Abstract
Stably Expressed Genes (SEGs) whose expression varies within a narrow range may be involved in core cellular processes necessary for basic functions. To identify such genes, we re-analyzed existing RNA-Seq gene expression profiles across 11 organs at 4 developmental stages (from immature to old age) in both sexes of F344 rats (n = 4/group; 320 samples). Expression changes (calculated as the maximum expression / minimum expression for each gene) of >19000 genes across organs, ages, and sexes ranged from 2.35 to >109-fold, with a median of 165-fold. The expression of 278 SEGs was found to vary ≤4-fold and these genes were significantly involved in protein catabolism (proteasome and ubiquitination), RNA transport, protein processing, and the spliceosome. Such stability of expression was further validated in human samples where the expression variability of the homologous human SEGs was significantly lower than that of other genes in the human genome. It was also found that the homologous human SEGs were generally less subject to non-synonymous mutation than other genes, as would be expected of stably expressed genes. We also found that knockout of SEG homologs in mouse models was more likely to cause complete preweaning lethality than non-SEG homologs, corroborating the fundamental roles played by SEGs in biological development. Such stably expressed genes and pathways across life-stages suggest that tight control of these processes is important in basic cellular functions and that perturbation by endogenous (e.g., genetics) or exogenous agents (e.g., drugs, environmental factors) may cause serious adverse effects.
Introduction
The development of genome-wide transcriptional profiling techniques has enabled measurement of the expression of tens of thousands of genes in parallel [1]. A consensus has formed that the status of a cell or an organ is, at least partially, a function of gene expression levels [2]. Frequently, investigations using transcriptomics data identify the genes whose expression levels differ between distinct biological conditions; for example, normal verses diseased tissues [3], or perturbed verses unperturbed samples [4]. Genes that are mostly responsible for the difference between the biological states are defined as differentially expressed genes (DEGs), which aid in the development of biomarkers for clinical diagnosis [5] and the understanding of diverse biological mechanisms of diseases [6, 7].
Genes whose expression remains relatively constant across multiple conditions are important in research methodologies and in understanding gene regulation. For example, quantification of often subtle changes in gene expression by PCR is performed through comparison with the quantity of endogenous controls, such as housekeeping genes [8], whose expression does not change under the experimental conditions. Housekeeping genes are involved in basic cell maintenance and, therefore, are expected to be present in all cells and maintain relatively constant expression levels under different experimental conditions. Identification of these genes increases understanding of various structural genomic features and facilitates the accurate measurement of expression of target genes [9]. Recently, a series of studies demonstrated that variability in gene expression can provide important insights into gene regulatory control related to development [10] and pathogenesis [11]. In an analysis of single cell RNA-seq data derived from embryos, those genes with low expression variability across developmental stages were found to have a functional impact on the regulation of basic cellular processes, and less likely to be associated with loss-of-function variants or copy number variation deletions inducing recessive diseases [12]. In another study, mRNA targets of miRNAs were found to be significantly enriched in those genes with highly stable expression in response to external perturbations, thus validating the hypothesis that miRNAs can buffer mRNA expression fluctuation [13].
The above studies encouraged us to further investigate the importance of stable gene expression in a postnatal healthy untreated animal model. We hypothesized that there are genes whose expression is relatively stable across various organs, ages, and sexes and that these genes are involved in core cellular processes indispensable for survival and growth [14]. A large dataset of gene expression measurements of various organs across the lifespan and in both sexes from a well-controlled model system is required to thoroughly and quantitatively test this hypothesis. The rat has been extensively used by the academic community and pharmaceutical industry as an animal model to identify gene functions, evaluate drug responses and understand human diseases. Recently, Yu et al. [15] used RNA-seq to comprehensively profile gene expression levels in 11 organs at 4 ages (2wk, immature; 6 wk, adolescence; 21 wk, adult; and 104 wk, aged) and both sexes of the untreated Fischer 344 rat. The organs include representatives of most of the major organ systems, including the nervous system (brain), respiratory system (lungs), cardiovascular system (heart), digestive system (liver), endocrine system (adrenal glands), lymphatic system (thymus and spleen), excretory system (kidney), muscular system (muscle), and reproductive system (testes and uterus). There were four biological replicates per organ, age, sex combination resulting in 320 samples. An annotated database has been created as a web-based open-access resource (The Rat BodyMap database; http://pgx.fudan.edu.cn/ratbodymap/) and we have used it to identify stably expressed genes (SEGs). Additional large-scale genomics databases derived from mouse and human were used to explore and confirm the biological importance of these SEGs, including pathway analysis, cross-species consistency, effects of gene knock-out, and extent of disease-causing mutations. Observing the action of their homologs in other biological systems enabled us to elucidate the biological importance of these SEGs. The pathways involving SEGs were found to be core processes, which are important to maintain basic cellular functions. Tight control of these core processes is essential to prevent damage to cells, and any perturbation by endogenous (e.g., genetics) or exogenous agents (e.g., drugs, environmental factors) may cause serious adverse effects.
Materials and Methods
Rat tissue RNA-seq data
The normalized RNA-sequencing data of 320 organ samples (as measured by fragments per kilobase of transcript per million mapped reads, FPKM) from male and female rats of 4 ages (2, 6, 21 and 104 weeks) were downloaded from the Rat BodyMap website (http://pgx.fudan.edu.cn/ratbodymap/). The downloaded RNA-seq data consisted of 40064 transcript expression measurements for 11 organs (adrenal, brain, heart, kidney, liver, lung, muscle, spleen, testis, thymus and uterus). Transcripts without corresponding Entrez Gene ID and transcripts that were not expressed (i.e., FPKM value constantly equals zero) in all samples were excluded. For redundant transcripts (i.e., multiple transcripts corresponding to the same Entrez Gene ID), only the one with the highest average FPKM value across all samples was retained. The minimum non-zero FPKM value was substituted for all remaining zero values. A total of 19343 transcripts, each of which corresponded to a unique rat gene, were selected for subsequent analysis.
Human tissue RNA-seq data
The raw RNA-sequencing data of human samples were retrieved from the NIH common fund's Genotype-Tissue Expression (GTEx) project [16] website (http://www.gtexportal.org/). The current version (V6) of GTEx contains transcriptomics data for 11983 organ samples from 570 subjects belonging to 6 different age brackets (20 yrs and older). The mode of death of these subjects is divided into 6 categories, of which only one category (violent and fast death) did not die of a disease process. Therefore, to avoid major pathological impact on gene expression, only the samples from subjects of violent and fast death were used in this study. To match the spectrum of human organs to the rat organs described above, only 10 types of human tissues were examined, i.e., adrenal gland, brain (cortex), heart (left ventricle), kidney (cortex), liver, lung, muscle (skeletal), spleen, testis and uterus (thymus is absent in adults). Sample characteristics are given in S1 Dataset. Following a procedure similar to the rat gene screening described above, those unnamed, non-expressed and redundant transcripts were excluded. A total of 47495 transcripts, each of which corresponded to a unique human gene, was selected for subsequent analysis.
Genetic damage index
The Genetic Damage Index (GDI) values of 19558 genes were provided in the supplementary data of [17] and downloaded from the Proceedings of the National Academy of Sciences website (http://www.pnas.org/content/112/44/13615/suppl/DCSupplemental). This data was derived from all alleles with a minor allele frequency less than 0.5 in the 1000 Genomes Project [18, 19] and GDI represents the mutational damage in each protein coding gene accumulated in the general human population.
Phenotype information of knockout mouse model
The phenotype information of knockout mouse models was collected from the International Mouse Phenotyping Consortium [20, 21] website (http://www.mousephenotype.org/). A total of 1357 mouse genes were categorized into genes observed or not observed to cause ‘complete preweaning lethality’ in knockout mice.
Pathways from KEGG (Kyoto Encyclopedia of Genes and Genomes)
The association between rat genes and KEGG pathway annotations was built by querying all the genes in ArrayTrack [22], an FDA-developed genomic tool for managing, analyzing, and interpreting gene expression data (http://www.fda.gov/ScienceResearch/BioinformaticsTools/Arraytrack/). Pathway analysis performed by ArrayTrack provided a tabular list of all the queried genes in corresponding KEGG pathways.
Determination of SEGs
For each candidate gene, the expression values across 320 rat samples were examined for expression stability. To minimize the impact of extreme expression values caused by random error, the highest and the lowest FPKM values were ignored. The expression fold change (FC) was then calculated as
where fpkmmax−1 and fpkmmin−1 represented the second highest and the second lowest expression value across 320 samples for each gene, respectively. Those genes showing FC ≤4.0 were defined as SEGs in the Rat BodyMap model. All the rat SEGs were mapped to human and mouse homolog genes according to the homology groups retrieved from the HomoloGene database (http://www.ncbi.nlm.nih.gov/homologene) of the National Center for Biotechnology Information (NCBI).
Statistical tests
A non-parametric Mann-Whitney U test (also known as Wilcoxon rank-sum test) was performed to examine the association between (1) the expression stability of rat genes and human homolog genes, (2) the expression stability of rat genes and the Genetic Damage Index of human homolog genes. All the human genes were ranked by the statistic of interest (i.e., expression stability in GTEx data or Genetic Damage Index). The null hypothesis was that the ranks of the homolog genes of rat SEGs were not significantly different from those of other genes. A p-value was calculated to determine whether the null hypothesis was valid.
In addition, Fisher’s exact test was performed to determine whether the SEGs were enriched in a certain category (i.e., a KEGG pathway or the genes causing complete preweaning lethality in mouse knockout model). The statistical question was framed in terms of the following 2 × 2 contingency table:
| SEG Homologs | Other Genes | |
| In category | Na | Nc |
| Not in category | Nb | Nd |
Na and Nb represented the numbers of SEG homologs in or not in the category. Nc and Nd were the numbers of other (non-SEG) genes in or not in the category. The odds ratio (OR) was calculated as OR = (Na × Nd)/(Nb × Nc). The null hypothesis was formulated as:
, where Prop1 = Na / (Na + Nb) and Prop2 = Nc / (Nc + Nd). For KEGG pathway analysis, the crude p-values were adjusted due to testing on multiple pathways, so as to control the false discovery rate (FDR) under 5%.
Results
Identification of Stably Expressed Genes (SEGs)
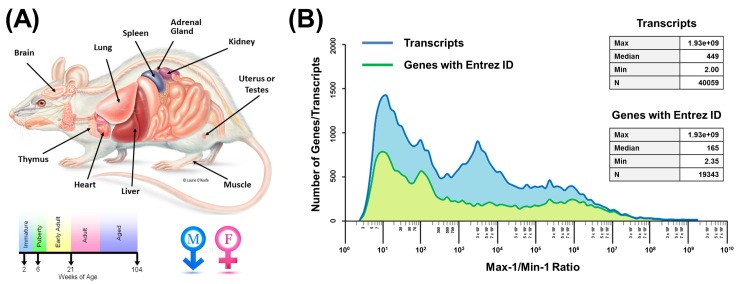
RNA-seq transcriptomics data were retrieved from the Rat BodyMap website. The samples consisted of 11 organs (adrenal gland, brain, heart, kidney, liver, lung, muscle, spleen, thymus, and testes or uterus) for 4 ages (2, 6, 21 and 104 weeks of age) and both sexes (Fig 1A). For each combination of age, sex and organ, there were 4 rats as biological replicates, resulting in 320 total samples. After excluding unnamed (without Entrez ID), non-expressed and redundant transcripts (see Materials and Methods), a total of 19343 unique genes (each corresponding to one transcript) remained for analysis of expression change in rats. The expression change for each gene was calculated as the maximum expression divided by the minimum expression for each gene across all sample. Initial inspection of the gene expression database showed an occasional expression value for some genes that was much higher or much lower than the other 320 samples (S1 Fig), suggesting that these outlier values may be technical artifacts of the measurement method. Removal of such values may allow better estimation of the expression change. Because this method of calculating gene expression change is sensitive to extreme outliers, the highest and lowest expression values of each gene were, therefore, removed before forming the ratio (see next paragraph for additional explanation). The distribution of the expression changes for these 19343 genes across these 320 samples is shown in Fig 1B (green), and ranged from 2.35 to >109-fold, with a median of 165-fold. The expression changes of the complete set of 40059 transcripts (excluding 5 genes not expressed in any sample) is shown in Fig 1B (blue), and ranged from 2.0 to >109-fold, with a median of 449. The distributions appear similar with the exception of a large peak of transcripts with expression changes of 1000–10000-fold in the complete transcript set.
Fig 1. Study design and gene expression stability.
(A) 320 organ samples were collected from rats of different ages and sexes. (B) A total of 19343 unique genes with Entrez ID were refined from 40059 transcripts. The distribution of expression changes demonstrated the rarity of genes with low variation across samples.
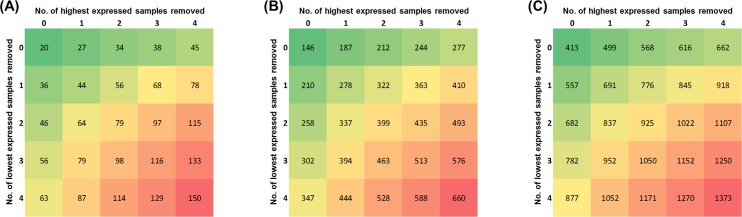
Our primary interest was identifying those genes showing relatively low expression variability across 11 organs, 4 ages, and both sexes. Two criteria were explored to set the cut-off threshold: (1) the ratio of the highest expression level of a gene to its lowest expression level (expression change) across all samples and (2) the number of highest and lowest expressing samples to be excluded. Adjusting these 2 factors allowed the selection of an appropriately low level of expression change with minimal interference by possibly false high or low expression values. Fig 2 shows matrix tables for the number of genes with expression changes of ≤3-fold, ≤4-fold, and ≤5-fold with removal of from 0–4 highest and lowest expression values. There were 20, 146 and 413 genes whose expression change was ≤3-fold, ≤4-fold, and ≤5-fold, respectively, and this number could be substantially increased by removing small numbers of highest and lowest expression values. As a trade-off for identifying genes with low expression variability without removing excessive numbers of samples, we focused on those genes whose expression varied by ≤4-fold after removal of the single highest and single lowest expressing samples. This resulted in 278 stably expressed genes (SEGs, S2 Dataset), which accounted for about 1% of all studied genes.
Fig 2. Numbers of stably expressed genes at different thresholds of expression fold-change and numbers of highest and lowest expressed samples removed.
(A) Cutoff criteria of expression changes of ≤3-fold. (B) Cutoff criteria of expression changes of ≤4-fold. (C) Cutoff criteria of expression changes of ≤5-fold.
Pathways enriched in SEGs
An unsupervised pathway analysis was performed to understand the associations between the 278 SEGs and Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway [23] terms (see Materials and Methods). One hundred six of these 278 SEGs were associated with at least one KEGG pathway (S2 Dataset). Pathways that were significantly enriched in SEGs included proteasome, ubiquitin mediated proteolysis, RNA transport, Epstein-Barr virus infection, aminoacyl-tRNA biosynthesis, protein processing in endoplasmic reticulum, legionellosis, and spliceosome (Table 1). The protein degradation pathway was notably represented with adjusted p-values of 1.8 x 10−16 and 6.4 x 10−6 for proteasome (16 genes) and ubiquitin mediated proteolysis (13 genes), respectively. And the odds ratio of SEGs enrichment was 41.52 for proteasome pathway and 8.01 for ubiquitin mediated proteolysis pathway. Most of the significant pathways were closely related to the processing of protein and RNA molecules, which are basic functions for all tissues and organs. The Epstein Barr virus infection pathway shared 10 genes (Psmc4, Psmd11, Psmd13, Psmd3, Psmd6, Psmd4, Psmd12, Psmc1, Psmd1, and Psmd7) with the proteasome pathway and 1 gene (RGD1561926) with the spliceosome pathway, suggesting that this pathway was significant because of the presence of protein degradation genes. The legionellosis pathway may have reached significance because of the presence of the genes Sar1a and Vcp that are also present in the protein processing in endoplasmic reticulum pathway.
Table 1. KEGG pathways significantly enriched in SEGs.
| KEGG Pathway Term | SEGs Associated with the Pathway | OR | Adjusted P-value |
|---|---|---|---|
| Proteasome | Psmc4; Psma4; Psma3l; Psmd11; Psmd13; Psmb1; Psmd3; Psmd6; Psmd4; Psma1; Psmd12; Psmb4; Psmc1; Psmd1; Psmd7; Psma5 | 41.52 | 1.78 x 10−16 |
| Ubiquitin mediated proteolysis | Ube2d3; Uba3; Cul1; Rbx1; Ube4a; Ube3c; Anapc5; Ddb1; Ube3a; Itch; Birc6; Keap1; Klhl9 | 8.01 | 6.41 x 10−6 |
| RNA transport | Nmd3; Snupn; Eif3s10; Eif4g1; Eif3c; Elac2; Eif2b5; Eif4g2_predicted; Eif4b; Ranbp2; Eif3h | 5.91 | 5.10 x 10−4 |
| Epstein-Barr virus infection | Psmc4; RGD1561926; Psmd11; Psmd13; Psmd3; Psmd6; Psmd4; Psmd12; Psmc1; Polr2b; Psmd1; Psmd7 | 4.79 | 1.17 x 10−3 |
| Aminoacyl-tRNA biosynthesis | Nars2; Farsb; Tars2; Zmat2; Sars; Lars | 11.66 | 1.35 x 10−3 |
| Protein processing in endoplasmic reticulum | Sar1a; Rad23b; Ube2d3; LOC685144; Nsfl1c; Cul1; Rbx1; Edem3; Dnajc10; Vcp | 4.90 | 3.25 x 10−3 |
| Legionellosis | Sar1a; Arf1; Sec22b; Rab1; Vcp | 7.41 | 3.15 x 10−2 |
| Spliceosome | Cwc15; Cdc5l; RGD1561926; Syf2; Prpf8; Plrg1; Prpf6 | 4.53 | 4.35 x 10−2 |
To understand the effect of changing the cutoff values on the types of genes and pathways identified, we adjusted the number of highest and lowest expressed genes to remove. Using an expression change of ≤4-fold and removing no samples, the highest and lowest expressed 2 samples, 3 samples, and 4 samples, resulted in 146, 399, 513, and 660 genes (along the diagonal in Fig 2B, FC≤4-fold panel). KEGG pathway analysis identified the same pathways as with the 278 SEGs, generally with increased significance level (Table 2; S1–S5 Tables). Examination of the significant pathways identified with cutoff criteria of expression changes of ≤3-fold (44 SEGs; S6 Table) and ≤5-fold (691 SEGs; S7 Table) calculated by using second highest and lowest samples (with exclusion of the single highest and single lowest expressed samples) also identified similar pathways involving protein degradation and processing, although with the expression change of ≤3-fold, only 17 of the 44 SEGs were associated with any KEGG pathway. Thus, our selection criteria appear to be robust and making small adjustments in them identify the same major pathways involved in protein degradation and processing.
Table 2. SEGs with expression changes of ≤4-fold–the number of genes in stably expressed pathways increases with relaxed selection criteria.
| Number of Genes in KEGG Pathways at Expression Change of ≤4-fold Calculated after Removal of N Highest and Lowest Expressed Samples | ||||||
|---|---|---|---|---|---|---|
| KEGG Pathway Term | N = 0 | N = 1 | N = 2 | N = 3 | N = 4 | Adjusted p-values |
| Proteasome | 8 | 16 | 19 | 20 | 24 | 3.5 x 10−7–10−21 |
| Ubiquitin mediated proteolysis | 8 | 13 | 15 | 15 | 18 | 8.7 x 10−3–6.4 x 10−6 |
| RNA transport | 8 | 11 | 12 | 15 | 16 | 1.2 x 10−3–3 x 10−4 |
| Aminoacyl-tRNA biosynthesis | 6 | 8 | 9 | 15 | 1.4 x 10−3–3 x 10−10 | |
| Protein processing in endoplasmic reticulum | 10 | 12 | 17 | 19 | 3 x 10−3–6 x 10−5 | |
| Spliceosome | 7 | 12 | 13 | 4 x 10−2–2 x 10−3 | ||
| Protein export | 5 | 6 | 8 | 3 x 10−3–2 x 10−5 | ||
| Legionellosis | 5 | 6 | 6 | 7 | >10−2 | |
| mTOR signaling pathway | 6 | 8 | 8 | >10−2 | ||
| Nucleotide excision repair | 5 | >10−2 | ||||
| Renal cell carcinoma | 6 | >10−2 | ||||
| AMPK signaling pathway | 10 | 11 | >10−2 | |||
| SNARE interactions in vesicular transport | 5 | 6 | >10−2 | |||
| Insulin signaling pathway | 10 | >10−2 | ||||
| Regulation of autophagy | 5 | >10−2 | ||||
Expression stability of human homologs of SEGs
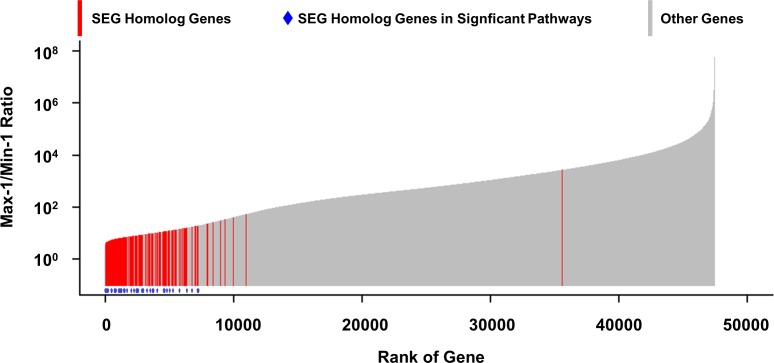
The expression stability of these SEGs identified in the rat was examined in human tissues. The human homologs were identified and used to query the human RNA-sequence data retrieved from the Genotype-Tissue Expression (GTEx) database as described in Materials and Methods. Gene expression data was available for 98 human tissue samples from 10 tissue types that matched the rat (all except thymus) in both sexes and at ages from 20–69 years from individuals who did not die of a disease process (S1 Dataset). The expression variation was calculated for these genes (S3 Dataset; see Materials and Methods) across the 98 samples and is displayed in Fig 3. The expression fold change varied from 3.2 to 5.6 x 107, with a median of 470. Those human genes homologous to the 278 rat SEGs are marked in red and the subset that fall into KEGG pathways are marked in blue in Fig 3. As can be verified by a non-parametric Mann-Whitney U test (see Materials and Methods), the human homologs of these rat SEGs are over-represented in the most stably expressed human genes with a highly significant p-value (2.49 x 10−104). Thus, the stability of expression of the rat SEGs is conserved across species, supporting the generalization of the findings.
Fig 3. Expression stability of human homologs of SEGs.
Each bar represented a gene, with the height corresponding to the expression change (maximum expression–minimum expression). The SEG homologs are red and those associated with significant pathways identified in Table 1 are marked with a blue dot.
Lethality of SEG knockout
The cross-species consistency of SEGs suggested that expression stability of these genes may be of such importance that absence of the SEGs may result in major systemic disturbance and/or lethal outcomes. The International Mouse Phenotyping Consortium (IMPC) is producing knockout mouse strains for every protein coding gene and carrying out high-throughput phenotyping of each line [24], which enabled us to examine the association between SEG knockout and lethal phenotype (S4 Dataset; see Materials and Methods). Of the 1357 genes examined thus far by IMPC, 16 are homologous to the 278 rat SEGs and knocking out of 15 of these (94%) produced complete preweaning lethality, defined by IMPC as death anytime between fertilization and weaning age (approximately 3–4 weeks of age). Knockout of one of the genes (Dnpep) did not result in preweaning lethality but did result in a changed phenotype that included reduced T cell numbers. Of the remaining 1341 genes, knockout of 421 produced complete preweaning lethality (31%). Thus, absence of mouse genes homologous to the rat SEGs resulted in a significantly higher risk of lethal phenotype than absence of non-SEGs (P = 3.84×10−7; Table 3). Characteristics of the 15 SEGs with lethal knockout phenotypes are shown in Table 4. Thus, the strong association of SEGs with a lethal phenotype supports the interpretation of a fundamental role of SEGs in biological development.
Table 3. The association between SEGs knockout and lethal phenotypes.
| SEG Homologs | Other Genes | |
|---|---|---|
| Lethal Phenotypes Observed | 15 | 421 |
| Lethal Phenotypes Not Observed | 1 | 920 |
| Odds Ratio (95% CI) | 32.78 (4.32 ~ 248.97) | |
| P-valuea | 3.84 × 10−7 | |
a P-value was determined with two-tailed Fisher’s exact test.
Table 4. Summary of SEGs with lethal knockout phenotypes.
| Lethal Knockout Mouse Gene | Corresponding Rat SEG | Gene Name | NCBI Gene Summary |
|---|---|---|---|
| Alg10b | Alg10b | ALG10, alpha-1,2-glucosyltransferase | Regulatory component of non-inactivating K+ channels |
| Chd4 | Chd4 | Chromodomain helicase DNA binding protein 4 | Member of the CHD protein family; may play a role in chromatin reorganization; may play a role in osmosignalling |
| Cog3 | Cog3 | Component of oligomeric golgi complex 3 | |
| Eif4g2 | Eif4g2 (predicted) | Eukaryotic translation initiation factor 4, gamma 2 | This gene product functions as a general repressor of translation by forming translationally inactive complexes |
| Exoc8 | Exoc8 | Exocyst complex component 8 | Encodes a subunit of the exocyst complex, an exocytosis-associated complex specifically located at sites of vesicle fusion |
| Lztr1 | Lztr1 | Leucine-zipper-like transcription regulator 1 | |
| Maea | Maea | Macrophage erythroblast attacher | |
| Mapkap1 | Mapkap1 | Mitogen-activated protein kinase associated protein 1 | |
| Ppp4r2 | Ppp4r2 | Protein phosphatase 4, regulatory subunit 2 | |
| Raf1 | Raf1 | V-raf-leukemia viral oncogene 1 | Acts as a mitogenic protein kinase; mutant forms may play a role in transformation |
| Rint1 | RGD1560433 | RAD50 interactor 1 | |
| Sec22b | Sec22b | SEC22 homolog B, vesicle trafficking protein | |
| Usp36 | Usp36 | Ubiquitin specific peptidase 36 | |
| Usp5 | Usp5 | Ubiquitin specific peptidase 5 | |
| Vps13d | Vps13d | Vacuolar protein sorting 13 homolog D |
Genetic damage index of SEGs
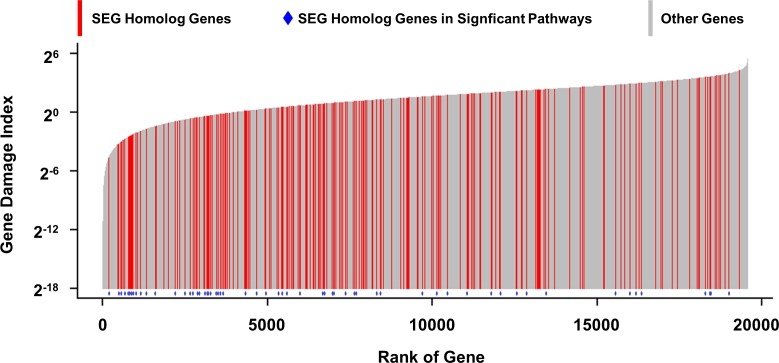
Recently, the frequency of non-synonymous mutations in each protein coding gene of the general human population was examined and a metric was developed (gene damage index, GDI) to quantify the mutational damage accumulated for each gene [17]. The GDI was proposed as a genome-wide and gene-level metric of evolutionarily accumulated mutational damage, with higher values associated with a higher non-synonymous mutational load. Because non-synonymous DNA mutations can have profound impact on the function and expression of human protein-coding genes by changing the DNA codon and correspondent amino acid sequences, it would be expected that SEGs would have lower GDI scores than non-SEGs. GDI scores for human genes (S5 Dataset) are displayed from lowest to highest in Fig 4 and the homologs of the rat SEGs are highlighted in red. While the overall average GDI was 4.35 (ranging from 0 to 42.91), the average GDI of SEG homologs was lower at 3.25 (ranging from 0.04 to 19.08). Using a non-parametric Mann-Whitney U test, the ranks of human homologs of the rat SEGs were compared with those of other genes (see Materials and Methods) and found to be significantly (p = 1.92×10−5) different. It was noted by Itan et al [17] that genes with low GDI were found to be enriched in proteasome and spliceosome pathways, which were also highlighted in our KEGG pathways analysis on SEGs (as shown in Table 1). Thus, SEGs appear to be subjected to less DNA sequence variation than the bulk of protein coding genes suggesting that expression stability is reflected in evolutionary DNA sequence stability in this set of genes.
Fig 4. Human homolog genes of SEGs showed significantly lower Genetic Damage Index (GDI).
Each bar represented a gene, with the height corresponding to the GDI value. The SEG homologs are red and those associated with significant pathways identified in Table 1 are marked with a blue dot.
Discussion
In the present study, we identified genes whose expression varied within a relatively narrow range across the life span, and between sexes and organs in F344 rats. The proteins encoded by these SEGs were found to play important roles in basic cellular functions, including protein catabolism (proteasome and ubiquitination), RNA transport, protein processing, and the spliceosome, and suggested stable expression of these genes is fundamental to good health. Several lines of evidence support the validity of these findings. First, the low expression variability of these genes was conserved across species. Examination of the expression of the human homologs of the rat SEGs in a database of tissues derived from humans (GTEx) showed that these genes exhibited much less expression variability across tissue, age and sex than other genes (p< 2.5 x 10−104). Second, we hypothesized that the transcriptomic stability of the SEGs played a pivotal role in normal physiological function and that dysregulation would result in disease or lethality. Examination of the effect of knocking out individual SEGs in mouse models developed in the International Mouse Phenotyping Consortium showed a lethal phenotype at the prenatal stage for a high proportion of SEGs. Of the 16 strains with knock-out of the mouse homolog of a rat SEG, 15 resulted in complete preweaning lethality (>93%); knocking out the mouse homologs of rat non-SEGs (1341 genes) resulted in complete preweaning lethality only 31% of the time, supporting the importance of these SEGs in development (p< 3.8 x 10−7). Third, we hypothesized that SEGs would be under evolutionary pressure to maintain stable expression and this would be reflected in the accumulation of fewer mutations in these genes. Recently, a human gene damage index (GDI) summarizing the non-synonymous mutational load of each protein-coding gene in the general human population was developed [17]. Examination of this database showed that the human homologs of the rat SEGs were less frequently mutated in healthy populations and exhibited lower GDI (p = 1.92×10−5). Thus, from multiple independent perspectives, stably expressed genes encoding proteins involved in protein catabolism, RNA transport, protein processing, and the spliceosome, appear to be of fundamental importance.
KEGG pathways analysis was performed to further interpret the pathological and pharmacological importance of SEGs. The results indicated that these genes were significantly involved in protein catabolism, such as proteasome components and ubiquitination. The ubiquitin-proteasome system degrades intracellular proteins in a highly regulated manner and is essential to a wide range of cellular processes [25]. Previous studies have shown that dysregulation and instability of the ubiquitin-proteasome system can result in multiple diseases [26], including aging [27]. For instance, elevated proteasomal activity has been detected in many types of cancers [28]. Because tumor growth and division can be halted by tumor suppressor proteins [29], tumor cells rely heavily on proteasomal function for the degradation of suppressor proteins [30]. Another example is the overactivity of the ubiquitin-proteasome system in rheumatoid arthritis. As a master regulator of many inflammatory cytokines involved in the pathogenesis of rheumatoid arthritis [31], NF-κB protein is normally inhibited when bound to IκB protein [32]. However, in the case of excessive ubiquitination of IκB followed by proteasomal degradation, NF-κB can be chronically active to cause inflammatory arthritis [33].
Since the 278 SEGs identified in our analysis are highly enriched in the ubiquitin-proteasome system, this may provide an opportunity to discover novel disease liability genes among SEGs. First, the expression levels can be monitored in disease models to confirm whether the expression stability of a certain SEG is maintained or altered in pathological conditions. Second, by using knockdown [34] or overexpression [35] techniques, gene expression can be purposely reduced or increased in particular age, sex or organ, to identify potential disease liabilities. It can then be determined if restoring the stability of expression will lead to beneficial outcomes in such disease models (e.g., suppression of tumor growth or anti-inflammatory effect). Third, because gene expression is an indirect measurement of biological function, direct examination of protein levels and pathway functions [36] are needed to confirm the biological significance of these findings. Finally, there have been wide discussions regarding the selection of stable reference genes for quantitative real-time PCR (qRT-PCR) [37]. The SEGs described here may offer better stable candidate reference genes for such pRT-PCR studies. Further validation of the proposed SEGs will be important for improving the accuracy of qRT-PCR.
Taken together, we identified a set of genes with a relatively narrow range of expression change across organs throughout the life span and between the sexes in a rat model. The proteins encoding by these genes are significantly involved in a series of housekeeping functions, including protein catabolism, RNA transport, protein processing, and the spliceosome. The fundamental biological importance of the stable expression of these genes was supported by multiple external data sources, including human organ transcriptomics, mouse knockout models, and human protein-coding gene mutations. Additional exploration of such stable gene expression, at the protein and pathway level, as well as in other gene expression datasets (including microarray data), may provide opportunities for disease gene identification, as well as potential drug targets for restoration of function.
Supporting Information
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
The expression value for each of the 320 samples is shown for four genes.
(PPTX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
The findings and conclusions presented in this article are the authors’ and do not necessarily reflect those of the Food and Drug Administration.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The authors received no specific funding for this work.
References
- 1.Shendure J, Ji H. Next-generation DNA sequencing. Nat Biotechnol. 2008;26: 1135–1145. 10.1038/nbt1486 [DOI] [PubMed] [Google Scholar]
- 2.Peixoto A, Monteiro M, Rocha B, Veiga-Fernandes H. Quantification of multiple gene expression in individual cells. Genome Res. 2004;14: 1938–1947. 10.1101/gr.2890204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu LX, Liu ZH, Jiang HC, Qu X, Zhang WH, Wu LF, et al. Profiling of differentially expressed genes in human gastric carcinoma by cDNA expression array. World J Gastroenterol. 2002;8: 580–585. 10.3748/wjg.v8.i4.580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Escrich E, Moral R, Garcia G, Costa I, Sanchez JA, Solanas M. Identification of novel differentially expressed genes by the effect of a high-fat n-6 diet in experimental breast cancer. Mol Carcinog. 2004;40: 73–78. 10.1002/mc.20028 [DOI] [PubMed] [Google Scholar]
- 5.Altintas DM, Allioli N, Decaussin M, de Bernard S, Ruffion A, Samarut J, et al. Differentially expressed androgen-regulated genes in androgen-sensitive tissues reveal potential biomarkers of early prostate cancer. PLoS One. 2013;8: e66278 10.1371/journal.pone.0066278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Myers JS, von Lersner AK, Robbins CJ, Sang QX. Differentially Expressed Genes and Signature Pathways of Human Prostate Cancer. PLoS One. 2015;10: e0145322 10.1371/journal.pone.0145322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shen Y, Wang X, Jin Y, Lu J, Qiu G, Wen X. Differentially expressed genes and interacting pathways in bladder cancer revealed by bioinformatic analysis. Mol Med Rep. 2014;10: 1746–1752. 10.3892/mmr.2014.2396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eisenberg E, Levanon EY. Human housekeeping genes, revisited. Trends Genet. 2013;29: 569–574. 10.1016/j.tig.2013.05.010 [DOI] [PubMed] [Google Scholar]
- 9.Janssens N, Janicot M, Perera T, Bakker A. Housekeeping genes as internal standards in cancer research. Mol Diagn. 2004;8: 107–113. [DOI] [PubMed] [Google Scholar]
- 10.Mason EA, Mar JC, Laslett AL, Pera MF, Quackenbush J, Wolvetang E, et al. Gene expression variability as a unifying element of the pluripotency network. Stem Cell Reports. 2014;3: 365–377. 10.1016/j.stemcr.2014.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mar JC, Matigian NA, Mackay-Sim A, Mellick GD, Sue CM, Silburn PA, et al. Variance of gene expression identifies altered network constraints in neurological disease. PLoS Genet. 2011;7: e1002207 10.1371/journal.pgen.1002207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hasegawa Y, Taylor D, Ovchinnikov DA, Wolvetang EJ, de Torrente L, Mar JC. Variability of Gene Expression Identifies Transcriptional Regulators of Early Human Embryonic Development. PLoS Genet. 2015;11: e1005428 10.1371/journal.pgen.1005428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang Z, Dong D, Zhang Z, Crabbe MJ, Wang L, Zhong Y. Preferential regulation of stably expressed genes in the human genome suggests a widespread expression buffering role of microRNAs. BMC Genomics. 2012;13 Suppl 7: S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Onken MD, Winkler AE, Kanchi KL, Chalivendra V, Law JH, Rickert CG, et al. A surprising cross-species conservation in the genomic landscape of mouse and human oral cancer identifies a transcriptional signature predicting metastatic disease. Clin Cancer Res. 2014;20: 2873–2884. 10.1158/1078-0432.CCR-14-0205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu Y, Fuscoe JC, Zhao C, Guo C, Jia M, Qing T, et al. A rat RNA-Seq transcriptomic BodyMap across 11 organs and 4 developmental stages. Nat Commun. 2014;5: 3230 10.1038/ncomms4230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45: 580–585. 10.1038/ng.2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Itan Y, Shang L, Boisson B, Patin E, Bolze A, Moncada-Velez M, et al. The human gene damage index as a gene-level approach to prioritizing exome variants. Proc Natl Acad Sci U S A. 2015;112: 13615–13620. 10.1073/pnas.1518646112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Genomes Project C, Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491: 56–65. 10.1038/nature11632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38: e164 10.1093/nar/gkq603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brown SD, Moore MW. Towards an encyclopaedia of mammalian gene function: the International Mouse Phenotyping Consortium. Dis Model Mech. 2012;5: 289–292. 10.1242/dmm.009878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bradley A, Anastassiadis K, Ayadi A, Battey JF, Bell C, Birling MC, et al. The mammalian gene function resource: the International Knockout Mouse Consortium. Mamm Genome. 2012;23: 580–586. 10.1007/s00335-012-9422-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fang H, Harris SC, Su Z, Chen M, Qian F, Shi L, et al. ArrayTrack: an FDA and public genomic tool. Methods Mol Biol. 2009;563: 379–398. 10.1007/978-1-60761-175-2_20 [DOI] [PubMed] [Google Scholar]
- 23.Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012;40: D109–114. 10.1093/nar/gkr988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koscielny G, Yaikhom G, Iyer V, Meehan TF, Morgan H, Atienza-Herrero J, et al. The International Mouse Phenotyping Consortium Web Portal, a unified point of access for knockout mice and related phenotyping data. Nucleic Acids Res. 2014;42: D802–809. 10.1093/nar/gkt977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kleiger G, Mayor T. Perilous journey: a tour of the ubiquitin-proteasome system. Trends Cell Biol. 2014;24: 352–359. 10.1016/j.tcb.2013.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang M, Kaufman RJ. Protein misfolding in the endoplasmic reticulum as a conduit to human disease. Nature. 2016;529: 326–335. 10.1038/nature17041 [DOI] [PubMed] [Google Scholar]
- 27.Kaushik S, Cuervo AM. Proteostasis and aging. Nat Med. 2015;21: 1406–1415. 10.1038/nm.4001 [DOI] [PubMed] [Google Scholar]
- 28.Chen D, Dou QP. The ubiquitin-proteasome system as a prospective molecular target for cancer treatment and prevention. Curr Protein Pept Sci. 2010;11: 459–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sherr CJ. Principles of tumor suppression. Cell. 2004;116: 235–246. [DOI] [PubMed] [Google Scholar]
- 30.Shen M, Schmitt S, Buac D, Dou QP. Targeting the ubiquitin-proteasome system for cancer therapy. Expert opinion on therapeutic targets. 2013;17: 1091–1108. 10.1517/14728222.2013.815728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Agarwal S, Deschner J, Long P, Verma A, Hofman C, Evans CH, et al. Role of NF-kappaB transcription factors in antiinflammatory and proinflammatory actions of mechanical signals. Arthritis Rheum. 2004;50: 3541–3548. 10.1002/art.20601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jacobs MD, Harrison SC. Structure of an IkappaBalpha/NF-kappaB complex. Cell. 1998;95: 749–758. [DOI] [PubMed] [Google Scholar]
- 33.Wang J, Maldonado MA. The ubiquitin-proteasome system and its role in inflammatory and autoimmune diseases. Cellular & molecular immunology. 2006;3: 255–261. [PubMed] [Google Scholar]
- 34.Kleinhammer A, Wurst W, Kuhn R. Gene knockdown in the mouse through RNAi. Methods Enzymol. 2010;477: 387–414. 10.1016/S0076-6879(10)77020-8 [DOI] [PubMed] [Google Scholar]
- 35.Prelich G. Gene overexpression: uses, mechanisms, and interpretation. Genetics. 2012;190: 841–854. 10.1534/genetics.111.136911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hipp MS, Bersuker K, Kopito RR. Live-cell imaging of ubiquitin-proteasome system function. Methods Mol Biol. 2012;832: 463–472. 10.1007/978-1-61779-474-2_33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kozera B, Rapacz M. Reference genes in real-time PCR. J Appl Genet. 2013;54: 391–406. 10.1007/s13353-013-0173-x [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
The expression value for each of the 320 samples is shown for four genes.
(PPTX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.