Abstract
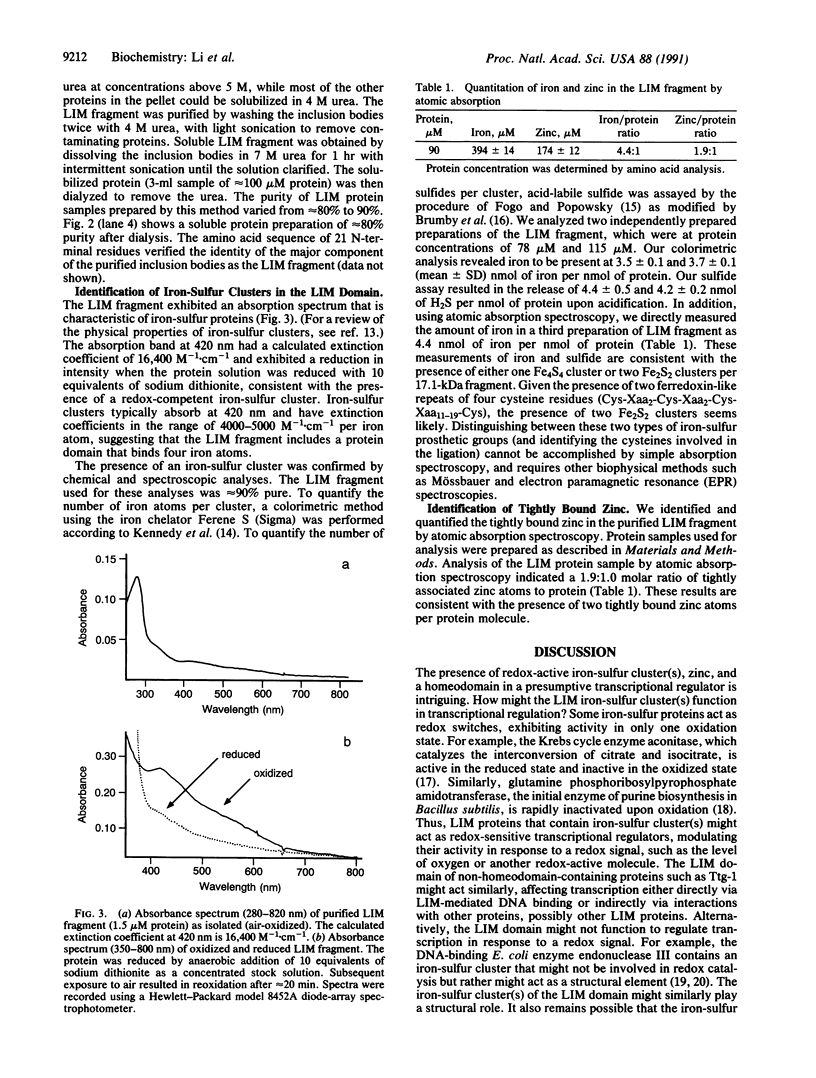
The cysteine-rich LIM motif is highly conserved between invertebrates and mammals. This motif shows similarity both to proteins that bind zinc and to ferredoxins, which contain iron-sulfur clusters. Two tandem copies of the LIM motif are found in a number of presumptive transcription factors, including the protein product of the Caenorhabditis elegans cell-lineage gene lin-11. To investigate the possible metal-binding properties of the LIM region of the lin-11 protein, we expressed and purified a 151-amino acid peptide containing the tandem LIM motifs. The purified peptide binds both zinc (two atoms per protein molecule) and iron (as a redox-active iron-sulfur cluster, with four atoms of iron and four atoms of inorganic sulfide per protein molecule). These observations suggest that the LIM motif is a metallodomain that might function in a redox-sensitive regulation of transcription.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Asahara H., Wistort P. M., Bank J. F., Bakerian R. H., Cunningham R. P. Purification and characterization of Escherichia coli endonuclease III from the cloned nth gene. Biochemistry. 1989 May 16;28(10):4444–4449. doi: 10.1021/bi00436a048. [DOI] [PubMed] [Google Scholar]
- BRUMBY P. E., MILLER R. W., MASSEY V. THE CONTENT AND POSSIBLE CATALYTIC SIGNIFICANCE OF LABILE SULFIDE IN SOME METALLOFLAVOPROTEINS. J Biol Chem. 1965 May;240:2222–2228. [PubMed] [Google Scholar]
- Bernlohr D. A., Switzer R. L. Reaction of Bacillus subtilis glutamine phosphoribosylpyrophosphate amidotransferase with oxygen: chemistry and regulation by ligands. Biochemistry. 1981 Sep 29;20(20):5675–5681. doi: 10.1021/bi00523a006. [DOI] [PubMed] [Google Scholar]
- Boehm T., Foroni L., Kaneko Y., Perutz M. F., Rabbitts T. H. The rhombotin family of cysteine-rich LIM-domain oncogenes: distinct members are involved in T-cell translocations to human chromosomes 11p15 and 11p13. Proc Natl Acad Sci U S A. 1991 May 15;88(10):4367–4371. doi: 10.1073/pnas.88.10.4367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm T., Greenberg J. M., Buluwela L., Lavenir I., Forster A., Rabbitts T. H. An unusual structure of a putative T cell oncogene which allows production of similar proteins from distinct mRNAs. EMBO J. 1990 Mar;9(3):857–868. doi: 10.1002/j.1460-2075.1990.tb08183.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bopp D., Burri M., Baumgartner S., Frigerio G., Noll M. Conservation of a large protein domain in the segmentation gene paired and in functionally related genes of Drosophila. Cell. 1986 Dec 26;47(6):1033–1040. doi: 10.1016/0092-8674(86)90818-4. [DOI] [PubMed] [Google Scholar]
- Cunningham R. P., Asahara H., Bank J. F., Scholes C. P., Salerno J. C., Surerus K., Münck E., McCracken J., Peisach J., Emptage M. H. Endonuclease III is an iron-sulfur protein. Biochemistry. 1989 May 16;28(10):4450–4455. doi: 10.1021/bi00436a049. [DOI] [PubMed] [Google Scholar]
- Edman P., Begg G. A protein sequenator. Eur J Biochem. 1967 Mar;1(1):80–91. doi: 10.1007/978-3-662-25813-2_14. [DOI] [PubMed] [Google Scholar]
- Freyd G., Kim S. K., Horvitz H. R. Novel cysteine-rich motif and homeodomain in the product of the Caenorhabditis elegans cell lineage gene lin-11. Nature. 1990 Apr 26;344(6269):876–879. doi: 10.1038/344876a0. [DOI] [PubMed] [Google Scholar]
- Greenberg J. M., Boehm T., Sofroniew M. V., Keynes R. J., Barton S. C., Norris M. L., Surani M. A., Spillantini M. G., Rabbitts T. H. Segmental and developmental regulation of a presumptive T-cell oncogene in the central nervous system. Nature. 1990 Mar 8;344(6262):158–160. doi: 10.1038/344158a0. [DOI] [PubMed] [Google Scholar]
- Herr W., Sturm R. A., Clerc R. G., Corcoran L. M., Baltimore D., Sharp P. A., Ingraham H. A., Rosenfeld M. G., Finney M., Ruvkun G. The POU domain: a large conserved region in the mammalian pit-1, oct-1, oct-2, and Caenorhabditis elegans unc-86 gene products. Genes Dev. 1988 Dec;2(12A):1513–1516. doi: 10.1101/gad.2.12a.1513. [DOI] [PubMed] [Google Scholar]
- Ingraham H. A., Chen R. P., Mangalam H. J., Elsholtz H. P., Flynn S. E., Lin C. R., Simmons D. M., Swanson L., Rosenfeld M. G. A tissue-specific transcription factor containing a homeodomain specifies a pituitary phenotype. Cell. 1988 Nov 4;55(3):519–529. doi: 10.1016/0092-8674(88)90038-4. [DOI] [PubMed] [Google Scholar]
- Karlsson O., Thor S., Norberg T., Ohlsson H., Edlund T. Insulin gene enhancer binding protein Isl-1 is a member of a novel class of proteins containing both a homeo- and a Cys-His domain. Nature. 1990 Apr 26;344(6269):879–882. doi: 10.1038/344879a0. [DOI] [PubMed] [Google Scholar]
- Kennedy M. C., Kent T. A., Emptage M., Merkle H., Beinert H., Münck E. Evidence for the formation of a linear [3Fe-4S] cluster in partially unfolded aconitase. J Biol Chem. 1984 Dec 10;259(23):14463–14471. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- MacFerrin K. D., Terranova M. P., Schreiber S. L., Verdine G. L. Overproduction and dissection of proteins by the expression-cassette polymerase chain reaction. Proc Natl Acad Sci U S A. 1990 Mar;87(5):1937–1941. doi: 10.1073/pnas.87.5.1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire E. A., Hockett R. D., Pollock K. M., Bartholdi M. F., O'Brien S. J., Korsmeyer S. J. The t(11;14)(p15;q11) in a T-cell acute lymphoblastic leukemia cell line activates multiple transcripts, including Ttg-1, a gene encoding a potential zinc finger protein. Mol Cell Biol. 1989 May;9(5):2124–2132. doi: 10.1128/mcb.9.5.2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabbitts T. H., Boehm T. LIM domains. Nature. 1990 Aug 2;346(6283):418–418. doi: 10.1038/346418a0. [DOI] [PubMed] [Google Scholar]
- Robbins A. H., Stout C. D. Structure of activated aconitase: formation of the [4Fe-4S] cluster in the crystal. Proc Natl Acad Sci U S A. 1989 May;86(10):3639–3643. doi: 10.1073/pnas.86.10.3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruvkun G., Finney M. Regulation of transcription and cell identity by POU domain proteins. Cell. 1991 Feb 8;64(3):475–478. doi: 10.1016/0092-8674(91)90227-p. [DOI] [PubMed] [Google Scholar]
- Scott M. P., Tamkun J. W., Hartzell G. W., 3rd The structure and function of the homeodomain. Biochim Biophys Acta. 1989 Jul 28;989(1):25–48. doi: 10.1016/0304-419x(89)90033-4. [DOI] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treisman J., Harris E., Desplan C. The paired box encodes a second DNA-binding domain in the paired homeo domain protein. Genes Dev. 1991 Apr;5(4):594–604. doi: 10.1101/gad.5.4.594. [DOI] [PubMed] [Google Scholar]
- Vallee B. L., Coleman J. E., Auld D. S. Zinc fingers, zinc clusters, and zinc twists in DNA-binding protein domains. Proc Natl Acad Sci U S A. 1991 Feb 1;88(3):999–1003. doi: 10.1073/pnas.88.3.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Way J. C., Chalfie M. mec-3, a homeobox-containing gene that specifies differentiation of the touch receptor neurons in C. elegans. Cell. 1988 Jul 1;54(1):5–16. doi: 10.1016/0092-8674(88)90174-2. [DOI] [PubMed] [Google Scholar]




