Abstract
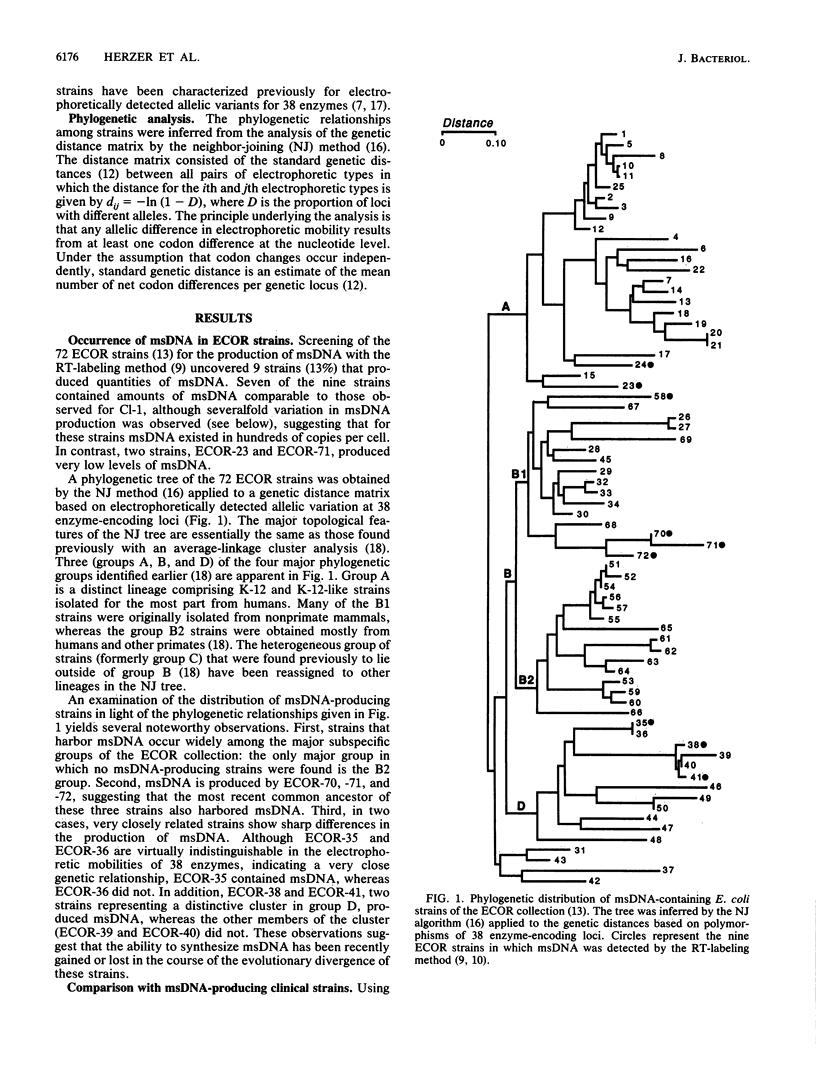
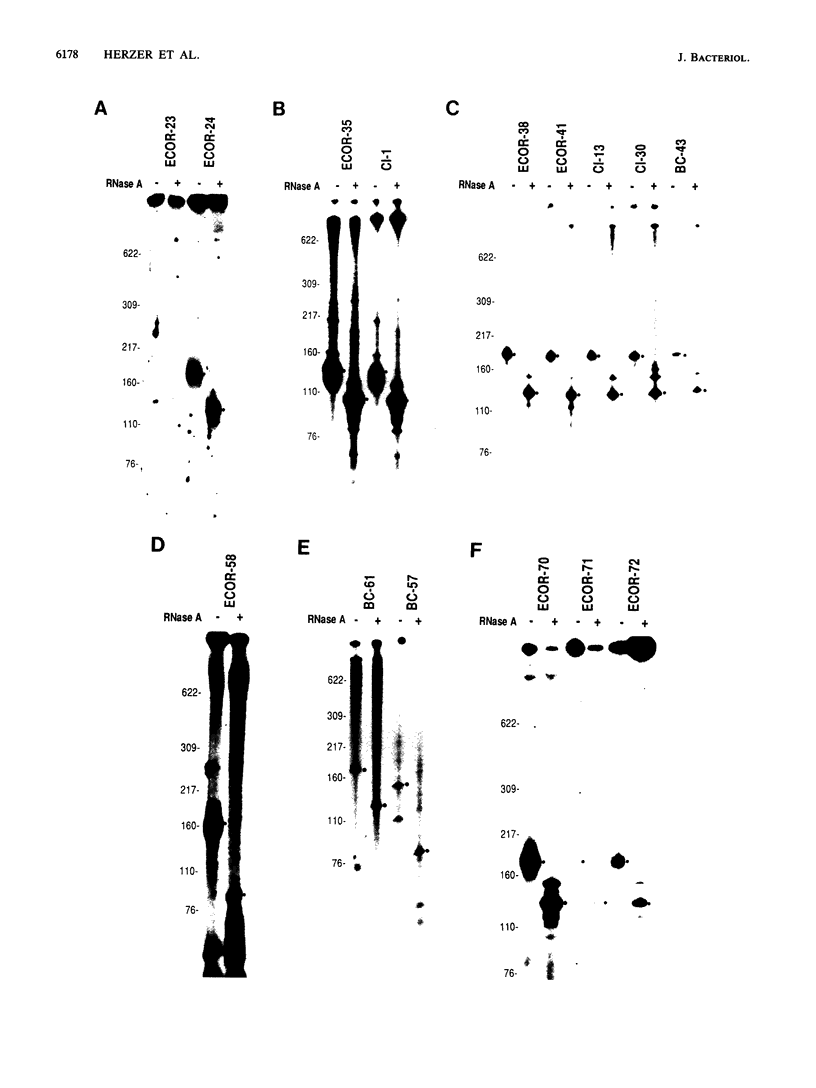
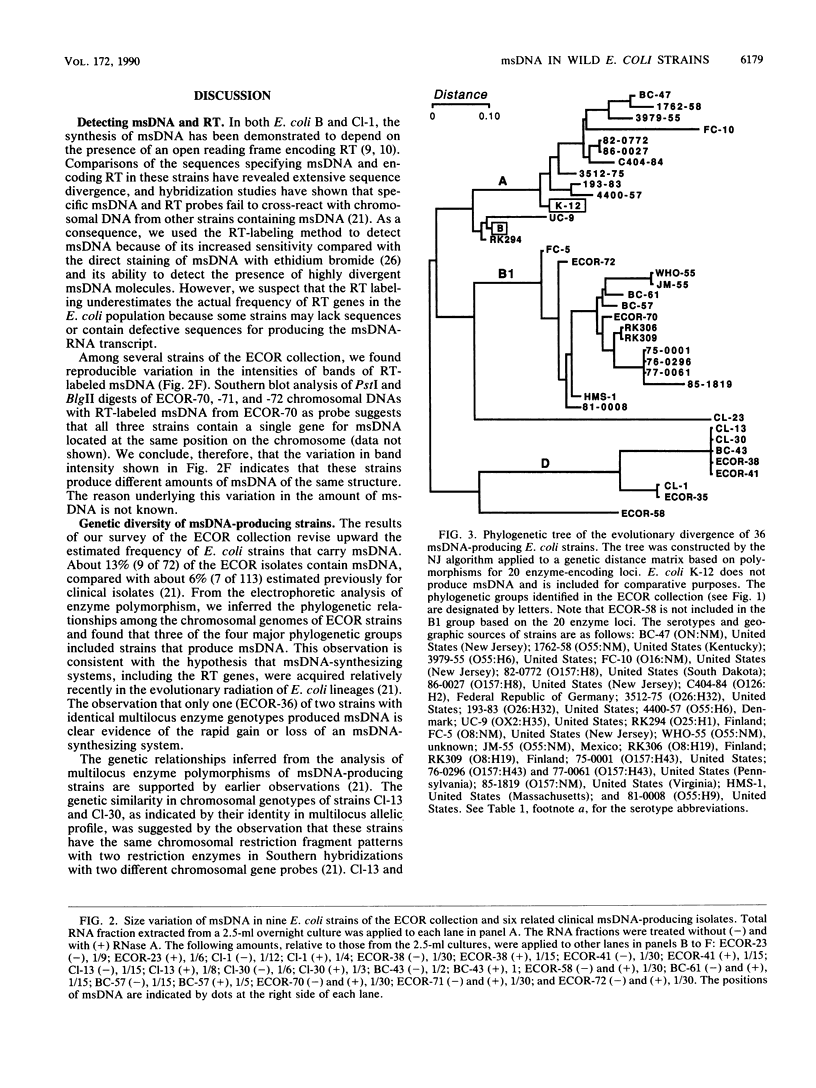
Multicopy single-stranded DNA (msDNA), a branched DNA-RNA molecule, has been shown in Escherichia coli B and clinical strain Cl-1 to be synthesized by reverse transcriptase. We report that 13% of the strains of the ECOR collection, a sample of 72 E. coli isolates representing the breadth of genetic variation of the species, produce msDNA. Three of the four major subspecific groups include msDNA-producing strains. Screening of 25 isolates that are genetically related to msDNA-producing clinical strains uncovered 22 additional msDNA-producing strains. A phylogenetic tree based on allelic variation detected electrophoretically at 20 enzyme-encoding loci revealed two major clusters and several deep branches composed of strains that synthesize msDNA. Although E. coli K-12 does not harbor msDNA, other closely related strains of the K-12 family do. The results support the hypothesis that msDNA-synthesizing systems, including reverse transcriptase genes, were acquired recently and independently in different lineages of E. coli.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Cairns J., Overbaugh J., Miller S. The origin of mutants. Nature. 1988 Sep 8;335(6186):142–145. doi: 10.1038/335142a0. [DOI] [PubMed] [Google Scholar]
- Chomczynski P., Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987 Apr;162(1):156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Dhundale A., Inouye M., Inouye S. A new species of multicopy single-stranded DNA from Myxococcus xanthus with conserved structural features. J Biol Chem. 1988 Jun 25;263(18):9055–9058. [PubMed] [Google Scholar]
- Dhundale A., Lampson B., Furuichi T., Inouye M., Inouye S. Structure of msDNA from Myxococcus xanthus: evidence for a long, self-annealing RNA precursor for the covalently linked, branched RNA. Cell. 1987 Dec 24;51(6):1105–1112. doi: 10.1016/0092-8674(87)90596-4. [DOI] [PubMed] [Google Scholar]
- Furuichi T., Dhundale A., Inouye M., Inouye S. Branched RNA covalently linked to the 5' end of a single-stranded DNA in Stigmatella aurantiaca: structure of msDNA. Cell. 1987 Jan 16;48(1):47–53. doi: 10.1016/0092-8674(87)90354-0. [DOI] [PubMed] [Google Scholar]
- Furuichi T., Inouye S., Inouye M. Biosynthesis and structure of stable branched RNA covalently linked to the 5' end of multicopy single-stranded DNA of Stigmatella aurantiaca. Cell. 1987 Jan 16;48(1):55–62. doi: 10.1016/0092-8674(87)90355-2. [DOI] [PubMed] [Google Scholar]
- Goullet P., Picard B. Comparative electrophoretic polymorphism of esterases and other enzymes in Escherichia coli. J Gen Microbiol. 1989 Jan;135(1):135–143. doi: 10.1099/00221287-135-1-135. [DOI] [PubMed] [Google Scholar]
- Inouye S., Hsu M. Y., Eagle S., Inouye M. Reverse transcriptase associated with the biosynthesis of the branched RNA-linked msDNA in Myxococcus xanthus. Cell. 1989 Feb 24;56(4):709–717. doi: 10.1016/0092-8674(89)90593-x. [DOI] [PubMed] [Google Scholar]
- Lampson B. C., Sun J., Hsu M. Y., Vallejo-Ramirez J., Inouye S., Inouye M. Reverse transcriptase in a clinical strain of Escherichia coli: production of branched RNA-linked msDNA. Science. 1989 Feb 24;243(4894 Pt 1):1033–1038. doi: 10.1126/science.2466332. [DOI] [PubMed] [Google Scholar]
- Lim D., Maas W. K. Reverse transcriptase-dependent synthesis of a covalently linked, branched DNA-RNA compound in E. coli B. Cell. 1989 Mar 10;56(5):891–904. doi: 10.1016/0092-8674(89)90693-4. [DOI] [PubMed] [Google Scholar]
- Mittler J. E., Lenski R. E. New data on excisions of Mu from E. coli MCS2 cast doubt on directed mutation hypothesis. Nature. 1990 Mar 8;344(6262):173–175. doi: 10.1038/344173a0. [DOI] [PubMed] [Google Scholar]
- Ochman H., Selander R. K. Standard reference strains of Escherichia coli from natural populations. J Bacteriol. 1984 Feb;157(2):690–693. doi: 10.1128/jb.157.2.690-693.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman H., Whittam T. S., Caugant D. A., Selander R. K. Enzyme polymorphism and genetic population structure in Escherichia coli and Shigella. J Gen Microbiol. 1983 Sep;129(9):2715–2726. doi: 10.1099/00221287-129-9-2715. [DOI] [PubMed] [Google Scholar]
- Orskov F., Whittam T. S., Cravioto A., Orskov I. Clonal relationships among classic enteropathogenic Escherichia coli (EPEC) belong to different O groups. J Infect Dis. 1990 Jul;162(1):76–81. doi: 10.1093/infdis/162.1.76. [DOI] [PubMed] [Google Scholar]
- Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987 Jul;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Selander R. K., Caugant D. A., Ochman H., Musser J. M., Gilmour M. N., Whittam T. S. Methods of multilocus enzyme electrophoresis for bacterial population genetics and systematics. Appl Environ Microbiol. 1986 May;51(5):873–884. doi: 10.1128/aem.51.5.873-884.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selander R. K., Korhonen T. K., Väisänen-Rhen V., Williams P. H., Pattison P. E., Caugant D. A. Genetic relationships and clonal structure of strains of Escherichia coli causing neonatal septicemia and meningitis. Infect Immun. 1986 Apr;52(1):213–222. doi: 10.1128/iai.52.1.213-222.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. M., Li W. H. The codon Adaptation Index--a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987 Feb 11;15(3):1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Herzer P. J., Weinstein M. P., Lampson B. C., Inouye M., Inouye S. Extensive diversity of branched-RNA-linked multicopy single-stranded DNAs in clinical strains of Escherichia coli. Proc Natl Acad Sci U S A. 1989 Sep;86(18):7208–7212. doi: 10.1073/pnas.86.18.7208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varmus H. E. Reverse transcription in bacteria. Cell. 1989 Mar 10;56(5):721–724. doi: 10.1016/0092-8674(89)90673-9. [DOI] [PubMed] [Google Scholar]
- Whittam T. S., Ochman H., Selander R. K. Multilocus genetic structure in natural populations of Escherichia coli. Proc Natl Acad Sci U S A. 1983 Mar;80(6):1751–1755. doi: 10.1073/pnas.80.6.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittam T. S., Wilson R. A. Genetic relationships among pathogenic Escherichia coli of serogroup O157. Infect Immun. 1988 Sep;56(9):2467–2473. doi: 10.1128/iai.56.9.2467-2473.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittam T. S., Wilson R. A. Genetic relationships among pathogenic strains of avian Escherichia coli. Infect Immun. 1988 Sep;56(9):2458–2466. doi: 10.1128/iai.56.9.2458-2466.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yee T., Furuichi T., Inouye S., Inouye M. Multicopy single-stranded DNA isolated from a gram-negative bacterium, Myxococcus xanthus. Cell. 1984 Aug;38(1):203–209. doi: 10.1016/0092-8674(84)90541-5. [DOI] [PubMed] [Google Scholar]