Abstract
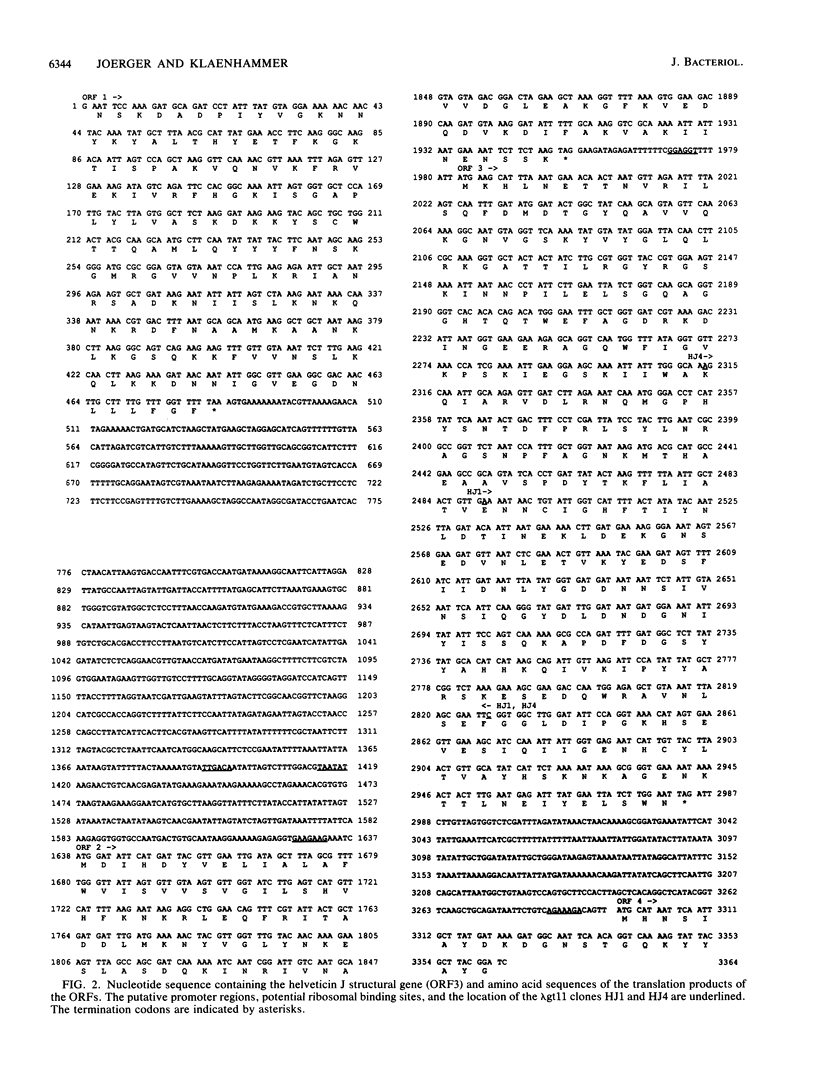
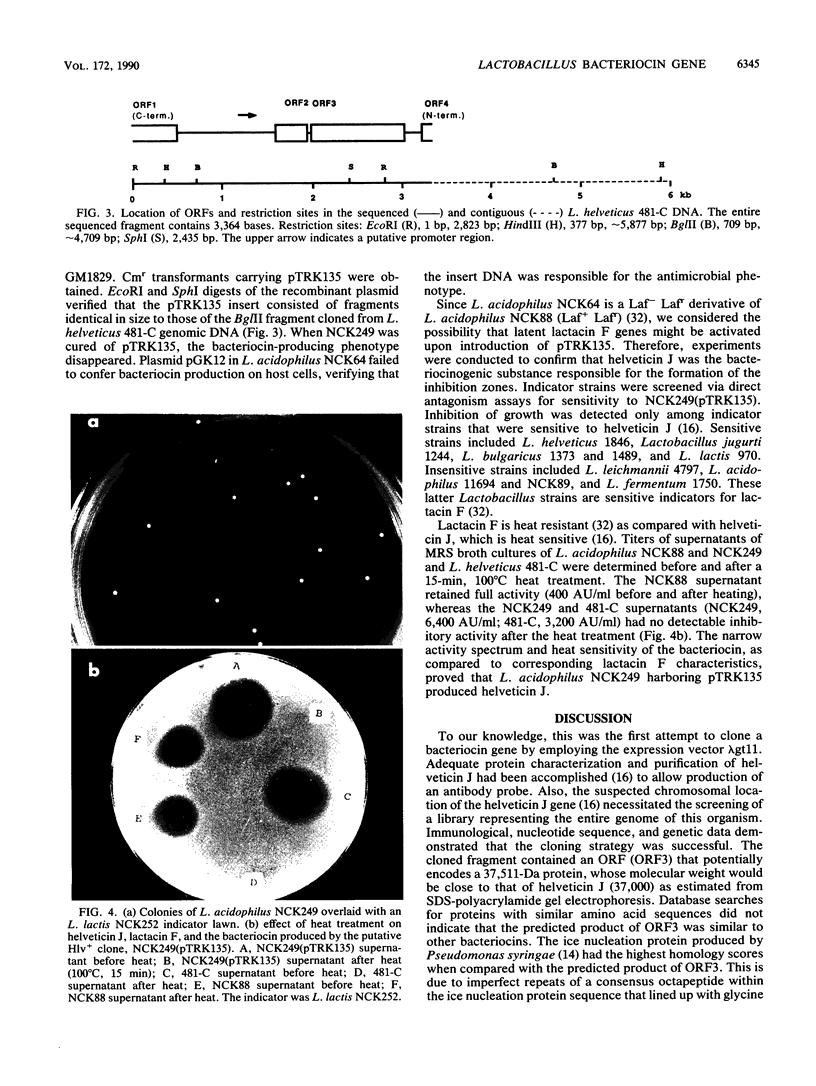
Lactobacillus helveticus 481 produces a 37-kDa bacteriocin called helveticin J. Libraries of chromosomal DNA from L. helveticus were prepared in lambda gt11 and probed for phage-producing fusion proteins that could react with polyclonal helveticin J antibody. Two recombinant phage, HJ1 and HJ4, containing homologous inserts of 350 and 600 bp, respectively, produced proteins that reacted with antibody. These two phage clones specifically hybridized to L. helveticus 481 total genomic DNA but not to DNA from strains that did not produce helveticin J or strains producing unrelated bacteriocins. HJ1 and HJ4 lysogens produced beta-galactosidase fusion proteins that shared similar epitopes with each other and helveticin J. The intact helveticin J gene (hlv) was isolated by screening a library of L. helveticus chromosomal DNA in lambda EMBL3 with the insert DNA from phage HJ4 as a probe. The DNA sequence of a contiguous 3,364-bp region was determined. Two complete open reading frames (ORF), designated ORF2 and ORF3, were identified within the sequenced fragment. The 3' end of another open reading frame, ORF1, was located upstream of ORF2. A noncoding region and a putative promoter were located between ORF1 and ORF2. ORF2 could encode an 11,808-Da protein. The L. helveticus DNA inserts of the HJ1 and HJ4 clones reside within ORF3, which begins 30 bp downstream from the termination codon of ORF2. ORF3 could encode a 37,511-Da protein. Downstream from ORF3, the 5' end of another ORF (ORF4) was found. A Bg/II fragment containing ORF2 and ORF3 was cloned into pGK12, and the recombinant plasmid, pTRK135, was transformed into Lactobacillus acidophilus via electroporation. Transformants carrying pTRK135 produced a bacteriocin that was heat labile and exhibited an acitivity spectrum that was the same as that of helveticin J.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson D. G., McKay L. L. Simple and rapid method for isolating large plasmid DNA from lactic streptococci. Appl Environ Microbiol. 1983 Sep;46(3):549–552. doi: 10.1128/aem.46.3.549-552.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barefoot S. F., Klaenhammer T. R. Detection and activity of lactacin B, a bacteriocin produced by Lactobacillus acidophilus. Appl Environ Microbiol. 1983 Jun;45(6):1808–1815. doi: 10.1128/aem.45.6.1808-1815.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchman G. W., Banerjee S., Hansen J. N. Structure, expression, and evolution of a gene encoding the precursor of nisin, a small protein antibiotic. J Biol Chem. 1988 Nov 5;263(31):16260–16266. [PubMed] [Google Scholar]
- Burnette W. N. "Western blotting": electrophoretic transfer of proteins from sodium dodecyl sulfate--polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radioiodinated protein A. Anal Biochem. 1981 Apr;112(2):195–203. doi: 10.1016/0003-2697(81)90281-5. [DOI] [PubMed] [Google Scholar]
- Conrad B., Mount D. W. Microcomputer programs for DNA sequence analysis. Nucleic Acids Res. 1982 Jan 11;10(1):31–38. doi: 10.1093/nar/10.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig R. J., Arraj J. A., Marinus M. G. Induction of damage inducible (SOS) repair in dam mutants of Escherichia coli exposed to 2-aminopurine. Mol Gen Genet. 1984;194(3):539–540. doi: 10.1007/BF00425572. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dower W. J., Miller J. F., Ragsdale C. W. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 1988 Jul 11;16(13):6127–6145. doi: 10.1093/nar/16.13.6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fickett J. W. Recognition of protein coding regions in DNA sequences. Nucleic Acids Res. 1982 Sep 11;10(17):5303–5318. doi: 10.1093/nar/10.17.5303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnier T., Cole S. T. Characterization of a bacteriocinogenic plasmid from Clostridium perfringens and molecular genetic analysis of the bacteriocin-encoding gene. J Bacteriol. 1986 Dec;168(3):1189–1196. doi: 10.1128/jb.168.3.1189-1196.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold L., Pribnow D., Schneider T., Shinedling S., Singer B. S., Stormo G. Translational initiation in prokaryotes. Annu Rev Microbiol. 1981;35:365–403. doi: 10.1146/annurev.mi.35.100181.002053. [DOI] [PubMed] [Google Scholar]
- Harmon K. S., McKay L. L. Restriction enzyme analysis of lactose and bacteriocin plasmids from Streptococcus lactis subsp. diacetylactis WM4 and cloning of BclI fragments coding for bacteriocin production. Appl Environ Microbiol. 1987 May;53(5):1171–1174. doi: 10.1128/aem.53.5.1171-1174.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joerger M. C., Klaenhammer T. R. Characterization and purification of helveticin J and evidence for a chromosomally determined bacteriocin produced by Lactobacillus helveticus 481. J Bacteriol. 1986 Aug;167(2):439–446. doi: 10.1128/jb.167.2.439-446.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston R. E., Mackenzie J. M., Jr, Dougherty W. G. Assembly of overlapping DNA sequences by a program written in BASIC for 64K CP/M and MS-DOS IBM-compatible microcomputers. Nucleic Acids Res. 1986 Jan 10;14(1):517–527. doi: 10.1093/nar/14.1.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaletta C., Entian K. D. Nisin, a peptide antibiotic: cloning and sequencing of the nisA gene and posttranslational processing of its peptide product. J Bacteriol. 1989 Mar;171(3):1597–1601. doi: 10.1128/jb.171.3.1597-1601.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaenhammer T. R., McKay L. L., Baldwin K. A. Improved lysis of group N streptococci for isolation and rapid characterization of plasmid deoxyribonucleic acid. Appl Environ Microbiol. 1978 Mar;35(3):592–600. doi: 10.1128/aem.35.3.592-600.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleeman E. G., Klaenhammer T. R. Adherence of Lactobacillus species to human fetal intestinal cells. J Dairy Sci. 1982 Nov;65(11):2063–2069. doi: 10.3168/jds.S0022-0302(82)82462-4. [DOI] [PubMed] [Google Scholar]
- Kok J., van der Vossen J. M., Venema G. Construction of plasmid cloning vectors for lactic streptococci which also replicate in Bacillus subtilis and Escherichia coli. Appl Environ Microbiol. 1984 Oct;48(4):726–731. doi: 10.1128/aem.48.4.726-731.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kékessy D. A., Piguet J. D. New method for detecting bacteriocin production. Appl Microbiol. 1970 Aug;20(2):282–283. doi: 10.1128/am.20.2.282-283.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Luchansky J. B., Muriana P. M., Klaenhammer T. R. Application of electroporation for transfer of plasmid DNA to Lactobacillus, Lactococcus, Leuconostoc, Listeria, Pediococcus, Bacillus, Staphylococcus, Enterococcus and Propionibacterium. Mol Microbiol. 1988 Sep;2(5):637–646. doi: 10.1111/j.1365-2958.1988.tb00072.x. [DOI] [PubMed] [Google Scholar]
- Manfioletti G., Schneider C. A new and fast method for preparing high quality lambda DNA suitable for sequencing. Nucleic Acids Res. 1988 Apr 11;16(7):2873–2884. doi: 10.1093/nar/16.7.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J., Gronenborn B., Müller-Hill B., Hans Hopschneider P. Filamentous coliphage M13 as a cloning vehicle: insertion of a HindII fragment of the lac regulatory region in M13 replicative form in vitro. Proc Natl Acad Sci U S A. 1977 Sep;74(9):3642–3646. doi: 10.1073/pnas.74.9.3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mount D. W., Conrad B. Microcomputer programs for graphic analysis of nucleic acid and protein sequences. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 2):811–817. doi: 10.1093/nar/12.1part2.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muriana P. M., Klaenhammer T. R. Conjugal Transfer of Plasmid-Encoded Determinants for Bacteriocin Production and Immunity in Lactobacillus acidophilus 88. Appl Environ Microbiol. 1987 Mar;53(3):553–560. doi: 10.1128/aem.53.3.553-560.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray N. E., Brammar W. J., Murray K. Lambdoid phages that simplify the recovery of in vitro recombinants. Mol Gen Genet. 1977 Jan 7;150(1):53–61. doi: 10.1007/BF02425325. [DOI] [PubMed] [Google Scholar]
- Oppenheimer C. L., Eckhardt A. E., Hill R. L. The nonidentity of porcine N-acetylglucosaminyltransferases I and II. J Biol Chem. 1981 Nov 25;256(22):11477–11482. [PubMed] [Google Scholar]
- Pugsley A. P. The ins and outs of colicins. Part I: Production, and translocation across membranes. Microbiol Sci. 1984 Oct;1(7):168–175. [PubMed] [Google Scholar]
- Rosenberg M., Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramm E., Mende J., Braun V., Kamp R. M. Nucleotide sequence of the colicin B activity gene cba: consensus pentapeptide among TonB-dependent colicins and receptors. J Bacteriol. 1987 Jul;169(7):3350–3357. doi: 10.1128/jb.169.7.3350-3357.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Tagg J. R., Dajani A. S., Wannamaker L. W. Bacteriocins of gram-positive bacteria. Bacteriol Rev. 1976 Sep;40(3):722–756. doi: 10.1128/br.40.3.722-756.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upreti G. C., Hinsdill R. D. Production and mode of action of lactocin 27: bacteriocin from a homofermentative Lactobacillus. Antimicrob Agents Chemother. 1975 Feb;7(2):139–145. doi: 10.1128/aac.7.2.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson M. E. Compilation of published signal sequences. Nucleic Acids Res. 1984 Jul 11;12(13):5145–5164. doi: 10.1093/nar/12.13.5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilbur W. J., Lipman D. J. Rapid similarity searches of nucleic acid and protein data banks. Proc Natl Acad Sci U S A. 1983 Feb;80(3):726–730. doi: 10.1073/pnas.80.3.726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada M., Ebina Y., Miyata T., Nakazawa T., Nakazawa A. Nucleotide sequence of the structural gene for colicin E1 and predicted structure of the protein. Proc Natl Acad Sci U S A. 1982 May;79(9):2827–2831. doi: 10.1073/pnas.79.9.2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- van Belkum M. J., Hayema B. J., Geis A., Kok J., Venema G. Cloning of two bacteriocin genes from a lactococcal bacteriocin plasmid. Appl Environ Microbiol. 1989 May;55(5):1187–1191. doi: 10.1128/aem.55.5.1187-1191.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. Signal sequences. The limits of variation. J Mol Biol. 1985 Jul 5;184(1):99–105. doi: 10.1016/0022-2836(85)90046-4. [DOI] [PubMed] [Google Scholar]