Abstract
Background
The scarcity of tissues from racial and ethnic minorities at biobanks poses a scientific constraint to research addressing health disparities in minority populations.
Methods
To address this gap, the Minority Biospecimen/Biobanking Geographic Management Program for region 3 (BMaP-3) established a working infrastructure for a “biobanking” hub in the southeastern United States and Puerto Rico. Herein we describe the steps taken to build this infrastructure, evaluate the feasibility of collecting formalin-fixed, paraffin-embedded tissue blocks and associated data from a single cancer type (breast), and create a web-based database and tissue microarrays (TMAs).
Results
Cancer registry data from 6 partner institutions were collected, representing 12,408 entries from 8,279 unique patients with breast cancer (years 2001–2011). Data were harmonized and merged, and deidentified information was made available online. A TMA was constructed from formalin-fixed, paraffin-embedded samples of infiltrating ductal carcinoma representing 427 patients with breast cancer (147 African Americans, 168 Hispanics, and 112 non-Hispanic whites) and was annotated according to biomarker status and race/ethnicity. Biomarker analysis of the TMA was consistent with the literature.
Conclusions
Contributions from participating institutions have facilitated a robust research tool. TMAs of infiltrating ductal breast carcinoma have now been released for 5 projects at 5 different institutions.
Keywords: biobank, tissue microarray, FFPE, tissue blocks, breast cancer, disparities, ethnicity, race, scarcity
Introduction
The burden of cancer disparities on African American and Hispanic populations is well documented.1–8 Discoveries in precision medicine and basic science that have the potential to improve the prognosis or treatment of cancer in affected individuals may be limited by the scarcity of research biospecimens from racial/ethnic minority populations. The availability of diverse samples is key to inclusive precision medicine.9 For example, the triple-negative breast cancer subtype, defined by the absence of ER, PR, and ERBB2 (formerly HER2 or HER2/neu) expression, accounts for 10% to 20% of all breast cancer cases, with an uneven distribution among ethnicities.10 In African American women, triple-negative breast cancer is nearly twice as prevalent as it is in European Americans, with some studies reporting triple negative subtypes in up to 40% of all breast cancer cases among premenopausal African Americans.10,11 Consistent with this observation, epidemiological studies have shown substantial differences in rates of breast cancer mortality between racial groups.5,8,12 For example, premenopausal African American women have a lower 5-year breast cancer–related survival rate than European American women (79.0% vs 91.0%, respectively), placing them at a higher risk of mortality despite an overall lower incidence of breast cancer.12 This disparity may be due to the increased frequency of triple-negative breast cancer observed in African American women who typically present at a later disease stage and are more likely to have lymph-node metastases at similar tumor sizes.13 A recent study has also shown that Hispanic women have a higher prevalence of ER-negative tumors compared with non-Hispanic women (36.2% vs 22.7%; P = .05) and an unexpectedly high proportion of ERBB2-positive tumors compared with non-Hispanic women (31.9% vs 14.3%; P < .01).14 These findings, coupled with more recent data, suggest that considerable molecular differences in cancers may exist in various ethnic groups.10 Because molecular features can be used for predicting cancer prognosis and therapeutic response, research focused on understanding the molecular features of cancers occurring in ethnic minorities is important.
Purpose
To address the lack of tissue-based studies for ethnic minorities with cancer, the Biospecimen/Biobanking Geographic Management Program (BMaP) was initiated by the National Cancer Institute’s Center to Reduce Cancer Health Disparities. BMaP coordinated regional efforts focused on health disparities in cancer research, training, and care and was organized into 6 geographical regions within the United States. BMaP region 3 (BMaP-3), which makes up the southeastern region, consists of Alabama, Florida, Georgia, Louisiana, Mississippi, and Puerto Rico, and it included 9 partner institutions that created a collaborative network. A key goal for BMaP-3 was to develop a state-of-the-art network that could provide a foundation and infrastructure to adequately and continuously supply high-quality, multiethnic, human biospecimens for cancer research. Since 2015, the National Cancer Institute has reorganized and consolidated the regions into 6 regions, and the institutions previously in region 3 are now, at the time of publication, in Geographical Management of Cancer Health Disparities (GMaP) region 2.15
In 2014, members of the participating BMaP-3 institutions completed a comprehensive assessment tool that revealed a diversity of samples by race/ethnicity and cancer site were available as formalin-fixed, paraffin-embedded (FFPE) tumor samples or fresh frozen samples (from 6 of 9 institutions).16 In addition, 5 of the institutions had a tissue advisory/steering committee, thus providing the necessary infrastructure and expertise to build a network approach. This provided the basis for the design of a BMaP-3 pilot study to test the feasibility of developing and implementing a regional, web-based database of available biospecimens and collecting FFPE blocks to create a unique tissue microarray (TMA) for health disparities in cancer research.
This pilot study represents a regional team, science-oriented approach for constructing and validating a multi-institutional breast cancer TMA from a racially and ethnically diverse population. This work was made possible by the infrastructure of the BMaP, and it can serve as a model for other multi-institutional networks focused on research utilizing human biospecimens. Using the construction and validation of a breast cancer TMA as an example, we illustrate how a regional team, science-oriented approach can effectively leverage the tissue and database resources of multiple institutions to facilitate tissue-based studies for a minority cohort — a difficult challenge for single institutions alone.
Materials and Methods
The research conducted in this study was performed with tissue samples and patient data unlinked from patient identifiers. Approval or waiver of informed consent was obtained by the Institutional Review Boards of the 6 institutions contributing FFPE biospecimens and data to support this pilot study.
Collection and Management of Data
Cancer registry coding discrepancies were harmonized by adopting a standardized coding for each variable and systematically replacing prior coding with current coding using software R v2.15.2 (R Foundation for Statistical Computing, Vienna, Austria). Cases were removed from the datasets for any of the following reasons: (a) patient was younger than 18 years of age at diagnosis (n = 1), (b) tumor marker and site-specific factor did not agree for either ER or PR status (n = 157), (c) patient had a Spanish surname and unknown “Spanish Hispanic” variable (n = 17), or (d) patient was assigned a study identification number that already existed (n = 1). The original site-specific factor 16 was captured after the year 2010; therefore, the site-specific factor was manually derived from the tumor marker 1 ER assay, tumor marker 2 PR assay, and ERBB2 variables, with assignments of either HR+/ERBB2+, HR+/ERBB2−, PR−/ER−/ERBB2+, or PR−/ER−/ERBB2−. If the original site-specific factor 16 disagreed with the derived site-specific factor, then the entry was deemed unreliable and was deleted (n = 19).
Multi-institutional, Web-based Database
To ensure no changes were made to the dataset originally merged by the biostatistician, the web-based dataset was exported and a comparison of the web-based dataset against the original merged dataset was executed. Counts were also verified between both datasets. The aggregated data were posted on a website (http://labpages.moffitt.org/bmap3/Cancer%20Database/Cancer_Database.html) in the form of an output of descriptive statistics generated by SPSS (IBM, Armonk, New York). BMaP-3 public access database (PAD) has logging features enabled so that, in the event of improper data modification, the user account responsible, the data that were changed, and the time of the change can be identified and corrected in a timely manner. The governance model for the BMaP-3 PAD consists of 3 hierarchical user-access levels (administrator, data manager, and investigator), ranging from full to search access.
Archival Case Selection and Tissue Microarray Construction
Each archival tissue sample was categorized according to tumor expression of ER, PR, and ERBB2. For each case, optimal areas for inclusion as cores in the TMA were marked on the slides stained with hematoxylin and eosin by pathologists. When sufficient tumor tissue was available, 3 or fewer 1-mm cores from each selected donor block were punched and put into 8 TMAs 60 × 10 mm in size using Tissue Arrayer (Beecher Instruments, Sun Prairie, Wisconsin). Each TMA also included 1 sample of normal breast tissue. Cores from kidney, spleen, and tonsil, as well as melanoma, human breast cancer lines (MCF-7 and MDA231), human prostate adenocarcinoma cells (LNCaP), and human ovarian carcinoma cells (SK-OV-3), were included as reference cells/tissues.
Immunohistochemical Staining and Evaluation
All TMA slides were stained with hematoxylin and eosin and processed for whole-slide imaging. Each TMA slide was scanned using ScanScope XT (Aperio, Vista, California) with a 20×/0.8 numerical aperture objective lens at a rate of 10 minutes per slide. Image analyses for stained TMAs were performed using Nuclear v9.1 (Aperio) to segment nuclei of various intensities. The captured hematoxylin and eosin–stained images were made available to investigators at all participating institutions. The biomarker status obtained from cancer registry data was validated with immunostains for ER, PR, and ERBB2 using the following antibodies from Ventana Medical Systems (Tucson, Arizona): 790-4324 CONFIRM anti-ER (SP1), 790-2223 CONFIRM anti-PR (1E2), and 790-100 PATHWAY anti-HER2/Neu (4B5). Antigen retrieval and incubation times were optimized for each antibody as follows: 60-minute retrieval for estrogen receptor (ER) and progesterone (PR) antibodies and 30-minute retrieval for erb-b2 receptor tyrosine kinase 2 (ERBB2) antibodies; 32-minute incubation for the ER antibody and 16-minute incubation for the PR and ERBB2 antibodies.
The Ventana Benchmark XT (Tucson, Arizona) platform was used for all immunohistochemistry analyses. The Allred scoring system was used to score the ER and PR stains.17 ERBB2 stains were scored in accordance with guidelines from the College of American Pathologists, which defines ERBB2-positive status by immunohistochemistry as protein overexpression.18 The HR rules for the categorization of cases were: if either ER or PR is positive, then HR+; if both are negative, then HR−; and if both ER and PR are missing, or if one is missing and the other is negative, then it was labeled as “missing.”
Results
Building an Infrastructure
To ensure equitable governance for issues concerning human biospecimen research sponsored by BMaP-3, a Tissue Advisory Board was formed with representatives from all 9 BMaP-3 institutions. A collaboration agreement, signed by members of each of the 9 partner institutions, allowed data and tissue sharing between the partner institutions regardless of the ability of any individual institution to contribute samples. Once the infrastructure was established, the goal was to evaluate the feasibility of collecting FFPE tissue blocks and associated data and creating a web-based database and TMAs.
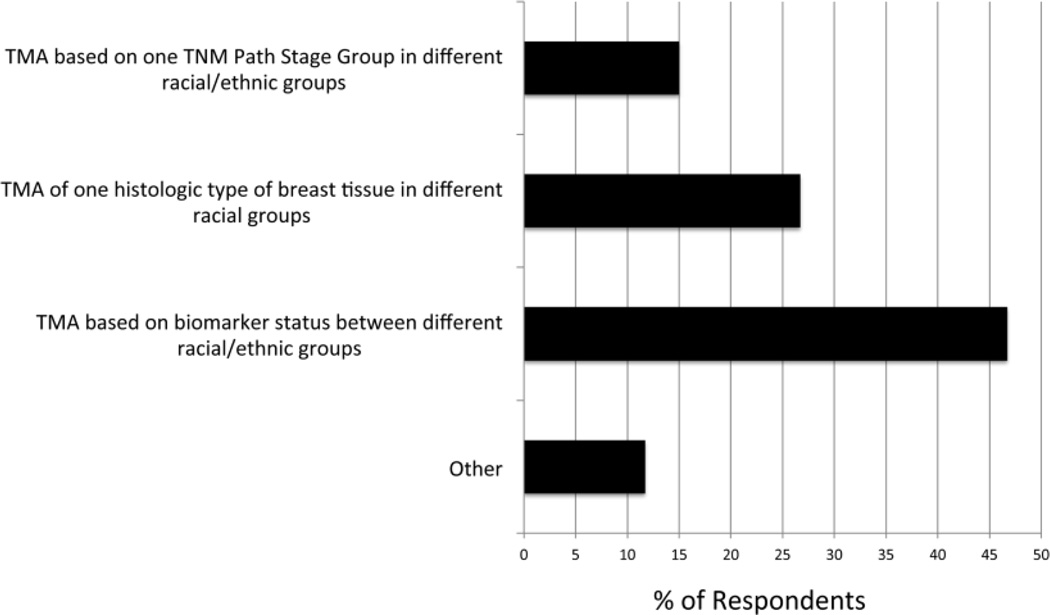
The first step in this pilot study was to choose a tumor type for the TMA. The BMaP-3 investigators chose breast cancer as the highest priority to address health disparities related to cancer in the southeastern United States and Puerto Rico. This decision was based on a variety of factors, including the scarcity of breast TMAs from diverse populations described in the literature, lack of commercially available breast TMAs with associated patient data on race and ethnicity, the observed rates of incidence and mortality associated with breast cancer at the 6 contributing cancer sites, and the results of a multi-institutional survey of researcher needs for TMAs.16 In addition, researchers specializing in breast cancer were surveyed to determine what type of breast cancer TMA is most needed for research. As shown in Fig 1, 29 of the 61 respondents (47.5%) expressed greatest need for a TMA based on biomarker status between different racial/ethnic groups, followed by a TMA based on 1 histological type of breast tissue in different racial/ethnic groups (26%), and a TMA based on 1 tumor, node, and metastasis pathology stage group in different racial/ethnic groups (14%).
Fig 1. Results of breast cancer researcher survey.
A multi-institutional, Web-based poll was distributed to investigators in region 3 (currently region 2) institutions to assess the research interest regarding different kinds of breast TMAs. Survey responses were anonymous. As can be seen, the greatest need was for a TMA based on biomarker status between different racial/ethnic groups. TMA = tissue microarray.
Collection and Management of Cancer Registry Data
Deidentified American College of Surgeons cancer registry data from each institution served as the basis for the web-based database and for the selection of cases to be included in the TMA. Requests for data from cancer registries at each of the 9 partner institutions specified the following criteria for selecting cases: date range, 2001 to 2011 (to avoid using biospecimens that might be needed for clinical care), age at diagnosis (≥ 18 years of age), primary breast cancer only (no metastasis), and first-course surgery at the institution. In addition, because cases from minority populations were a priority for inclusion in the TMA, members at partnering institutions were asked to specifically retrieve patient data from African Americans and Hispanics/Latinos. The 17 data elements requested are described in Table 1.
Table 1.
Data Elements and Their Descriptions
| Data Element | Description |
|---|---|
| Study identifier | Unique code used by honest broker for reidentification |
| Year first seen | Year of first patient contact (inpatient or outpatient), with the reporting facility for the diagnosis of the tumor, it treatment, or both |
| Site primary | Code for the primary site of the tumor being reported using either ICD-O (2nd ed. or 3rd ed.) |
| Age at diagnosis | Records the age of the patient in complete years at his or her last birthday before diagnosis |
| Sex | Identifies the sex of the patient |
| Race | Identifies the primary race of the patient |
| Spanish/Hispanic origin | Identifies patients of Spanish or Hispanic origin |
| Histology (ICD-O [3rd ed.]) | First 4 digits of the morphology code that describe the tumor/cell type |
| Behavior (ICD-O [3rd ed.])a | Code for the behavior of the tumor being reported (MCC Dictionary) |
| Year of surgery | Year in which a surgical procedure was performed |
| Surgery of primary site, at hospital | Type of surgery performed to breast aimed at modifying, controlling, removing, or destroying cancerous tissue |
| Systemic/surgery sequence | Records the sequencing of systemic treatment and surgical procedures given as part of first-line therapy |
| Surgery/radiation sequence | Records the sequencing of radiation and surgical procedures provided as part of first-line therapy |
| Collaborative stage site-specific factor 16 | Combinations of ER, PR, and ERBB2 results |
| Tumor marker 1 | ER assay |
| Tumor marker 2 | PR assay |
| ERBB2 | Interpretation of results (not the same as the laboratory value) for the ERBB2 test used to determine the status of ERBB2 |
| TNM, pathology, stage group | The anatomic extent of disease based on tumor, node, and metastasis elements of the tumor following surgery |
Behavior is the 5th digit of the ICD-O (3rd ed.) morphology code.
ICD-O = International Classification of Diseases for Oncology, TNM = tumor, node, metastasis.
Cancer registry datasets were retrieved from 6 institutions, and a biostatistician harmonized the datasets. A total of 12,408 entries from 8,279 unique patients with breast cancer pathology were confirmed after merging the datasets. Deidentified information about breast cancer cases collected from the different cancer registries is available in BMaP-3 PAD (https://apps.mathbiol.org/bmap [access restricted]).
Archival Case Selection and Tissue Microarray Construction
The dataset was further analyzed for cases with infiltrating ductal carcinoma (histology selected by the Tissue Advisory Board) and then cross-tabulated by race/ethnicity and biomarker status (triple negative, HR+/ERBB2−, HR+/ERBB2+, ER−/PR−/ERBB2+). The demographical and clinical characteristics of the data from the breast cancer cases obtained from the 6 institutions are presented in Table 2. Because cancer registry standards for ERBB2 were largely adopted from 2006 onward, many of the archival cases (collected from 2001 to 2005) were categorized as missing for ERBB2 status. Therefore, the Tissue Advisory Board decided that the 3 biomarker categories for selecting cases would be as follows: (1) triple negative, (2) HR+/ERBB2−, and (3) ERBB2+ (HR+/ERBB2+ and ER−/PR–/ERBB2+ grouped together). All unique cases of female infiltrating ductal carcinomas were tabulated according to biomarker status (site-specific factor 16) and by race/ethnicity (Table 3). Although a large portion of cases were missing data, this analysis revealed that, in this cohort, African American women have a higher incidence of triple-negative breast cancer compared with white women, and Hispanic women have a higher incidence of HR-negative breast cancer compared with non-Hispanic women. These data, as well as data on breast cancer cases not included in the TMA, were incorporated into the BMaP-3 PAD web-based database.
Table 2.
Characteristics of Patients at the 6 Contributing Institutions (N = 8,279)
| Variable | No. of Patients, n (%) |
|---|---|
| Sex | |
| Male | 59 (0.72) |
| Female | 8,219 (99.30) |
| Race | |
| American Indian | 2 (0.02) |
| Asian Indian | 1 (0.01) |
| Asian Other | 1 (0.01) |
| Black/African American | 2,012 (24.30) |
| Filipino | 2 (0.02) |
| Pacific Islander | 3 (0.04) |
| Vietnamese | 3 (0.04) |
| White | 5,904 (71.30) |
| Other | 80 (0.97) |
| Unknown | 271 (3.30) |
| Ethnicity | |
| Cuban | 43 (0.52) |
| Dominican Republic | 7 (0.08) |
| Mexican | 24 (0.29) |
| Puerto Rican | 248 (3.00) |
| South/Central American | 66 (0.80) |
| Spanish (not otherwise specified) | 368 (4.40) |
| Other Spanish | 9 (0.12) |
| Non-Spanish | 7,370 (89.00) |
| Spanish surname only | 2 (0.02) |
| Unknown | 142 (1.70) |
| Age, y | |
| < 25 | 23 (0.28) |
| 25–34 | 222 (2.70) |
| 35–44 | 1,126 (13.60) |
| 45–54 | 2,217 (26.80) |
| 55–64 | 2,267 (27.40) |
| 65–74 | 1,588 (19.20) |
| ≥ 75 | 836 (10.10) |
Table 3.
Site-Specific Factor 16 for Infiltrating Ductal Carcinoma by Race and Ethnicity for Women in All Institutions Combined
| HR+/ERBB2− | HR+/ERBB2+ |
PR−/ER− /ERBB2− |
PR−/ER− /ERBB2+ |
Missing | Totals | ||
|---|---|---|---|---|---|---|---|
| Race, n (%) | |||||||
| African American |
169 (15) | 58 (5) | 130 (11) | 33 (3) | 755 (66) | 1,145 | |
| White | 585 (18) | 91 (3) | 163 (5) | 43 (2) | 2,295 (72) | 3,177 | |
| Other/missing | 99 (36) | 15 (6) | 58 (21) | 26 (9) | 77 (28) | 275 | |
| Total | 853 (19) | 164 (3) | 351 (8) | 102 (2) | 3,127 (68) | 4,597 | |
| Ethnicity, n (%) | |||||||
| Hispanic | 128 (25) | 21 (4) | 97 (19) | 28 (6) | 234 (46) | 508 | |
| Non-Hispanic | 711 (18) | 139 (3) | 251 (6) | 73 (2) | 2,835 (71) | 4,009 | |
| Other/missing | 14 (17) | 4 (5) | 3 4) | 1 (1) | 58 (73) | 80 | |
| Total | 853 (19) | 164 (3) | 351 (8) | 102 (2) | 3,127 (68) | 4,597 | |
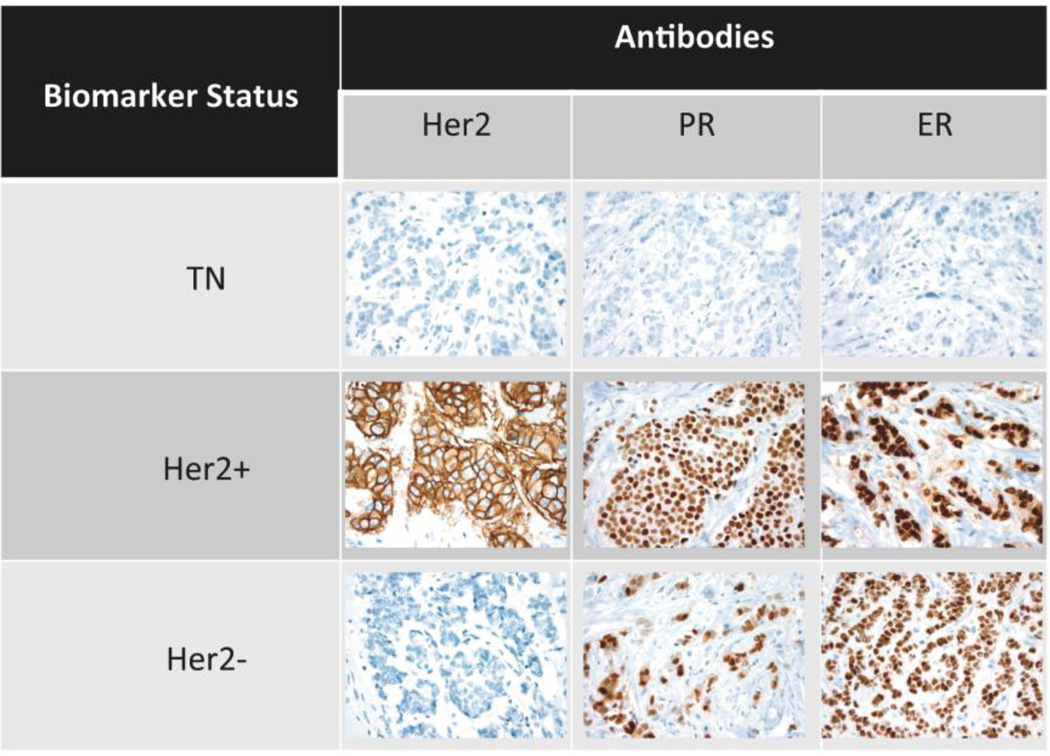
FFPE blocks were requested from participating institutions, and 427 blocks from 427 unique patients with breast cancer were obtained. Tumor expressions of ER, PR, and ERBB2 were annotated based on cancer registry data in the following manner: (1) HR+ (ER+, PR+, or both) and ERBB2−, (2) HR+ (ER+, PR+, or both) and ERBB2+, and (3) HR− (ER− and PR−) and ERBB2−. These 427 FFPEs were used to construct the TMA blocks, of which 8 blocks were constructed. TMA slides from each of the blocks were stained for ER, PR, and ERBB2 to confirm the biomarker status of the samples. Fig 2 shows a sample representation of tumors from each of the 3 types of breast TMAs (ERBB2+, ERBB2−, and triple negative) for each stain. Thereafter, the cancer registry data were reviewed, and 4 samples originally classified as triple negative were reclassified as either ERBB2+ or HR+.
Fig 2. Sample representation of immunostaining according to biomarker status.
TMA slides were immunostained with anti–estrogen receptor, anti–progesterone receptor, and anti–erb-b2 receptor tyrosine kinase 2 antibodies. TMA = tissue microarray.
Several discrepancies were found between the cancer registry data and the TMA immunohistochemistry results. Specifically, for all racial/ethnic groups, the biomarker status obtained from the cancer registry data was 100% in concordance to the staining results for ERBB2− cases. However, this was not true for the triple-negative cases (86.8% and 85.7% rates of accuracy for African Americans/whites and Hispanics, respectively), and concordance was much lower in the case of ERBB2+ staining (43.6% and 30.6% for African Americans/whites and Hispanics, respectively). This finding could be due to different factors such as tumor heterogeneity or loss of antigenicity (eg, some donor blocks were several years old).15 These stains were scanned and quantified, and links to the images were made available upon request. Detailed information on the samples included on the TMA is posted on http://labpages.moffitt.org/bmap3/Cancer%20Database/Cancer_Database.html.
Discussion
This pilot study successfully accomplished the following goals:
Creation of a regional infrastructure with an organizing framework for communications, processes for memorandums of understanding and Institutional Review Board approval from multiple institutions, and an established Tissue Advisory Board and related guidelines
Processes for retrieval and synchronization of data from multiple cancer registries into a single dataset
Development of a collaborative model for optimizing TMA design considerations
Development and implementation of a web-based database of cancer registry cases
All of these accomplishments demonstrate the overall feasibility of creating a unique, centralized, public resource for multiethnic biobanking and the collection of biospecimens. Given the large number of cases included in the web-based database and the amount of TMAs, this model demonstrates that no institution could have accomplished these outcomes alone. A primary benefit of this approach is that members of all the partner institutions are eligible to access the data and tissue resources regardless of their capabilities to contribute to the database or TMA.
The first step in this pilot study was to select a tumor type for creation of the TMAs. We chose to focus on breast cancer as the highest priority to address health disparities related to cancer in the southeastern United States and Puerto Rico for a variety of reasons.16 This decision was based on1,10,13,14,20–22:
Analysis of the literature on TMAs from patients with different racial/ethnic backgrounds showing a scarcity of breast TMAs from diverse populations
Lack of commercially available breast TMAs that provide associated data on race and ethnicity
Observed incidence and mortality rates associated with breast cancer at each of the 6 contributing cancer sites by race/ethnicity
Data from the comprehensive assessment tool on FFPE biospecimens for 6 cancer types available for study at the 9 study institutions
Survey results of researcher needs for TMAs
This pilot study demonstrated the feasibility of collecting cancer registry data for more than 12,000 breast cancer cases and creating a web-based database. We were successful in creating a racially/ethnically diverse TMA from FFPEs representing 427 breast cancer cases. For validation of the TMAs, we focused on studying the characteristics of infiltrating ductal carcinoma among different racial/ethnic groups because the investigators from the BMaP-3 institutions identified these as an important area of investigation, given the known differences in incidence of triple-negative ductal carcinomas among diverse populations.
The results of the biomarker analysis showed a higher incidence rate of triple-negative breast cancer among African American women compared with white women and a higher incidence rate of HR-negative breast cancer among Hispanic women compared with non-Hispanic women. These observations are consistent with the literature.10,14 However, our analysis is compromised by a large amount of missing data. Recent work from Hines et al14 showed that breast cancer among Hispanic women comprises a distinct spectrum of tumor subtypes compared with non-Hispanic white women. In that study, a diverse TMA based on biomarkers was created, allowing researchers to study potential molecular differences in triple-negative breast cancer in different racial/ethnic groups.14
Although most institutions had compatible bioinformatics systems for the management of biospecimen data, these systems were used for fresh frozen samples. No single institution had a direct database for associated FFPE tissue data; thus, cancer registry data and pathology reports were the sources of data for cases likely to have tissues stored at the respective institution. When needed, data would have had to be abstracted from pathology reports — which was beyond the resources of this project. Our findings suggest that the idea of synchronizing pathology reports with cancer registry data should be pursued because pathology reports offer more complete and accurate information on biomarker data than is currently provided by a cancer registry.
Conclusions
We have created a model for collaboration. Through the contribution of FFPE blocks from the BMaP-3 institutions, this project facilitated the creation of a robust research tool that would not be readily available if each institution worked independently. Moreover, each institution benefited from the collaboration and from the team approach to tackle a research/clinical issue common to all team members. It is expected that investigators at the partner institutions will utilize the web-based database on an ongoing basis for data analysis. This database is freely available with open access for the BMaP-3 investigators. We fully expect that the model and process for the work described herein could be applied and exported to other multi-institutional networks focused on a participatory approach to biobanking.
We also demonstrated the feasibility of creating TMAs from racial and ethnically diverse populations through the BMaP-3 partnership, and this project can serve as a model for collaborations of multiple institutions. To date, these TMAs of infiltrating ductal breast carcinoma have been released for 5 projects at 5 different institutions.
Lead line: The Minority Biospecimen/Biobanking Geographic Management Program for region 3 successfully created a race and ethnicity diverse database with 12,408 entries and constructed a tissue microarray representing 427 patients with breast cancer (147 African Americans, 168 Hispanics, and 112 non-Hispanic whites).
Acknowledgments
This work was supported in part by the Tissue Core Facility and Bioinformatics and Biostatistics Core at Moffitt Cancer Center, a National Cancer Institute–designated Comprehensive Cancer Center (P30-CA076292). Substantial support was provided by Region 3 GMaP/BMaP partner institutions: Emory Winship Cancer Center, Moffitt Cancer Center, Morehouse School of Medicine, Ponce Health Sciences University, Tulane University, Tuskegee University, University of Alabama at Birmingham, University of Mississippi Medical Center, and Xavier University of Louisiana. This work was supported in part by National Cancer Institute grants 3U56 CA118809-14S2; 3U54 CA153509-02S3 and 3U54 CA153509-03S1 (Moffitt Cancer Center).
The authors would like to thank Ivana Sehovic and Zena Sayegh for managing and constructing the BMaP-3 TMA blocks. Marithea Goberville, PhD, provided editorial support.
Footnotes
The Web-based database is maintained at https://apps.mathbiol.org/bmap (access restricted). The list of data elements captured is maintained in the public domain at https://apps.mathbiol.org/bmap/data_descriptions/pub.
References
- 1.Lisovicz N, Johnson RE, Higginbotham J, et al. The Deep South Network for cancer control. Building a community infrastructure to reduce cancer health disparities. Cancer. 2006;107(8 suppl):1971–1979. doi: 10.1002/cncr.22151. [DOI] [PubMed] [Google Scholar]
- 2.Ooi SL, Martinez ME, Li CI. Disparities in breast cancer characteristics and outcomes by race/ethnicity. Breast Cancer Res Treat. 2011;127(3):729–738. doi: 10.1007/s10549-010-1191-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Partridge EE, Fouad MN, Hinton AW, et al. The deep South network for cancer control: eliminating cancer disparities through community-academic collaboration. Fam Community Health. 2005;28(1):6–19. doi: 10.1097/00003727-200501000-00004. [DOI] [PubMed] [Google Scholar]
- 4.Ward E, Jemal A, Cokkinides V, et al. Cancer disparities by race/ethnicity and socioeconomic status. CA Cancer J Clin. 2004;54(2):78–93. doi: 10.3322/canjclin.54.2.78. [DOI] [PubMed] [Google Scholar]
- 5.Ademuyiwa FO, Edge SB, Erwin DO, et al. Breast cancer racial disparities: unanswered questions. Cancer Res. 2011;71(3):640–444. doi: 10.1158/0008-5472.CAN-10-3021. [DOI] [PubMed] [Google Scholar]
- 6.Evens AM, Antillón M, Aschebrook-Kilfoy B, et al. Racial disparities in Hodgkin’s lymphoma: a comprehensive population-based analysis. Ann Oncol. 2012;23(8):2128–2137. doi: 10.1093/annonc/mdr578. [DOI] [PubMed] [Google Scholar]
- 7.Fedewa SA, Ward EM, Stewart AK, et al. Delays in adjuvant chemotherapy treatment among patients with breast cancer are more likely in African American and Hispanic populations: a national cohort study 2004–2006. J Clin Oncol. 2010;28(27):4135–4141. doi: 10.1200/JCO.2009.27.2427. [DOI] [PubMed] [Google Scholar]
- 8.Warner ET, Tamimi RM, Hughes ME, et al. Racial and ethnic differences in breast cancer survival: mediating effect of tumor characteristics and sociodemographic and treatment factors. J Clin Oncol. 2015;33(20):2254–2261. doi: 10.1200/JCO.2014.57.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tian Q, Price ND, Hood L. Systems cancer medicine: towards realization of predictive, preventive, personalized and participatory (P4) medicine. J Intern Med. 2012;271(2):111–121. doi: 10.1111/j.1365-2796.2011.02498.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carey LA, Perou CM, Livasy CA, et al. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. JAMA. 2006;295(21):2492–2502. doi: 10.1001/jama.295.21.2492. [DOI] [PubMed] [Google Scholar]
- 11.Kohler BA, Sherman RL, Howlader N, et al. Annual Report to the Nation on the Status of Cancer, 1975–2011, Featuring Incidence of Breast Cancer Subtypes by Race/Ethnicity, Poverty, and State. J Natl Cancer Inst. 2015;107(6):djv048. doi: 10.1093/jnci/djv048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.U.S. Department of Health and Human Services. National Institutes of Health; National Cancer Institute. [Accessed March 24, 2016];Surveillance, Epidemiology, and End Results (SEER) 2010 http://seer.cancer.gov/
- 13.McBride R, Hershman D, Tsai WY, et al. Within-stage racial differences in tumor size and number of positive lymph nodes in women with breast cancer. Cancer. 2007;110(6):1201–1208. doi: 10.1002/cncr.22884. [DOI] [PubMed] [Google Scholar]
- 14.Hines LM, Risendal B, Byers T, et al. Ethnic disparities in breast tumor phenotypic subtypes in Hispanic and non-Hispanic white women. J Womens Health (Larchmt) 2011;20(10):1543–1550. doi: 10.1089/jwh.2010.2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.National Cancer Institute. Geographical Management of Cancer Health Dispartieis (GMaP) [Accessed August 19, 2016]; Updated August 1, 2016. http://www.cancer.gov/about-nci/organization/crchd/inp/gmap.
- 16.Wells KJ, Lima DS, Meade CD, et al. Assessing needs and assets for building a regional network infrastructure to reduce cancer related health disparities. Eval Program Plann. 2014;44:14–25. doi: 10.1016/j.evalprogplan.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Allred DC, Carlson RW, Berry DA, et al. NCCN task force report: estrogen receptor and progesterone receptor testing in breast cancer by immunohistochemistry. J Natl Compr Canc Netw. 2009;(7 suppl 6):S1–S21. doi: 10.6004/jnccn.2009.0079. [DOI] [PubMed] [Google Scholar]
- 18.Wolff AC, Hammond ME, Schwartz JN, et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. J Clin Oncol. 2007;25(1):118–145. doi: 10.1200/JCO.2006.09.2775. [DOI] [PubMed] [Google Scholar]
- 19.Miller RT, Swanson PE, Wick MR. Fixation and epitope retrieval in diagnostic immunohistochemistry: a concise review with practical considerations. Appl Immunohistochem Mol Morphol. 2000;8(3):228–235. doi: 10.1097/00129039-200009000-00010. [DOI] [PubMed] [Google Scholar]
- 20.Wu Y, Sarkissyan M, Elshimali Y, et al. Triple negative breast tumors in African-American and Hispanic/Latina women are high in CD44+, low in CD24+, and have loss of PTEN. PLoS One. 2013;8(10):e78259. doi: 10.1371/journal.pone.0078259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lindner R, Sullivan C, Offor O, et al. Molecular phenotypes in triple negative breast cancer from African American patients suggest targets for therapy. PLoS One. 2013;8(11):e71915. doi: 10.1371/journal.pone.0071915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Winter JL, Stackhouse BL, Russell GB, et al. Measurement of PTEN expression using tissue microarrays to determine a race-specific prognostic marker in breast cancer. Arch Pathol Lab Med. 2007;131(5):767–772. doi: 10.5858/2007-131-767-MOPEUT. [DOI] [PubMed] [Google Scholar]