Abstract
Epigenetic regulation in myeloid cells is crucial for cell differentiation and activation in response to developmental and environmental cues. Epigenetic control involves posttranslational modification of DNA or chromatin, and is also coupled to upstream signaling pathways and transcription factors. In this review, we summarize key epigenetic events and how dynamics in the epigenetic landscape of myeloid cells shape the development, immune activation, and innate immune memory.
INTRODUCTION
Epigenetics in the context of cell differentiation and activation refers to mechanisms that regulate and potentially stabilize gene expression in response to developmental and environmental cues. Epigenetic regulation is mediated by posttranslational modification of DNA or chromatin and by noncoding RNAs. In myeloid cells, the predominant focus of research on epigenetic mechanisms has been chromatin-mediated regulation of macrophages, which will be the focus of this review (1–3). Differentiation of macrophages from myeloid precursors is regulated by developmental signals and pioneer transcription factors that impart an “epigenetic landscape” that helps determine macrophage phenotype and how cells respond to environmental challenges. Macrophages protect the host from pathogenic microorganisms and other environmental insults, providing a rapid response as an initial line of defense. Accordingly, recognition of pathogen-associated molecular patterns through germ line-encoded receptors initiates and subsequently amplifies the adaptive immune response through cytokine production and antigen presentation. Importantly, macrophage phenotype is plastic, and macrophages carry out their distinct roles while maintaining the ability to adapt to local or systemic environmental changes (4–7).
Macrophages generate a transcriptional response that is both cell type and stimulus specific, helping the host develop specific innate and adaptive responses to successfully control various infections. In addition, macrophages must coordinate responses to, and have the ability to “remember,” many stimuli, including signaling cues from other cells, the extracellular matrix, hormones, and active components of bacteria or viruses. Based on their repertoire of pattern recognition receptors, receptor-mediated signaling events, or their underlying differentiation phenotype, macrophages exhibit diverse responses to numerous stimuli and also drive rapid and appropriate immune responses. For example, a number of functionally distinct macrophage subsets has been described that can be broadly categorized in terms of ontogeny, homeostatic role, or maturation status (5, 8). Recent studies have highlighted the complex ontogeny of macrophages and dendritic cells and the unexpected complexity of the myeloid system, with highly specialized function and distribution in various tissues (6, 7, 9). Furthermore, during inflammatory processes, macrophages exhibit extensive plasticity of their phenotypes, including polarization and activation, and such characteristics are controlled by both changes at the transcriptional level and an unanticipated degree of epigenetic control (1, 2, 9). Epigenetic regulation is not only coupled with transcription factor-mediated regulation but also linked with upstream signaling pathways that connect external signals to gene function to shape the identity and function of macrophages during differentiation and activation. Chromatin-mediated epigenetic mechanisms also participate in innate memory-like phenomena that can either promote tolerance to a stimulus or prime cells for a more robust response.
In this article, we first discuss myeloid lineage decisions during development, with an emphasis on the emerging role of pioneer transcription factors and distal regulatory elements (enhancers) in regulating cell lineage and cell-type-specific responses in a chromatin-regulated manner. We then discuss how epigenetic regulation affects the response of macrophages to activation and classify stimulus response genes in concert with signaling pathways and transcription factors. Furthermore, we illustrate how chromatin can provide a memory of prior stimuli, and discuss the basis for extensive plasticity in heterogeneous macrophage populations present in different tissues.
EPIGENETIC PRINCIPLES IN MYELOID CELLS
The traditional definition of the term “epigenetics” refers to stably inherited changes in gene expression and phenotype that do not involve changes to the underlying DNA sequence. More recently, within the context of cell differentiation and activation, the term epigenetics is often used to connote mechanisms that stabilize gene expression even after environmental signals that regulate gene expression have resolved. These changes are not necessarily heritable or propagated across cell mitosis, and mechanistically are mediated by histone modifications and other chromatin changes and modification of DNA such a (hydroxyl)methylation (1–3). In this review, epigenetic changes typically refer to stable, chromatin-mediated alterations in the transcriptional potential of a cell.
In myeloid cells, as in all mammalian cells, nuclear DNA is wrapped around histones to form nucleosomes, which are compacted into chromatin. These interactions not only enable the marked compaction of DNA required for packaging in the nucleus but also impose a barrier to transcription. This organization generates a default state of inaccessibility, and hence, the most fundamental issue of epigenetic regulation is how to ensure access of transcription machinery to DNA despite the compact and protective chromatin organization (10). Epigenetic mechanisms are typically mediated by posttranslational modifications of histones and DNA methylation, ATP-dependent chromatin-remodeling complexes, and noncoding RNA, and function at gene regulatory regions such as promoters and enhancers (11–14). The sum total and pattern of epigenetic modifications determine accessibility of DNA, linking epigenetic regulation with transcription.
Another issue is that epigenetic marks have traditionally been considered to be stable and are a suitable mechanism to explain stable phenotypes in various differentiated cell types that express different patterns of gene expression (15). At the cellular level, signaling cascades initiated at the cell surface relay messages via effector proteins, ultimately culminating in the nucleus, where transcription factors are targeted to induce or shut down a particular gene expression signature. This event may be transient in nature, for example, when a cell needs to respond acutely to an event and then return to its previous steady state. However, the development, health, and/or adaptation of an organism often require an environmental signal to be converted into a long-lived phenotypic change. Stabilization of cell phenotypes by epigenetic mechanisms includes changes in chromatin landscapes in response to external stimuli that control gene expression (10, 14, 15). Epigenetic modifications that persist after the original stimulus could provide a mechanism for extending transient, short-lived signals into a more stable and sustained cellular response lasting several hours or days. This epigenetic landscape, in turn, can be reprogrammed in response to subsequent stimulation for activation and polarization. Such reprogramming of the epigenetic landscape helps integrate signaling over time and maintains distinct cell fates to respond rapidly and properly to subsequent stimuli (10, 14, 15).
Investigation of the epigenetics of macrophages to date has focused primarily on posttranslational modification of histones. More than 60 different residues on histone tails are known to be posttranslationally modified by various covalent modifications such as acetylation, methylation, phosphorylation, ubiquitination, sumoylation, ADP-ribosylation, and others, and there are more than 100 possible histone modifications identified to date (16). These histone marks are “written” and “erased” by enzymes that exhibit specificity for particular histone amino acid residues and marks. Some histone marks, such as H4 lysine 16 acetylation (H4K16ac) and H2B ubiquitination (H2Bub), can directly alter nucleosome structure, and these marks generally increase nucleosome mobility along the DNA strand and accessibility of DNA to transcription factors, which correlate with gene expression (17, 18). Other histone marks, such as H3 lysine 4 methylation (H3K4me), contribute to a sort of “code” that is “read” and interpreted by additional chromatin regulators that recognize and bind to these marks and serve as “signaling platforms” for recruitment of additional regulatory proteins (14). DNA sequence-specific transcription factors and interacting coactivators/corepressors function in concert with chromatin regulators to determine the rates of transcription initiation and elongation, and orchestrate changes in the transcriptional program. However, it is often not clear whether these histone modifications play a causal role in regulating transcription or are passive by-products or markers of transcription, although it has been shown that H3K4me3 or histone H3 lysine 27 acetylation (H3K27ac) at promoters directly facilitates RNA polymerase (Pol) II recruitment and polymerase initiation complex formation in vitro in cell-free systems (19, 20).
In addition to histone modifications, chromatin remodelers, such as the SWI/SNF family of proteins in humans, can “open” chromatin through nucleosome sliding, eviction, or breathing where the associated DNA is unwrapped to expose regulatory sites (12). In this way, remodeling of nucleosomes can regulate the accessibility of DNA to transcription factors to control gene expression. Whereas analysis of histone modifications reveals overwhelming complexity and leaves mechanistic questions unaddressed, DNA accessibility provides a simple testable paradigm for understanding the role of nucleosomes in gene regulatory processes (13). Many high-throughput assays have been developed to take advantage of next-generation sequencing technologies in assessing the genome-wide chromatin state of mammalian cells. These include DNase I digestion followed by high-throughput sequencing (DNase-seq), formaldehyde-assisted isolation of regulatory elements (FAIRE-seq), and assay for transposase-accessible chromatin using sequencing (ATAC-seq), which reveal nucleosome-depleted regions known as open chromatin. Chromatin immunoprecipitation using specific antibodies for histone marks or transcription factors followed by sequencing (ChIP-seq) reveals genome-wide patterns of histone marks and transcription factor occupancy, known as the “cistrome” (21). On a finer scale, DNase-seq data can be used to identify binding sites of transcription factors by analyzing the distribution of the signal or “footprint” within a DNase-hypersensitive site (21, 22). ChIP experiments can be used for unbiased profiling of chromatin remodelers, transcription factors, and other proteins that are associated with DNA as well as histone modifications through enriching for bound sequences by means of a specific antibody (21). Taken together, the positioning and modifications of nucleosomes throughout the genome define the chromatin state of a cell, and genome-wide data in macrophages have rapidly accumulated. We discuss the current understanding of epigenetic regulation in macrophages and how these epigenetic principles can be applied to define the identity and function of macrophages during differentiation and activation while the cells remember previous experiences.
EPIGENETICS IN DEVELOPMENT OF MACROPHAGES
Macrophages can be classified by cell surface marker expression, ontogeny, or differential dependence on lineage-defining transcription factors and growth factor signaling, which sharply distinguish specific populations in different tissues. During cellular differentiation, common myeloid precursors undergo a series of progressive choices that eventually change their transcriptional program and chromatin, which play a critical part in the selective activation and repression of genes important in various macrophage homeostatic and stress-related functions (2, 5, 6, 23).
It is thought that expression of transcription factors called pioneer factors or master regulators determines myeloid cell lineage commitment, and the composition and function of those are likely to vary between cell types (24–26). High levels of the lineage-determining transcription factor PU.1 are important for differentiation of macrophages and neutrophils (27, 28). PU.1 instructs progenitors to upregulate myeloid-specific cell surface antigens and to downregulate other cell-specific markers and transcription factors (29). By definition, pioneer factors can bind target DNA sequences even in a silent or native chromatin environment, differing from other transcription factors, which readily bind only to already accessible DNA (30). Indeed, recent genome-wide studies in macrophages and dendritic cells (24, 31–33) indicate that a small set of myeloid lineage factors such as PU.1 and members of the CCAAT/enhancer-binding protein (C/EBP) and activator protein 1 (AP1) families function in a collaborative manner to shape the genome and establish a large fraction of the enhancer-like elements. Their binding to tens of thousands of chromatin sites is established during differentiation, and PU.1 genomic distribution in differentiated macrophages and B cells shows highly distinct patterns (31, 34–36). Accordingly, PU.1 is associated with nearly all genomic enhancers marked by H3K4me1 (31, 35, 36). Moreover, reexpression of PU.1 in PU.1-deficent myeloid precursors, or even in fibroblasts, leads to the local deposition of H3K4me1 and increased accessibility of the underlying DNA at PU.1-bound genomic regions (31, 36). Another example of pioneer factors in myeloid cells is the C/EBP family. C/EBPα can switch B cells into macrophages by increasing accessibility of relevant chromatin (37), and depending on the order of expression, C/EBPα and GATA-2 regulate the differentiation of different types of myeloid cells such as neutrophils and mast cells (26). C/EBPα-mediated conversion of primary pre-B cells into macrophages also results in the downregulation of histone deacetylase 7 (HDAC7), which inhibits the induction of key genes for macrophage function, such as inflammatory response and phagocytosis (38). In addition, natural genetic variation between inbred strains of mice in the binding sites of the PU.1 and C/EBPα impairs not only the binding of pioneer factors but also that of closely bound NF-κB in response to signals, which is consistent with a model in which pioneer factors form enhancers and set up the epigenetic landscape, which then determines the binding profiles of signaling-dependent transcription factors (35).
The enhancer landscape, established by pioneer factors, is unique to a particular cell lineage. The defining characteristic of enhancers is their ability to drive gene expression at a distance (39), and modularity of enhancer elements permits a given gene to be transcribed in multiple tissues, at varied levels, in response to different signaling cascades, and at different states during development. Recent studies of the ENCODE consortium revealed that the great majority of binding sites for transcription factors are in enhancers rather than in promoters, suggesting that enhancers are likely to be specific to the lineage and represent future regulatory potential for the lineage (40–42). Many different enhancer states can be defined by enrichment for specific combinations of histone modifications, low nucleosomal occupancy (open chromatin), co-occupancy by multiple transcription factors, binding of coactivators and RNA Pol II, and production of enhancer RNAs (eRNAs) (43–46). In contrast to promoters, which exhibit relatively high levels of H3K4me3 as compared to H3K4me1 or H3K4me2, enhancers exhibit relatively low H3K4me3 and high H3K4me1 (43–45). H3K27ac in H3K4me1-associated regions separates active from poised enhancers (47), and many transcription coregulators, such as myeloid/lymphoid or mixed-lineage leukemia (MLL) proteins, p300 and CREB-binding protein, the mammalian SWI/SNF complexes, and the Mediator complex, can promote enhancer activity (48–51). Importantly, enhancer landscapes vary between cell types and predict developmental states (52). For example, recent genome-wide studies in macrophages and dendritic cells found that enhancer landscape, defined by mapping histone modifications and a comprehensive panel of myeloid transcription factors, is unique to the cell lineage and is correlated with lineage-specific gene expression (24). In addition, tissue-resident macrophages from mouse have a large repertoire of tissue-specific enhancers, which drive tissue-specific transcriptional and functional responses by inducing the expression of divergent secondary transcription factors that collaborate with PU.1 (53, 54). In peritoneal macrophages, it has been shown that local tissue-derived retinoic acid promotes the peritoneal macrophage phenotype through the reversible induction of GATA-6 (55), implying that the local microenvironment may reprogram the tissue-dependent characteristics of enhancers and transcription factors.
Although many studies identify and/or predict enhancer position and activity using a combination of histone marks, coactivator and Pol II occupancy, and eRNA expression, major challenges are to connect enhancers with their target genes and to validate the physiological function of enhancers. Techniques that measure physical interactions between DNA loci, using variations of chromosome conformation capture (3C), allow inference of DNA looping and have been helpful in linking enhancers with their target genes. A newly developed variant of 3C, termed Hi-C, and chromatin interaction analysis by paired-end tag sequencing (ChIA-PET) allows for genome-wide, unbiased mapping of long-range interactions (56, 57). Most recent resolution of the Hi-C technology is ~0.1 to 1 Mb for mammalian genomes (58), making it possible to map enhancer-promoter interactions genome-wide. It has been shown that chromatin compartments called topologically associated domains (TADs) are largely conserved in their position between cell types and developmental states (59, 60). However, TADs can be further substratified into smaller domains, and inside TADs the chromatin structure of regions <100 kb does differ in a cell-type-specific manner (56), which implies that different enhancer-promoter interactions within TADs can be dynamically regulated in a context-dependent manner. Accordingly, a recent ChIA-PET study in embryonic stem cells revealed that large genomic domains with a high density of enhancer-associated marks, called super-enhancers, exist within TADs and interact with promoters of adjacent genes within the TAD (61). Furthermore, genome editing of putative enhancers mediated by CRISPR (clustered regularly interspaced short palindromic repeats) or TALEN (transcription activator-like effector nuclease) provides insights into the functional importance of these elements (57, 61). Finally, eRNAs have been shown to modulate the levels of nearby gene expression, potentially by facilitating enhancer-promoter interactions through chromatin looping, recruitment of cofactors, the release of the negative elongation factor complex (44, 62), and transcription-associated changes in histone marks. RNA interference-mediated inhibition of eRNAs results in reduced expression of nearby mRNAs, suggesting a direct role of these eRNAs in enhancer function and providing potential tools for validating the physiological function of enhancers (63–65).
Finally, DNA methylation has been shown to play a central role in development and function of myeloid cells. Methylation of cytosines generally silences certain genomic regions by modulating the binding of particular transcription factors to their binding sites or the binding of other factors that display different affinity for methylated cytosines. Terminally differentiated human myeloid cells show a marked hypomethylation pattern compared to lymphoid cells (66). DNAmethylation levels decrease sharply as myeloid differentiation progresses, while they increase upon lymphoid commitment (66, 67). For instance, the in vitro differentiation of monocytes into macrophages or dendritic cells involves active DNA demethylation. Genes encoding transcription factors that are important for myeloid cell differentiation, such as C/EBPα, display low levels of DNA methylation and are enriched in both activating H3K4me3 and repressive H3K27me3 marks, known as bivalent domains (68, 69). Recent studies revealed that ten-eleven translocation (TET) enzymes catalyze the oxidation of 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC) and to further oxidation products, and these oxidized nucleotides can lead to active/passive demethylation in many different ways (70, 71). Enrichment of 5hmC at the transcription start site (TSS) and gene bodies indicates the potential role of such modification in transcriptional control, and TET proteins contribute to histone modification by interacting with chromatin-modifying enzymes. TET proteins can contribute to both activating and repressive effects on gene expression, probably depending on the context and location of binding (72–74). For example, PU.1 binds TET2 and recruits it to the promoters of genes that become demethylated. PU.1 is also able to recruit DNA methyltransferase DNMT3B to target the deposition of de novo DNA methylation (75). Although genome-wide analysis of 5hmC has not yet been described for myeloid cells, it seems that dynamic regulation of the methylation and hydroxymethylation of cytosine at gene-specific loci during differentiation and activation of myeloid cells may contribute to modulation of the immune responses.
EPIGENETICS IN MACROPHAGE ACTIVATION
Macrophages respond to external stimulation by activating hundreds of response genes. Different transcriptional responses of cytokine and inflammatory/antiviral genes define unique macrophage biological functions. In a classic model of stimulus-dependent activation, each stimulus elicits a characteristic transcriptional response solely by activating a unique combination of signaling pathways (76, 77). Although signal transduction pathways and transcription factors are clearly central mediators of transcriptional activation, the possible roles of chromatin must also be considered. For example, nucleosome remodeling, as detected by DNase I hypersensitivity and restriction enzyme accessibility, was initially observed at the promoters of the IL12B and IFNB1 genes (78, 79). Inducible histone modifications relevant to an immune response were first observed by ChIP at the human IFNB1 gene (80). Like most plastic cells of the myeloid system, macrophages are responsive to a wide spectrum of regulatory molecules and provide a robust model system for investigation of chromatin regulation for transcriptional responses at a genome-wide level. Most data in this area have been generated by analyses of the macrophage in response to prototypical inflammatory stimuli such as Toll-like receptor (TLR) agonists.
Epigenetics in Resting States
The resting state of a macrophage is associated with its development history. An epigenetic landscape is established during differentiation, and pioneer factors bind to and open the regulatory regions that are the future binding sites of transcription factors in response to stimuli. For example, PU.1 is associated not only with macrophage-specific enhancers but also with most lipopolysaccharide (LPS)-induced enhancers in both unstimulated and stimulated macrophages (Fig. 1) (36). This finding suggests that epigenetic landscapes for inducible genes are directly targeted by PU.1, rendering the genes they control susceptible to transcriptional induction when the mature cell encounters a stimulus (31). In addition to pioneer factors, H3K4me1 enrichment as well as binding of p300 to inducible gene enhancers was usually observed in both unstimulated and stimulated macrophages (31, 36). Promoters also are marked by basal permissive histone marks such as H3K4me3 and a nucleosome-depleted region upstream of the TSS. There is often basal low-level transcription of TLR-inducible genes, and the promoters of primary response genes are occupied at baseline by Pol II that is paused in the vicinity of the TSS (81, 82). Basal transcription rates can be set by prebound primer factors, including JunB, activating transcription factor 3 (ATF3), and interferon regulatory factor 4 (IRF4), which may also recruit additional factors after cell stimulation (24). The negative histone marks, such as H3K9me3 (83), H3K27me3 (84), and H4K20me3 (85), also are distributed at promoters of inflammatory genes. Histone H3K9me, which generally correlates with transcriptional repression, is readily apparent at the promoters of some but not all inducible genes in unstimulated human dendritic cells (86). In turn, the activation of cells was associated with the rapid loss of H3K9me at these promoters. However, macrophages and dendritic cells display relatively very low levels of H3K9me2 at antiviral and antimicrobial genes as compared to fibroblasts, myocytes, or neurons (83), suggesting a potential explanation of their professional role in immune defense. H3K27me, another repressive histone modification, which acts by recruiting polycomb complexes, has also been identified at a selective subset of inducible genes prior to their activation (84, 87). H3K27me3 is downregulated at immune response genes after stimulation by the action of Jmjd3, a JmjC family histone demethylase, which is required for activation of a subset of TLR4-dependent genes (84). Finally, many corepressors that act prior and/or subsequent to stimulus responses restrain inflammatory cytokines in the basal state. These include occupancy of gene loci by repressors such as B-cell lymphoma 6 (BCL-6) and nuclear receptors that recruit corepressor complexes that contain HDACs and histone demethylases that limit the amount of positive histone marks (88). BCL-6 has been shown to corepress many LPS-inducible genes, such that its loss results in hypersensitivity to proinflammatory signals (88). The Nuclear receptor corepressor (NCoR1)-HDAC3 corepressor contributes to maintaining low H3K9/14 acetylation levels under resting conditions (89) and contains the histone methyltransferase SET and MYND domain-containing protein 5 (SMYD5), which contributes to repression by catalyzing H4K20me3, inhibiting the expression of TLR4 target genes (85). The binding of these corepressors to distinct subsets of target genes in resting cells has the potential to selectively regulate their activation.
FIGURE 1.
Epigenetic features of promoters and enhancers of inflammatory genes under basal and acute activated conditions. During macrophage differentiation, PU.1 binds to promoters and enhancers and facilitates the opening of chromatin and an increase in H3K4me3 at promoters and H3K4me1 at enhancers. The negative histone marks/corepressors also are distributed at both promoters and enhancers. There is often low or nonproductive basal transcription. Inflammatory stimulation of macrophages leads to release and loss of corepressors and negative histone marks, increased histone acetylation, additional nucleosome remodeling by BRG1, and recruitment of signaling transcription factors such as NF-κB. The subsequent recruitment of histone acetyltransferases (HATs) results in histone acetylation, which is subsequently bound by Brd4/P-TEFb complex. This results in successive rounds of Pol II elongation and active transcription. Enhancers of active genes are characterized by occupancy by HATs, H3K27ac, and low levels of transcription of eRNA. Interaction between enhancers and promoters involves structural connections that include the Mediator complex and cohesions to promote formation of the preinitiation complex, to initiate transcription. Substantial data support that many interactions between enhancers and promoters (“looping”) are constitutive, although it is possible that such interactions may be enhanced by stimulation.
NFR: Nucleosome Free Region
Epigenetics in Acute Activation
In macrophages, TLR stimulation results in the release of the aforementioned epigenetic brakes, such as dismissal of BCL-6 and corepressors from gene loci and concomitant induction or activation of demethylases that erase the negative histone marks H3K27me3, H3K9me3, and H4K20me3 (Fig. 1) (84, 85, 87, 88). In addition to histone modifications, induction of a subset of genes that includes IL6 and IL12B requires nucleosome remodeling by the SWI/SNF complexes (82, 90). This epigenetic remodeling facilitates recruitment of signaling transcription factors such as NF-κB, an increase in positive histone marks such H4ac and H3K4me3, and release of paused Pol II (81) to promote transcription elongation. Although low levels of precursor transcripts are constitutively produced at these genes, NF-κB and possibly other inducible factors are needed to enhance the efficiency of transcription elongation and pre-mRNA processing, in addition to enhancing the frequency of transcription initiation (81, 91). These inducible factors promote H4 acetylation, which is then recognized by the bromodomain-containing adaptor protein Brd4. Brd4 recruits the positive transcription elongation factor (P-TEFb), which promotes elongation and pre-mRNA processing through its ability to phosphorylate the C-terminal domain of RNA Pol II (81).
Enhancers also are activated, as demonstrated by recruitment of p300, increased histone acetylation, binding of signaling transcription factors, and transcription of eRNA (Fig. 1) (31, 36, 92, 93). Although some enhancers can be activated solely by pioneer factors and/or lineage-dependent transcription factors, signal-dependent transcription factors will be required for other enhancers to be fully activated (35, 94, 95). Signal-dependent transcription factors frequently activate common sets of genes in different cell types but can also regulate gene expression in a cell-type-specific manner. For example, a large set of enhancers appears to be specifically activated upon LPS stimulation, which is commonly associated with p300 and its homolog CBP (36). They promote transcriptional activation by acetylating histones and transcription factors and possibly also by mechanisms unrelated to acetylation that have not yet been clarified. Interestingly, ~ 90% of the binding of the p65 subunit of NF-κB occurs at enhancers that are already poised (where chromatin is in an accessible state), whereas the remainder is associated with the de novo induction of latent enhancers in collaboration with PU.1 and C/EBPα (35, 95). In addition, modulation of the enhancer repertoire and transcriptional induction in response to gamma interferon (IFN-γ) or interleukin-4 (IL-4) was almost completely abolished in STAT1 or STAT6 knockout macrophages (94), implicating signal-activated transcription factors such as STATs in enhancer remodeling. However, little is known about the mechanisms by which the enhancer-bound transcription factors and the chromatin state of enhancers influence the recruitment of the general transcription machinery and RNA Pol II to the promoter.
Furthermore, early loss-of-function studies have revealed variable requirements of nucleosome remodeling by the SWI/SNF complexes at distinct LPS-induced genes in mouse macrophages (90). A majority of primary response genes were induced in a SWI/SNF-independent manner, whereas other primary response genes and most secondary response genes required signal-induced SWI/SNF complexes for their transcriptional induction (82). Interestingly, a substantial subset of SWI/SNF-dependent, LPS-induced primary response genes was found to require IRF3 for activation, and the promoters for these genes usually contain consensus IRF3-binding sites (82). Inducible nucleosome remodeling at these promoters, analyzed by restriction enzyme accessibility, was absent in LPS-stimulated macrophages from IRF3 knockout mice, demonstrating that IRF3 promotes nucleosome remodeling, either directly or indirectly, at this select subset of LPS-induced primary response genes. Taken together, the above results suggest that the nucleosomes found at the promoters of selectively regulated genes serve as a barrier to transcriptional activation, such that these genes will be activated only by stimuli capable of inducing factors that bind nucleosome-embedded sites and recruit SWI/SNF complexes to their promoters. Although it seems clear that enhancers determine gene selectivity, little is known about the role of promoters in determining gene selectivity. Interestingly, recent genome-wide studies showed that broad H3K4me3 domains (the top 5%) preferentially mark cell identity and function genes in a given cell type. The broadest H3K4me3 domains also have more paused Pol II at their promoters and enhanced transcriptional consistency rather than increased transcriptional levels (96). On promoters of active genes, H3K4me1, a well-established feature of enhancers, was associated with transcriptional silencing in macrophages, embryonic fibroblasts, and embryonic stem cells together with H3K27me3 and H4K20me1, suggesting an unexpected role of H3K4me1 in restricting the recruitment of chromatin modifiers to defined regions within promoters (97). Cap analysis of gene expression (CAGE) across a large collection of primary cell types has revealed that many mammalian promoters are composite entities composed of multiple closely separated TSSs, with independent cell-type-specific expression profiles (98). In addition, ChIA-PET results show that some promoters were actually acting like enhancers (99), suggesting a more complex layer of promoter-dependent regulation.
EPIGENETICS IN MAINTAINING DISTINCT FUNCTIONAL STATES UPON ACTIVATION
With respect to distinct transcriptional responses in the innate immune system, the chromatin-dependent mechanisms described above that contribute to macrophage activation and development may reflect an ability to adapt to different environments and maintain functional states in order to properly respond to subsequent challenges. Activation of macrophages plays a key role in tissue homeostasis, disease pathogenesis, and resolving and nonresolving inflammation and is involved in the outcome of many diseases, including autoimmune diseases, metabolic diseases, and bacterial and viral infections (4, 5, 8, 100). It has been well established that macrophages change their activation states in response to growth factors (e.g., colony-stimulating factor-1 [CSF-1] and granulocyte-macrophage CSF [GM-CSF]) and external cues, such as cytokines, microbes, microbial products, and other modulators. Activated macrophages polarize toward various functional phenotypes depending on the pathogen and cytokines expressed in the microenvironment, with a spectrum of activation states commonly observed (4, 5, 8, 100). Recently it has become clear that the signaling pathways and transcription factors important for distinct macrophage activation states induces epigenetic changes, as exemplified by alterations in histone modifications and chromatin accessibility (101). Epigenetic regulation could provide a suitable mechanism to explain how transient signals are transformed into more sustained patterns of functional distinct gene expression, and how the epigenetic landscape of the macrophage “remembers” its history of differentiation and previous environmental stimulation. Epigenetically conferred transcriptional memory provides the molecular basis for integration of various polarizing signals into functional distinct states and for reprogramming of macrophages for altered responses to subsequent environmental challenges.
Epigenetics in Macrophage Polarization
Macrophage polarization states are defined by the inducing stimulus and by the ensuing patterns of gene expression, which determine function. In vivo, macrophage phenotype is heterogeneous, and multiple polarization states have been described. These states exist on a spectrum of overlapping phenotypes and gene expression patterns related to the original classification of M1 and M2 macrophages (4, 5, 10). Although it is unclear how the epigenetic landscape differently regulates macrophage polarization to explain spectrum of activation, recent studies have revealed that epigenetic changes contribute to macrophage polarization in classical M1 and M2 models, two extreme ends of a functional spectrum.
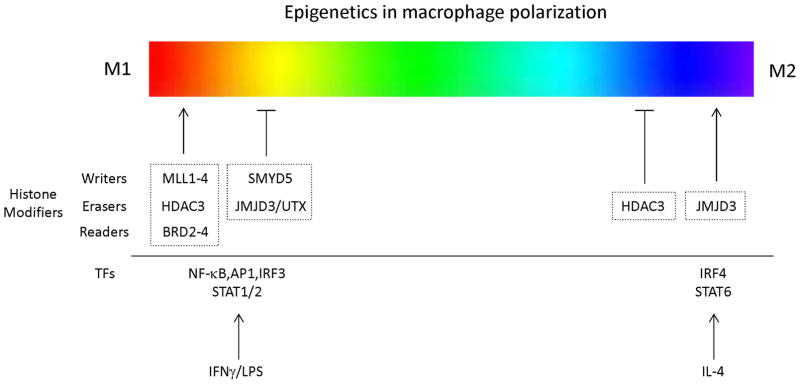
Notably, macrophage polarization has been shown to be associated with histone modifiers such as writers, erasers, and readers of histone marks (Fig. 2). The histone methyltransferase MLL was recently shown to be a regulator of M1 activation (102). MLL expression increases upon stimulation with IFN-γ/LPS, M1 polarizing factors, and inhibition of MLL reduces the induction of the M1 gene CXCL10. In addition, H3K4 methylation at de novo enhancers is primarily dependent on the histone methyltransferases MLL1, MLL2/4, and MLL3, and H4K20me3 by SET and MYND domain-containing protein 5 (SMYD5) restricts the expression of TLR4 target genes in macrophages (95). Two histone mark erasers, Jmjd3 and HDAC3, also have been shown to regulate M1/M2 polarization. Expression of a subset of LPS-induced genes is strongly decreased in Jmjd3-deficient macrophages (87), and Jmjd3/UTX inhibition reduces LPS-induced inflammatory cytokine production in human macrophages (103). Interestingly, Jmjd3 is also induced by IL-4 (104) and was shown to be crucial for M2 polarization and immune responses against helminth infections through IRF4 (105). In addition, H3K27me3 demethylation in the Nfatc1 locus by Jmjd3 promotes receptor activator of NF-κB ligand (RANKL)-induced osteoclast differentiation (106), suggesting that Jmjd3 allows various responses to different external stimuli and regulates macrophage polarization in a context-dependent manner. Another histone eraser, HDAC3, promotes M1 responses and at the same time acts as a brake for M2 polarization. Macrophages lacking HDAC3 show an M2-like phenotype in the absence of external stimuli and are hyperresponsive to IL-4 (107). HDAC3 is required for the activation of hundreds of mainly STAT1-dependent, inflammatory genes in M1 macrophages, and this was shown to be dependent on defective IFN-β responses in macrophages (108). In contrast to histone deacetylation by HDAC3, bromodomain and extraterminal domain family (BET) proteins, such as the Brd2 to -4 readers of histone acetylation, are crucial for inducing inflammatory genes (81, 109), and the BET protein inhibitors I-BET and JQ1 block macrophage inflammatory responses (109, 110). A more recent study showed that I-BET also suppresses RANKL-induced osteoclastogenesis by inhibiting the MYC-NFAT axis (111), suggesting that epigenetic modulation broadly regulates macrophage polarization and differentiation.
FIGURE 2.
Epigenetic regulators of macrophage polarization. A color spectrum illustrates different polarized macrophage populations, showing the linear scale of two macrophage designations, M1 and M2. Histone modifiers implicated in M1 and M2 macrophage polarization are indicated. TF, transcription factor.
TLR-induced expression of core M1 inflammatory cytokines is transient, and gene expression is rapidly repressed to near baseline levels (112). In contrast to activation, less is known about mechanisms of gene repression. Nuclear receptors, TLR-induced transcriptional repressors ATF3 and hairy and enhancer of split 1 (Hes1), feedback inhibitors induced by IL-10, and the p50 NF-κB subunit can recruit corepressor complexes that contain HDACs and histone demethylases and decrease gene expression (113, 114). However, the precise mechanisms of action of these repressors and how chromatin states are regulated during deactivation of M1 inflammatory genes are not known. Histone marks can be long-lived, and the extent to which positive marks are removed, and whether negative marks are installed, during gene repression is not clear. In addition, there is evidence for a repressive role for the nucleosome remodeling and deacetylase (NuRD), which presumably shifts nucleosomes to a configuration that limits access of gene loci to transcription factors and general transcriptional machinery (90, 115, 116). Interestingly, deactivation of cytokine gene expression is delayed by the M1-promoting cytokines IFN-γ and GM-CSF, which work in part by suppressing expression of transcriptional repressors such as Hes1 (117).
EPIGENETICS IN PRIMING MACROPHAGE ACTIVATION STATES
Of note, priming by IFNs enables macrophages to be fully activated with augmented and sustained expression of inflammatory cytokine genes in response to pathogen-associated molecular patterns and inflammatory cytokines. Basal IFN-β production under homeostatic conditions can systemically calibrate type I IFN response and maintain macrophages in a primed state of increased readiness to respond rapidly and strongly to infectious challenges (118). Recent reports show that in mice the microbiome, comprising commensal microorganisms that colonize body surfaces, promotes a partial and low-grade M1-like phenotype in macrophages throughout the body, including those in lymphoid organs (119, 120). This M1-like priming of macrophages induces chromatin remodeling with increased H3K4me3 marks at Ifnb, Il6, and Tnf promoters, which is associated with increased binding of NF-κB p65, IRF3, and Pol II upon cell stimulation (119, 120). Interestingly, poising of IFN response genes is targeted by the influenza A protein NS1, which mimics H3K4-containing peptides and thereby suppresses the positive functions of H3K4me3 by blocking interactions with the human Pol II-associated factor 1 (PAF1) transcription elongation complex (hPAF1C), the reader of this epigenetic mark (121).
IFN-γ is the most potent M1-activating cytokine and promotes a TLR-induced classical inflammatory activation state. IFN-γ and STAT1 have been implicated in nucleosome remodeling and opening of chromatin (122). Recent genome-wide studies of human macrophages revealed that IFN-γ induced stable and coordinated recruitment of STAT1, IRF1, and associated histone acetylation to enhancers and promoters of genes that are synergistically activated by IFN-γ and LPS, such as TNF, IL6, and IL12B (123, 124). Importantly, CRISPR-mediated deletion established the functional importance of IFN-γ-primed enhancer at the TNF locus. In T cells, p300 recruitment to a large percentage of enhancers in T-bet-deficient cells was dependent on the STAT1/4 proteins, indicating the potential role of STAT1 in shaping enhancer landscape independently of pioneer factors (125). Over all, these results suggest that the IFN-mediated STAT pathway may create a primed chromatin environment to augment TLR-induced gene transcription to explain a synergy mechanism in concert with signaling pathways.
EPIGENETICS IN MEMORY-LIKE BEHAVIOR OF MONOCYTES/MACROPHAGES
It has long been known that monocytes exposed to endotoxins can enter a refractory functional state, characterized by incapacity to produce proinflammatory cytokines (126). More recently, another state termed “trained immunity” was introduced to describe how an initial infection or vaccinations could increase the responsiveness of monocytes to a secondary infection (Fig. 3) (127). Both tolerance and training represent clinically relevant functional states, such as the immune paralysis encountered during bacterial sepsis or endotoxic shock or the nonspecific protective effects of live microorganism vaccination that strongly influence susceptibility to secondary infections (126–129). Importantly, tolerance and training states are persistent, but fully reversible, after the initial challenge. They affect responses to subsequent stimulation with similar or different stimuli by “remembering” prior environmental stimulation. Thus, the essential features of such phenomena are their persistence beyond the initial stimulation and their decay over time, and in this way they can be considered short-term memories of environmental exposure. Tolerance and training are likely initial examples of a more pervasive phenomenon of the conditioning of biological systems by exposure to environmental changes. This conditioning reflects the need to adapt to the external milieu and to maintain such an adapted state after stimulus termination to avoid the adverse consequences of exaggerated, chronic, or repeated stimulation.
FIGURE 3.
Epigenetic regulation of endotoxin tolerance and trained immunity in macrophages. A proposed model of endotoxin tolerance and trained immunity is depicted in the graph. The red line indicates tolerized genes that remain refractory to a second stimulus, and the blue line represents trained genes that show enhanced expression in response to subsequent stimulation. Innate immune responses during and after first stimuli can lead to epigenetic reprogramming, which translates into decreased or increased immune response to subsequent stimulation.
EPIGENETICS IN TOLERANCE
Exposure to a potent inflammatory stimulus can lead to acquired refractoriness to induction of genes encoding inflammatory molecules after subsequent stimulation (Fig. 3). This observation was first made with LPS as the stimulus and is referred to as LPS tolerance or endotoxin tolerance (126). However, tumor necrosis factor (TNF) has similarly been shown to induce tolerance (130). Tolerant macrophages exhibit a selective defect in the induction of a subset of genes, including inflammatory cytokine genes, called tolerized genes. By contrast, nontolerized genes, for example, those encoding antimicrobial products and some chemokines, are expressed (131). Many molecular mechanisms seem to contribute to tolerance, ranging from mechanisms for suppressing the transduction of inflammatory signals, to expression of microRNAs, to active repression of genes encoding inflammatory molecules through chromatin regulation (126).
Earlier studies of endotoxin-tolerant human mono-cytic cell lines reported RelB to direct deposition of histone H3K9me2, resulting in impaired p65 NF-κB transactivation of the IL1B promoter (132). Similarly, binding of high-mobility group box-1 protein (HMGB1) and histone H1 linker at the promoters of TNF and IL1B genes also suppresses chromatin remodeling (133). Recent works have clarified that epigenetic mechanisms likely explain the gene-specific nature of tolerance. LPS-induced, TNF-induced, and sepsis-induced tolerance are governed by changes in chromatin accessibility and histone modification, more specifically at the level of H3K4me3, H3K27me2, and histone methyltransferase complexes (122, 130, 131, 134, 135). Tolerized genes exhibit decreased chromatin accessibility, as assessed by restriction enzyme accessibility assays, and diminished recruitment of transcription factors such as p65. The molecular explanation for diminished accessibility involves decreased TLR-induced recruitment of Brahma-related gene 1 (BRG1)-containing nucleosome-remodeling complexes and complex changes in histone acetylation and methylation. The signals to explain gene-specific regulation in tolerance are not clear, although newly transcribed gene products are important to establish tolerance (131), and glycogen synthase kinase 3 (GSK3) plays a key role in TNF-induced tolerance (130). Recent motif-based analyses of TLR4-induced genes revealed that NF-κB motifs were the key regulatory elements significantly enriched in tolerizable genes, by facilitating the formation of an NCoR-HDAC3-p50 repressosome (136). Interestingly, IFN-γ prevents tolerance by preserving expression of the receptor-interacting protein 140 (RIP140) coactivator and promoting TLR-induced chromatin accessibility upon secondary TLR challenge (122, 137). By contrast, nontolerized genes maintain an open chromatin state. Nontolerized genes exhibit more H4 acetylation and maintain H3K4me3 after restimulation (131). Like the genes in naive macrophages, these genes are capable of recruiting the BRG1 and chromodomain helicase DNA-binding protein 4 (CHD4) chromatin-remodeling complexes to their promoters. Interestingly, nontolerized genes are induced with enhanced kinetics and magnitude in tolerant macrophages. Differences in their sensitivity to inhibitors of protein synthesis, such as cycloheximide, showed that a subset of nontolerized genes that were secondary response genes in naive macrophages was converted into primary response genes in tolerant macrophages (131). IFN-γ also altered the transcriptional requirements for secondary tolerized IL6 gene expression, and converted IL6 into a primary response gene in tolerant monocytes by increasing IL6 promoter accessibility in the absence of new protein synthesis (122). These results suggest that removal of a requirement for new protein synthesis for chromatin remodeling represents one mechanism by which a secondary tolerized gene can become converted into a primary nontolerized gene by IFN-γ. Epigenetic mechanisms that regulate polarization of macrophages to a tolerized state need to be further clarified by genome-wide analysis of chromatin landscape and by identification of key transcription factors and chromatin-remodeling complexes that regulate the tolerization process. It will be interesting to determine whether defects in establishing tolerance states contribute to chronic inflammation and disease.
EPIGENETICS IN TRAINED IMMUNITY
Conversely, trained macrophages have been described as cells responsible for nonspecific protection against reinfection independently of adaptive immunity, and increased production of proinflammatory cytokines is characteristic of trained macrophages (Fig. 3) (127). It has been shown that infections with bacteria, yeasts, or viruses (e.g., Candida albicans, BCG, or herpesvirus) are capable of conferring a beneficial protection against reinfection with a second, unrelated pathogen, due to the upregulation of the basal level of innate immunity such as macrophage activation, as well as heightened TNF and IFN-γ production (138, 139). Interestingly, two recent studies have shown that helminth coinfection resulted in impaired antiviral immunity and was associated with STAT6-dependent, helminth-induced alternative activation of macrophages (140, 141), suggesting that infection with one pathogen predisposes the host to respond to subsequent infection, and macrophage phenotypes and the local cytokine environment may have a role in immunomodulation for protective immune responses.
It has been shown that C. albicans- and β-glucan-dependent functional reprogramming of monocytes requires the C-type lectin receptor Dectin-1 and the noncanonical Raf-1 pathway (142), whereas protection associated with BCG vaccination/training is induced through NOD2 (nucleotide-binding oligomerization domain-containing protein 2) recognition of peptidoglycans (143). In vivo, training of both human and murine monocytes/macrophages was associated with enhanced H3K4me3 levels but not changes in H3K27me3. These genome-wide modifications in H3K4me3 correlate with changes in gene expression. Of interest, the H3K4me3 signature is already apparent after 24 h of β-glucan incubation and H3K4me3 patterns do not diminish after 7 days. More recently, it has been demonstrated that β-glucan priming specifically induced about 3,000 distal regulatory elements with distinct motif profiles for transcription factors in DNase I-hypersensitive sites at dynamic epigenomic regions (144). Notably, β-glucan-mediated training of monocytes also leads to epigenetic remodeling at genes involved in the mTOR pathway and glycolysis. Accordingly, trained immunity could not be induced in monocytes from patients deficient in Dectin-1 or in monocytes treated with inhibitors of AKT, mTOR, or hypoxia-inducible factor-1α (HIF1α) (145). β-Glucan-trained monocytes displayed reduced oxygen consumption and increased glucose consumption, which is consistent with a switch from oxidative metabolism to glycolysis.
THE ROLE OF LATENT ENHANCERS FOR MEMORY-LIKE BEHAVIOR
In line with memory-like behavior for selective gene expression, another mechanistic insight recently emerged from an analysis of enhancers for LPS-induced genes in macrophages that do not exhibit an H3K4me1 mark in the basal state (94). Although most LPS-induced enhancers possess the properties described above, with PU.1 binding, nucleosome remodeling and open chromatin, and H3K4me1 deposition, a significant subset of enhancers appears to remain unmarked prior to LPS exposure. LPS stimulation induces the acquisition of H3K4me1/H3K4me3 and H3K27ac at several thousand regulatory regions that are completely unmarked in naive cells (94). Importantly, when the stimulus has ceased, H3K27ac and H3K4me3 disappear, whereas H3K4me1 persists at a new class of enhancers, termed latent enhancers, despite the release of PU.1 and the partner transcription factors that contributed to initial activation. The maintenance of this modification correlates with more rapid induction of enhancer acetylation in response to a subsequent stimulus. Stimulation of macrophages by a variety of stimuli such as IFN-γ and IL-4 also induces the acquisition of different patterns of latent enhancers, suggesting that stimulus-specific latent enhancers may modulate gene expression in response to a subsequent stimulus. Similar to tolerance and training, these chromatin features may represent a chromatin-mediated memory mechanism, whereby the H3K4me1 mark is maintained in the absence of the transcription factors responsible for its initial deposition.
CONCLUDING REMARKS
Macrophages continuously sample the environment and quickly react to it. Thus, the differentiation and functional specialization of macrophages must be tightly regulated to ensure that these cells execute their proper function. In this review, we have attempted to emphasize the dynamic role of epigenetic regulation in macrophages with respect to differentiation, activation, and memory-like behaviors on many levels and over broad time frames. Recent genome-wide studies provide chromatin dynamics at extraordinary resolution and testify that chromatin is far more dynamic than was appreciated previously. We described how individual or combinations of pioneer factors establish the resting enhancer landscape throughout differentiation and determine various responses to activation. Genome-wide mapping of diverse enhancer features, their functional relationship with promoters, and their ultimate transcriptional outputs have provided a number of striking discoveries, ranging from the identification of the large number of enhancers to the widespread production of eRNAs. We discussed that macrophage function, upon activation, is coordinated by regulation of the chromatin landscape in the context of enhancers, promoters, and rapid response genes. Preexisting chromatin marks deposited during differentiation interpret, calibrate, and transmit environmental signals to determine the magnitude and specificity of gene expression. In addition, epigenetic control mechanisms may have a role in providing a flexible mechanism for the stable preactivation of cells that ensures macrophages mount the correct level of response under inflammatory conditions. Finally, we presented the epigenetic mechanisms governing the polarization of macrophages and the alternative perspectives of chromatin state mediating memory in trained or tolerant states. Epigenetic regulation provides stable, yet reversible, marks that bestow flexibility on these processes. Evidence reviewed here suggests that epigenetic changes fundamentally reprogram macrophages to exhibit altered gene expression programs in response to environmental stimuli. Such reprogramming would allow transcriptional memory to shape macrophage phenotype in response to environmental changes and may contribute to the complex macrophage phenotypes. Thus, the epigenetic mechanisms that are supposed to control the memory of the environmental impact may also contribute to the persistence of disease-associated phenotypes, particularly in sustaining chronic inflammation and in mediating the interactions of genes and environment that lead to disease. Of note, many single nucleotide polymorphisms associated with autoimmune/inflammatory diseases, affecting chromatin states, are concentrated in regulatory regions that are subjected to epigenetic regulation (45, 146–148). In this context, it would be interesting to consider the possibility of treating chronic inflammatory states by targeting specific epigenetic modifications. Investigation of epigenetic regulation in myeloid cells is still at an early stage, and there are many exciting areas for future research. As advances in technology rapidly increase the complexity of interacting transcription factors, cis-regulatory elements, chromatin regulators, and posttranslational modifications of histones and DNA methylation, it is a major challenging issue to decipher global regulatory processes on a mechanistic level and integrate new findings with known genomic features. Finally, despite many features conserved between the human and mouse systems (149, 150), there are important known species differences in myeloid cells such as macrophages and dendritic cells (149–153). Systematically determining the similarities and differences between mouse and human regulatory networks in myeloid cells will help to interpret biomedical insights derived from research performed on mouse models.
References
- 1.Smale ST, Tarakhovsky A, Natoli G. Chromatin contributions to the regulation of innate immunity. Annu Rev Immunol. 2014;32:489–511. doi: 10.1146/annurev-immunol-031210-101303. [DOI] [PubMed] [Google Scholar]
- 2.Winter DR, Amit I. The role of chromatin dynamics in immune cell development. Immunol Rev. 2014;261:9–22. doi: 10.1111/imr.12200. [DOI] [PubMed] [Google Scholar]
- 3.Heinz S, Romanoski CE, Benner C, Glass CK. The selection and function of cell type-specific enhancers. Nat RevMol Cell Biol. 2015;16:144–154. doi: 10.1038/nrm3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gordon S, Martinez FO. Alternative activation of macrophages: mechanism and functions. Immunity. 2010;32:593–604. doi: 10.1016/j.immuni.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 5.Murray PJ, Allen JE, Biswas SK, Fisher EA, Gilroy DW, Goerdt S, Gordon S, Hamilton JA, Ivashkiv LB, Lawrence T, Locati M, Mantovani A, Martinez FO, Mege JL, Mosser DM, Natoli G, Saeij JP, Schultze JL, Shirey KA, Sica A, Suttles J, Udalova I, van Ginderachter JA, Vogel SN, Wynn TA. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity. 2014;41:14–20. doi: 10.1016/j.immuni.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Merad M, Sathe P, Helft J, Miller J, Mortha A. The dendritic cell lineage: ontogeny and function of dendritic cells and their subsets in the steady state and the inflamed setting. Annu Rev Immunol. 2013;31:563–604. doi: 10.1146/annurev-immunol-020711-074950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ginhoux F, Jung S. Monocytes and macrophages: developmental pathways and tissue homeostasis. Nat Rev Immunol. 2014;14:392–404. doi: 10.1038/nri3671. [DOI] [PubMed] [Google Scholar]
- 8.Sica A, Mantovani A. Macrophage plasticity and polarization: in vivo veritas. J Clin Invest. 2012;122:787–795. doi: 10.1172/JCI59643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xue J, Schmidt SV, Sander J, Draffehn A, Krebs W, Quester I, De Nardo D, Gohel TD, Emde M, Schmidleithner L, Ganesan H, Nino-Castro A, Mallmann MR, Labzin L, Theis H, Kraut M, Beyer M, Latz E, Freeman TC, Ulas T, Schultze JL. Transcriptome-based network analysis reveals a spectrum model of human macrophage activation. Immunity. 2014;40:274–288. doi: 10.1016/j.immuni.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 11.Sadeh R, Allis CD. Genome-wide “re”-modeling of nucleosome positions. Cell. 2011;147:263–266. doi: 10.1016/j.cell.2011.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Becker PB, Workman JL. Nucleosome remodeling and epigenetics. Cold Spring Harb Perspect Biol. 2013;5:a017905. doi: 10.1101/cshperspect.a017905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Henikoff S, Shilatifard A. Histone modification: cause or cog? Trends Genet. 2011;27:389–396. doi: 10.1016/j.tig.2011.06.006. [DOI] [PubMed] [Google Scholar]
- 14.Badeaux AI, Shi Y. Emerging roles for chromatin as a signal integration and storage platform. Nat Rev Mol Cell Biol. 2013;14:211–224. doi: 10.1038/nrm3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Monticelli S, Natoli G. Short-term memory of danger signals and environmental stimuli in immune cells. Nat Immunol. 2013;14:777–784. doi: 10.1038/ni.2636. [DOI] [PubMed] [Google Scholar]
- 16.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 17.Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- 18.Fierz B, Chatterjee C, McGinty RK, Bar-Dagan M, Raleigh DP, Muir TW. Histone H2B ubiquitylation disrupts local and higher-order chromatin compaction. Nat Chem Biol. 2011;7:113–119. doi: 10.1038/nchembio.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lauberth SM, Nakayama T, Wu X, Ferris AL, Tang Z, Hughes SH, Roeder RG. H3K4me3 interactions with TAF3 regulate preinitiation complex assembly and selective gene activation. Cell. 2013;152:1021–1036. doi: 10.1016/j.cell.2013.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stasevich TJ, Hayashi-Takanaka Y, Sato Y, Maehara K, Ohkawa Y, Sakata-Sogawa K, Tokunaga M, Nagase T, Nozaki N, McNally JG, Kimura H. Regulation of RNA polymerase II activation by histone acetylation in single living cells. Nature. 2014;516:272–275. doi: 10.1038/nature13714. [DOI] [PubMed] [Google Scholar]
- 21.Meyer CA, Liu XS. Identifying and mitigating bias in next-generation sequencing methods for chromatin biology. Nat Rev Genet. 2014;15:709–721. doi: 10.1038/nrg3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thurman RE, Rynes E, Humbert R, Vierstra J, Maurano MT, Haugen E, Sheffield NC, Stergachis AB, Wang H, Vernot B, Garg K, John S, Sandstrom R, Bates D, Boatman L, Canfield TK, Diegel M, Dunn D, Ebersol AK, Frum T, Giste E, Johnson AK, Johnson EM, Kutyavin T, Lajoie B, Lee BK, Lee K, London D, Lotakis D, Neph S, Neri F, Nguyen ED, Qu H, Reynolds AP, Roach V, Safi A, Sanchez ME, Sanyal A, Shafer A, Simon JM, Song L, Vong S, Weaver M, Yan Y, Zhang Z, Zhang Z, Lenhard B, Tewari M, Dorschner MO, Hansen RS, Navas PA, Stamatoyannopoulos G, Iyer VR, Lieb JD, Sunyaev SR, Akey JM, Sabo PJ, Kaul R, Furey TS, Dekker J, Crawford GE, Stamatoyannopoulos JA. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Geissmann F, Manz MG, Jung S, Sieweke MH, Merad M, Ley K. Development of monocytes, macrophages, and dendritic cells. Science. 2010;327:656–661. doi: 10.1126/science.1178331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garber M, Yosef N, Goren A, Raychowdhury R, Thielke A, Guttman M, Robinson J, Minie B, Chevrier N, Itzhaki Z, Blecher-Gonen R, Bornstein C, Amann-Zalcenstein D, Weiner A, Friedrich D, Meldrim J, Ram O, Cheng C, Gnirke A, Fisher S, Friedman N, Wong B, Bernstein BE, Nusbaum C, Hacohen N, Regev A, Amit I. A high-throughput chromatin immunoprecipitation approach reveals principles of dynamic gene regulation in mammals. Mol Cell. 2012;47:810–822. doi: 10.1016/j.molcel.2012.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mullen AC, Orlando DA, Newman JJ, Lovén J, Kumar RM, Bilodeau S, Reddy J, Guenther MG, DeKoter RP, Young RA. Master transcription factors determine cell-type-specific responses to TGF-β signaling. Cell. 2011;147:565–576. doi: 10.1016/j.cell.2011.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Graf T, Enver T. Forcing cells to change lineages. Nature. 2009;462:587–594. doi: 10.1038/nature08533. [DOI] [PubMed] [Google Scholar]
- 27.Scott EW, Simon MC, Anastasi J, Singh H. Requirement of transcription factor PU.1 in the development of multiple hematopoietic lineages. Science. 1994;265:1573–1577. doi: 10.1126/science.8079170. [DOI] [PubMed] [Google Scholar]
- 28.Scott EW, Fisher RC, Olson MC, Kehrli EW, Simon MC, Singh H. PU.1 functions in a cell-autonomous manner to control the differentiation of multipotential lymphoid-myeloid progenitors. Immunity. 1997;6:437–447. doi: 10.1016/s1074-7613(00)80287-3. [DOI] [PubMed] [Google Scholar]
- 29.Nerlov C, Graf T. PU.1 induces myeloid lineage commitment in multipotent hematopoietic progenitors. Genes Dev. 1998;12:2403–2412. doi: 10.1101/gad.12.15.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zaret KS, Carroll JS. Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 2011;25:2227–2241. doi: 10.1101/gad.176826.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pham TH, Minderjahn J, Schmidl C, Hoffmeister H, Schmidhofer S, Chen W, Längst G, Benner C, Rehli M. Mechanisms of in vivo binding site selection of the hematopoietic master transcription factor PU.1. Nucleic Acids Res. 2013;41:6391–6402. doi: 10.1093/nar/gkt355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schonheit J, Kuhl C, Gebhardt ML, Klett FF, Riemke P, Scheller M, Huang G, Naumann R, Leutz A, Stocking C, Priller J, Andrade-Navarro MA, Rosenbauer F. PU.1 level-directed chromatin structure remodeling at the 8 gene drives dendritic cell commitment. Cell Rep. 2013;3:1617–1628. doi: 10.1016/j.celrep.2013.04.007. [DOI] [PubMed] [Google Scholar]
- 34.Escoubet-Lozach L, Benner C, Kaikkonen MU, Lozach J, Heinz S, Spann NJ, Crotti A, Stender J, Ghisletti S, Reichart D, Cheng CS, Luna R, Ludka C, Sasik R, Garcia-Bassets I, Hoffmann A, Subramaniam S, Hardiman G, Rosenfeld MG, Glass CK. Mechanisms establishing TLR4-responsive activation states of inflammatory response genes. PLoS Genet. 2011;7:e1002401. doi: 10.1371/journal.pgen.1002401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heinz S, Romanoski CE, Benner C, Allison KA, Kaikkonen MU, Orozco LD, Glass CK. Effect of natural genetic variation on enhancer selection and function. Nature. 2013;503:487–492. doi: 10.1038/nature12615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ghisletti S, Barozzi I, Mietton F, Polletti S, De Santa F, Venturini E, Gregory L, Lonie L, Chew A, Wei CL, Ragoussis J, Natoli G. Identification and characterization of enhancers controlling the inflammatory gene expression program in macrophages. Immunity. 2010;32:317–328. doi: 10.1016/j.immuni.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 37.Di Stefano B, Sardina JL, van Oevelen C, Collombet S, Kallin EM, Vicent GP, Lu J, Thieffry D, Beato M, Graf T. C/EBPα poises B cells for rapid reprogramming into induced pluripotent stem cells. Nature. 2014;506:235–239. doi: 10.1038/nature12885. [DOI] [PubMed] [Google Scholar]
- 38.Barneda-Zahonero B, Román-González L, Collazo O, Rafati H, Islam AB, Bussmann LH, di Tullio A, De Andres L, Graf T, López-Bigas N, Mahmoudi T, Parra M. HDAC7 is a repressor of myeloid genes whose downregulation is required for transdifferentiation of pre-B cells into macrophages. PLoS Genet. 2013;9:e1003503. doi: 10.1371/journal.pgen.1003503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Calo E, Wysocka J. Modification of enhancer chromatin: what, how, and why? Mol Cell. 2013;49:825–837. doi: 10.1016/j.molcel.2013.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gerstein MB, Kundaje A, Hariharan M, Landt SG, Yan KK, Cheng C, Mu XJ, Khurana E, Rozowsky J, Alexander R, Min R, Alves P, Abyzov A, Addleman N, Bhardwaj N, Boyle AP, Cayting P, Charos A, Chen DZ, Cheng Y, Clarke D, Eastman C, Euskirchen G, Frietze S, Fu Y, Gertz J, Grubert F, Harmanci A, Jain P, Kasowski M, Lacroute P, Leng J, Lian J, Monahan H, O’Geen H, Ouyang Z, Partridge EC, Patacsil D, Pauli F, Raha D, Ramirez L, Reddy TE, Reed B, Shi M, Slifer T, Wang J, Wu L, Yang X, Yip KY, Zilberman-Schapira G, Batzoglou S, Sidow A, Farnham PJ, Myers RM, Weissman SM, Snyder M. Architecture of the human regulatory network derived from ENCODE data. Nature. 2012;489:91–100. doi: 10.1038/nature11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Neph S, Vierstra J, Stergachis AB, Reynolds AP, Haugen E, Vernot B, Thurman RE, John S, Sandstrom R, Johnson AK, Maurano MT, Humbert R, Rynes E, Wang H, Vong S, Lee K, Bates D, Diegel M, Roach V, Dunn D, Neri J, Schafer A, Hansen RS, Kutyavin T, Giste E, Weaver M, Canfield T, Sabo P, Zhang M, Balasundaram G, Byron R, MacCoss MJ, Akey JM, Bender MA, Groudine M, Kaul R, Stamatoyannopoulos JA. An expansive human regulatory lexicon encoded in transcription factor footprints. Nature. 2012;489:83–90. doi: 10.1038/nature11212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, Ku M, Durham T, Kellis M, Bernstein BE. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Andersson R, Gebhard C, Miguel-Escalada I, Hoof I, Bornholdt J, Boyd M, Chen Y, Zhao X, Schmidl C, Suzuki T, Ntini E, Arner E, Valen E, Li K, Schwarzfischer L, Glatz D, Raithel J, Lilje B, Rapin N, Bagger FO, Jørgensen M, Andersen PR, Bertin N, Rackham O, Burroughs AM, Baillie JK, Ishizu Y, Shimizu Y, Furuhata E, Maeda S, Negishi Y, Mungall CJ, Meehan TF, Lassmann T, Itoh M, Kawaji H, Kondo N, Kawai J, Lennartsson A, Daub CO, Heutink P, Hume DA, Jensen TH, Suzuki H, Hayashizaki Y, Müller F, Forrest AR, Carninci P, Rehli M, Sandelin A FANTOM Consortium. An atlas of active enhancers across human cell types and tissues. Nature. 2014;507:455–461. doi: 10.1038/nature12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roadmap Epigenomics Consortium. Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, Ziller MJ, Amin V, Whitaker JW, Schultz MD, Ward LD, Sarkar A, Quon G, Sandstrom RS, Eaton ML, Wu YC, Pfenning AR, Wang X, Claussnitzer M, Liu Y, Coarfa C, Harris RA, Shoresh N, Epstein CB, Gjoneska E, Leung D, Xie W, Hawkins RD, Lister R, Hong C, Gascard P, Mungall AJ, Moore R, Chuah E, Tam A, Canfield TK, Hansen RS, Kaul R, Sabo PJ, Bansal MS, Carles A, Dixon JR, Farh KH, Feizi S, Karlic R, Kim AR, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang P, Xue Y, Han Y, Lin L, Wu C, Xu S, Jiang Z, Xu J, Liu Q, Cao X. The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science. 2014;344:310–313. doi: 10.1126/science.1251456. [DOI] [PubMed] [Google Scholar]
- 47.Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, Boyer LA, Young RA, Jaenisch R. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Herz HM, Hu D, Shilatifard A. Enhancer malfunction in cancer. Mol Cell. 2014;53:859–866. doi: 10.1016/j.molcel.2014.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang Z, Zang C, Cui K, Schones DE, Barski A, Peng W, Zhao K. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell. 2009;138:1019–1031. doi: 10.1016/j.cell.2009.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Morris SA, Baek S, Sung MH, John S, Wiench M, Johnson TA, Schiltz RL, Hager GL. Overlapping chromatin-remodeling systems collaborate genome wide at dynamic chromatin transitions. Nat Struct Mol Biol. 2014;21:73–81. doi: 10.1038/nsmb.2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, Taatjes DJ, Dekker J, Young RA. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lara-Astiaso D, Weiner A, Lorenzo-Vivas E, Zaretsky I, Jaitin DA, David E, Keren-Shaul H, Mildner A, Winter D, Jung S, Friedman N, Amit I. Immunogenetics. Chromatin state dynamics during blood formation. Science. 2014;345:943–949. doi: 10.1126/science.1256271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lavin Y, Winter D, Blecher-Gonen R, David E, Keren-Shaul H, Merad M, Jung S, Amit I. Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell. 2014;159:1312–1326. doi: 10.1016/j.cell.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gosselin D, Link VM, Romanoski CE, Fonseca GJ, Eichenfield DZ, Spann NJ, Stender JD, Chun HB, Garner H, Geissmann F, Glass CK. Environment drives selection and function of enhancers controlling tissue-specific macrophage identities. Cell. 2014;159:1327–1340. doi: 10.1016/j.cell.2014.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Okabe Y, Medzhitov R. Tissue-specific signals control reversible program of localization and functional polarization of macrophages. Cell. 2014;157:832–844. doi: 10.1016/j.cell.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jin F, Li Y, Dixon JR, Selvaraj S, Ye Z, Lee AY, Yen CA, Schmitt AD, Espinoza CA, Ren B. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature. 2013;503:290–294. doi: 10.1038/nature12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kieffer-Kwon KR, Tang Z, Mathe E, Qian J, Sung MH, Li G, Resch W, Baek S, Pruett N, Grøntved L, Vian L, Nelson S, Zare H, Hakim O, Reyon D, Yamane A, Nakahashi H, Kovalchuk AL, Zou J, Joung JK, Sartorelli V, Wei CL, Ruan X, Hager GL, Ruan Y, Casellas R. Interactome maps of mouse gene regulatory domains reveal basic principles of transcriptional regulation. Cell. 2013;155:1507–1520. doi: 10.1016/j.cell.2013.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rao SS, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, Aiden EL. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ghavi-Helm Y, Klein FA, Pakozdi T, Ciglar L, Noordermeer D, Huber W, Furlong EE. Enhancer loops appear stable during development and are associated with paused polymerase. Nature. 2014;512:96–100. doi: 10.1038/nature13417. [DOI] [PubMed] [Google Scholar]
- 61.Dowen JM, Fan ZP, Hnisz D, Ren G, Abraham BJ, Zhang LN, Weintraub AS, Schuijers J, Lee TI, Zhao K, Young RA. Control of cell identity genes occurs in insulated neighborhoods in mammalian chromosomes. Cell. 2014;159:374–387. doi: 10.1016/j.cell.2014.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schaukowitch K, Joo JY, Liu X, Watts JK, Martinez C, Kim TK. Enhancer RNA facilitates NELF release from immediate early genes. Mol Cell. 2014;56:29–42. doi: 10.1016/j.molcel.2014.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lam MT, Cho H, Lesch HP, Gosselin D, Heinz S, Tanaka-Oishi Y, Benner C, Kaikkonen MU, Kim AS, Kosaka M, Lee CY, Watt A, Grossman TR, Rosenfeld MG, Evans RM, Glass CK. Rev-Erbs repress macrophage gene expression by inhibiting enhancer-directed transcription. Nature. 2013;498:511–515. doi: 10.1038/nature12209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li W, Notani D, Ma Q, Tanasa B, Nunez E, Chen AY, Merkurjev D, Zhang J, Ohgi K, Song X, Oh S, Kim HS, Glass CK, Rosenfeld MG. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature. 2013;498:516–520. doi: 10.1038/nature12210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ilott NE, Heward JA, Roux B, Tsitsiou E, Fenwick PS, Lenzi L, Goodhead I, Hertz-Fowler C, Heger A, Hall N, Donnelly LE, Sims D, Lindsay MA. Long non-coding RNAs and enhancer RNAs regulate the lipopolysaccharide-induced inflammatory response in human monocytes. Nat Commun. 2014;5:3979. doi: 10.1038/ncomms4979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ji H, Ehrlich LI, Seita J, Murakami P, Doi A, Lindau P, Lee H, Aryee MJ, Irizarry RA, Kim K, Rossi DJ, Inlay MA, Serwold T, Karsunky H, Ho L, Daley GQ, Weissman IL, Feinberg AP. Comprehensive methylome map of lineage commitment from haematopoietic progenitors. Nature. 2010;467:338–342. doi: 10.1038/nature09367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zilbauer M, Rayner TF, Clark C, Coffey AJ, Joyce CJ, Palta P, Palotie A, Lyons PA, Smith KG. Genome-wide methylation analyses of primary human leukocyte subsets identifies functionally important cell-type-specific hypomethylated regions. Blood. 2013;122:e52–e60. doi: 10.1182/blood-2013-05-503201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Klug M, Schmidhofer S, Gebhard C, Andreesen R, Rehli M. 5-Hydroxymethylcytosine is an essential intermediate of active DNA demethylation processes in primary human monocytes. Genome Biol. 2013;14:R46. doi: 10.1186/gb-2013-14-5-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang X, Ulm A, Somineni HK, Oh S, Weirauch MT, Zhang HX, Chen X, Lehn MA, Janssen EM, Ji H. DNA methylation dynamics during ex vivo differentiation and maturation of human dendritic cells. Epigenetics Chromatin. 2014;7:21. doi: 10.1186/1756-8935-7-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ficz G, Branco MR, Seisenberger S, Santos F, Krueger F, Hore TA, Marques CJ, Andrews S, Reik W. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011;473:398–402. doi: 10.1038/nature10008. [DOI] [PubMed] [Google Scholar]
- 73.Williams K, Christensen J, Pedersen MT, Johansen JV, Cloos PA, Rappsilber J, Helin K. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature. 2011;473:343–348. doi: 10.1038/nature10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wu H, D’Alessio AC, Ito S, Wang Z, Cui K, Zhao K, Sun YE, Zhang Y. Genome-wide analysis of 5-hydroxymethylcytosine distribution reveals its dual function in transcriptional regulation in mouse embryonic stem cells. Genes Dev. 2011;25:679–684. doi: 10.1101/gad.2036011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.de la Rica L, Rodríguez-Ubreva J, García M, Islam AB, Urquiza JM, Hernando H, Christensen J, Helin K, Gómez-Vaquero C, Ballestar E. PU.1 target genes undergo Tet2-coupled demethylation and DNMT3b-mediated methylation in monocyte-to-osteoclast differentiation. Genome Biol. 2013;14:R99. doi: 10.1186/gb-2013-14-9-r99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Medzhitov R, Horng T. Transcriptional control of the inflammatory response. Nat Rev Immunol. 2009;9:692–703. doi: 10.1038/nri2634. [DOI] [PubMed] [Google Scholar]
- 77.Smale ST, Natoli G. Transcriptional control of inflammatory responses. Cold Spring Harb Perspect Biol. 2014;6:a016261. doi: 10.1101/cshperspect.a016261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Weinmann AS, Plevy SE, Smale ST. Rapid and selective remodeling of a positioned nucleosome during the induction of IL-12 p40 transcription. Immunity. 1999;11:665–675. doi: 10.1016/s1074-7613(00)80141-7. [DOI] [PubMed] [Google Scholar]
- 79.Agalioti T, Lomvardas S, Parekh B, Yie J, Maniatis T, Thanos D. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-β promoter. Cell. 2000;103:667–678. doi: 10.1016/s0092-8674(00)00169-0. [DOI] [PubMed] [Google Scholar]
- 80.Parekh BS, Maniatis T. Virus infection leads to localized hyperacetylation of histones H3 and H4 at the IFN-β promoter. Mol Cell. 1999;3:125–129. doi: 10.1016/s1097-2765(00)80181-1. [DOI] [PubMed] [Google Scholar]
- 81.Hargreaves DC, Horng T, Medzhitov R. Control of inducible gene expression by signal-dependent transcriptional elongation. Cell. 2009;138:129–145. doi: 10.1016/j.cell.2009.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ramirez-Carrozzi VR, Braas D, Bhatt DM, Cheng CS, Hong C, Doty KR, Black JC, Hoffmann A, Carey M, Smale ST. A unifying model for the selective regulation of inducible transcription by CpG islands and nucleosome remodeling. Cell. 2009;138:114–128. doi: 10.1016/j.cell.2009.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fang TC, Schaefer U, Mecklenbrauker I, Stienen A, Dewell S, Chen MS, Rioja I, Parravicini V, Prinjha RK, Chandwani R, MacDonald MR, Lee K, Rice CM, Tarakhovsky A. Histone H3 lysine 9 di-methylation as an epigenetic signature of the interferon response. J Exp Med. 2012;209:661–669. doi: 10.1084/jem.20112343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.De Santa F, Totaro MG, Prosperini E, Notarbartolo S, Testa G, Natoli G. The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell. 2007;130:1083–1094. doi: 10.1016/j.cell.2007.08.019. [DOI] [PubMed] [Google Scholar]
- 85.Stender JD, Pascual G, Liu W, Kaikkonen MU, Do K, Spann NJ, Boutros M, Perrimon N, Rosenfeld MG, Glass CK. Control of proinflammatory gene programs by regulated trimethylation and demethylation of histone H4K20. Mol Cell. 2012;48:28–38. doi: 10.1016/j.molcel.2012.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Saccani S, Natoli G. Dynamic changes in histone H3 Lys 9 methylation occurring at tightly regulated inducible inflammatory genes. Genes Dev. 2002;16:2219–2224. doi: 10.1101/gad.232502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.De Santa F, Narang V, Yap ZH, Tusi BK, Burgold T, Austenaa L, Bucci G, Caganova M, Notarbartolo S, Casola S, Testa G, Sung WK, Wei CL, Natoli G. Jmjd3 contributes to the control of gene expression in LPS-activated macrophages. EMBO J. 2009;28:3341–3352. doi: 10.1038/emboj.2009.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Barish GD, Yu RT, Karunasiri M, Ocampo CB, Dixon J, Benner C, Dent AL, Tangirala RK, Evans RM. Bcl-6 and NF-κB cistromes mediate opposing regulation of the innate immune response. Genes Dev. 2010;24:2760–2765. doi: 10.1101/gad.1998010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ogawa S, Lozach J, Jepsen K, Sawka-Verhelle D, Perissi V, Sasik R, Rose DW, Johnson RS, Rosenfeld MG, Glass CK. A nuclear receptor corepressor transcriptional checkpoint controlling activator protein 1-dependent gene networks required for macrophage activation. Proc Natl Acad Sci U S A. 2004;101:14461–14466. doi: 10.1073/pnas.0405786101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ramirez-Carrozzi VR, Nazarian AA, Li CC, Gore SL, Sridharan R, Imbalzano AN, Smale ST. Selective and antagonistic functions of SWI/SNF and Mi-2β nucleosome remodeling complexes during an inflammatory response. Genes Dev. 2006;20:282–296. doi: 10.1101/gad.1383206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Amir-Zilberstein L, Ainbinder E, Toube L, Yamaguchi Y, Handa H, Dikstein R. Differential regulation of NF-κB by elongation factors is determined by core promoter type. Mol Cell Biol. 2007;27:5246–5259. doi: 10.1128/MCB.00586-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jin F, Li Y, Ren B, Natarajan R. PU.1 and C/EBPα synergistically program distinct response to NF-κB activation through establishing monocyte specific enhancers. Proc Natl Acad Sci U S A. 2011;108:5290–5295. doi: 10.1073/pnas.1017214108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.De Santa F, Barozzi I, Mietton F, Ghisletti S, Polletti S, Tusi BK, Muller H, Ragoussis J, Wei CL, Natoli G. A large fraction of extragenic RNA Pol II transcription sites overlap enhancers. PLoS Biol. 2010;8:e1000384. doi: 10.1371/journal.pbio.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ostuni R, Piccolo V, Barozzi I, Polletti S, Termanini A, Bonifacio S, Curina A, Prosperini E, Ghisletti S, Natoli G. Latent enhancers activated by stimulation in differentiated cells. Cell. 2013;152:157–171. doi: 10.1016/j.cell.2012.12.018. [DOI] [PubMed] [Google Scholar]
- 95.Kaikkonen MU, Spann NJ, Heinz S, Romanoski CE, Allison KA, Stender JD, Chun HB, Tough DF, Prinjha RK, Benner C, Glass CK. Remodeling of the enhancer landscape during macrophage activation is coupled to enhancer transcription. Mol Cell. 2013;51:310–325. doi: 10.1016/j.molcel.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Benayoun BA, Pollina EA, Ucar D, Mahmoudi S, Karra K, Wong ED, Devarajan K, Daugherty AC, Kundaje AB, Mancini E, Hitz BC, Gupta R, Rando TA, Baker JC, Snyder MP, Cherry JM, Brunet A. H3K4me3 breadth is linked to cell identity and transcriptional consistency. Cell. 2014;158:673–688. doi: 10.1016/j.cell.2014.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cheng J, Blum R, Bowman C, Hu D, Shilatifard A, Shen S, Dynlacht BD. A role for H3K4 monomethylation in gene repression and partitioning of chromatin readers. Mol Cell. 2014;53:979–992. doi: 10.1016/j.molcel.2014.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Consortium F, the RP, Clst. Forrest AR, Kawaji H, Rehli M, Baillie JK, de Hoon MJ, Haberle V, Lassman T, Kulakovskiy IV, Lizio M, Itoh M, Andersson R, Mungall CJ, Meehan TF, Schmeier S, Bertin N, Jorgensen M, Dimont E, Arner E, Schmidl C, Schaefer U, Medvedeva YA, Plessy C, Vitezic M, Severin J, Semple C, Ishizu Y, Young RS, Francescatto M, Alam I, Albanese D, Altschuler GM, Arakawa T, Archer JA, Arner P, Babina M, Rennie S, Balwierz PJ, Beckhouse AG, Pradhan-Bhatt S, Blake JA, Blumenthal A, Bodega B, Bonetti A, Briggs J, Brombacher F, Burroughs AM, Califano A, et al. A promoter-level mammalian expression atlas. Nature. 2014;507:462–470. doi: 10.1038/nature13182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Li G, Ruan X, Auerbach RK, Sandhu KS, Zheng M, Wang P, Poh HM, Goh Y, Lim J, Zhang J, Sim HS, Peh SQ, Mulawadi FH, Ong CT, Orlov YL, Hong S, Zhang Z, Landt S, Raha D, Euskirchen G, Wei CL, Ge W, Wang H, Davis C, Fisher-Aylor KI, Mortazavi A, Gerstein M, Gingeras T, Wold B, Sun Y, Fullwood MJ, Cheung E, Liu E, Sung WK, Snyder M, Ruan Y. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell. 2012;148:84–98. doi: 10.1016/j.cell.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mosser DM, Edwards JP. Exploring the full spectrum of macrophage activation. Nat Rev Immunol. 2008;8:958–969. doi: 10.1038/nri2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lawrence T, Natoli G. Transcriptional regulation of macrophage polarization: enabling diversity with identity. Nat Rev Immunol. 2011;11:750–761. doi: 10.1038/nri3088. [DOI] [PubMed] [Google Scholar]
- 102.Kittan NA, Allen RM, Dhaliwal A, Cavassani KA, Schaller M, Gallagher KA, Carson WF, IV, Mukherjee S, Grembecka J, Cierpicki T, Jarai G, Westwick J, Kunkel SL, Hogaboam CM. Cytokine induced phenotypic and epigenetic signatures are key to establishing specific macrophage phenotypes. PLoS One. 2013;8:e78045. doi: 10.1371/journal.pone.0078045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kruidenier L, Chung CW, Cheng Z, Liddle J, Che K, Joberty G, Bantscheff M, Bountra C, Bridges A, Diallo H, Eberhard D, Hutchinson S, Jones E, Katso R, Leveridge M, Mander PK, Mosley J, Ramirez-Molina C, Rowland P, Schofield CJ, Sheppard RJ, Smith JE, Swales C, Tanner R, Thomas P, Tumber A, Drewes G, Oppermann U, Patel DJ, Lee K, Wilson DM. A selective jumonji H3K27 demethylase inhibitor modulates the proinflammatory macrophage response. Nature. 2012;488:404–408. doi: 10.1038/nature11262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ishii M, Wen H, Corsa CA, Liu T, Coelho AL, Allen RM, Carson WF, IV, Cavassani KA, Li X, Lukacs NW, Hogaboam CM, Dou Y, Kunkel SL. Epigenetic regulation of the alternatively activated macrophage phenotype. Blood. 2009;114:3244–3254. doi: 10.1182/blood-2009-04-217620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Satoh T, Takeuchi O, Vandenbon A, Yasuda K, Tanaka Y, Kumagai Y, Miyake T, Matsushita K, Okazaki T, Saitoh T, Honma K, Matsuyama T, Yui K, Tsujimura T, Standley DM, Nakanishi K, Nakai K, Akira S. The Jmjd3-Irf4 axis regulates M2 macrophage polarization and host responses against helminth infection. Nat Immunol. 2010;11:936–944. doi: 10.1038/ni.1920. [DOI] [PubMed] [Google Scholar]
- 106.Yasui T, Hirose J, Tsutsumi S, Nakamura K, Aburatani H, Tanaka S. Epigenetic regulation of osteoclast differentiation: possible involvement of Jmjd3 in the histone demethylation of Nfatc1. J Bone Miner Res. 2011;26:2665–2671. doi: 10.1002/jbmr.464. [DOI] [PubMed] [Google Scholar]
- 107.Mullican SE, Gaddis CA, Alenghat T, Nair MG, Giacomin PR, Everett LJ, Feng D, Steger DJ, Schug J, Artis D, Lazar MA. Histone deacetylase 3 is an epigenomic brake in macrophage alternative activation. Genes Dev. 2011;25:2480–2488. doi: 10.1101/gad.175950.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Chen X, Barozzi I, Termanini A, Prosperini E, Recchiuti A, Dalli J, Mietton F, Matteoli G, Hiebert S, Natoli G. Requirement for the histone deacetylase Hdac3 for the inflammatory gene expression program in macrophages. Proc Natl Acad Sci U S A. 2012;109:E2865–E2874. doi: 10.1073/pnas.1121131109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Nicodeme E, Jeffrey KL, Schaefer U, Beinke S, Dewell S, Chung CW, Chandwani R, Marazzi I, Wilson P, Coste H, White J, Kirilovsky J, Rice CM, Lora JM, Prinjha RK, Lee K, Tarakhovsky A. Suppression of inflammation by a synthetic histone mimic. Nature. 2010;468:1119–1123. doi: 10.1038/nature09589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Belkina AC, Nikolajczyk BS, Denis GV. BET protein function is required for inflammation: Brd2 genetic disruption and BET inhibitor JQ1 impair mouse macrophage inflammatory responses. J Immunol. 2013;190:3670–3678. doi: 10.4049/jimmunol.1202838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Park-Min KH, Lim E, Lee MJ, Park SH, Giannopoulou E, Yarilina A, van der Meulen M, Zhao B, Smithers N, Witherington J, Lee K, Tak PP, Prinjha RK, Ivashkiv LB. Inhibition of osteoclastogenesis and inflammatory bone resorption by targeting BET proteins and epigenetic regulation. Nat Commun. 2014;5:5418. doi: 10.1038/ncomms6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ivashkiv LB. Inflammatory signaling in macrophages: transitions from acute to tolerant and alternative activation states. Eur J Immunol. 2011;41:2477–2481. doi: 10.1002/eji.201141783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ivashkiv LB. Epigenetic regulation of macrophage polarization and function. Trends Immunol. 2013;34:216–223. doi: 10.1016/j.it.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Murray PJ, Smale ST. Restraint of inflammatory signaling by interdependent strata of negative regulatory pathways. Nat Immunol. 2012;13:916–924. doi: 10.1038/ni.2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Roger T, Lugrin J, Le Roy D, Goy G, Mombelli M, Koessler T, Ding XC, Chanson AL, Reymond MK, Miconnet I, Schrenzel J, François P, Calandra T. Histone deacetylase inhibitors impair innate immune responses to Toll-like receptor agonists and to infection. Blood. 2011;117:1205–1217. doi: 10.1182/blood-2010-05-284711. [DOI] [PubMed] [Google Scholar]
- 116.Shimizu-Hirota R, Xiong W, Baxter BT, Kunkel SL, Maillard I, Chen XW, Sabeh F, Liu R, Li XY, Weiss SJ. MT1-MMP regulates the PI3Kδ·Mi-2/NuRD-dependent control of macrophage immune function. Genes Dev. 2012;26:395–413. doi: 10.1101/gad.178749.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hu X, Ivashkiv LB. Cross-regulation of signaling pathways by interferon-γ: implications for immune responses and autoimmune diseases. Immunity. 2009;31:539–550. doi: 10.1016/j.immuni.2009.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ivashkiv LB, Donlin LT. Regulation of type I interferon responses. Nat Rev Immunol. 2014;14:36–49. doi: 10.1038/nri3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Abt MC, Osborne LC, Monticelli LA, Doering TA, Alenghat T, Sonnenberg GF, Paley MA, Antenus M, Williams KL, Erikson J, Wherry EJ, Artis D. Commensal bacteria calibrate the activation threshold of innate antiviral immunity. Immunity. 2012;37:158–170. doi: 10.1016/j.immuni.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ganal SC, Sanos SL, Kallfass C, Oberle K, Johner C, Kirschning C, Lienenklaus S, Weiss S, Staeheli P, Aichele P, Diefenbach A. Priming of natural killer cells by nonmucosal mononuclear phagocytes requires instructive signals from commensal microbiota. Immunity. 2012;37:171–186. doi: 10.1016/j.immuni.2012.05.020. [DOI] [PubMed] [Google Scholar]
- 121.Marazzi I, Ho JS, Kim J, Manicassamy B, Dewell S, Albrecht RA, Seibert CW, Schaefer U, Jeffrey KL, Prinjha RK, Lee K, García-Sastre A, Roeder RG, Tarakhovsky A. Suppression of the antiviral response by an influenza histone mimic. Nature. 2012;483:428–433. doi: 10.1038/nature10892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Chen J, Ivashkiv LB. IFN-γ abrogates endotoxin tolerance by facilitating Toll-like receptor-induced chromatin remodeling. Proc Natl Acad Sci U S A. 2010;107:19438–19443. doi: 10.1073/pnas.1007816107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Qiao Y, Giannopoulou EG, Chan CH, Park SH, Gong S, Chen J, Hu X, Elemento O, Ivashkiv LB. Synergistic activation of inflammatory cytokine genes by interferon-γ-induced chromatin remodeling and Toll-like receptor signaling. Immunity. 2013;39:454–469. doi: 10.1016/j.immuni.2013.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Chow NA, Jasenosky LD, Goldfeld AE. A distal locus element mediates IFN-γ priming of lipopolysaccharide-stimulated TNF gene expression. Cell Rep. 2014;9:1718–1728. doi: 10.1016/j.celrep.2014.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Vahedi G, Takahashi H, Nakayamada S, Sun HW, Sartorelli V, Kanno Y, O’Shea JJ. STATs shape the active enhancer landscape of T cell populations. Cell. 2012;151:981–993. doi: 10.1016/j.cell.2012.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Biswas SK, Lopez-Collazo E. Endotoxin tolerance: new mechanisms, molecules and clinical significance. Trends Immunol. 2009;30:475–487. doi: 10.1016/j.it.2009.07.009. [DOI] [PubMed] [Google Scholar]
- 127.Netea MG, Quintin J, van der Meer JW. Trained immunity: a memory for innate host defense. Cell Host Microbe. 2011;9:355–361. doi: 10.1016/j.chom.2011.04.006. [DOI] [PubMed] [Google Scholar]
- 128.Shalova IN, Lim JY, Chittezhath M, Zinkernagel AS, Beasley F, Hernández-Jiménez E, Toledano V, Cubillos-Zapata C, Rapisarda A, Chen J, Duan K, Yang H, Poidinger M, Melillo G, Nizet V, Arnalich F, López-Collazo E, Biswas SK. Human monocytes undergo functional re-programming during sepsis mediated by hypoxia-inducible factor-1α. Immunity. 2015;42:484–498. doi: 10.1016/j.immuni.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 129.Benn CS, Netea MG, Selin LK, Aaby P. A small jab—a big effect: nonspecific immunomodulation by vaccines. Trends Immunol. 2013;34:431–439. doi: 10.1016/j.it.2013.04.004. [DOI] [PubMed] [Google Scholar]
- 130.Park SH, Park-Min KH, Chen J, Hu X, Ivashkiv LB. Tumor necrosis factor induces GSK3 kinase-mediated cross-tolerance to endotoxin in macrophages. Nat Immunol. 2011;12:607–615. doi: 10.1038/ni.2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Foster SL, Hargreaves DC, Medzhitov R. Gene-specific control of inflammation by TLR-induced chromatin modifications. Nature. 2007;447:972–978. doi: 10.1038/nature05836. [DOI] [PubMed] [Google Scholar]
- 132.Chen X, El Gazzar M, Yoza BK, McCall CE. The NF-κB factor RelB and histone H3 lysine methyltransferase G9a directly interact to generate epigenetic silencing in endotoxin tolerance. J Biol Chem. 2009;284:27857–27865. doi: 10.1074/jbc.M109.000950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.El Gazzar M, Yoza BK, Chen X, Garcia BA, Young NL, McCall CE. Chromatin-specific remodeling by HMGB1 and linker histone H1 silences proinflammatory genes during endotoxin tolerance. Mol Cell Biol. 2009;29:1959–1971. doi: 10.1128/MCB.01862-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Carson WF, Cavassani KA, Dou Y, Kunkel SL. Epigenetic regulation of immune cell functions during post-septic immunosuppression. Epigenetics. 2011;6:273–283. doi: 10.4161/epi.6.3.14017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wen H, Dou Y, Hogaboam CM, Kunkel SL. Epigenetic regulation of dendritic cell-derived interleukin-12 facilitates immunosuppression after a severe innate immune response. Blood. 2008;111:1797–1804. doi: 10.1182/blood-2007-08-106443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Yan Q, Carmody RJ, Qu Z, Ruan Q, Jager J, Mullican SE, Lazar MA, Chen YH. Nuclear factor-κB binding motifs specify Toll-like receptor-induced gene repression through an inducible repressosome. Proc Natl Acad Sci U S A. 2012;109:14140–14145. doi: 10.1073/pnas.1119842109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ho PC, Tsui YC, Feng X, Greaves DR, Wei LN. NF-κB-mediated degradation of the coactivator RIP140 regulates inflammatory responses and contributes to endotoxin tolerance. Nat Immunol. 2012;13:379–386. doi: 10.1038/ni.2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Barton ES, White DW, Cathelyn JS, Brett-McClellan KA, Engle M, Diamond MS, Miller VL, Virgin HW., IV Herpesvirus latency confers symbiotic protection from bacterial infection. Nature. 2007;447:326–329. doi: 10.1038/nature05762. [DOI] [PubMed] [Google Scholar]
- 139.Bistoni F, Vecchiarelli A, Cenci E, Puccetti P, Marconi P, Cassone A. Evidence for macrophage-mediated protection against lethal Candida albicans infection. Infect Immun. 1986;51:668–674. doi: 10.1128/iai.51.2.668-674.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Osborne LC, Monticelli LA, Nice TJ, Sutherland TE, Siracusa MC, Hepworth MR, Tomov VT, Kobuley D, Tran SV, Bittinger K, Bailey AG, Laughlin AL, Boucher JL, Wherry EJ, Bushman FD, Allen JE, Virgin HW, Artis D. Coinfection. Virus-helminth coinfection reveals amicrobiota-independent mechanism of immunomodulation. Science. 2014;345:578–582. doi: 10.1126/science.1256942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Reese TA, Wakeman BS, Choi HS, Hufford MM, Huang SC, Zhang X, Buck MD, Jezewski A, Kambal A, Liu CY, Goel G, Murray PJ, Xavier RJ, Kaplan MH, Renne R, Speck SH, Artyomov MN, Pearce EJ, Virgin HW. Coinfection. Helminth infection reactivates latent gamma-herpesvirus via cytokine competition at a viral promoter. Science. 2014;345:573–577. doi: 10.1126/science.1254517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Quintin J, Saeed S, Martens JH, Giamarellos-Bourboulis EJ, Ifrim DC, Logie C, Jacobs L, Jansen T, Kullberg BJ, Wijmenga C, Joosten LA, Xavier RJ, van der Meer JW, Stunnenberg HG, Netea MG. Candida albicans infection affords protection against reinfection via functional reprogramming of monocytes. Cell Host Microbe. 2012;12:223–232. doi: 10.1016/j.chom.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Kleinnijenhuis J, Quintin J, Preijers F, Joosten LA, Ifrim DC, Saeed S, Jacobs C, van Loenhout J, de Jong D, Stunnenberg HG, Xavier RJ, van der Meer JW, van Crevel R, Netea MG. Bacille Calmette-Guerin induces NOD2-dependent nonspecific protection from reinfection via epigenetic reprogramming of monocytes. Proc Natl Acad Sci U S A. 2012;109:17537–17542. doi: 10.1073/pnas.1202870109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Saeed S, Quintin J, Kerstens HH, Rao NA, Aghajanirefah A, Matarese F, Cheng SC, Ratter J, Berentsen K, van der Ent MA, Sharifi N, Janssen-Megens EM, Ter Huurne M, Mandoli A, van Schaik T, Ng A, Burden F, Downes K, Frontini M, Kumar V, Giamarellos-Bourboulis EJ, Ouwehand WH, van der Meer JW, Joosten LA, Wijmenga C, Martens JH, Xavier RJ, Logie C, Netea MG, Stunnenberg HG. Epigenetic programming of monocyte-to-macrophage differentiation and trained innate immunity. Science. 2014;345:1251086. doi: 10.1126/science.1251086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Cheng SC, Quintin J, Cramer RA, Shepardson KM, Saeed S, Kumar V, Giamarellos-Bourboulis EJ, Martens JH, Rao NA, Aghajanirefah A, Manjeri GR, Li Y, Ifrim DC, Arts RJ, van der Veer BM, Deen PM, Logie C, O’Neill LA, Willems P, van de Veerdonk FL, van der Meer JW, Ng A, Joosten LA, Wijmenga C, Stunnenberg HG, Xavier RJ, Netea MG. mTOR- and HIF-1α-mediated aerobic glycolysis as metabolic basis for trained immunity. Science. 2014;345:1250684. doi: 10.1126/science.1250684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Farh KK, Marson A, Zhu J, Kleinewietfeld M, Housley WJ, Beik S, Shoresh N, Whitton H, Ryan RJ, Shishkin AA, Hatan M, Carrasco-Alfonso MJ, Mayer D, Luckey CJ, Patsopoulos NA, De Jager PL, Kuchroo VK, Epstein CB, Daly MJ, Hafler DA, Bernstein BE. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature. 2015;518:337–343. doi: 10.1038/nature13835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.1000 Genomes Project Consortium. Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, McVean GA. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.McVicker G, van de Geijn B, Degner JF, Cain CE, Banovich NE, Raj A, Lewellen N, Myrthil M, Gilad Y, Pritchard JK. Identification of genetic variants that affect histone modifications in human cells. Science. 2013;342:747–749. doi: 10.1126/science.1242429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Cheng Y, Ma Z, Kim BH, Wu W, Cayting P, Boyle AP, Sundaram V, Xing X, Dogan N, Li J, Euskirchen G, Lin S, Lin Y, Visel A, Kawli T, Yang X, Patacsil D, Keller CA, Giardine B, Mouse EC, Kundaje A, Wang T, Pennacchio LA, Weng Z, Hardison RC, Snyder MP. Principles of regulatory information conservation between mouse and human. Nature. 2014;515:371–375. doi: 10.1038/nature13985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Lin S, Lin Y, Nery JR, Urich MA, Breschi A, Davis CA, Dobin A, Zaleski C, Beer MA, Chapman WC, Gingeras TR, Ecker JR, Snyder MP. Comparison of the transcriptional landscapes between human and mouse tissues. Proc Natl Acad Sci U S A. 2014;111:17224–17229. doi: 10.1073/pnas.1413624111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Shay T, Jojic V, Zuk O, Rothamel K, Puyraimond-Zemmour D, Feng T, Wakamatsu E, Benoist C, Koller D, Regev A ImmGen Consortium. Conservation and divergence in the transcriptional programs of the human and mouse immune systems. Proc Natl Acad Sci U S A. 2013;110:2946–2951. doi: 10.1073/pnas.1222738110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Schmidt D, Wilson MD, Ballester B, Schwalie PC, Brown GD, Marshall A, Kutter C, Watt S, Martinez-Jimenez CP, Mackay S, Talianidis I, Flicek P, Odom DT. Five-vertebrate ChIP-seq reveals the evolutionary dynamics of transcription factor binding. Science. 2010;328:1036–1040. doi: 10.1126/science.1186176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Seok J, Warren HS, Cuenca AG, Mindrinos MN, Baker HV, Xu W, Richards DR, McDonald-Smith GP, Gao H, Hennessy L, Finnerty CC, López CM, Honari S, Moore EE, Minei JP, Cuschieri J, Bankey PE, Johnson JL, Sperry J, Nathens AB, Billiar TR, West MA, Jeschke MG, Klein MB, Gamelli RL, Gibran NS, Brownstein BH, Miller-Graziano C, Calvano SE, Mason PH, Cobb JP, Rahme LG, Lowry SF, Maier RV, Moldawer LL, Herndon DN, Davis RW, Xiao W, Tompkins RG Inflammation and Host Response to Injury, Large Scale Collaborative Research Program. Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc Natl Acad Sci U S A. 2013;110:3507–3512. doi: 10.1073/pnas.1222878110. [DOI] [PMC free article] [PubMed] [Google Scholar]