Abstract
Background
Escherichia coli producing ESBL/AmpC enzymes are unwanted in animal production chains as they may pose a risk to human and animal health. Molecular characterization of plasmids and strains carrying genes that encode these enzymes is essential to understand their local and global spread.
Objectives
To investigate the diversity of genes, plasmids and strains in ESBL/AmpC-producing E. coli from the Colombian poultry chain isolated within the Colombian Integrated Program for Antimicrobial Resistance Surveillance (Coipars).
Methods
A total of 541 non-clinical E. coli strains from epidemiologically independent samples and randomly isolated between 2008 and 2013 within the Coipars program were tested for antimicrobial susceptibility. Poultry isolates resistant to cefotaxime (MIC ≥ 4 mg/L) were screened for ESBL/AmpC genes including blaCTX-M, blaSHV, blaTEM, blaCMY and blaOXA. Plasmid and strain characterization was performed for a selection of the ESBL/AmpC-producing isolates. Plasmids were purified and transformed into E. coli DH10B cells or transferred by conjugation to E. coli W3110. When applicable, PCR Based Replicon Typing (PBRT), plasmid Multi Locus Sequence Typing (pMLST), plasmid Double Locus Sequence Typing (pDLST) and/or plasmid Replicon Sequence Typing (pRST) was performed on resulting transformants and conjugants. Multi Locus Sequence Typing (MLST) was used for strain characterization.
Results
In total, 132 of 541 isolates were resistant to cefotaxime and 122 were found to carry ESBL/AmpC genes. Ninety-two harboured blaCMY-2 (75%), fourteen blaSHV-12 (11%), three blaSHV-5 (2%), five blaCTX-M-2 (4%), one blaCTX-M-15 (1%), one blaCTX-M-8 (1%), four a combination of blaCMY-2 and blaSHV-12 (4%) and two a combination of blaCMY-2 and blaSHV-5 (2%). A selection of 39 ESBL/AmpC-producing isolates was characterized at the plasmid and strain level. ESBL/AmpC genes from 36 isolates were transferable by transformation or conjugation of which 22 were located on IncI1 plasmids. These IncI1 plasmids harboured predominantly blaCMY-2 (16/22), and to a lesser extend blaSHV-12 (5/22) and blaCTX-M-8 (1/22). Other plasmid families associated with ESBL/AmpC-genes were IncK (4/33), IncHI2 (3/33), IncA/C (2/33), IncΒ/O (1/33) and a non-typeable replicon (1/33). Subtyping of IncI1 and IncHI2 demonstrated IncI1/ST12 was predominantly associated with blaCMY-2 (12/16) and IncHI2/ST7 with blaCTX-M-2 (2/3). Finally, 31 different STs were detected among the 39 selected isolates.
Conclusions
Resistance to extended spectrum cephalosporins in E. coli from Colombian poultry is mainly caused by blaCMY-2 and blaSHV-12. The high diversity of strain Sequence Types and the dissemination of homogeneous IncI1/ST12 plasmids suggest that spread of the resistance is mainly mediated by horizontal gene transfer.
Introduction
Infections caused by isolates of Escherichia coli resistant to extended spectrum cephalosporins (ESC), can result in antimicrobial treatment failure and increased health expenditures in humans [1–3], livestock [4] and companion animals [5]. Isolates resistant to ESC produce enzymes capable of hydrolyzing the β-lactam ring of these drugs, and exhibit reduced susceptibility to third generation cephalosporins and monobactams. These ESC hydrolyzing enzymes are mostly plasmid mediated and they include Extended Spectrum Beta-Lactamases (ESBLs), and AmpC beta-lactamases [6,7]. ESBL/AmpC-producing E. coli have been isolated from humans as well as from livestock, animal meat products, companion animals [8], and vegetables [9]. ESBL/AmpC-producing E. coli have been frequently reported in poultry and therefore poultry production and poultry meat are considered a reservoir of ESBL/AmpC-producing E. coli potentially causing infections in humans [10–15].
Previous studies have compared the occurrence and characteristics of genes, plasmids and strains of ESBL/AmpC-producing E. coli from humans, poultry and poultry products [11,16,17]. Some of these studies have found genetic relatedness of strains, genes and/or plasmids, suggesting transmission of ESBL/AmpC-producing E. coli between poultry and humans. Some studies suggest the transmission of bacterial strains, with plasmids and genes through clonal spread [12,16] while others suggest the transmission of plasmids conferring the resistance through horizontal gene transfer between different E. coli strains [13,17]. Whether this transmission happens by means of clonal spread or dissemination of mobile genetic elements, likely corresponds to different conditions within the analysed geographical areas [18].
In Colombia, available literature from human clinical isolates shows that the most frequent ESBL- gene variants are blaCTX-M-12, blaCTX-M-15, blaSHV-12 and blaSHV-5 [19–24]. More recently, some of these studies included AmpC-genes on their genotypic screening, and demonstrated the association of blaCMY-2 to community onset infections [22,23]. Nevertheless, reports that describe the genetic determinants of ESBL/AmpC-producing E. coli from Colombian poultry are not available. This information is essential to know whether a proportion of the mobile genetic elements harbouring ESBL/AmpC—producing E.coli, in particular plasmids, identified in isolates from poultry and humans are genetically related. Therefore, the aim of this study was to characterize the genes, plasmids and strains of a collection of ESBL/AmpC-producing E. coli from baseline studies in the Colombian poultry chain using sequence-based characterization that enables comparisons regardless of time, place and source of isolates.
Materials and Methods
Origin of samples and isolation of E. coli
Non-clinical samples were obtained from three different sources from the Colombian poultry production chain: i) broiler farms ii) broilers at slaughter and iii) raw chicken meat at retail. The samples from farms (n = 1097), slaughterhouses (n = 1566), and retail (n = 1203) were taken as part of a pilot project for the establishment of the Colombian Integrated Program for Antimicrobial Resistance Surveillance (Coipars) [25]. The samples were collected between 2008 and 2013 and originated from i) feces, drag swabs and cloacal swabs from broiler farms ii) cecal content and carcass rinse from slaughterhouses iii) chicken thighs (homogenized with stomacher method) from independent stores and a large retail distribution centre. Carcass rinse samples were obtained from independent stores and supermarkets. Three out of 32 departments of Colombia (that is, provinces) responsible for more than 65% of Colombian chicken production, were represented at the farm level [26]. At the slaughterhouse and retail level, samples were collected from the most populated departments, 18/32 and 23/32, respectively. After collection, samples were stored in insulated containers at 4°C and transported to the laboratory within 24 hours.
Isolates were recovered from samples from the three sources using the methodology previously described by the Canadian Integrated Program for Antimicrobial Resistance Surveillance (CIPARS) [27]. Briefly, samples were mixed in a 1:10 ratio (w/v) with buffered peptone water (BD, USA) and one loop of the mixture was streaked on a MacConkey agar plate (BD, USA) and incubated overnight at 35°C. For chicken thighs samples, 50 ml of EC broth (7% w/v) (BD, USA) was added to the mixture and incubated overnight at 45°C for 24 hours. Next, one loop of the mixture was streaked on a MacConkey agar plate (BD, USA) and incubated overnight at 35°C.
From MacConkey agar plates, one typical lactose-fermenting colony was further plated on BHI agar and incubated overnight at 35°C. One colony of the pure culture was screened with Sulphur Indole and Motility medium (SIM) (Difco, USA). Subsequently, presumptive E. coli strains (negative H2S production and positive tryptophan degradation) were stored at -80°C on sterile Skim Milk (20% w/w) (Difco, USA).
Analyzed strains, antimicrobial resistance screening and species confirmation
From the stored isolates from farms and slaughterhouses, a convenient selection was made. Given the country-wide scope of an integrated surveillance program like Coipars, and the absence of previous reports of ESC resistance from poultry in Colombia, our strategy aimed to represent as much levels, years of production and departments as possible. A sum of 155 and 182 isolates representing 14% and 12% of the total number of samples, respectively, was made in a way that at least one isolate originated from every sampled department. In order to avoid overrepresentation of genes, plasmids and strains due to sample duplicity, repeated isolates from the same production flocks (that is, epidemiologically related isolates) were excluded. In contrast to samples from farms and slaughter, each isolate from retail originated from epidemiologically unrelated flocks and were therefore all included to assess the level of exposure to retail consumers and widen the comparisons with isolates from humans in Colombia. Based on this selection, 541 epidemiologically independent isolates were considered for this study, 45 from farms, 66 from slaughterhouses and 430 from retail.
Isolates were tested using the BD Phoenix automated system and the NMIC/ID-121 panels (BD, USA). Drugs included in the panel and the tested concentrations are available in S1 Table. Minimal Inhibitory concentrations (MIC) were interpreted using CLSI 2013 guidelines [28]. Strains resistant to cefotaxime (MIC ≥ 4 mg/L) were screened for the presence of ESBL/AmpC genes. Additional species confirmation was performed with matrix-assisted laser desorption ionization—time of flight mass spectrometry (MALDI-TOF MS) (Bruker, Delft, the Netherlands). The isolates originated from baseline studies that were financed for specific years only during the development of Coipars (Table 1). As a result for some years no samples were included.
Table 1. Distribution of cefotaxime-resistant E. coli in Colombian poultry (A) and ESBL/AmpC genes and isolates selected for plasmid and strain characterization (B).
| Source | Farms | Slaughter | Retail | Total | |
|---|---|---|---|---|---|
| (A) | 2008 | 8 / 29a | 13 / 29 | - | 21 / 58 |
| 2009 | 4 / 9 | - | 20 / 165 | 24 / 174 | |
| 2010 | - | - | 44 / 168 | 44 / 168 | |
| 2011 | - | - | 30 / 97 | 30 / 97 | |
| 2012 | 1 / 7 | 6 / 20 | - | 7 / 27 | |
| 2013 | - | 6 / 17 | - | 6 / 17 | |
| 2008–2013 | 13 / 45 | 25 / 66 | 94 / 430 | 132 / 541 | |
| (B) | blaCMY-2 | 8 (2)b | 14 (6) | 70 (13) | 92 (21) |
| blaSHV-12 | 2 (1) | 5 (2) | 7 (1) | 14 (4) | |
| blaSHV-5 | - (0) | 1 (1) | 2 (0) | 3 (1) | |
| blaCMY-2-blaSHV-12c | - | 1 | 3 | 4 | |
| blaCMY-2-blaSHV-5 | - | - | 2 | 2 | |
| blaCTX-M-15 | - | - | 1 | 1 | |
| blaCTX-M-2 | 1 | 2 | 2 | 5 | |
| blaCTX-M-8 | - | 1 | - | 1 | |
| Total | 11 (4) | 24 (13) | 87 (22) | 122 (39) |
a Cefotaxime-resistant / Tested isolates
b Numbers of isolates selected for plasmid and strain characterization are presented in brackets
c All isolates positive for combinations of blaCMY-blaSHV and blaCTX-M were selected for plasmid and strain characterization
ESBL/AmpC gene characterization
Polymerase chain reaction (PCR) with previously described primers was used to screen for ESBL [29] and plasmidic AmpC [30]. The genes analysed included blaCTX-M, blaCMY, blaSHV, blaTEM, blaOXA-1-like, blaOXA-2-like and blaOXA-10-like. PCR products were purified using ExoSAP-IT according to manufacturer’s protocol (Affymetrix, USA) and sent to BaseClear (the Netherlands) for sequencing. Sequence results were analysed with BioNumerics v7.5 software (Applied Maths, Belgium) and compared with reference sequences deposited at http://lahey.org/studies/.
Plasmid and strain characterization
A selection of strains based on the distribution of ESBL/AmpC variants was made and further characterized. Due to its low prevalence, the selection included all isolates positive for blaCTX-M (n = 7) and all combinations of blaCMY-blaSHV (n = 6). For the highly prevalent blaCMY and blaSHV genes, a random selection was made using the Random function in Microsoft® Excel to assign random numbers to isolates positive for blaCMY and blaSHV. The size of selection was fixed to a maximum of 30% for both genes. Finally, strains with the largest 21 and 5 numbers, representing 23% and 29% of the isolates, were selected for blaCMY and blaSHV respectively.
Plasmids were purified from the original donor strain using the Qiagen Plasmid Midi-kit (Qiagen, Germany). Plasmid DNA was transformed into ElectroMAX DH10B cells through electroporation (Invitrogen, USA) by mixing 5μl of isolated plasmid DNA with 20μl of electro-competent cells. Electroporation conditions were 25 μF, 200 Ω, and 2.0 Kv. Cells were recovered for 1 hour at 37°C in SOC-medium (Invitrogen, USA). Subsequently, transformants were selected on Luria-Bertani agar (LΒ-agar) supplemented with 2 mg/L cefotaxime (Sigma, USA). Conjugation experiments were performed when electroporation did not yield transformants. Donor strain and recipient rifampicin-resistant strain E. coli W3110, were grown separately overnight in 5 ml of LΒ-Broth. Volumes of 0.1 ml of donor and 0.9 ml of recipient strains were added to 9 ml of LB broth and cultured overnight. Conjugants were selected on LΒ-agar supplemented with 2 mg/L cefotaxime (Sigma, USA) and 75 mg/L rifampicin (Sigma, USA). Lysates of DNA for confirmation of ESBL/AmpC genes, PCR Based Replicon Typing (PBRT), plasmid Multi Locus Sequence Typing (pMLST), plasmid Double Locus Sequence Typing (pDLST) and plasmid Replicon Sequence Typing (pRST) were obtained from transformants and transconjugants. PBRT was performed using the PBRT-kit (Diatheva, Italy) following the manufacturer instructions. Noteworthy, the kit did not differentiate between IncI1 or IncIγ plasmids. For readability all IncI1-Iγ plasmids are here designated as IncI1. Plasmid MLST, pDLST and pRST were performed as previously described for IncI1, IncHI2 and IncF plasmids, respectively [31–33].
For Multi Locus Sequence Typing (MLST), bacterial lysates were obtained after boiling the original donor strain and PCR protocols were performed as previously described at http://mlst.warwick.ac.uk/mlst/dbs/Ecoli/. PCR products were purified and sequenced as described above.
Allele and profile analysis for pMLST, pDLST and pRST was done at http://pubmlst.org/plasmid/. Sequences and minimal spanning trees for MLST were analysed with BioNumerics v7.5 software (Applied Maths, Belgium).
Results
Screening of resistance, ESBL/AmpC genes and selection of isolates for molecular characterization
In total 132 of 541 strains were phenotypically resistant to cefotaxime (MIC≥ 4mg/L), 13 from farms, 25 from slaughterhouses and 94 from retail (Table 1). From these, 122 were positive for ESBL and/or AmpC genes, 11 from farms, 24 from slaughterhouses and 87 from retail (Table 1). The most prevalent gene variant at the farm, slaughterhouse and retail level was blaCMY-2, present in 73% (n = 8/11), 58% (n = 14/24) and 80% (n = 70/87), respectively. Second most prevalent was blaSHV-12, present in 18% (n = 2/11), 21% (n = 5/24) and 8% (n = 7/87), respectively (Table 1). In addition to these gene variants we found blaSHV-5 (n = 3), blaCTX-M-15 (n = 1), blaCTX-M-2 (n = 5), blaCTX-M-8 (n = 1) and combinations of blaCMY-2-blaSHV-12 (n = 4) and blaCMY-2-blaSHV-5 (n = 2) distributed in the three levels. After sequencing, all positive reactions for blaTEM PCR, were identified as blaTEM-1 or blaTEM-1b and were considered as wild-type beta-lactamase. None of the isolates were positive for blaOXA-1-like, -2-like or -10-like genes.
Based on the distribution of gene variants, 39 isolates were selected and further characterized at the plasmid level using PBRT, pMLST, pDLST and pRST (if applicable) and at the strain level using MLST. The selection was aimed to investigate the diversity of genetic determinants and cover as much plasmid and strain STs as possible. Accordingly, all strains positive for blaCTX-M (n = 7) and all for blaCMY-blaSHV (n = 6) were included to cover the diversity or homogeneity of plasmids and strains associated to their spread. In contrast, a random selection was performed for the strains positive for blaCMY (n = 92) and blaSHV (n = 17) (Table 1).
Plasmid typing
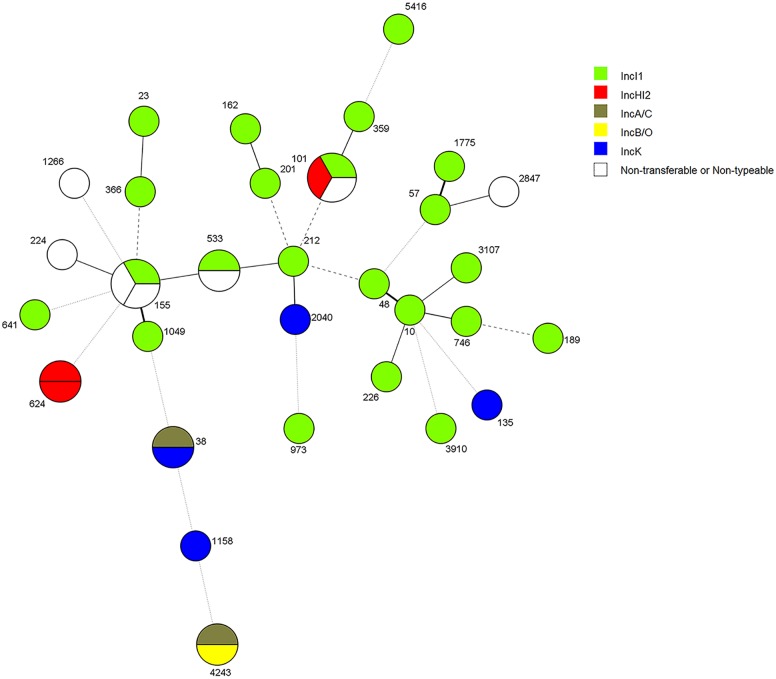
ESBL/AmpC genes from 36 strains were transferable using electroporation or conjugation. In the remaining 3 strains (EC1.6, EC10PP62457 and UGCAR489EC) the genes were non-transferable (Table 2). The transferred genes of 33 strains were located on a single plasmid replicon in the recipient strain after selective isolation. For 1 strain the plasmids remained non-typeable (Fig 1). IncI1 plasmids were most abundant in isolates from all years, sources and department of origin of the samples (22/33), except for the department of Bolivar (Table 2). IncK (4/33), IncHI2 (3/33), IncA/C (2/33), IncΒ/O (1/33) and a non-typeable replicon (strain FBOG54) were also associated with ESBL/AmpC-genes at a low frequency (Fig 1 and Table 2). Overall, IncI1 plasmids were associated with carriage of blaCMY-2, blaSHV-12 and blaCTX-M-8 genes, IncHI2 with carriage of blaCTX-M-2. Further plasmid typing, pMLST for IncI1 and pDLST for IncHI2, demonstrated IncI1/ST12 (13/22) and the new IncHI2/ST7 (2/3) as the most abundant plasmid Sequence Types, respectively (Table 2). In addition to IncHI2/ST7, two new IncI1 STs were observed, designated as ST229 and ST230 (Table 2). Three IncI1 and one IncHI2 plasmid remained non-typeable due to missing amplification of one of the alleles in the pMLST and pDLST schemes (Table 2). Information relating to the year, source and origin of the strains with their respective gene, ST, incompatibility group (PBRT) and plasmid ST is shown in Table 2.
Table 2. Strain and plasmid typing on selected strains after ESBL/AmpC genes characterization.
| Source | Year | Location (Department) | Strain | ESBL/AmpC enzymes in original isolatesa | Plasmid replicons | Plasmid ST/FAB formula | Strain ST | Clonal Complex |
|---|---|---|---|---|---|---|---|---|
| FARM | 2008 | Santander | ECSLV.10.C1 | SHV-12 | I1b | Non-typeablee | 366 | - |
| Santander | ECSXXXIV.1.C | CMY-2 | I1b | 12 | 57 | 350 | ||
| 2009 | Cundinamarca | EC1.6 | CTX-M-2 | - | - | 1266 | - | |
| Cundinamarca | EC4.5 | CMY-2 | I1b | Non-typeablee | 101 | 101 | ||
| SLAUGHTER | 2008 | Santander | ECSIIIL.9.C1 | CMY-2 | I1-Fd | 12-F24:A-:B1 | 155 | 155 |
| Santander | ECSIIL.18.C2 | SHV-12 | I1b | 12 | 3107 | - | ||
| Santander | ECSIVL.9 | CTX-M-2 | HI2b | Non-typeablee | 101 | 101 | ||
| Santander | ECSLII.8.C3 | CMY-2 | I1b | 12 | 155 | 155 | ||
| Santander | ECSLIV.27.C2 | CMY-2 | I1b | 12 | 10 | 10 | ||
| Santander | ECSLIV.7.C1 | CMY-2 | I1b | Non-typeablee | 3910 | - | ||
| Santander | ECSVIL.11.C1 | CMY-2/SHV-12 | I1b | 12 | 359 | - | ||
| Santander | ECSVIL.21.C3 | CMY-2 | I1b | 12 | 212 | - | ||
| Santander | ECSXXXII.7 | CTX-M-2 | A/Cb | 38 | 38 | |||
| 2012 | Cundinamarca | FBOG54 | SHV-5 | Non-typeableb | 155 | 155 | ||
| Santander | FSAN126 | CMY-2 | I1b | 231 | 1775 | - | ||
| 2013 | Sucre | FSUC314 | CTX-M-8 | I1b | 114 | 641 | 86 | |
| Magdalena | FMAG347 | SHV-12 | I1c | 230 g | 226 | 226 | ||
| RETAIL | 2009 | Cundinamarca | EC102.1 | CMY-2 | I1c | 229 g | 23 | 23 |
| Cundinamarca | EC107.1 | CTX-M-2 | HI2b | 7 g | 624 | 648 | ||
| Cundinamarca | EC108.1 | CTX-M-2 | HI2b | 7 g | 624 | 648 | ||
| Cundinamarca | EC114.1 | CMY-2/SHV-12 | B/Ob | 4243 | - | |||
| Cundinamarca | EC10PP62457 | CMY-2 | - | - | 101 | 101 | ||
| Cundinamarca | EC9PP62328 | SHV-12 | I1b | 230 g | 973 | - | ||
| 2010 | Cundinamarca | UGBOG166EC | CMY-2 | Kb | 2040 | - | ||
| Cundinamarca | UGBOG204EC | CMY-2/SHV-5 | I1b | 12 | 189 | 165 | ||
| Cundinamarca | UGBOG301EC | CMY-2 | I1b | 12 | 201 | 469 | ||
| Cundinamarca | UGBOG304EC | CMY-2/SHV-12 | Kb/ I1bf | 26 | 746 | - | ||
| Cundinamarca | UGBOG34EC | CMY-2 | I1b | 12 | 48 | 10 | ||
| Meta | UGVIL369EC | CMY-2/SHV-12 | I1b | 12 | 1049 | - | ||
| Meta | UGVIL380EC | CMY-2 | Kc | 135 | - | |||
| 2011 | Atlántico | UGBAR389EC | CMY-2 | I1b | 12 | 5416 | - | |
| Atlántico | UGBAR425EC | CMY-2 | I1b | 12 | 533 | - | ||
| Atlántico | UGBAR428EC | CMY-2 | I1-Fd | 12-F29:A-:B- | 533 | - | ||
| Bolívar | UGCAR489EC | CTX-M-15 | - | - | 224 | - | ||
| Bolívar | UGCAR500EC | CMY-2 | Kb | 1158 | - | |||
| Cundinamarca | UGCAR511EC | CMY-2 | A/Cc | 4243 | - | |||
| Córdoba | UGMON457EC | CMY-2/SHV-5 | I1b | 12 | 162 | 469 | ||
| Boyacá | UGSAN546EC | CMY-2 | I1-Fd | Non-typeablee-F36:A1:B1 | 2847 | - | ||
| Boyacá | UGTUN878EC | CMY-2 | Kb | 38 | 38 |
a Underlined genes were transferable to recipient strains by electroporation or conjugation of plasmids
b Single replicon transferred by electroporation or
c by conjugation
d Both replicons were transferred together by conjugation. It is uncertain in which of the plasmids the ESBL/AmpC gene was transferred
e No PCR amplification product for one of the alleles
f blaCMY-2 and blaSHV-12 were transferred in IncK and IncI1 plasmids respectively
g New plasmid sequence types
Fig 1. Distribution of plasmid families in ESBL/AmpC-producing E. coli from different Sequence Types in the Colombian poultry chain.
Strain typing
Thirty-one different Sequence Types (STs) were detected among the 39 strains selected (Fig 1 and Table 2). Sequence Types ST38 (n = 2), ST101 (n = 3), ST155 (n = 3), ST533 (n = 2), ST624 (n = 2) and ST4243 (n = 2) were found more than once. ST155 and ST4243 were found in slaughterhouses and retail, respectively. ST533 and ST624 were found both in retail but ST533 in 2011 and ST624 in 2009. ST38 was encountered in different years and sources. Finally, ST101 was found in samples from farms (n = 1), slaughterhouses (n = 1) and retail (n = 1) in different years.
Discussion
We have provided the molecular characterization of ESBL/AmpC-producing E. coli from different base-line studies within the Colombian poultry production chain. Our selection aimed to reflect the characteristics of resistance in different Colombian departments in order to comply with the country-wide scope of an integrated program like Coipars. We have found that the most prevalent genes and plasmids were present in highly heterogeneous strain STs, which may well be indicative for non-clonal spread of resistance within the poultry chain. In addition, our results suggest that dissemination of the most prevalent genes, blaCMY-2 and blaSHV-12, is mainly through horizontal transfer of plasmids belonging to the IncI1 group.
Among the systems providing automation to the phenotypic screening of ESC resistance, Vitek 2 (bioMérieux) and BD Phoenix have been used in the investigation of ESBL-producing E. coli in animal isolates [34–37]. Although Vitek 2 is most commonly used, no dedicated assessment of their performance for isolates from animal origin is available in the literature. When testing human isolates in studies from Europe and the USA, Phoenix has been found to provide a better sensitivity and specifity in comparison with other commercial systems [38–41]. In our study, Phoenix was used for detection of resistance to cefotaxime regardless of its ESBL evaluation. In general, this drug is preferred among other cephalosporins given its advantage to cover both ESBL and plasmidic AmpC phenotypes [42]. In this respect, the automated phenotypic screening of cefotaxime resistance enabled the analysis of a large number of isolates with good discriminatory power. In total 122 out of 132 resistant strains were found to carry ESBL/AmpC genes, showing good performance of the automated system to screen large bacterial collections. The remaining isolates may be chromosomal ampC-promoter mutants or harbour a gene we did not screen for.
Our study has some limitations related to the limited number of isolates from farms and slaughterhouses from which a convenient selection of isolates was available. The exclusion of epidemiologically related isolates resulted in a marked reduction of samples, from 155 to 45 at farms and from 182 to 66 at slaughter. This exclusion may have influenced the prevalence of genes, plasmids and strains we encountered since it resulted in the uneven distribution of samples between the 3 levels of production. Additionally it may have resulted in loss of coverage of the diversity of genetic determinants within same production flocks. Nevertheless, this was a requisite to avoid general overrepresentation of genes, plasmids and strains due to sample duplicity. Thus, the characteristics of resistance were reported individually for the 3 different levels and our results served principally as a base line for the Colombian poultry chain. In addition, we included all isolates from retail and finally found that the proportion of the two most prevalent genes, blaCMY-2 and blaSHV-12, was equal in the three sources. This strongly suggests that in our collection of E. coli the distribution of ESBL/AmpC genes in the 3 levels of production we sampled are comparable.
Gene frequencies
Other studies of ESBL/AmpC-producing E. coli from poultry in Latin America are scarce. However, available reports from Brazil have showed that together with blaCMY-2 [43], blaCTX-M-2 and blaCTX-M-8 are important mediators of ESC-resistance in E. coli from poultry and chicken meat [44–48]. It is important to note that some of these studies only screened for blaCTX-M and did not include detection of plasmidic AmpC genes in their PCR screening. This will affect the accuracy of a comparison with the prevalences described in this study. Furthermore, studies from the USA and Canada demonstrated that resistance to third generation cephalosporins in samples from chicken meat is predominantly mediated by blaCMY-2 [49–54]. In our study, the prevalence of blaCMY-2 was higher than other genes within the three sources. We found a proportion of over 58% in ESBL/AmpC positive strains from farms, slaughter and retail. In our case, the encountered prevalence was based on random isolation of E. coli (that is, without selection using antimicrobials during enrichment). Therefore, the frequencies of other genes than blaCMY-2 may be underestimated and comparisons with reports using selective isolation with cephalosporins during enrichment have to be interpreted carefully. Our finding of this gene as the main mediator of ESC resistance, is in accordance with previous studies (using selective isolation) from European countries like Denmark, Sweden, and the Netherlands where blaCMY-2 was found to be highly prevalent at grandparent and parent level of the broiler production pyramid [55–57].
This is the first publication from Colombia in relation to ESBL/AmpC—producing E coli from poultry and chicken meat. In reports from human isolates, blaCTX-M-12, blaCTX-M-15, blaSHV-12 and blaSHV-5 [19–22] were most prevalent. More recently, blaCMY-2 has been included as part of the genotypic screening and associated to community onset infections caused by E. coli ST131 [23]. Although previous studies did not characterize the plasmids mediating the resistance, our findings suggest a possible relationship with some of the ESBL/AmpC genes found in humans in Colombia, namely blaCMY-2, blaSHV-12, blaSHV-5 and blaCTX-M-15. Further studies using a one health approach, simultaneous collection of samples from human and animal sources and sequence-based molecular characterization of plasmids and strains are necessary to confirm this relationship.
ESBL/AmpC carrying plasmids
Various plasmid families, namely IncI1, IncK, IncF, IncHI2, IncA/C and IncΒ/O were found harbouring ESBL/AmpC genes. However, we have shown that the association of blaCMY-2 and blaSHV-12 was mainly to IncI1 plasmids. Both the diversity of the analysed strains (year, source and place of isolation) and the diversity in E. coli STs, in contrast to the homogeneity in plasmid types, support the hypothesis that spread of ESBL/AmpC in the Colombian poultry chain is mainly mediated by horizontal gene transfer of plasmids. A minimal spanning tree representing the genetic relatedness of E. coli STs and plasmid lineages is shown in Fig 1.
In contrast to strain Sequence Types, there is evidence that supports the association between specific plasmid Sequence Types and the spread of particular ESBL/AmpC genes. In a previous study, Smith and colleagues [58] reported the combination of IncI1/ST12 and blaCMY-2 in more than 80% of the plasmids they analysed. Among our isolates, blaCMY-2 was the most frequently encountered gene, and IncI1/ST12 was most frequently associated to its spread. In particular, we found 12 isolates carrying blaCMY-2 in IncI1/ST12 plasmids out of 16 carrying blaCMY-2 in overall IncI1 plasmids. It is likely that this combination of plasmid ST-gene is widespread in Colombian poultry since blaCMY-2 was the most prevalent gene in our study and IncI1 accounted for most of its occurrence (16/24). The features associated to this plasmid ST and its interactions with E. coli hosts are of further interest to assess the factors influencing the successful spread of blaCMY-2 in poultry.
Sequence types of ESBL/AmpC producers
As a result of our selection, different samples from all years and levels of production were characterized. Some departments were represented with more than one sample, namely Cundinamarca (n = 15) Santander (12), Atlántico (3), Meta (2), Bolívar (2) and Boyacá (2). In addition single samples from Sucre, Magdalena and Córdoba were included. In general, we observed high heterogeneity of strain STs harbouring same combinations of gene variants and plasmids (Fig 1 and Table 2). In addition, but to a lesser extent, samples originating from the same years, levels of production and departments carried the same genes, plasmids and strain STs, as observed for ST155 (Santander), ST624 (Cundinamarca) and ST533 (Atlántico).
From a plasmid perspective, IncI1 harbouring blaCMY was distributed among highly heterogeneous E. coli STs from different years, levels of production and departments. However, it was also present among the closely related E. coli ST155 and ST533, in both cases carrying blaCMY-2 on an IncI1/ST12 plasmid (Table 2). In this respect, we conclude our selection of strains was sufficient to detect inter-department diversity and intra-department diversity to a limited level, and also the likelihood that in one geographical area E. coli belonging to the same ST exist. Nonetheless, an in-depth analysis for the spread within departments was out of the scope of the present study. This would require a much more intensive sampling scheme.
At a broader scale, seven out of 31 STs have been previously reported as ESBL/AmpC producers in samples from both humans and poultry, namely ST10, ST23, ST38, ST48, ST57, ST155 and ST624 [11,14,59–64]. One has only been found as ESBL/AmpC producer in poultry, ST641 [11,60,65,66] and 3 in humans, ST101, ST162 and ST746 [63,67]. Additionally, ST224, ST359 and ST973 have been previously reported in other livestock and companion animals [65,68,69]. To our knowledge this is the first report in which ESBL/AmpC producers are associated with ST135, ST189, ST201, ST212, ST226, ST366, ST533, ST1049, ST1158, ST1266, ST1775, ST2040, ST2847, ST 3107, ST3910, ST4243 and ST5416. At the level of isolates from human clinical isolates in Colombia, only two strains belonging to ST38 coincide in our selection. Different than blaCTX-M-15 [24] our strains carried blaCMY-2 or blaCTX-M-2 and were not considered as an evident link for the transmission of resistance.
In conclusion, the molecular characterization of ESBL/AmpC-producing E. coli has identified the genetic determinants mediating the spread of resistance to ESC in Colombian poultry and chicken meat. The differences in distribution of genes, plasmids and strains between our study and other reports may be related to different practices of farming and supply of chickens and chicken meat in the country. Further characterization of ESBL/AmpC-producing E. coli from human and poultry sources in Colombia is necessary to understand the potential transmission of resistance determinants. This approach could be enhanced by the use of next generation sequencing of isolates and plasmids.
Supporting Information
(XLSX)
Acknowledgments
The authors wish to thank Carolina Duarte from Instituto Nacional de Salud and Yamile Celis from Secretaría Distrital de Salud for valuable input from the human health sector in Colombia, Massiel Rivera and María Valencia from Corpoica for laboratory assistance, Marta Rozwandowicz for information of genes, plasmids and Sequence Types of ESBL/AmpC-producing E. coli and Alessandra Carattoli for assignment of novel Sequence Types to IncI1 and IncHI2 plasmids. WHO-AGISAR is acknowledged for facilitating the exchange of researchers and knowledge between research groups.
Data Availability
All relevant data are within the paper and its Supporting Information file.
Funding Statement
The authors received no specific funding for this work.
References
- 1.Leistner R, Gürntke S, Sakellariou C, Denkel LA, Bloch A, Gastmeier P, et al. Bloodstream infection due to extended-spectrum beta-lactamase (ESBL)-positive K. pneumoniae and E. coli: an analysis of the disease burden in a large cohort. Infection. 2014;42. [DOI] [PubMed] [Google Scholar]
- 2.Macvane SH, Tuttle LO, Nicolau DP. Impact of extended-spectrum β-lactamase-producing organisms on clinical and economic outcomes in patients with urinary tract infection. J Hosp Med. 2014;9: 232–238. 10.1002/jhm.2157 [DOI] [PubMed] [Google Scholar]
- 3.Jean S-S, Ko W-C, Xie Y, Pawar V, Zhang D, Prajapati G, et al. Clinical characteristics of patients with community-acquired complicated intra-abdominal infections: A prospective, multicentre, observational study. Int J Antimicrob Agents. 2014;44: 222–228. 10.1016/j.ijantimicag.2014.05.018 [DOI] [PubMed] [Google Scholar]
- 4.Meunier D, Jouy E, Lazizzera C, Kobisch M, Madec JY. CTX-M-1- and CTX-M-15-type β-lactamases in clinical Escherichia coli isolates recovered from food-producing animals in France. Int J Antimicrob Agents. 2006;28: 402–407. 10.1016/j.ijantimicag.2006.08.016 [DOI] [PubMed] [Google Scholar]
- 5.Teshager T, Dominguez L, Moreno MA, Saenz Y, Torres C, Cardenosa S. Isolation of an SHV-12 beta -Lactamase-Producing Escherichia coli Strain from a Dog with Recurrent Urinary Tract Infections. Antimicrob Agents Chemother. 2000;44: 3483–3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jacoby GA, Han P. Detection of extended-spectrum beta-lactamases in clinical isolates of Klebsiella pneumoniae and Escherichia coli. J Clin Microbiol. 1996;34: 908–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jacoby GA. AmpC beta-lactamases. Clin Microbiol Rev. 2009;22: 161–82. 10.1128/CMR.00036-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ewers C, Bethe A, Semmler T, Guenther S, Wieler LH. Extended-spectrum β-lactamase-producing and AmpC-producing Escherichia coli from livestock and companion animals, and their putative impact on public health: A global perspective. Clin Microbiol Infect. 2012. pp. 646–655. [DOI] [PubMed] [Google Scholar]
- 9.Zurfluh K, Nüesch-Inderbinen M, Morach M, Berner AZ, Hächler H, Stephan R. Extended-spectrum-β-lactamase-producing Enterobacteriaceae isolated from vegetables imported from the Dominican Republic, India, Thailand, and Vietnam. Appl Environ Microbiol. 2015;81: 3115–3120. 10.1128/AEM.00258-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hasman H, Mevius D, Veldman K, Olesen I, Aarestrup FM. β-Lactamases among extended-spectrum β-lactamase (ESBL)-resistant Salmonella from poultry, poultry products and human patients in The Netherlands. J Antimicrob Chemother. 2005;56: 115–121. 10.1093/jac/dki190 [DOI] [PubMed] [Google Scholar]
- 11.Leverstein-van Hall MA, Dierikx CM, Cohen Stuart J, Voets GM, van den Munckhof MP, van Essen-Zandbergen A, et al. Dutch patients, retail chicken meat and poultry share the same ESBL genes, plasmids and strains. Clin Microbiol Infect. 2011;17: 873–880. 10.1111/j.1469-0691.2011.03497.x [DOI] [PubMed] [Google Scholar]
- 12.Kluytmans JAJW, Overdevest ITMA, Willemsen I, Kluytmans-Van Den Bergh MFQ, Van Der Zwaluw K, Heck M, et al. Extended-spectrum β-lactamase-producing Escherichia coli from retail chicken meat and humans: Comparison of strains, plasmids, resistance genes, and virulence factors. Clin Infect Dis. 2013;56: 478–487. 10.1093/cid/cis929 [DOI] [PubMed] [Google Scholar]
- 13.Voets GM, Fluit AC, Scharringa J, Schapendonk C, van den Munckhof T, Leverstein-van Hall MA, et al. Identical plasmid AmpC beta-lactamase genes and plasmid types in E. coli isolates from patients and poultry meat in the Netherlands. Int J Food Microbiol. 2013;167: 359–362. 10.1016/j.ijfoodmicro.2013.10.001 [DOI] [PubMed] [Google Scholar]
- 14.Dierikx C, van der Goot J, Fabri T, van Essen-Zandbergen A, Smith H, Mevius D. Extended-spectrum-β-lactamase- and AmpC-β-lactamase-producing Escherichia coli in Dutch broilers and broiler farmers. J Antimicrob Chemother. 2013;68: 60–67. 10.1093/jac/dks349 [DOI] [PubMed] [Google Scholar]
- 15.Huijbers PMC, Graat EAM, Haenen APJ, van Santen MG, van Essen-Zandbergen A, Mevius DJ, et al. Extended-spectrum and AmpC beta-lactamase-producing Escherichia coli in broilers and people living and/or working on broiler farms: prevalence, risk factors and molecular characteristics. J Antimicrob Chemother. 2014;69: 2669–2675. 10.1093/jac/dku178 [DOI] [PubMed] [Google Scholar]
- 16.Overdevest I, Willemsen I, Rijnsburger M, Eustace A, Xu L, Hawkey P, et al. Extended-spectrum β-lactamase genes of Escherichia coli in chicken meat and humans, the Netherlands. Emerg Infect Dis. 2011;17: 1216–1222. 10.3201/eid1707.110209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Been M, Lanza VF, de Toro M, Scharringa J, Dohmen W, Du Y, et al. Dissemination of Cephalosporin Resistance Genes between Escherichia coli Strains from Farm Animals and Humans by Specific Plasmid Lineages. PLoS Genet. 2014;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lazarus B, Paterson DL, Mollinger JL, Rogers BA. Do human extraintestinal Escherichia coli infections resistant to expanded-spectrum cephalosporins originate from food-producing animals? A systematic review. Clin Infect Dis. 2015. pp. 439–452. 10.1093/cid/ciu785 [DOI] [PubMed] [Google Scholar]
- 19.Villegas MV, Correa A, Perez F, Miranda MC, Zuluaga T, Quinn JP. Prevalence and characterization of extended-spectrum β-lactamases in Klebsiella pneumoniae and Escherichia coli isolates from Colombian hospitals. Diagn Microbiol Infect Dis. 2004;49: 217–222. 10.1016/j.diagmicrobio.2004.03.001 [DOI] [PubMed] [Google Scholar]
- 20.Espinal P, Garza-Ramos U, Reyna F, Rojas-Moreno T, Sanchez-Perez A, Carrillo B, et al. Identification of SHV-type and CTX-M-12 extended-spectrum beta-lactamases (ESBLs) in multiresistant Enterobacteriaceae from Colombian Caribbean hospitals. J Chemotherapy. 2010. pp. 160–4. [DOI] [PubMed] [Google Scholar]
- 21.Ruiz SJ, Montealegre MC, Ruiz-Garbajosa P, Correa A, Briceño DF, Martinez E, et al. First characterization of CTX-M-15-producing Escherichia coli ST131 and ST405 clones causing community-onset infections in South America. J Clin Microbiol. 2011;49: 1993–1996. 10.1128/JCM.00045-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martinez P, Garzón D, Mattar S. CTX-M-producing Escherichia coli and Klebsiella pneumoniae isolated from community-acquired urinary tract infections in Valledupar, Colombia. Brazilian J Infect Dis. 2012;16: 420–425. [DOI] [PubMed] [Google Scholar]
- 23.Leal AL, Cortés JA, Arias G, Ovalle MV, Saavedra SY, Buitrago G, et al. Emergence of resistance to third generation cephalosporins by Enterobacteriaceae causing community-onset urinary tract infections in hospitals in Colombia. Enferm Infecc Microbiol Clín. 2013;31: 298–303. 10.1016/j.eimc.2012.04.007 [DOI] [PubMed] [Google Scholar]
- 24.Blanco VM, Maya JJ, Correa A, Perenguez M, Munoz JS, Motoa G, et al. Prevalence and risk factors for extended-spectrum beta-lactamase-producing Escherichia coli causing community-onset urinary tract infections in Colombia. Enferm Infecc Microbiol Clin. 2016; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Donado-Godoy P, Castellanos R, León M, Arevalo A, Clavijo V, Bernal J, et al. The establishment of the Colombian Integrated Program for Antimicrobial Resistance Surveillance (COIPARS): A pilot project on poultry farms, slaughterhouses and retail market. Zoonoses Public Health. 2015;62: 58–69. 10.1111/zph.12192 [DOI] [PubMed] [Google Scholar]
- 26.Donado-Godoy P, Gardner I, Byrne BA, León M, Perez-Gutierrez E, Ovalle MV, et al. Prevalence, Risk Factors, and Antimicrobial Resistance Profiles of Salmonella from Commercial Broiler Farms in Two Important Poultry-Producing Regions of Colombia. J Food Prot. 2012;75: 874–883. 10.4315/0362-028X.JFP-11-458 [DOI] [PubMed] [Google Scholar]
- 27.Canadian Integrated program for Antimicrobial Resistance Surveillance (CIPARS). Annual Report. Appendix A—Methods. In: http://www.phac-aspc.gc.ca/cipars-picra/2008/6-eng.php. 2008.
- 28.Clinical and Laboratory Standards Institute (CLSI), 2013. Performance Standards for Antimicrobial Susceptibility Testing; Twenty-third Informational Supplement CLSI document M100-S23, Clinical and Laboratory Standards Institute, Wayne, PA, USA. [Google Scholar]
- 29.Dierikx CM, van Duijkeren E, Schoormans AHW, van Essen-Zandbergen A, veldman K, Kant A, et al. Occurrence and characteristics of extended-spectrum-β-lactamase- and AmpC-producing clinical isolates derived from companion animals and horses. J Antimicrob Chemother. 2012;67: 1368–1374. 10.1093/jac/dks049 [DOI] [PubMed] [Google Scholar]
- 30.Dierikx C, van Essen-Zandbergen A, Veldman K, Smith H, Mevius D. Increased detection of extended spectrum beta-lactamase producing Salmonella enterica and Escherichia coli isolates from poultry. Vet Microbiol. 2010;145: 273–278. 10.1016/j.vetmic.2010.03.019 [DOI] [PubMed] [Google Scholar]
- 31.García-Fernández A, Chiaretto G, Bertini A, Villa L, Fortini D, Ricci A, et al. Multilocus sequence typing of IncI1 plasmids carrying extended-spectrum β-lactamases in Escherichia coli and Salmonella of human and animal origin. J Antimicrob Chemother. 2008;61: 1229–1233. 10.1093/jac/dkn131 [DOI] [PubMed] [Google Scholar]
- 32.García-Fernández A, Carattoli A. Plasmid double locus sequence typing for IncHI2 plasmids, A subtyping scheme for the characterization of IncHI2 plasmids carrying extended-spectrum β-lactamase and quinolone resistance genes. J Antimicrob Chemother. 2010;65: 1155–1161. 10.1093/jac/dkq101 [DOI] [PubMed] [Google Scholar]
- 33.Villa L, García-Fernández A, Fortini D, Carattoli A. Replicon sequence typing of IncF plasmids carrying virulence and resistance determinants. J Antimicrob Chemother. 2010;65: 2518–2529. 10.1093/jac/dkq347 [DOI] [PubMed] [Google Scholar]
- 34.El-Jade MR, Parcina M, Schmithausen RM. ESBL Detection: Comparison of a Commercially Available Chromogenic Test for Third Generation Cephalosporine Resistance and Automated Susceptibility Testing in Enterobactericeae. PLoS One. 2016; 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fadlelmula A, Al-hamam NA, Al-dughaym AM. A potential camel reservoir for extended-spectrum β-lactamase-producing Escherichia coli causing human infection in Saudi Arabia. Trop Anim Health Prod. 2016; 427–433. 10.1007/s11250-015-0970-9 [DOI] [PubMed] [Google Scholar]
- 36.Abreu R, Castro B, Espigares E, Rodríguez-Álvarez C, et al. Prevalence of CTX-M-Type Extended-Spectrum β-Lactamases in Escherichia coli Strains. Foodborne Pathog Dis. 2014;11: 868–874. 10.1089/fpd.2014.1796 [DOI] [PubMed] [Google Scholar]
- 37.Braun SD, Ahmed MFE, El-Adawy H, Hotzel H, Engelmann I, Weiß D, et al. Surveillance of Extended-Spectrum Beta-Lactamase-Producing Escherichia coli in Dairy Cattle Farms in the Nile Delta, Egypt. Front Microbiol. 2016;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hall MAL, Fluit AC, Paauw A, Box ATA, Brisse S, Verhoef J. Evaluation of the Etest ESBL and the BD Phoenix, VITEK 1, and VITEK 2 Automated Instruments for Detection of Extended-Spectrum Beta-Lactamases in Multiresistant Escherichia coli and Klebsiella spp. J Clin Microbiol. 2002;40: 3703–3711. 10.1128/JCM.40.10.3703-3711.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hall MAL, Waar K, Muilwijk J, Stuart JC. Consequences of switching from a fixed 2: 1 ratio of amoxicillin/clavulanate (CLSI) to a fixed concentration of clavulanate (EUCAST) for susceptibility testing of Escherichia coli. J Antimicrob Chemother. 2013; 2636–2640. 10.1093/jac/dkt218 [DOI] [PubMed] [Google Scholar]
- 40.Thomson KS, Cornish NE, Hong SG, Hemrick K, Herdt C, Moland ES. Comparison of Phoenix and VITEK 2 Extended-Spectrum-β-Lactamase Detection Tests for Analysis of Escherichia coli and Klebsiella Isolates with Well-Characterized β-Lactamases. J Clin Microbiol. 2007;45: 2380–2384. 10.1128/JCM.00776-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Winstanley T, Courvalin P. Expert Systems in Clinical Microbiology. Clin Microbiol Rev. 2011;24: 515–556. 10.1128/CMR.00061-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aarestrup FM, Hasman H, Veldman K, Mevius D. Evaluation of eight different cephalosporins for detection of cephalosporin resistance in Salmonella enterica and Escherichia coli. Microb Drug Resist. 2010;16: 253–261. 10.1089/mdr.2010.0036 [DOI] [PubMed] [Google Scholar]
- 43.Botelho LAB, Kraychete GB, e Silva JLC, Regis DVV, Picão RC, Moreira BM, et al. Widespread distribution of CTX-M and plasmid-mediated AmpC β-lactamases in Escherichia coli from Brazilian chicken meat. Mem Inst Oswaldo Cruz. 2015;110: 249–254. 10.1590/0074-02760140389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Koga VL, Scandorieiro S, Vespero EC, Oba A, De Brito BG, De Brito KCT, et al. Comparison of Antibiotic Resistance and Virulence Factors among Escherichia coli Isolated from Conventional and Free-Range Poultry. Biomed Res Int. 2015;2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Koga VL, Rodrigues GR, Scandorieiro S, Vespero EC, Oba A, de Brito BG, et al. Evaluation of the Antibiotic Resistance and Virulence of Escherichia coli Strains Isolated from Chicken Carcasses in 2007 and 2013 from Paraná, Brazil. Foodborne Pathog Dis. 2015;12: 479–85. 10.1089/fpd.2014.1888 [DOI] [PubMed] [Google Scholar]
- 46.Ferreira JC, Penha Filho RAC, Andrade LN, Berchieri Junior A, Darini ALC. IncI1/ST113 and IncI1/ST114 conjugative plasmids carrying blaCTX-M-8 in Escherichia coli isolated from poultry in Brazil. Diagn Microbiol Infect Dis. 2014;80: 304–306. 10.1016/j.diagmicrobio.2014.09.012 [DOI] [PubMed] [Google Scholar]
- 47.Ferreira JC, Penha Filho RAC, Andrade LN, Berchieri A, Darini ALC. Detection of chromosomal blaCTX-M-2 in diverse Escherichia coli isolates from healthy broiler chickens. Clin Microbiol Infect. 2014;20: O623–O626. 10.1111/1469-0691.12531 [DOI] [PubMed] [Google Scholar]
- 48.Ferreira JC, Penha Filho RAC, Andrade LN, Berchieri Junior A, Darini ALC. Evaluation and characterization of plasmids carrying CTX-M genes in a non-clonal population of multidrug-resistant Enterobacteriaceae isolated from poultry in Brazil. Diagn Microbiol Infect Dis. 2016; [DOI] [PubMed] [Google Scholar]
- 49.Doi Y, Paterson DL, Egea P, Pascual A, López-Cerero L, Navarro MD, et al. Extended-spectrum and CMY-type β-lactamase-producing Escherichia coli in clinical samples and retail meat from Pittsburgh, USA and Seville, Spain. Clin Microbiol Infect. 2010;16: 33–38. 10.1111/j.1469-0691.2009.03001.x [DOI] [PubMed] [Google Scholar]
- 50.Zhao S, Blickenstaff K, Bodeis-Jones S, Gaines SA, Tong E, McDermott PF. Comparison of the prevalences and antimicrobial resistances of Escherichia coli isolates from different retail meats in the United States, 2002 to 2008. Appl Environ Microbiol. 2012;78: 1701–1707. 10.1128/AEM.07522-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Park YS, Adams-Haduch JM, Rivera JI, Curry SR, Harrison LH, Doi Y. Escherichia coli producing CMY-2 β-lactamase in retail chicken, pittsburgh, pennsylvania, USA. Emerg Infect Dis. 2012. pp. 515–516. 10.3201/eid1803.111434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Glenn LM, Englen MD, Lindsey RL, Frank JF, Turpin JE, Berrang ME, et al. Analysis of antimicrobial resistance genes detected in multiple-drug-resistant Escherichia coli isolates from broiler chicken carcasses. Microb Drug Resist. 2012;18: 453–63. 10.1089/mdr.2011.0224 [DOI] [PubMed] [Google Scholar]
- 53.Mollenkopf DF, Cenera JK, Bryant EM, King CA, Kashoma I, Kumar A, et al. Organic or antibiotic-free labeling does not impact the recovery of enteric pathogens and antimicrobial-resistant Escherichia coli from fresh retail chicken. Foodborne Pathog Dis. 2014;11: 920–929. 10.1089/fpd.2014.1808 [DOI] [PubMed] [Google Scholar]
- 54.Sheikh AA, Checkley S, Avery B, Chalmers G, Bohaychuk V, Boerlin P, et al. Antimicrobial resistance and resistance genes in Escherichia coli isolated from retail meat purchased in Alberta, Canada. Foodborne Pathog Dis. 2012;9: 625–31. 10.1089/fpd.2011.1078 [DOI] [PubMed] [Google Scholar]
- 55.Agersø Y, Jensen JD, Hasman H, Pedersen K. Spread of extended spectrum cephalosporinase-producing Escherichia coli clones and plasmids from parent animals to broilers and to broiler meat in a production without use of cephalosporins. Foodborne Pathog Dis. 2014;11: 740–746. 10.1089/fpd.2014.1742 [DOI] [PubMed] [Google Scholar]
- 56.Nilsson O, Börjesson S, Landén A, Bengtsson B. Vertical transmission of Escherichia coli carrying plasmid-mediated AmpC (pAmpC) through the broiler production pyramid. J Antimicrob Chemother. 2014;69: 1497–1500. 10.1093/jac/dku030 [DOI] [PubMed] [Google Scholar]
- 57.Dierikx CM, Van Der Goot JA, Smith HE, Kant A, Mevius DJ. Presence of ESBL/AmpC-producing Escherichia coli in the broiler production pyramid: A descriptive study. PLoS One. 2013;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Smith H, Bossers A, Harders F, Wu G, Woodford N, Schwarz S, et al. Characterization of epidemic IncI1-Iγ plasmids harboring ambler class A and C genes in Escherichia coli and Salmonella enterica from animals and humans. Antimicrob Agents Chemother. 2015;59: 5357–5365. 10.1128/AAC.05006-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ben Sallem R, Ben Slama K, Rojo-Bezares B, Porres-Osante N, Jouini A, Klibi N, et al. IncI1 plasmids carrying blaCTX-M-1 or blaCMY-2 genes in Escherichia coli from healthy humans and animals in Tunisia. Microb Drug Resist. 2014;20: 495–500. 10.1089/mdr.2013.0224 [DOI] [PubMed] [Google Scholar]
- 60.Börjesson S, Jernberg C, Brolund A, Edquist P, Finn M, Landén A, et al. Characterization of plasmid-mediated ampc-producing E. coli from swedish broilers and association with human clinical isolates. Clin Microbiol Infect. 2013;19. [DOI] [PubMed] [Google Scholar]
- 61.Correia S, Pacheco R, Radhouani H, Diniz JC, Ponce P, Jones-Dias D, et al. High prevalence of ESBL-producing Escherichia coli isolates among hemodialysis patients in Portugal: Appearance of ST410 with the blaCTX-M-14 gene. Diagn Microbiol Infect Dis. 2012;74: 423–425. 10.1016/j.diagmicrobio.2012.08.017 [DOI] [PubMed] [Google Scholar]
- 62.Ellis C, Chung C, Tijet N, Patel SN, Desjardins M, Melano RG, et al. OXA-48-like carbapenemase-producing Enterobacteriaceae in Ottawa, Canada. Diagn Microbiol Infect Dis. 2013. pp. 399–400. 10.1016/j.diagmicrobio.2013.04.017 [DOI] [PubMed] [Google Scholar]
- 63.Potron A, Poirel L, Rondinaud E, Nordmann P. Intercontinental spread of OXA-48 beta-lactamase-producing Enterobacteriaceae over a 11-year period, 2001 to 2011. Eurosurveillance. 2013;18. [DOI] [PubMed] [Google Scholar]
- 64.Wang J, Stephan R, Power K, Yan Q, Hächler H, Fanning S. Nucleotide sequences of 16 transmissible plasmids identified in nine multidrug-resistant Escherichia coli isolates expressing an ESBL phenotype isolated from food-producing animals and healthy humans. J Antimicrob Chemother. 2014;69: 2658–2668. 10.1093/jac/dku206 [DOI] [PubMed] [Google Scholar]
- 65.Guo YF, Zhang WH, Ren SQ, Yang L, Lü DH, Zeng ZL, et al. IncA/C plasmid-mediated spread of CMY-2 in multidrug-resistant Escherichia coli from food animals in China. PLoS One. 2014;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vogt D, Overesch G, Endimiani A, Collaud A, Thomann A, Perreten V. Occurrence and genetic characteristics of third-generation cephalosporin-resistant Escherichia coli in Swiss retail meat. Microb Drug Resist. 2014;20: 485–494. 10.1089/mdr.2013.0210 [DOI] [PubMed] [Google Scholar]
- 67.Poirel L, Lagrutta E, Taylor P, Pham J, Nordmann P. Emergence of metallo- β-lactamase NDM-1-producing multidrug-resistant Escherichia coli in Australia. Antimicrob Agents Chemother. 2010;54: 4914–4916. 10.1128/AAC.00878-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dahmen S, Haenni M, Châtre P, Madec JY. Characterization of blaCTX-M IncFII plasmids and clones of Escherichia coli from pets in France. J Antimicrob Chemother. 2013;68: 2797–2801. 10.1093/jac/dkt291 [DOI] [PubMed] [Google Scholar]
- 69.Hordijk J, Mevius DJ, Kant A, Bos MEH, Graveland H, Bosman AB, et al. Within-farm dynamics of ESBL/AmpC-producing Escherichia coli in veal calves: A longitudinal approach. J Antimicrob Chemother. 2013;68: 2468–2476. 10.1093/jac/dkt219 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information file.