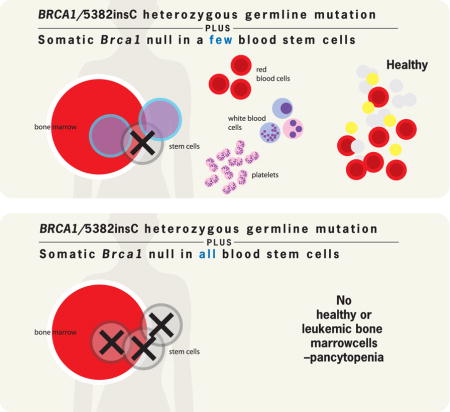
To assess the effects of the germline Brca15382insC mutant allele on hematopoiesis, we crossed the Brca15382insC/+ mice with the Vav1-iCre;Brca1F22-24/+ mice to generate Vav1-iCre; Brca1F22-24/5382insC biallelic mutant mice. This is a similar genetic configuration predicted to occur in many human cancers—a germline mutation (Brca15382insC) in one allele followed by somatic loss of heterozygosity as a result of deletion of the second allele. The biallelic Vav1-iCre; Brca1F22-24/5382insC mice were healthy at weaning. In contrast to the severe hematopoietic defects in Vav1-iCre;Brca1F22-24/F22-24 mice, average peripheral blood counts (Figure S5a–f) and bone marrow stem and progenitor cell frequencies were normal in adult Vav1-iCre; Brca1F22-24/5382insC mice (Figure S5h-p). However, the normal blood counts may be attributed to the main presence of cells that lack recombination of the floxed null allele (Figure S5g). In contrast to the presence of only non-recombined hematopoietic cells in Vav1-iCre; Brca1F22-24/5382insC mice (same amplification curve as obtained from DNA derived from Cre-negative Brca1F22-24/+ control bone marrow), hematopoietic cells from Vav1-iCre;Brca1F22-24/F22-24 mice exhibited significant recombination (Figure S5g). A possible explanation for the lack of somatic recombination of the Brca1F22-24 allele when the germline allele is Brca15382insC, is that the BRCA15382insC protein is more deleterious to hematopoietic cells than the simple null allele, and thus the only cells that survived into adulthood were those that were not somatically recombined.
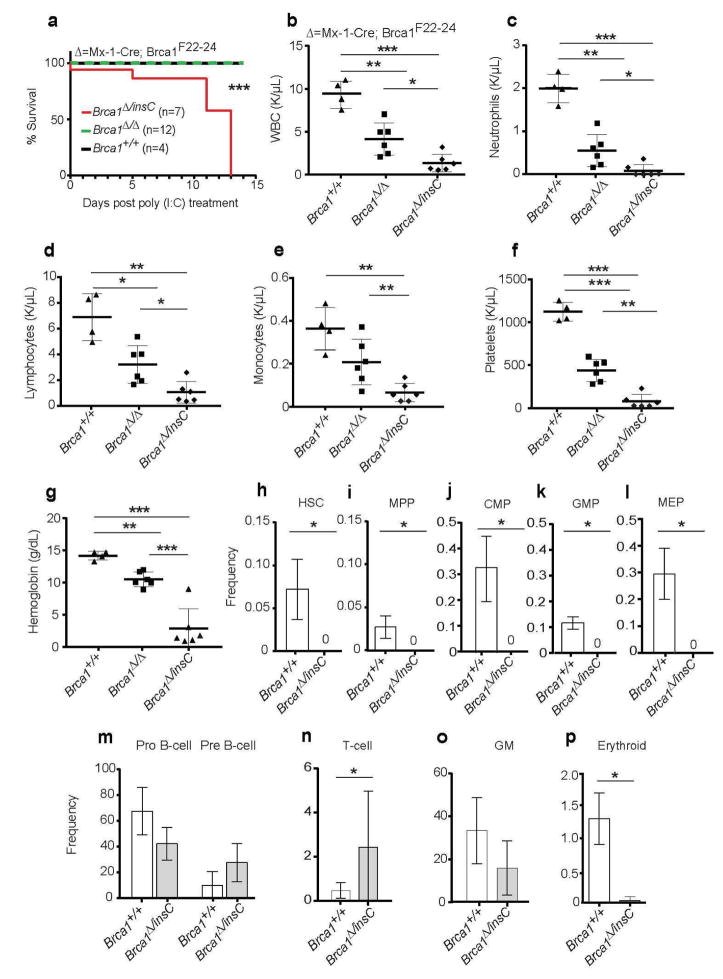
To test this, we used Mx1-Cre to conditionally recombine the floxed allele in the adult hematopoietic system. The un-induced Mx1-Cre;Brca1F22-24/F22-24 biallelic mice were healthy at weaning but when Mx1-Cre was induced with pIpC at 4 weeks of age, these mice experienced fully penetrant, rapid morbidity and mortality In contrast, Mx1-Cre;Brca1F22-24/F22-24 mice did not (Figure 5a). To confirm there were no differences in recombination we generated mice with the null mutation in the germline. To do this Brca1F22-24/F22-24 mice were mated with CMV deletor mice (Dupe et al., 1997). The progeny with recombination in the germline (Brca1Δ/+) were then used to generate Mx1-Cre;Brca1F22-24/Δ biallelic mice to compare to Mx1-Cre;Brca1F22-24/5382insC biallelic mutant mice. Again, the Mx1-Cre;Brca1F22-24/Δ biallelic mice survived while the Mx1-Cre;Brca1F22-24/5382insC biallelic mutant mice did not (Figure 5a). These data indicate that the Brca1Δ/5382insC genotype is more deleterious to hematopoietic cells than the Brca1Δ/Δ genotype.
Figure 5. Substitution of mouse Brca1 with the human BRCA1 5382insC generates a compound heterozygote with a more severe hematopoietic phenotype than homozygous Brca1 null mice.
(A) Kaplan-Meier survival curve of control (black line; Brca1+/+; n=4), Mx1-Cre;Brca1F22-24/ F22-24 (n=7) plus Mx1-Cre;Brca1F22-24/Δ (n=5) (green dashed line; Brca1Δ/ Δ) and Mx1-Cre;Brca1F22-24/5382insC (red line; Brca1Δ/insC; n=7) mice. (B–G) Complete blood cell counts from wild-type mice (Brca1+/+; n=4), Mx1-Cre;Brca1F22-24/F22-24 (n=4 mice) plus Mx1-Cre;Brca1F22-24/Δ (n=5) mice (Brca1Δ/Δ) and Mx1-Cre;Brca1F22-24/5382insC (Brca1Δ/insC; n=7 mice) (B) White blood cells (WBC), (C) neutrophils, (D) lymphocytes, (E) monocytes, (F) platelets, and (G) hemoglobin are shown. All data represent means ± standard deviation. (H) HSCs, (I) MPPs, (J) CMPs, (K) GMPs, (L) MEPs (M) B lineage cells, (N) T lineage cells, (O) myeloid cells, and (P) erythroid cells in the bone marrow of control (+/+; n=3; white bar) and Mx1-Cre;Brca1F22-24/5382insC (Δ/+; n=3; light shaded bar). Statistical significance was assessed using a two-tailed Student’s t-test except in (A) where a log-rank test was used (*P<0.05, **P<0.01, ***P<0.001).
The rapid mortality in pIpC-treated Mx1-Cre; Brca1F22-24/5382insC mice was associated with severe pancytopenia. The organs of the pIpC induced Mx1-Cre;Brca1F22-24/5382insC mice were pale (data not shown). As expected, for both the Mx1-Cre;Brca1F22-24/F22-24 and Mx1-Cre;Brca1F22-24/5382insC mutant mice in Figure 5a, there were significantly decreased absolute numbers of white blood cells, including neutrophils and lymphocytes, as well as significantly reduced numbers of platelets (Figure 5b–f). However, Mx1-Cre; Brca1F22-24/5382insC mice had a significantly more severe pancytopenia than Mx1-Cre;Brca1F22-24/Δ mice with the lowest absolute numbers of white blood cells, including neutrophils, monocytes and lymphocytes, as well as reduced numbers of platelets and hemoglobin (Figure 5b–g).
The severe anemia in the Mx1-Cre; Brca1F22-24/5382insC mice was the largest and most significant difference from the Mx1-Cre;Brca1F22-24/Δ mice (Figure 5g). Since the half-life of mouse red blood cells in wildtype mice has been measured at more than 20 days (Van Putten, 1958), it is possible there was bleeding secondary to the severe thrombocytopenia. In fact, in two mice that were necropsied immediately after death, we observed large pools of blood in the abdominal and thoracic cavities. Thrombocytopenia can occur quickly with loss of progenitors as mouse platelet half-life has been estimated to be 3.4 days (Jayachandran et al., 2010).
To further characterize pIpC-induced Mx1-Cre;Brca1F22-24/5382insC mice, we examined hematopoiesis in their bone marrow. Six-week old Mx1-Cre;Brca1F22-24/5382insC mice that had been treated with pIpC at 4 weeks of age had a severe reduction in HSCs and early progenitors compared to pIpC-treated controls (Figure 5h–l; P<0.01). In fact, the bone marrow of pIpC-treated Mx1-Cre;Brca1F22-24/5382insC mice had no detectable HSCs (Figure 5h). Hematopoietic progenitors including MPPs, CMPs, GMPs and MEPs were also not detectable in the bone marrow of Mx1-Cre;Brca1F22-24/5382insC (Figure 5i–l). When compared with hematopoiesis in Mx1-Cre;Brca1F22-24/F22-24 mice (Vasanthakumar et al., 2016) (data not shown), a more severe hematopoietic defect in Mx1-Cre;Brca1F22-24/5382insC mice is present, suggesting that the BRCA15382insC protein is more deleterious to hematopoietic stem and progenitor cells than the null allele.
DISCUSSION
In this manuscript, we show a cohort of patients with BRCA1 mutations experienced increased hematopoietic toxicity and complications after cancer chemotherapy. We also observed that Brca1 is required for HSC function and normal hematopoiesis in mice. When Brca1 was conditionally deleted from embryonic hematopoietic cells, young adult mice developed pancytopenia (Figure 2) and a loss of nearly all HSCs (Figure 3). Moreover, heterozygosity for a loss-of-function allele of Brca1 in mouse hematopoietic cells led to a slight but significant decrease in white blood cells and lymphocytes (Figure 2) as well as deficits in HSC reconstituting potential upon serial transplantation (Figure S3).
These results are consistent with a reduced hematopoietic regenerative capacity in BRCA1 heterozygous humans after chemotherapy, suggesting that even a partial loss of BRCA1 function reduces the capacity for hematopoietic recovery due to direct DNA damage or replication stress after myeloablation. The concept that replication stress leads to more chemotherapeutic toxicity for BRCA1 mutation carriers is also consistent with prior work that has suggested there is enhanced replication stress (due to decreased stalled fork repair) in BRCA1 heterozygous epithelial cells. This abnormality in heterozygous cells was hypothesized to enhance the formation of tumors in epithelial cells (Pathania et al., 2014).
These data suggest a “cell death or transformation” tissue specificity hypothesis and could explain why patients with germline BRCA1 mutations have a predisposition to epithelial cancers, but do not have a predisposition to hematological malignancies. The ultimate loss of BRCA1 heterozygosity, which is promoted by diminished DNA repair in the heterozygous state and is thought to be required for transformation of epithelial cells to cancer (Pathania et al., 2014), is not tolerated by hematopoietic stem cells.
This is the first report of generation and characterization a humanized Brca1 allele. The human BRCA1 cDNA was knocked into the mouse Brca1 locus to study its function. Humanization of mouse genes has proven useful for in vivo functional evaluation of human p53 mutations (Song et al., 2007). Like p53, the introduction of human mutations into the mouse Brca1 allele is advantageous as there are significant differences in amino acid sequence between mouse Brca1 and human BRCA1. The mouse protein is only 60% identical to the human BRCA1 protein (Sharan et al., 1995). Our finding of embryonic lethality for the Brca15382insC/5382insC genotype but not in un-recombined Brca1BRCA1/BRCA1 mice (Figure 4), confirms that human BRCA1 can perform many of the necessary functions of mouse Brca1 after being knocked into the mouse Brca1 locus.
Conditional deficiency for Brca1 using Mx1-Cre in adult mice has previously been reported to increase differentiation in MLL-AF9 induced leukemia (Santos et al., 2014), diminish hematopoietic cell proliferation in vitro, and lead to mild leukopenia and anemia (Vasanthakumar et al., 2016). However, neither of these studies reported a deleterious effect of Brca1 deficiency on HSC frequency or function. Use of the Vav1-iCre allele to delete Brca1 in embryonic and adult HSCs and use of the new human Brca15382insC allele were not part of the prior studies. The use of different Cre alleles suggests that deletion of Brca1 in the embryonic HSCs (Vav1-iCre) may be more deleterious than deletion in adult HSCs (Mx1-Cre).
A trivial explanation for why an HSC defect was only observed in mice using the Vav1-iCre allele for conditional deletion of Brca1 is that there was more recombination in HSCs with the Vav1-iCre allele compared to those with the Mx1-Cre allele. Although this explanation is not possible to confirm or refute without analysis of the original mice, because Vasanthakumar et al did provide evidence for full recombination in hematopoietic cells from their Mx1-Cre transgenic mice, it is an unlikely explanation (Vasanthakumar et al., 2016).
Different BRCA1 mutations have been shown to have distinct effects on cancer phenotypes. Humans with mutations at the extreme C- and N-terminus of BRCA1 experience more breast cancer and less ovarian cancer as compared to humans with mutations in the middle of the BRCA1 gene (Rebbeck et al., 2015). Our surprising observation that the Brca15382insC mutation led to a more severe adult hematopoietic phenotype than the Brca1 null mutation (Figure 5) suggests that distinct germline BRCA1 mutations may result in different degrees of chemotherapeutic toxicity, as well. More patients with these mutations are needed to make associations and more investigation into the mechanism of this increased toxicity of the Brca15382insC allele in the mice will be important.
Because the Brca15382insC allele, in contrast to the null allele, expresses a mutant protein (Figure S4c vs. Figure 2a), it could indeed have hypo- or hyper-morphic effects on cells. Expression of the BRCA1 5382insC mutant mRNA from humans heterozygous for the BRCA1 5382insC mutation is equivalent to the expression of the wild type mRNA from the other allele in primary human fibroblasts, induced pluripotent stem cells and teratomas. However, the expression of BRCA1 5382insC in these cells does not promote excessive cell death, differentiation, survival or growth (Soyombo et al., 2013). Further, heterologous expression of this mutant BRCA1 in cell lines does not lead to altered growth or survival (data not shown). These observations may be due to the expression from the normal BRCA1 allele. The mutant protein may only be detrimental in a completely deficient BRCA1 background. Further studies to understand why the BRCA1 5382insC allele leads to an in vivo phenotype distinct from the Brca1 null allele are necessary.
Stem cells are susceptible to DNA damage due to their longevity and self-renewal potential. HSCs from mice with mutations in DNA damage repair proteins that also lead to cancer susceptibility syndromes, such as Brca2 (Navarro et al., 2006) and Msh2 (Reese et al., 2003), have defects in their ability to reconstitute bone marrow in irradiated mice, and mice with mutant Rad50 exhibit hematopoietic failure (Bender et al., 2002). However, the hematopoietic phenotype we observed after Brca1 deletion is much more severe than the phenotypes reported in these studies.
Several mouse models have been generated to study BRCA1-mutant breast cancer (Dine and Deng, 2013; Drost et al., 2011; Drost and Jonkers, 2009; Evers and Jonkers, 2006; Shakya et al., 2011). These models confirm that Brca1 maintains genome stability in vivo, and that without normal Brca1 in breast epithelial tissues, breast tumorigenesis occurs. However, breast cancer develops in Brca1 knockout mice only after a long latency (even if p53 is also deficient). This is consistent with the fact that human BRCA1 mutation carriers are only diagnosed with cancer as adults, if ever.
Here we describe mice with different Brca1 alleles mutated specifically in the hematopoietic system that have distinct phenotypes, which, in contrast to the breast cancer phenotype, occur rapidly and are fully penetrant (for allele/phenotype summary see Table S2). In addition to the new information about the role of Brca1 in hematopoiesis, these allele combinations provide the field with powerful tools for rapid investigation of the pathogenicity of BRCA1 variants of unknown significance.
Finally, given the potent requirement for Brca1 in HSCs, an inherited BRCA1 mutation may be a marker to add to the list of patient risk factors, such as age and co-morbidities (Caggiano et al., 2005), that support the prophylactic use of growth factors and antibiotics, and close monitoring for chemotherapy-related hematopoietic complications. Preventative use of myeloid growth factor support may, however, be counterproductive if unrepaired replication-induced mutations are increased in BRCA1 heterozygotes. Prophylactic growth factor support and antibiotics should, therefore, be evaluated prospectively in BRCA1 mutation carriers who are receiving chemotherapy.
METHODS
Patients
A list of patients with BRCA1 or BRCA2 mutations treated between January 1, 2011 and October 31, 2014, were identified from the University of Texas Southwestern Medical Center’s Cancer Genetics database. Patients were categorized based on cancer type and chemotherapy treatments. A retrospective chart review was then conducted on these patients to collect information on patient characteristics (Table 1), as well as comorbidities and past medical/surgical histories, type of cancer, age of diagnosis, treatment, treatment complications (if applicable), and complete blood cell counts. For any patient who had at least one complete blood cell count recorded in their medical record, baseline complete blood cell count values were selected for each patient based on the following criteria: pre-treatment (but as close to beginning of therapy as possible within 5 years of cancer diagnosis), no active infection, no procedural context (e.g. post-biopsy or post-operative), and did not appear to be an outlier if other complete blood cell counts were available for comparison.
For the patients who underwent chemotherapy for their cancer, the most severe adverse hematopoietic event during chemotherapy and its associated toxicity score was recorded. Grades of blood cell count toxicity were assigned based on the National Cancer Institute Common Terminology Criteria for Adverse Events v3.0 guidelines (Trotti et al., 2003) (Table 2). Neutropenic fever was defined as an absolute neutrophil count < 500 cells/mm3 and fever. Fever was defined as a single oral temperature of >38.3°C (101°F) or a temperature of >38.0°C (100.4°F) sustained for more than one hour.
Following collection of these data, statistical analyses were conducted on de-identified data. Range, mean, and standard deviations of complete blood cell components (neutrophils, platelets, hemoglobin, etc.) in BRCA1/2 mutation carriers were compared to the normal ranges. Analysis of variations in complete blood cell components in response to different chemotherapy regimens were also evaluated for differences between the BRCA1 and BRCA2 mutant patients. This study (STU 072014-043; Analysis of Complete Blood Counts (CBCs) in BRCA Mutation Carriers) was approved by the University of Texas Southwestern Medical Center Institutional Review Board.
Mice
The Brca1F22-24 (McCarthy et al., 2007), Mx1-Cre (Rajewsky et al., 1996) and Vav1-iCre (Georgiades et al., 2002) alleles, all on a C57/BL6 pure background, have been previously described and were obtained from Jackson Labs. C57BL/Ka-Thy-1.2 (CD45.1) mice were used as transplant recipients. Both male and female mice between 6–14 weeks old were used in all studies. Mx1-Cre was induced as describe previously (Oravecz-Wilson et al., 2009).
The BRCA1 knock-in mice were generated as described in detail in the Extended Experimental Procedures and Figure S4. Briefly, the targeting vector was constructed to generate a knock-in allele that conditionally generated the BRCA1 5382insC mutation as well as constitutively humanized BRCA1 mice (Figure 4A and Figure S4).
All Brca1 mutant mice were genotyped from tail snips using real-time PCR assays designed by and available from Transnetyx. The assays were designed to detect the wild-type and mutant alleles in the presence or absence of recombination. Mice were housed in the Unit for Laboratory Animal Medicine at the University of Texas Southwestern Medical Center under specific pathogen-free conditions and were monitored regularly for evidence of disease and abnormal peripheral blood cell counts. The animal use protocol was approved by the University of Texas Southwestern Institutional Animal Care and Use Committee (APN 2011-0143).
Bone Marrow Transplantation
Adult recipient mice (CD45.1) were administered a minimum lethal dose of radiation using an XRAD 320 X-ray irradiator (Precision X-Ray Inc.) to deliver two doses of approximately 540 rad (1,080 rad in total) at least 3 hours apart. Cells were injected into the retro-orbital venous sinus of anesthetized recipients. For competitive bone marrow transplants 5 x 105 donor and 5 x 105 recipient cells were transplanted. Blood was obtained from the submandibular plexus of recipient mice at the indicated time points after transplantation. Red blood cells were lysed with ammonium chloride potassium buffer. The remaining cells were stained with antibodies (Tonbo Biosciences, Inc.) against CD45.2, CD45.1, CD45R (B220), CD11b, CD3, and Gr-1 to assess donor cell engraftment. Mice that died were omitted from the analyses.
Hematopoietic Analysis
Bone marrow cells were isolated by flushing the long bones (femurs and tibias) in Ca2+ and Mg2+ free Hank’s buffered salt solution (Corning) supplemented with 3% heat-inactivated bovine serum (Gibco). Spleens were prepared by crushing tissues between frosted slides. Cell number and viability were assessed by a Vi-CELL cell viability analyzer (Beckman Coulter) or by counting on a hemocytometer.
Flow cytometric analysis of specific hematopoietic progenitors was performed as previously described (Foley et al., 2013; Signer et al., 2014). Complete blood cell count analysis was performed on peripheral blood using the Hemavet 950 with MULTI-TROL Mouse as an equilibration control (Drew Scientific).
Western blot analysis
Mouse tissues were lysed in RIPA buffer (Cell Signaling Technology). 100 μg of total protein was electrophoresed on 6% SDS-PAGE gels, and transferred to a PVDF membrane. Proteins were detected with anti-Brca1 antiserum (1:100; GH118, kind gift of Dr. Jos Jonkers), mouse monoclonal anti-Hip1 (1:1000; 1B11, Ross laboratory (Ames et al., 2013)) and rabbit polyclonal anti-γH2AX (1:1000, Cell Signaling). Blots were developed with Supersignal West Pico chemiluminescence substrate (Pierce).
Statistical Analysis
Statistical significance was assessed using a two-tailed Student’s t-test with p values (*P<0.05, **P<0.01, ***P<0.001). A Fisher’s Exact test (*P<0.05) was used to asses statistical significance in Figure 1 (G, L–O). For Kaplan Meier curves depicting survival analyses, a log-rank test was used. All statistical analyses were performed using GraphPad Prism version 7.00 for Windows, GraphPad Software, La Jolla California USA, www.graphpad.com. All RNA sequencing expression data and accession codes can be found at the Gene Expression Omnibus: GSE91390.
Supplementary Material
HIGHLIGHTS.
Mouse hematopoietic stem cells (mHSCs) require normal Brca1 to survive.
Knocked-in wild-type human BRCA1 cDNA fully substitutes for mouse Brca1.
A BRCA1/5382insC mutation is more deleterious to mHSCs than a Brca1 null allele.
Acknowledgments
We are grateful to Dr. Martin Dietrich, Lesli Kiedrowski, Abigail Soyombo and other members of the Ross lab for their technical assistance and intellectual contributions. This work was supported by National Cancer Institute grants to TSR (R01 CA82363-03 and R01 CA098730-01), the Lucy and Henry Billingsley Fund and a Burroughs Wellcome Fund Clinical Scientist Award in Translational Research (TSR). VEM was supported by the NIH (5T32 CA124334-09) and the Cancer Prevention and Research institute of Texas (CPRIT; RP140110). SJM is a Howard Hughes Medical Institute (HHMI) Investigator, the Mary McDermott Cook Chair in Pediatric Genetics, the Kathryne and Gene Bishop Distinguished Chair in Pediatric Genetics, the director of the Hamon Laboratory for Stem Cells and Cancer, and a CPRIT Scholar. TSR holds the Jeanne Ann Plitt Professorship in Breast Cancer Research and the H. Ben and Isabelle T. Decherd Chair in Internal Medicine at UT Southwestern Medical Center.
Footnotes
AUTHOR CONTRIBUTIONS
VEM and RAS designed experiments, collected data, interpreted results and edited the manuscript. RW, and TL collected, interpreted results and edited the manuscript. SJM interpreted data and edited the manuscript. TSR designed experiments, interpreted data and wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akashi K, Traver D, Miyamoto T, Weissman IL. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 2000;404:193–197. doi: 10.1038/35004599. [DOI] [PubMed] [Google Scholar]
- Ames HM, Wang AA, Coughran A, Evaul K, Huang S, Graves CW, Soyombo AA, Ross TS. Huntingtin-interacting protein 1 phosphorylation by receptor tyrosine kinases. Mol Cell Biol. 2013;33:3580–3593. doi: 10.1128/MCB.00473-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baer R, Ludwig T. The BRCA1/BARD1 heterodimer, a tumor suppressor complex with ubiquitin E3 ligase activity. Curr Opin Genet Dev. 2002;12:86–91. doi: 10.1016/s0959-437x(01)00269-6. [DOI] [PubMed] [Google Scholar]
- Beerman I, Seita J, Inlay MA, Weissman IL, Rossi DJ. Quiescent hematopoietic stem cells accumulate DNA damage during aging that is repaired upon entry into cell cycle. Cell Stem Cell. 2014;15:37–50. doi: 10.1016/j.stem.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender CF, Sikes ML, Sullivan R, Huye LE, Le Beau MM, Roth DB, Mirzoeva OK, Oltz EM, Petrini JH. Cancer predisposition and hematopoietic failure in Rad50(S/S) mice. Genes Dev. 2002;16:2237–2251. doi: 10.1101/gad.1007902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caggiano V, Weiss RV, Rickert TS, Linde-Zwirble WT. Incidence, cost, and mortality of neutropenia hospitalization associated with chemotherapy. Cancer. 2005;103:1916–1924. doi: 10.1002/cncr.20983. [DOI] [PubMed] [Google Scholar]
- Carreau M, Gan OI, Liu L, Doedens M, Dick JE, Buchwald M. Hematopoietic compartment of Fanconi anemia group C null mice contains fewer lineage-negative CD34+ primitive hematopoietic cells and shows reduced reconstruction ability. Exp Hematol. 1999;27:1667–1674. doi: 10.1016/s0301-472x(99)00102-2. [DOI] [PubMed] [Google Scholar]
- Dine J, Deng CX. Mouse models of BRCA1 and their application to breast cancer research. Cancer Metastasis Rev. 2013;32:25–37. doi: 10.1007/s10555-012-9403-7. [DOI] [PubMed] [Google Scholar]
- Domchek SM, Tang J, Stopfer J, Lilli DR, Hamel N, Tischkowitz M, Monteiro AN, Messick TE, Powers J, Yonker A, et al. Biallelic deleterious BRCA1 mutations in a woman with early-onset ovarian cancer. Cancer Discov. 2013;3:399–405. doi: 10.1158/2159-8290.CD-12-0421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drost R, Bouwman P, Rottenberg S, Boon U, Schut E, Klarenbeek S, Klijn C, van der Heijden I, van der Gulden H, Wientjens E, et al. BRCA1 RING function is essential for tumor suppression but dispensable for therapy resistance. Cancer Cell. 2011;20:797–809. doi: 10.1016/j.ccr.2011.11.014. [DOI] [PubMed] [Google Scholar]
- Drost RM, Jonkers J. Preclinical mouse models for BRCA1-associated breast cancer. Br J Cancer. 2009;101:1651–1657. doi: 10.1038/sj.bjc.6605350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupe V, Davenne M, Brocard J, Dolle P, Mark M, Dierich A, Chambon P, Rijli FM. In vivo functional analysis of the Hoxa-1 3′ retinoic acid response element (3′RARE) Development. 1997;124:399–410. doi: 10.1242/dev.124.2.399. [DOI] [PubMed] [Google Scholar]
- Evers B, Jonkers J. Mouse models of BRCA1 and BRCA2 deficiency: past lessons, current understanding and future prospects. Oncogene. 2006;25:5885–5897. doi: 10.1038/sj.onc.1209871. [DOI] [PubMed] [Google Scholar]
- Foley SB, Hildenbrand ZL, Soyombo AA, Magee JA, Wu Y, Oravecz-Wilson KI, Ross TS. Expression of BCR/ABL p210 from a knockin allele enhances bone marrow engraftment without inducing neoplasia. Cell Rep. 2013;5:51–60. doi: 10.1016/j.celrep.2013.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiades P, Ogilvy S, Duval H, Licence DR, Charnock-Jones DS, Smith SK, Print CG. VavCre transgenic mice: a tool for mutagenesis in hematopoietic and endothelial lineages. Genesis. 2002;34:251–256. doi: 10.1002/gene.10161. [DOI] [PubMed] [Google Scholar]
- Haneline LS, Gobbett TA, Ramani R, Carreau M, Buchwald M, Yoder MC, Clapp DW. Loss of FancC function results in decreased hematopoietic stem cell repopulating ability. Blood. 1999;94:1–8. [PubMed] [Google Scholar]
- Howlett NG, Taniguchi T, Olson S, Cox B, Waisfisz Q, De Die-Smulders C, Persky N, Grompe M, Joenje H, Pals G, et al. Biallelic inactivation of BRCA2 in Fanconi anemia. Science. 2002;297:606–609. doi: 10.1126/science.1073834. [DOI] [PubMed] [Google Scholar]
- Ito K, Hirao A, Arai F, Matsuoka S, Takubo K, Hamaguchi I, Nomiyama K, Hosokawa K, Sakurada K, Nakagata N, et al. Regulation of oxidative stress by ATM is required for self-renewal of haematopoietic stem cells. Nature. 2004;431:997–1002. doi: 10.1038/nature02989. [DOI] [PubMed] [Google Scholar]
- Jayachandran M, Miller VM, Brunn GJ, Owen WG. Platelet response as a sentinel marker of toll-like receptor 4 activation in mice. Thromb Res. 2010;126:414–417. doi: 10.1016/j.thromres.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiel MJ, Yilmaz OH, Iwashita T, Terhorst C, Morrison SJ. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell. 2005;121:1109–1121. doi: 10.1016/j.cell.2005.05.026. [DOI] [PubMed] [Google Scholar]
- Kottemann MC, Smogorzewska A. Fanconi anaemia and the repair of Watson and Crick DNA crosslinks. Nature. 2013;493:356–363. doi: 10.1038/nature11863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouroukis CT, Chia S, Verma S, Robson D, Desbiens C, Cripps C, Mikhael J. Canadian supportive care recommendations for the management of neutropenia in patients with cancer. Curr Oncol. 2008;15:9–23. doi: 10.3747/co.2008.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy A, Savage K, Gabriel A, Naceur C, Reis-Filho JS, Ashworth A. A mouse model of basal-like breast carcinoma with metaplastic elements. J Pathol. 2007;211:389–398. doi: 10.1002/path.2124. [DOI] [PubMed] [Google Scholar]
- Mohrin M, Bourke E, Alexander D, Warr MR, Barry-Holson K, Le Beau MM, Morrison CG, Passegue E. Hematopoietic stem cell quiescence promotes error-prone DNA repair and mutagenesis. Cell Stem Cell. 2010;7:174–185. doi: 10.1016/j.stem.2010.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro S, Meza NW, Quintana-Bustamante O, Casado JA, Jacome A, McAllister K, Puerto S, Surralles J, Segovia JC, Bueren JA. Hematopoietic dysfunction in a mouse model for Fanconi anemia group D1. Mol Ther. 2006;14:525–535. doi: 10.1016/j.ymthe.2006.05.018. [DOI] [PubMed] [Google Scholar]
- Nijnik A, Woodbine L, Marchetti C, Dawson S, Lambe T, Liu C, Rodrigues NP, Crockford TL, Cabuy E, Vindigni A, et al. DNA repair is limiting for haematopoietic stem cells during ageing. Nature. 2007;447:686–690. doi: 10.1038/nature05875. [DOI] [PubMed] [Google Scholar]
- Oguro H, Ding L, Morrison SJ. SLAM family markers resolve functionally distinct subpopulations of hematopoietic stem cells and multipotent progenitors. Cell Stem Cell. 2013;13:102–116. doi: 10.1016/j.stem.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oravecz-Wilson KI, Philips ST, Yilmaz OH, Ames HM, Li L, Crawford BD, Gauvin AM, Lucas PC, Sitwala K, Downing JR, et al. Persistence of leukemia-initiating cells in a conditional knockin model of an imatinib-responsive myeloproliferative disorder. Cancer Cell. 2009;16:137–148. doi: 10.1016/j.ccr.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathania S, Bade S, Le Guillou M, Burke K, Reed R, Bowman-Colin C, Su Y, Ting DT, Polyak K, Richardson AL, et al. BRCA1 haploinsufficiency for replication stress suppression in primary cells. Nat Commun. 2014;5:5496. doi: 10.1038/ncomms6496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrin-Vidoz L, Sinilnikova OM, Stoppa-Lyonnet D, Lenoir GM, Mazoyer S. The nonsense-mediated mRNA decay pathway triggers degradation of most BRCA1 mRNAs bearing premature termination codons. Hum Mol Genet. 2002;11:2805–2814. doi: 10.1093/hmg/11.23.2805. [DOI] [PubMed] [Google Scholar]
- Prakash R, Zhang Y, Feng W, Jasin M. Homologous recombination and human health: the roles of BRCA1, BRCA2, and associated proteins. Cold Spring Harb Perspect Biol. 2015;7:a016600. doi: 10.1101/cshperspect.a016600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajewsky K, Gu H, Kuhn R, Betz UA, Muller W, Roes J, Schwenk F. Conditional gene targeting. J Clin Invest. 1996;98:600–603. doi: 10.1172/JCI118828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebbeck TR, Mitra N, Wan F, Sinilnikova OM, Healey S, McGuffog L, Mazoyer S, Chenevix-Trench G, Easton DF, Antoniou AC, et al. Association of type and location of BRCA1 and BRCA2 mutations with risk of breast and ovarian cancer. JAMA. 2015;313:1347–1361. doi: 10.1001/jama.2014.5985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese JS, Liu L, Gerson SL. Repopulating defect of mismatch repair-deficient hematopoietic stem cells. Blood. 2003;102:1626–1633. doi: 10.1182/blood-2002-10-3035. [DOI] [PubMed] [Google Scholar]
- Reid S, Schindler D, Hanenberg H, Barker K, Hanks S, Kalb R, Neveling K, Kelly P, Seal S, Freund M, et al. Biallelic mutations in PALB2 cause Fanconi anemia subtype FA-N and predispose to childhood cancer. Nat Genet. 2007;39:162–164. doi: 10.1038/ng1947. [DOI] [PubMed] [Google Scholar]
- Rossi DJ, Bryder D, Seita J, Nussenzweig A, Hoeijmakers J, Weissman IL. Deficiencies in DNA damage repair limit the function of haematopoietic stem cells with age. Nature. 2007;447:725–729. doi: 10.1038/nature05862. [DOI] [PubMed] [Google Scholar]
- Santos MA, Faryabi RB, Ergen AV, Day AM, Malhowski A, Canela A, Onozawa M, Lee JE, Callen E, Gutierrez-Martinez P, et al. DNA-damage-induced differentiation of leukaemic cells as an anti-cancer barrier. Nature. 2014;514:107–111. doi: 10.1038/nature13483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer SL, Tian L, Kahkonen M, Schwartzentruber J, Kircher M, Majewski J, Dyment DA, Innes AM, Boycott KM, Moreau LA, et al. Biallelic mutations in BRCA1 cause a new Fanconi anemia subtype. Cancer Discov. 2015;5:135–142. doi: 10.1158/2159-8290.CD-14-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scully R, Ganesan S, Vlasakova K, Chen J, Socolovsky M, Livingston DM. Genetic analysis of BRCA1 function in a defined tumor cell line. Mol Cell. 1999;4:1093–1099. doi: 10.1016/s1097-2765(00)80238-5. [DOI] [PubMed] [Google Scholar]
- Seal S, Thompson D, Renwick A, Elliott A, Kelly P, Barfoot R, Chagtai T, Jayatilake H, Ahmed M, Spanova K, et al. Truncating mutations in the Fanconi anemia J gene BRIP1 are low-penetrance breast cancer susceptibility alleles. Nat Genet. 2006;38:1239–1241. doi: 10.1038/ng1902. [DOI] [PubMed] [Google Scholar]
- Shakya R, Reid LJ, Reczek CR, Cole F, Egli D, Lin CS, deRooij DG, Hirsch S, Ravi K, Hicks JB, et al. BRCA1 tumor suppression depends on BRCT phosphoprotein binding, but not its E3 ligase activity. Science. 2011;334:525–528. doi: 10.1126/science.1209909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanley S, McReynolds K, Ardern-Jones A, Ahern R, Fernando I, Yarnold J, Evans G, Eccles D, Hodgson S, Ashley S, et al. Acute chemotherapy-related toxicity is not increased in BRCA1 and BRCA2 mutation carriers treated for breast cancer in the United Kingdom. Clin Cancer Res. 2006;12:7033–7038. doi: 10.1158/1078-0432.CCR-06-1246. [DOI] [PubMed] [Google Scholar]
- Sharan SK, Wims M, Bradley A. Murine Brca1: sequence and significance for human missense mutations. Hum Mol Genet. 1995;4:2275–2278. doi: 10.1093/hmg/4.12.2275. [DOI] [PubMed] [Google Scholar]
- Signer RA, Magee JA, Salic A, Morrison SJ. Haematopoietic stem cells require a highly regulated protein synthesis rate. Nature. 2014;509:49–54. doi: 10.1038/nature13035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H, Hollstein M, Xu Y. p53 gain-of-function cancer mutants induce genetic instability by inactivating ATM. Nat Cell Biol. 2007;9:573–580. doi: 10.1038/ncb1571. [DOI] [PubMed] [Google Scholar]
- Soyombo AA, Wu Y, Kolski L, Rios JJ, Rakheja D, Chen A, Kehler J, Hampel H, Coughran A, Ross TS. Analysis of induced pluripotent stem cells from a BRCA1 mutant family. Stem Cell Reports. 2013;1:336–349. doi: 10.1016/j.stemcr.2013.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotti A, Colevas AD, Setser A, Rusch V, Jaques D, Budach V, Langer C, Murphy B, Cumberlin R, Coleman CN, et al. CTCAE v3.0: development of a comprehensive grading system for the adverse effects of cancer treatment. Semin Radiat Oncol. 2003;13:176–181. doi: 10.1016/S1053-4296(03)00031-6. [DOI] [PubMed] [Google Scholar]
- Van Putten LM. The life span of red cells in the rat and the mouse as determined by labeling with DFP32 in vivo. Blood. 1958;13:789–794. [PubMed] [Google Scholar]
- Vasanthakumar A, Arnovitz S, Marquez R, Lepore J, Rafidi G, Asom A, Weatherly M, Davis EM, Neistadt B, Duszynski R, et al. Brca1 deficiency causes bone marrow failure and spontaneous hematologic malignancies in mice. Blood. 2016;127:310–313. doi: 10.1182/blood-2015-03-635599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia B, Dorsman JC, Ameziane N, de Vries Y, Rooimans MA, Sheng Q, Pals G, Errami A, Gluckman E, Llera J, et al. Fanconi anemia is associated with a defect in the BRCA2 partner PALB2. Nat Genet. 2007;39:159–161. doi: 10.1038/ng1942. [DOI] [PubMed] [Google Scholar]
- Xia B, Sheng Q, Nakanishi K, Ohashi A, Wu J, Christ N, Liu X, Jasin M, Couch FJ, Livingston DM. Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol Cell. 2006;22:719–729. doi: 10.1016/j.molcel.2006.05.022. [DOI] [PubMed] [Google Scholar]
- Zhang F, Ma J, Wu J, Ye L, Cai H, Xia B, Yu X. PALB2 links BRCA1 and BRCA2 in the DNA-damage response. Curr Biol. 2009;19:524–529. doi: 10.1016/j.cub.2009.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.