Supplemental Digital Content is available in the text
Keywords: genetic polymorphism, hepatitis B virus related hepatocellular carcinoma, meta-analysis, tumor necrosis factor-alpha
Abstract
Background:
To assess the association between tumor necrosis factor-alpha (TNF-α) G308A, G238A and C863T polymorphisms and hepatitis B virus related hepatocellular carcinoma (HBV-HCC) susceptibility.
Methods:
We interrogated the databases of Pubmed, Sciencedirect and Viley online library up to March 8, 2016. Odds ratios (ORs) and corresponding 95% confidence intervals (95%CIs) were calculated in a fixed-effects model or a random-effects model when appropriate.
Results:
In total, 12 case–control studies which containing 1580 HBV-HCC cases, 2033 HBV carrier controls, 395 HBV spontaneously recovered (SR) controls and 1116 healthy controls were included. Compared with GG genotype, the genotypes GA/AA of G308A were associated with a significantly increased HBV-HCC risk when the controls were all healthy individuals (AA vs. GG, OR 2.483, 95%CI 1.243 to 4.959; GA vs. GG, OR 1.383, 95%CI 1.028 to 1.860; GA/AA vs. GG, OR 1.381, 95%CI 1.048 to 1.820). Meanwhile, only the AA vs. GG model of G238A and HBV-HCC showed a statistic significance when the controls were healthy individuals (OR 4.776, 95%CI 1.280 to 17.819). CT genotype of TNF-α C863T could increase HBV-HCC risk whenever the controls were healthy individuals, HBV carriers or HBV recovers.
Conclusion:
This meta-analysis shows that AA genotype in TNF-α G308A and TNF-α G238A and CT genotype in TNF-α C863T may increase HBV-HCC risk. Therefore, HBV infection seemed to be a more important factor for tumorigenesis of HCC than genetic predisposition in G308A of TNF-α, and interaction between TNF-α C863T polymorphisms and HBV infection might be associated with increased HCC risk.
1. Introduction
Hepatocellular carcinoma (HCC), a type of hepatocyte epithelial tumor, is the sixth most common cancer and the third-leading cause of cancer related death worldwide.[1–3] And with lack of effective therapy in most HCC patients, a poor prognosis with a 5-year survival rate of 5% in developing countries is reported.[2,4] Etiologically, carcinogenesis of HCC is a complex, multistep and multifactor process, in which chronic infection with hepatitis B virus (HBV) or hepatitis C virus (HCV) is the most well-established environmental risk factor for HCC worldwide.[5] However, out of these 2 causative agents, HBV is regarded as the predominant causative factor of HCC. This is primarily due to its role in induction of chronic inflammation, which develops through the action of various inflammatory mediators.[6] Among inflammatory mediators, tumor necrosis factor alpha (TNF-α), a potent pleiotropic proinflammatory cytokine involved in the growth, differentiation and cellular function, plays an essential role and has been implicated in inflammation-associated tumors.[7–9]
Several polymorphisms (G308A, G238A, C863A, C857T, T1031C) in TNF-α promoter region are confirmed to influence the expression of TNF-α under genetic controls at the transcription and posttranscription levels.[10–13] And TNF-α polymorphisms have been reported to be associated with breast cancer,[14] non-Hodgkin lymphoma,[15] prostate cancer,[16] uterine endometrial cancer,[17] and gastric carcinoma.[18] In recent years, a number of studies investigated the possible association of TNF-α polymorphisms with HCC but controversies exist, especially between TNF-α polymorphisms and hepatitis B virus related hepatocellular carcinoma (HBV-HCC) risk. There are 2 meta-analyses conducted by Yang et al[19] and Wei et al,[20] and they all reported that GA/AA of G308A in TNF-α is correlated with increased risk of HCC; and Yang et al[19] also reported G238A of TNF-α polymorphism is not associated with HCC. While 2 meta-analyses conducted later by Tian et al[21] and Cheng et al[22] showed inconsistent results. As all these meta-analyses did not focus on the relationship between TNF-α polymorphism and HBV-HCC, and different controls in control group might also influence the results. We apply the methods of evidence-based medicine to evaluate and analyze the documented studies of G308A, G238A, C863A polymorphisms of TNF-α and HBV-HCC so as to provide a more systematic and comprehensive assessment of their associations.
2. Materials and methods
2.1. Inclusion criteria
All the studies included into this meta-analysis must meet the following criteria: case–control studies investigated the association of G308A, G238A, or C863T of TNF-α with HBV-HCC; original studies provided sufficient data that could be extracted and used to calculate odds ratios (ORs) and 95% confidence intervals (95% CI); the cases must be HBV-HCC and controls are HBV carriers, spontaneously recovers from HBV or healthy individuals, and Hardy–Weinberg equilibrium (HWE) in the control group should be confirmed. HBV carrier were the person with HBsAg positive for a period of 6 months or more but with normal levels of transaminases, were HBeAg negative or anti-HBe positive, had serum HBV DNA levels less than 105 copies/mL and had no clinical symptoms of liver disease along with no radiological evidence of cirrhosis or varices on endoscopy.
2.2. Exclusion criteria
The study where cases are not definitely restricted to HBV-HCC.
Repeat publications, sample size < 10 and studies were only reported superficially, such as in the form of an abstract.
The studies in which the full text or main data could not be obtained.
Gray literature that was unpublished.
2.3. Literature search strategy
We performed an electronic search of PubMed (1966 to September 2016), ScienceDirect Online (1995 to September 2016), Wiley online library (2010 to September 2016) for case–control studies evaluating the relationship of G308A, G238A, or C863A of TNF-α and HBV-HCC. The search keywords were used with different combinations with both medical subject headings terms and text words: “hepatitis B virus” or “HBV” or “liver cancer” or “hepatocellular carcinoma” or “HCC” or “hepatitis B virus related virus hepatocellular carcinoma” or “HBV-HCC” plus “tumor necrosis factor-alpha” or “TNF-α” or “polymorphism” or “G308A” or “G238A” or “C863T” or “variant.” Publication date was not restricted in our search. Reference lists of the included studies and supplemental materials were checked manually to further identify related studies. The approval by an institutional review board is not required because this study was based on published studies.
2.4. Selection of studies
Two reviewers (QX and PC) independently screened the title, abstract, and keywords of each article retrieved. Full-text papers were screened for further assessment if the information given suggested that the study fulfilled the inclusion criteria and did not meet the exclusion criteria. Discrepancies were settled by discussion and consensus with all the authors.
2.5. Data extraction
The following information was independently extracted from the identified studies by 3 reviewers (BQF, ZZL, and WW) using a standard form with first author's surname, year of publication, country, ethnicity, number of HBV-HCC cases and controls as well as individual genotype, method of genotype test, and HWE test. Ethnicities were stratified as Asians, Caucasians, and others. Controls were stratified as HBV carrier controls, HBV spontaneously recovered (SR) controls and healthy controls. The authors of original studies were consulted for missing information where necessary. Discrepancies were resolved by open discussion.
2.6. Hardy–Weinberg equilibrium test
HWE test was performed and the HWE significance of the control groups was calculated with StataSE12.0 (StataCorp LP, College Station, TX) when the original information was not provided.
2.7. Data synthesis and analysis
The significance for 5 genetic models (allele, dominant, recessive, codominant, and super-dominant genetic models) was evaluated for each study separately. All the associations were indicated as ORs with the corresponding 95% CI. Based on the individual ORs, a pooled OR was estimated. Subgroup analysis was also performed by stratifying control group and ethnicity. Fixed-effects model[23] or the random-effects model[24] of meta-analysis was chosen according to the results of heterogeneity tests among individual studies by StataSE12.0 (StataCorp LP). The significance of the pooled OR was determined using the Z test and P-value less than 0.05 was considered statistically significant.
Heterogeneity assumption was assessed by Cochran Q statistic[25] and I2 statistic. The heterogeneity was considered statistically significant if P < 0.10. The random-effects model (if P < 0.05 and I2 > 60%) or the fixed-effects model (if P ≥ 0.05 and I2 < 60%) was used to pool the ORs. Sensitivity analyses were made to determine whether the results were robust and evaluate the sources of heterogeneity. Egger test and Begg test were used to evaluate the publication bias, which was considered when P < 0.05.
3. Results
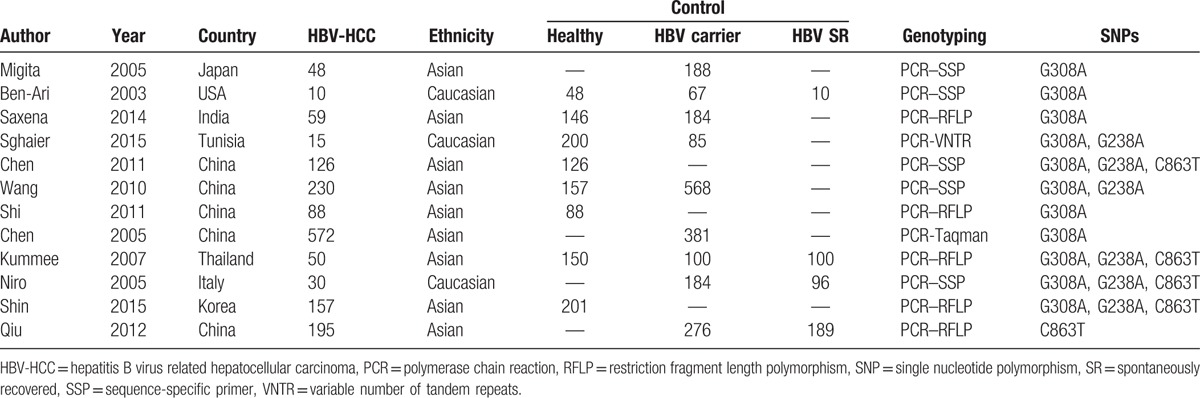
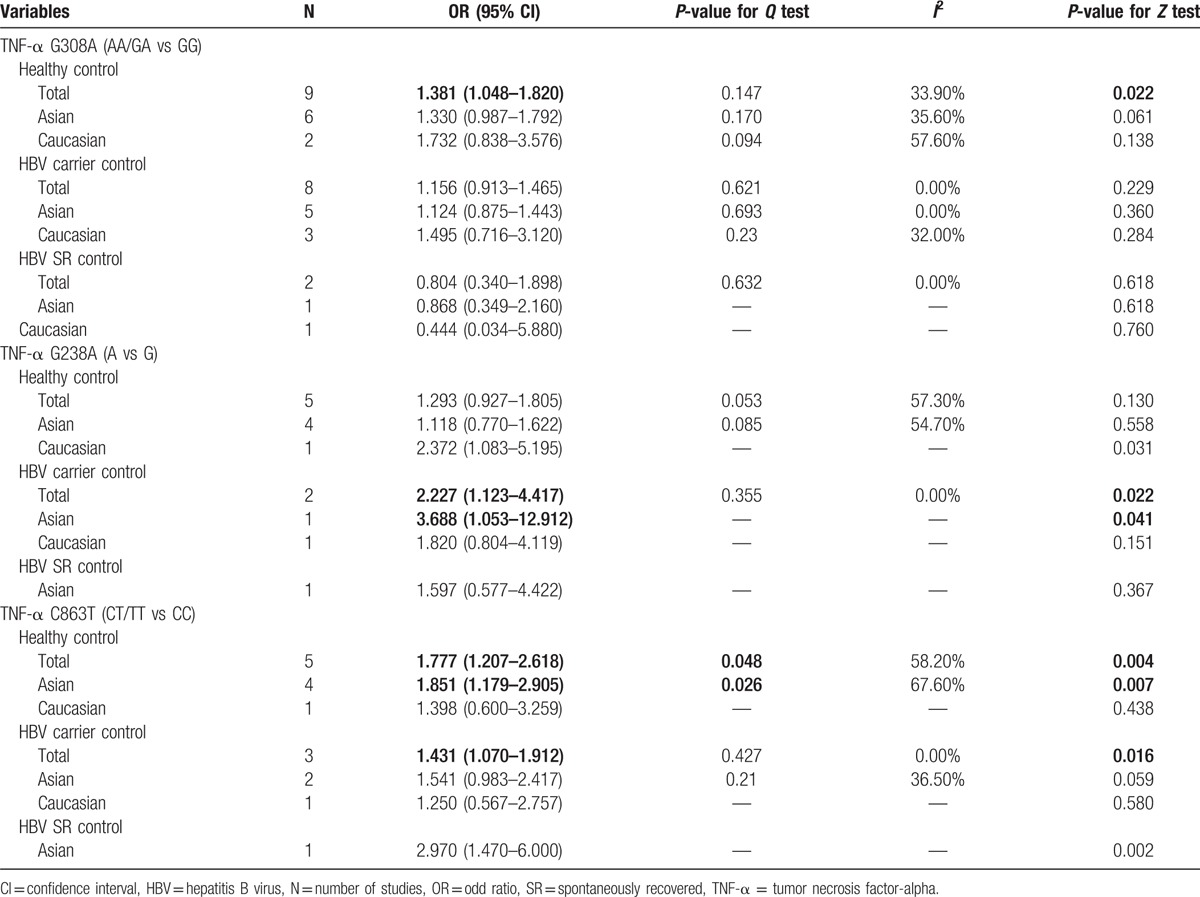
Figure 1 shows detailed information for study selection. Thirty-six case–control studies about HCC and G308A, G238A, or C863T of TNF-α were obtained. But only 12 case–control studies[26–37] focusing on HBV-HCC were included in this meta-analysis with 1580 HBV-HCC cases, 2033 HBV carrier controls, 395 HBV SR controls, and 1116 healthy controls included. The basic characteristics of all primary studies are listed in Table 1 and the detailed information of pooled OR values for all primary studies displayed in Supplementary 1. Of these studies, 10 studies with 1385 cases and 3079 controls reported an association between TNF-α G308A polymorphism and risk of HBV-HCC, and 6 studies with 608 cases and 1967 controls reported the effect of TNF-α G238A polymorphism on the risk of HBV-HCC, and 5 studies with 558 cases and 1422 controls reported the effect of TNF-α C863T polymorphism on the risk of HBV-HCC.
Figure 1.
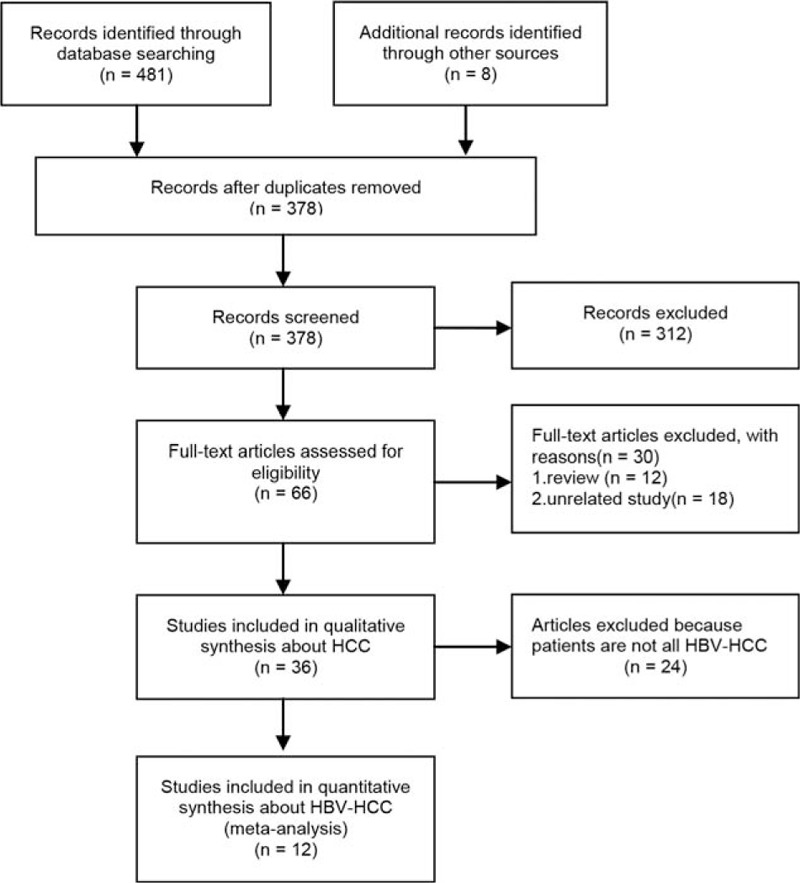
Flowchart of literature searches.
Table 1.
The basic characteristics of all primary studies.
3.1. TNF-α G308A polymorphism on risk of HBV-HCC
The number of cases for GA and AA genotypes was reported together in the study by Niro et al,[35] which could only be used for dominant-model analysis (GA/AA vs GG). Table 2 shows the results of pooled analyses. As I2 standing for the heterogeneity among studies for all models was less than 60% and P-value for the heterogeneity was more than 0.05, thus fixed-effects models were applied. When the controls were all healthy individuals, the A allele of TNF-α G308A polymorphism was significantly associated with risk of HBV-HCC (OR = 1.416, 95% CI = 1.108–1.810, P = 0.005) (Fig. 2). And the estimated ORs for AA versus GG and GA versus GG were 2.483 (P = 0.010) and 1.383 (P = 0.032), thus a dominant genetic model was suggested to be used. Therefore, the GA/AA genotypes of TNF-α G308A polymorphism was significantly associated with risk of HBV-HCC (OR = 1.381, 95% CI = 1.048–1.820, P = 0.022) (Fig. 3). But when the controls were all HBV carriers, only AA versus GG of TNF-α G308A was significantly associated with risk of HBV-HCC (OR = 2.773, 95% CI = 1.107–6.945, P = 0.030) and when compared to HBV SR controls, TNF-α G308A was not significantly associated with risk of HBV-HCC. As shown in Table 3, subgroup analyses stratified by ethnicity found GA/AA genotypes of TNF-α G308A polymorphism was not significantly associated with risk of HBV-HCC in Asian and Caucasian, but there was a trend for AA/GA genotypes of TNF-α G308A polymorphism to increase the incidence of HBV-HCC with a P-value of 0.061. Begg tests and Egger tests for publication bias revealed that there was no any obvious evidence of publication bias, which could be seen in Table 2.
Table 2.
Pooled risk estimates for TNF-α G308A, G238A, and C863T polymorphisms and HBV-HCC stratified by control group.
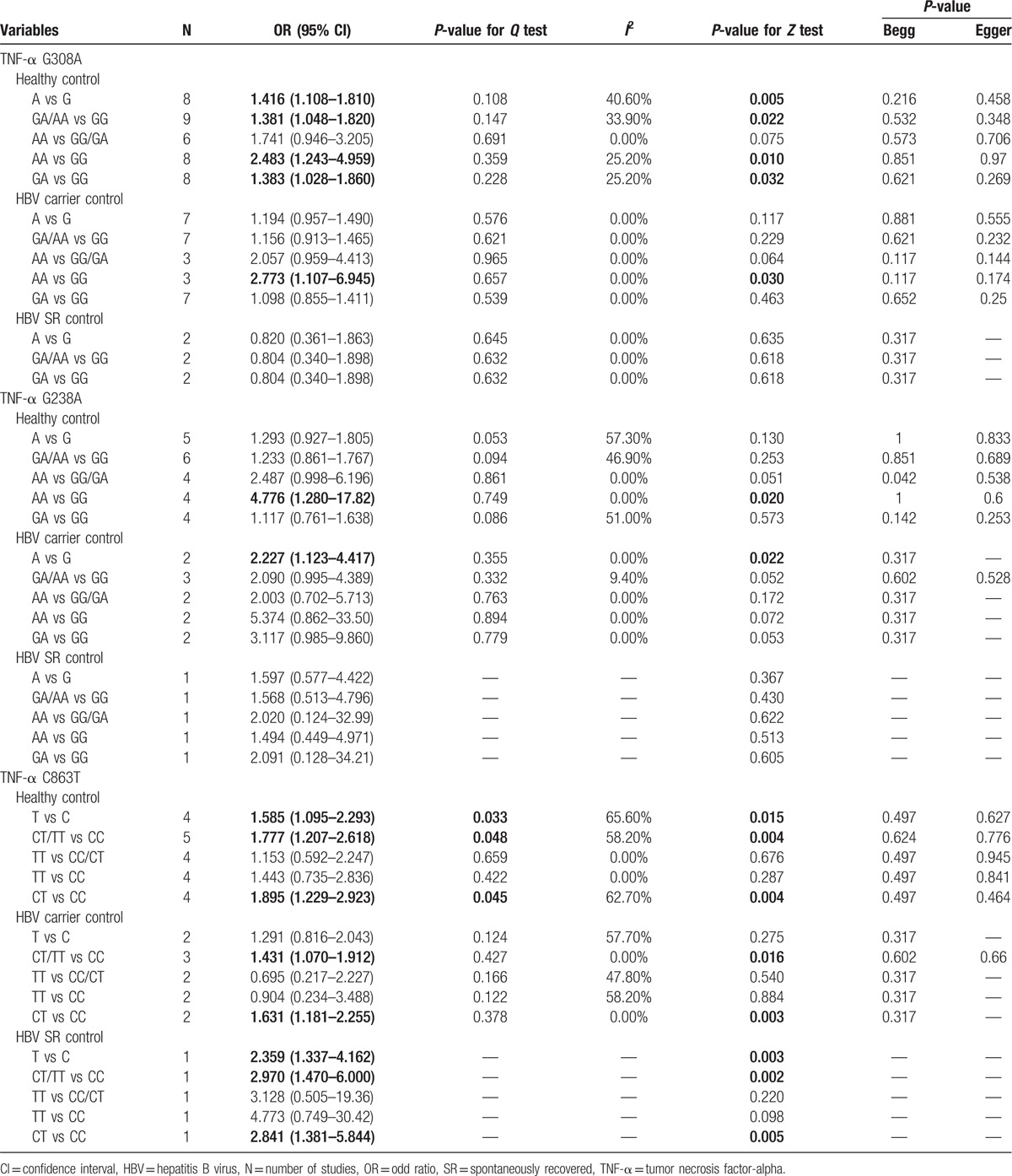
Figure 2.
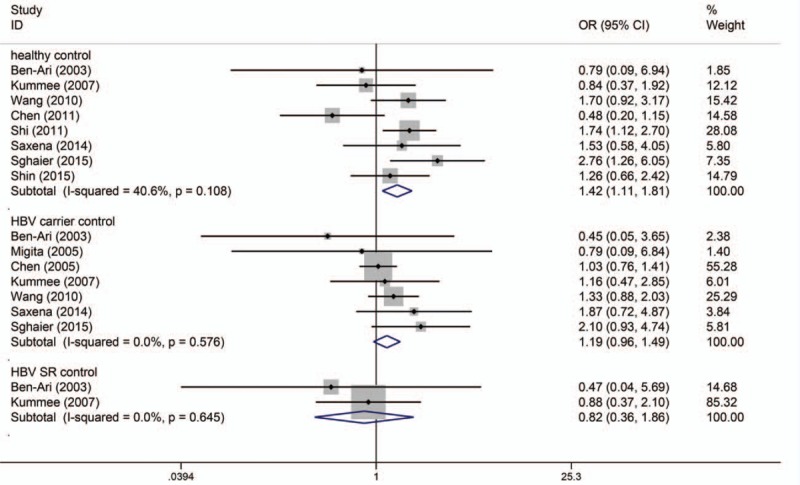
Allele model (A/G) of TNF-α G308A polymorphism on risk of HBV-HCC. The association was indicated as odds ratio (OR) estimate with the corresponding 95% confidence interval. The OR estimate of each study is marked with a solid black square. The size of the square represents the weight that the corresponding study exerts in the meta-analysis. The confidence intervals of pooled estimates are displayed as a horizontal line through the diamond. OR more than 1 indicates increased risk of HBV-HCC.
Figure 3.
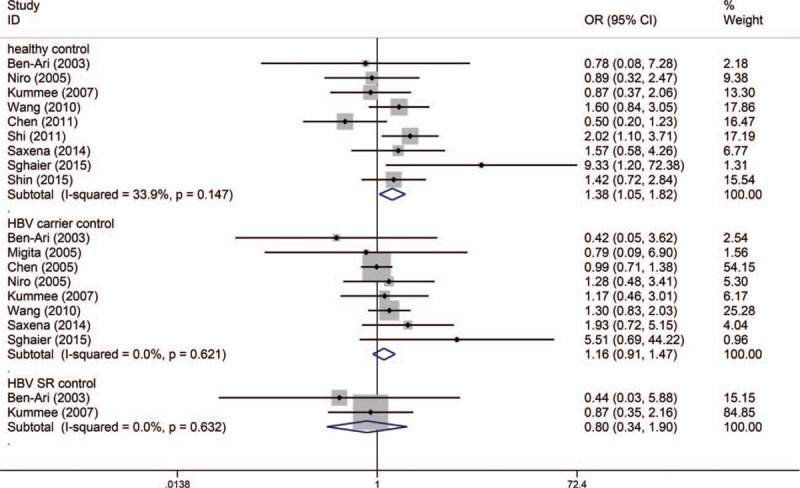
Dominant model (GA/AA vs GG) of TNF-α G308A polymorphism on risk of HBV-HCC. The association was indicated as odds ratio (OR) with the corresponding 95% confidence interval. The OR estimate of each study is marked with a solid black square. The size of the square represents the weight that the corresponding study exerts in the meta-analysis. The confidence intervals of pooled estimates are displayed as a horizontal line through the diamond. OR more than 1 indicates increased risk of HBV-HCC.
Table 3.
Pooled risk estimates for TNF-α G308A, G238A, and C863T polymorphisms and HBV-HCC stratified by ethnicity.
3.2. TNF-α G238A polymorphism on risk of HBV-HCC
Similar to the TNF-α G238A polymorphism, the numbers of cases for GA and AA genotypes were reported together in the study by Niro et al,[35] which could only be used for dominant-model analysis (GA/AA vs GG). Table 2 shows the results of the TNF-α G238A polymorphism on risk of HBV-HCC. As I2 standing for the heterogeneity among studies for all models was less than 60% and P-value for the heterogeneity was more than 0.05, thus fixed-effects models were applied. When the controls were all healthy individuals, only AA versus GG of TNF-α G238A polymorphism was significantly associated with risk of HBV-HCC (OR = 4.776, 95% CI = 1.280–17.819, P = 0.020) (Fig. 4). When the controls were all HBV carriers, only A versus G of TNF-α G238A was significantly associated with risk of HBV-HCC (OR = 2.227, 95% CI = 1.123–4.417, P = 0.022) and when compared to HBV SR controls, TNF-α G238A was not significantly associated with risk of HBV-HCC. As shown in Table 3, subgroup analyses stratified by ethnicity found A versus G of TNF-α G308A polymorphism was significantly associated with risk of HBV-HCC in Asian but not in Caucasian, while there was only 1 article in each subgroup. Begg tests and Egger tests for publication bias revealed that there was no any obvious evidence of publication bias, which could be seen in Table 2.
Figure 4.
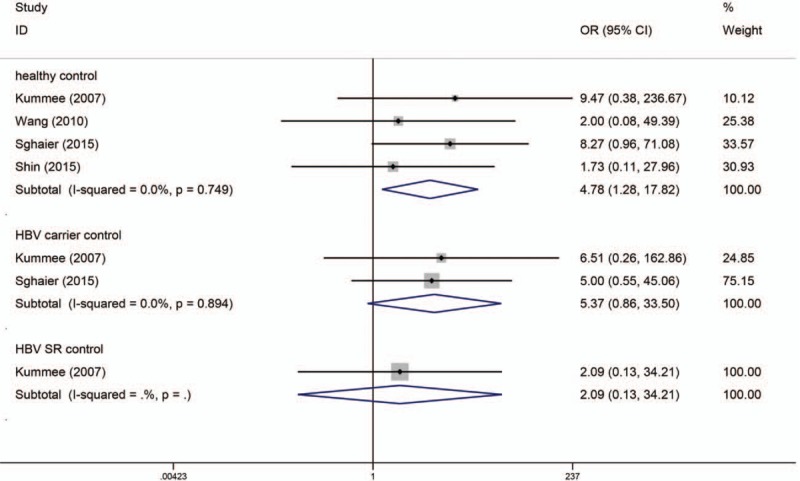
Codominant model (AA vs GG) of TNF-α G238A polymorphism on risk of HBV-HCC. The association was indicated as odds ratio (OR) estimate with the corresponding 95% confidence interval. The OR estimate of each study is marked with a solid black square. The size of the square represents the weight that the corresponding study exerts in the meta-analysis. The confidence intervals of pooled estimates are displayed as a horizontal line through the diamond. OR more than 1 indicates increased risk of HBV-HCC.
3.3. TNF-α C863T polymorphism on risk of HBV-HCC
As the numbers of cases for CT and TT genotypes were reported together in the study by Niro et al,[35] only dominant-model analysis (CT/TT vs CC) could be used for the study by Niro et al. Table 2 also shows the results of the TNF-α C863T polymorphism on risk of HBV-HCC. As I2 standing for the heterogeneity among studies for some models was more than 60% and P-value for the heterogeneity was less than 0.05, thus random-effects models were applied. The model of CT versus CC of TNF-α C863A polymorphism was significantly associated with risk of HBV-HCC. Whenever the controls were healthy individuals (OR = 1.895, 95% CI = 1.229–2.923, P = 0.004), HBV carriers (OR = 1.631, 95% CI = 1.181–2.255, P = 0.003) or HBV SR controls (OR = 2.841, 95% CI = 1.381–5.844, P = 0.005) (Fig. 5). Meanwhile the dominant-model analysis (CT/TT vs CC) also revealed significant association between the TNF-α C863T and risk of HBV-HCC (Fig. 6). As heterogeneity was found in the statistical analyses, we did sensitivity analyses to evaluate the sources of heterogeneity. We found that heterogeneity between studies was mainly caused by the study conducted by Kummee et al, and after this study was excluded, there was no longer any evidence of significant heterogeneity. As shown in Table 3, subgroup analyses found CT/TT versus CC of TNF-α G308A polymorphism and risk of HBV-HCC did not differ substantially according to ethnicity. Begg tests and Egger tests for publication bias revealed that there was no any obvious evidence of publication bias, which could be seen in Table 2.
Figure 5.
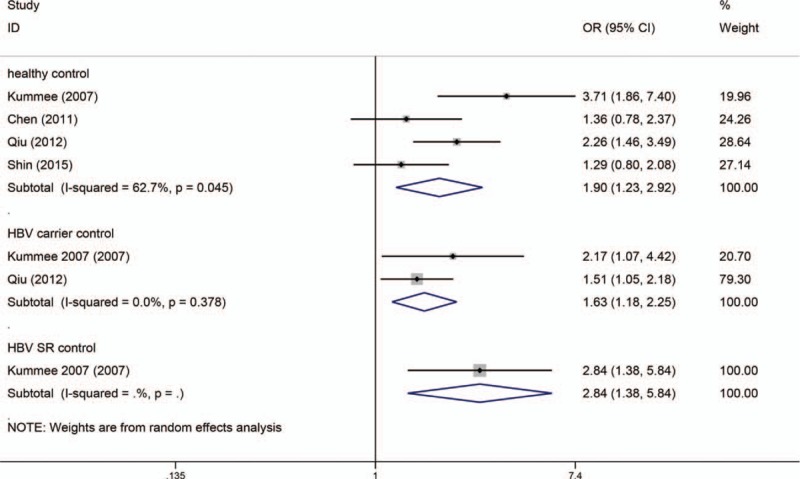
Codominant model (CA vs CC) of TNF-α C863A polymorphism on risk of HBV-HCC. The association was indicated as odds ratio (OR) with the corresponding 95% confidence interval. The OR estimate of each study is marked with a solid black square. The size of the square represents the weight that the corresponding study exerts in the meta-analysis. The confidence intervals of pooled estimates are displayed as a horizontal line through the diamond. OR more than 1 indicates increased risk of HBV-HCC.
Figure 6.
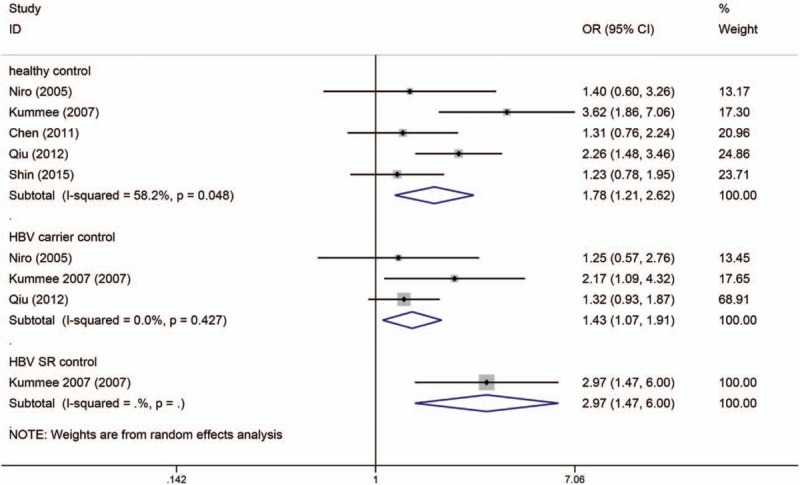
Dominant model (CA/AA vs CC) of TNF-α C863A polymorphism on risk of HBV-HCC. The association was indicated as odds ratio (OR) with the corresponding 95% confidence interval. The OR estimate of each study is marked with a solid black square. The size of the square represents the weight that the corresponding study exerts in the meta-analysis. The confidence intervals of pooled estimates are displayed as a horizontal line through the diamond. OR more than 1 indicates increased risk of HBV-HCC.
4. Discussion
The tumorigenesis of HCC, a complex and multifactor process, was found to correlate with many environmental factors.[38–40] Among which chronic infection with HBV is the most well-established environmental risk factor for HCC worldwide. However, a little part of HBsAg carriers eventually develop HCC.[41] And an interaction of environmental factors and genetic predisposition had been shown by Yu et al[42] in the development of HCC. Various studies have shown that TNF-α, a potent proinflammatory and immunomodulatory cytokine,[43] exerts an antiviral effect with profoundly suppressing HBV gene expression in infected hepatocytes noncytolytically. Literature has shown that several functional SNPs in the TNF-α promoter region were reported to influence the TNF-α constitutive and inducible expression levels.[44,45]
Until recently, there are many studies performed to explore the association between TNF-α polymorphisms and HCC risk and several meta-analyses were conducted to investigate the association between TNF-α polymorphisms (G308A, G238A, C863T) and HCC. While, all these meta-analyses did not focus on the relationship between TNF-α polymorphism and HBV-HCC patients or HBV infection recovered patients and controversies exist. Thus, clarifying the independent role of each polymorphism on HBV and HCC should be quite necessary.
Our data showed that the variant genotypes AA/AG of G308A were associated with an increased HBV-HCC risk when compared to all healthy individuals. And this result might be biologically credible considering the function of TNF-α in inflammation and tumor development. While, there was no obvious association in studies which the controls were HBV carriers (thought AA vs GG of G308A was correlated with increased HBV-HCC, credibility is not high because there were only 3 studies) and SR controls. Thus, HBV infection seemed to be a more important factor for tumorigenesis of HCC than genetic predisposition in G308A of TNF-α, and the association between G308A of TNF-α polymorphisms and HCC in HBV-free patients should be conducted to confirm our result.
The previous results conduct on G238A polymorphism of TNF-α and HCC risk were different. Contrary to the previous finding made by Yang et al,[19] which showed no significant, 3 meta-analyses conducted by Wei et al,[20] Cheng et al,[22] and Tian et al[21] showed that there was a significant association between TNF-α 238 G/A polymorphism and increased risk of liver cancer under A versus G, AG versus GG, and AA/AG versus GG models. As the meta-analysis of Yang et al[19] included only 5 studies for G238A polymorphism of TNF-α and HCC, the association between G238A polymorphism of TNF-α and increased HCC risk seems robust. In the present studies, 6 studies were included to investigate the association between G238A polymorphism of TNF-α and HBV-HCC risk, and our finding is almost different. Our result showed that only AA versus GG of G238A was correlated with increased HBV-HCC when controls were healthy individuals, and only A versus G of G238A was correlated with increased HBV-HCC when controls were HBV carriers. There were several factors might contribute to these discrepancies. Firstly, the HCC patients in our meta-analysis were all related to HBV. Secondly, the controls were classified to healthy individuals, HBV carriers and HBV SR controls, not just noncancers. Finally, the number of studies in which the controls were HBV carriers or HBV SR controls was too few to obtain credible Results.
We also found 5 studies that had examined the association between TNF-α C863T polymorphisms and HBV-HCC. The pooled result showed that TNF-α C863T polymorphisms were associated with increased HBV-HCC risk in a heterozygous comparison (CT vs CC) and dominant model (CT/TT vs CC) whenever the controls were healthy individuals, HBV carriers and HBV SR controls. Previous studies demonstrated that allele A of TNF-α C863T was associated with increased circulating TNF-a levels in response to HBV infection,[13,46] which could activate nuclear factor κB (NF-κB). NF-κB was a nuclear transcription factor that regulated expression of proinflammatory genes that were associated with hepatic inflammation and hepatic fibrosis.[47,48] Thus the conclusion was assumed that the interaction between TNF-α C863T polymorphisms and HBV infection might be associated with increased HCC risk. As studies investigating TNF-α C863T polymorphisms and HBV-HCC were not enough, this assumption might be interpreted with caution and more studies are needed, thought the statistical results for detecting bias did not show publication bias.
However, there were several inevitable study limitations should be interpreted after taking into consideration. Firstly, there was language limitation as current meta-analysis only contains English literature with some articles possibly published in other languages and not accessible to the international journals. Secondly, our results were all based on unadjusted ORs, while a more precise estimation should take into account the effect of multiple confounders such as specific environmental and lifestyle factors on the association. Thirdly, the number of studies and the number of subjects in the studies included in the meta-analysis were small. This might not have enough power to explore the associations between TNF-α C863T polymorphisms and HBV-HCC. So further studies estimating the effect of haplotypes, covariants of more risk polymorphisms, and gene–environment interactions about HCC should be conducted.
In summary, this meta-analysis and systemic review investigated the association of TNF-α G308A, G238A, and C863T polymorphisms and HBV-HCC susceptibility. Our novel data demonstrated that AA genotype in TNF-α G308A and TNF-α G238A and CA genotype in TNF-α C863T may increase HBV-HCC risk. Therefore, HBV infection seemed to be a more important factor for tumorigenesis of HCC than genetic predisposition in G308A of TNF-α, and interaction between TNF-α C863T polymorphisms and HBV infection might be associated with increased HCC risk.
Supplementary Material
Footnotes
Abbreviations: CIs = confidence intervals, HBV = hepatitis B virus, HBV-HCC = hepatitis B virus related hepatocellular carcinoma, HCC = hepatocellular carcinoma, HCV = hepatitis C virus, HWE = Hardy–Weinberg equilibrium, OR = odds ratio, SR = spontaneously recovered, TNF-α = tumor necrosis factor-alpha.
The authors have no conflicts of interest to disclose.
Supplemental Digital Content is available for this article.
References
- [1].Tsochatzis EA, Meyer T, Burroughs AK. Hepatocellular carcinoma. N Engl J Med 2012;366:92.author reply 92–93. [DOI] [PubMed] [Google Scholar]
- [2].Jemal A, Bray F, Center MM, et al. Global cancer statistics. CA Cancer J Clin 2011;61:69–90. [DOI] [PubMed] [Google Scholar]
- [3].Parkin DM. The global health burden of infection-associated cancers in the year 2002. Int J Cancer 2006;118:3030–44. [DOI] [PubMed] [Google Scholar]
- [4].Llovet JM, Bruix J. Molecular targeted therapies in hepatocellular carcinoma. Hepatology 2008;48:1312–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Rapisarda V, Loreto C, Malaguarnera M, et al. Hepatocellular carcinoma and the risk of occupational exposure. World J Hepatol 2016;8:573–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Coussens LM, Werb Z. Inflammation and cancer. Nature 2002;420:860–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Yoshimura A. Signal transduction of inflammatory cytokines and tumor development. Cancer Sci 2006;97:439–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Hajeer AH, Hutchinson IV. Influence of TNFalpha gene polymorphisms on TNFalpha production and disease. Hum Immunol 2001;62:1191–9. [DOI] [PubMed] [Google Scholar]
- [9].Hajeer AH, Hutchinson IV. TNF-alpha gene polymorphism: clinical and biological implications. Microsc Res Tech 2000;50:216–28. [DOI] [PubMed] [Google Scholar]
- [10].Kroeger KM, Carville KS, Abraham LJ. The −308 tumor necrosis factor-alpha promoter polymorphism effects transcription. Mol Immunol 1997;34:391–9. [DOI] [PubMed] [Google Scholar]
- [11].Wilson AG, Symons JA, McDowell TL, et al. Effects of a polymorphism in the human tumor necrosis factor alpha promoter on transcriptional activation. Proc Natl Acad Sci USA 1997;94:3195–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Wilson AG, di Giovine FS, Blakemore AI, et al. Single base polymorphism in the human tumour necrosis factor alpha (TNF alpha) gene detectable by NcoI restriction of PCR product. Hum Mol Genet 1992;1:353. [DOI] [PubMed] [Google Scholar]
- [13].Higuchi T, Seki N, Kamizono S, et al. Polymorphism of the 5′-flanking region of the human tumor necrosis factor (TNF)-alpha gene in Japanese. Tissue Antigens 1998;51:605–12. [DOI] [PubMed] [Google Scholar]
- [14].Mestiri S, Bouaouina N, Ahmed SB, et al. Genetic variation in the tumor necrosis factor-alpha promoter region and in the stress protein hsp70-2: susceptibility and prognostic implications in breast carcinoma. Cancer 2001;91:672–8. [DOI] [PubMed] [Google Scholar]
- [15].Warzocha K, Ribeiro P, Bienvenu J, et al. Genetic polymorphisms in the tumor necrosis factor locus influence non-Hodgkin's lymphoma outcome. Blood 1998;91:3574–81. [PubMed] [Google Scholar]
- [16].Oh BR, Sasaki M, Perinchery G, et al. Frequent genotype changes at −308, and 488 regions of the tumor necrosis factor-alpha (TNF-alpha) gene in patients with prostate cancer. J Urol 2000;163:1584–7. [PubMed] [Google Scholar]
- [17].Sasaki M, Nakajima K, Perinchery G, et al. Frequent genotype changes at −308 of the human tumor necrosis factor-alpha promoter region in human uterine endometrial cancer. Oncol Rep 2000;7:369–73. [PubMed] [Google Scholar]
- [18].Machado JC, Figueiredo C, Canedo P, et al. A proinflammatory genetic profile increases the risk for chronic atrophic gastritis and gastric carcinoma. Gastroenterology 2003;125:364–71. [DOI] [PubMed] [Google Scholar]
- [19].Yang Y, Luo C, Feng R, et al. The TNF-alpha, IL-1B and IL-10 polymorphisms and risk for hepatocellular carcinoma: a meta-analysis. J Cancer Res Clin Oncol 2011;137:947–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Wei Y, Liu F, Li B, et al. Polymorphisms of tumor necrosis factor-alpha and hepatocellular carcinoma risk: a HuGE systematic review and meta-analysis. Dig Dis Sci 2011;56:2227–36. [DOI] [PubMed] [Google Scholar]
- [21].Tian X, Ma P, Sui C, et al. Comprehensive assessment of the association between tumor necrosis factor alpha G238A polymorphism and liver cancer risk. Tumour Biol 2014;35:103–9. [DOI] [PubMed] [Google Scholar]
- [22].Cheng K, Zhao YJ, Liu L, et al. Tumor necrosis factor-alpha 238 G/A polymorphism and risk of hepatocellular carcinoma: evidence from a meta-analysis. Asian Pac J Cancer Prev 2013;14:3275–9. [DOI] [PubMed] [Google Scholar]
- [23].Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst 1959;22:719–48. [PubMed] [Google Scholar]
- [24].DerSimonian R, Kacker R. Random-effects model for meta-analysis of clinical trials: an update. Contemp Clin Trials 2007;28:105–14. [DOI] [PubMed] [Google Scholar]
- [25].Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med 2002;21:1539–58. [DOI] [PubMed] [Google Scholar]
- [26].Migita K, Miyazoe S, Maeda Y, et al. Cytokine gene polymorphisms in Japanese patients with hepatitis B virus infection--association between TGF-beta1 polymorphisms and hepatocellular carcinoma. J Hepatol 2005;42:505–10. [DOI] [PubMed] [Google Scholar]
- [27].Ben-Ari Z, Mor E, Papo O, et al. Cytokine gene polymorphisms in patients infected with hepatitis B virus. Am J Gastroenterol 2003;98:144–50. [DOI] [PubMed] [Google Scholar]
- [28].Saxena R, Chawla YK, Verma I, et al. IFN-gamma (+874) and not TNF-alpha (−308) is associated with HBV-HCC risk in India. Mol Cell Biochem 2014;385:297–307. [DOI] [PubMed] [Google Scholar]
- [29].Sghaier I, Zidi S, Mouelhi L, et al. The relationship between TNF alpha gene polymorphisms (−238/−308), TNF RII VNTR (p75) and outcomes of hepatitis B virus infection in Tunisian population. Gene 2015;568:140–5. [DOI] [PubMed] [Google Scholar]
- [30].Chen X, Zhang L, Chang Y, et al. Association of TNF-alpha genetic polymorphisms with hepatocellular carcinoma susceptibility: a case-control study in a Han Chinese population. Int J Biol markers 2011;26:181–7. [DOI] [PubMed] [Google Scholar]
- [31].Wang B, Wang J, Zheng Y, et al. A study of TNF-alpha-238 and −308 polymorphisms with different outcomes of persistent hepatitis B virus infection in China. Pathology 2010;42:674–80. [DOI] [PubMed] [Google Scholar]
- [32].Shi Z, Du C. Tumor necrosis factor alpha 308 G/A polymorphism and hepatocellular carcinoma risk in a Chinese population. Genet Test Mol Biomarkers 2011;15:569–72. [DOI] [PubMed] [Google Scholar]
- [33].Chen CC, Yang SY, Liu CJ, et al. Association of cytokine and DNA repair gene polymorphisms with hepatitis B-related hepatocellular carcinoma. Int J Epidemiol 2005;34:1310–8. [DOI] [PubMed] [Google Scholar]
- [34].Kummee P, Tangkijvanich P, Poovorawan Y, et al. Association of HLA-DRB1∗13 and TNF-alpha gene polymorphisms with clearance of chronic hepatitis B infection and risk of hepatocellular carcinoma in Thai population. J Viral Hepat 2007;14:841–8. [DOI] [PubMed] [Google Scholar]
- [35].Niro GA, Fontana R, Gioffreda D, et al. Tumor necrosis factor gene polymorphisms and clearance or progression of hepatitis B virus infection. Liver Int 2005;25:1175–81. [DOI] [PubMed] [Google Scholar]
- [36].Shin SP, Kim NK, Kim JH, et al. Association between hepatocellular carcinoma and tumor necrosis factor alpha polymorphisms in South Korea. World J Gastroenterol 2015;21:13064–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Qiu B, Wang X, Zhang P, et al. Association of TNF-alpha promoter polymorphisms with the outcome of persistent HBV infection in a northeast Chinese Han population. Acta Biochim Biophys Sin 2012;44:712–8. [DOI] [PubMed] [Google Scholar]
- [38].Zhu ZZ, Cong WM. Roles of hepatitis B virus and hepatitis C virus in hepato-carcinogenesis. Chin J Hepatol 2003;11:574–6. [PubMed] [Google Scholar]
- [39].Kuper H, Tzonou A, Kaklamani E, et al. Tobacco smoking, alcohol consumption and their interaction in the causation of hepatocellular carcinoma. Int J Cancer 2000;85:498–502. [PubMed] [Google Scholar]
- [40].Chen CJ, Wang LY, Lu SN, et al. Elevated aflatoxin exposure and increased risk of hepatocellular carcinoma. Hepatology 1996;24:38–42. [DOI] [PubMed] [Google Scholar]
- [41].Bowen DG, Walker CM. Adaptive immune responses in acute and chronic hepatitis C virus infection. Nature 2005;436:946–52. [DOI] [PubMed] [Google Scholar]
- [42].Yu MW, Yang SY, Chiu YH, et al. A p53 genetic polymorphism as a modulator of hepatocellular carcinoma risk in relation to chronic liver disease, familial tendency, and cigarette smoking in hepatitis B carriers. Hepatology 1999;29:697–702. [DOI] [PubMed] [Google Scholar]
- [43].Dragani TA. Risk of HCC: genetic heterogeneity and complex genetics. J Hepatol 2010;52:252–7. [DOI] [PubMed] [Google Scholar]
- [44].Partida-Rodriguez O, Torres J, Flores-Luna L, et al. Polymorphisms in TNF and HSP-70 show a significant association with gastric cancer and duodenal ulcer. Int J Cancer 2010;126:1861–8. [DOI] [PubMed] [Google Scholar]
- [45].Nieves-Ramirez ME, Partida-Rodriguez O, Alegre-Crespo PE, et al. Characterization of single-nucleotide polymorphisms in the tumor necrosis factor alpha promoter region and in lymphotoxin alpha in squamous intraepithelial lesions, precursors of cervical cancer. Transl Oncol 2011;4:336–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Sinha S, Mishra SK, Sharma S, et al. Polymorphisms of TNF-enhancer and gene for FcgammaRIIa correlate with the severity of falciparum malaria in the ethnically diverse Indian population. Malar J 2008;7:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Lu H, Lei X, Zhang Q. Moderate activation of IKK2-NF-κB in unstressed adult mouse liver induces cytoprotective genes and lipogenesis without apparent signs of inflammation or fibrosis. BMC Gastroenterol 2015;15:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Wilson CL, Jurk D, Fullard N, et al. NFkappaB1 is a suppressor of neutrophil-driven hepatocellular carcinoma. Nat Commun 2015;6:6818. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


