Abstract
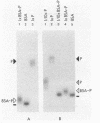
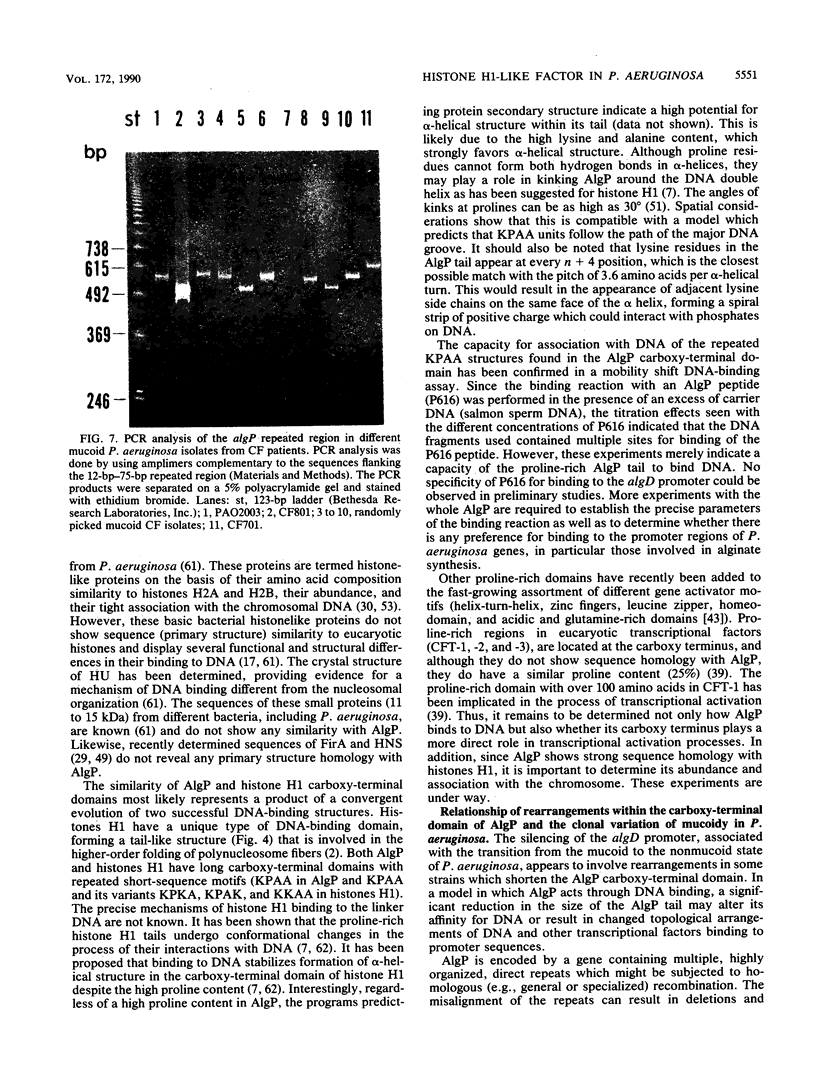
A novel procaryotic transcriptional regulatory element, AlgP, with a histone H1-like carboxy-terminal domain was identified in Pseudomonas aeruginosa. AlgP is required for transcription of the key biosynthetic gene algD, which is necessary for production of the exopolysaccharide alginate causing mucoidy in P. aeruginosa. Mucoidy is a critical virulence determinant of P. aeruginosa invariably associated with the respiratory infections causing high mortality in cystic fibrosis. Here we show that AlgP and histones H1 both have repeated units of the Lys-Pro-Ala-Ala motif (KPAA) and its variations within their long (over 100 amino acids) carboxy-terminal domains. This region of histone H1 tails has been shown to bind to the linker DNA in eucaryotic chromatin fibers. A synthetic 50-mer peptide consisting of repeats from the AlgP carboxy-terminal domain was found to bind DNA in a mobility shift DNA-binding assay. AlgP is encoded by a gene that contains multiple direct repeats organized as tandem, head-to-tail, 12-base-pair (bp) units overlapping with six highly conserved 75-bp units. The repetitive structure of the algP gene appears to participate in the processes underlying the metastable character of mucoidy in P. aeruginosa. Relatively large DNA rearrangements spanning the region with tandem direct repeats encoding the carboxy-terminal histone H1-like structure of AlgP were detected in several strains upon conversion from the mucoid to the nonmucoid phenotype. The frequency of the detectable algP rearrangements associated with the transition into the nonmucoid state varied from strain to strain and ranged from 0 to 50%. The nonmucoid derivatives with the clearly rearranged chromosomal copy of algP were complemented to mucoidy with plasmids containing algP from P. aeruginosa PAO. When a random collection of mucoid strains, isolated from different cystic fibrosis patients, was analyzed by using polymerase chain reaction, an additional level of strain-dependent sequence variation in algP was observed. Variations in the number of the 12-bp repeats were found; however, they did not appear to influence the mucoid status of the strains examined. Thus, the repeated region of algP appears to be a hot spot for DNA rearrangements and strain-dependent variability.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Albertini A. M., Hofer M., Calos M. P., Miller J. H. On the formation of spontaneous deletions: the importance of short sequence homologies in the generation of large deletions. Cell. 1982 Jun;29(2):319–328. doi: 10.1016/0092-8674(82)90148-9. [DOI] [PubMed] [Google Scholar]
- Allan J., Mitchell T., Harborne N., Bohm L., Crane-Robinson C. Roles of H1 domains in determining higher order chromatin structure and H1 location. J Mol Biol. 1986 Feb 20;187(4):591–601. doi: 10.1016/0022-2836(86)90337-2. [DOI] [PubMed] [Google Scholar]
- Aricó B., Miller J. F., Roy C., Stibitz S., Monack D., Falkow S., Gross R., Rappuoli R. Sequences required for expression of Bordetella pertussis virulence factors share homology with prokaryotic signal transduction proteins. Proc Natl Acad Sci U S A. 1989 Sep;86(17):6671–6675. doi: 10.1073/pnas.86.17.6671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunier D., Michel B., Ehrlich S. D. Copy choice illegitimate DNA recombination. Cell. 1988 Mar 25;52(6):883–892. doi: 10.1016/0092-8674(88)90430-8. [DOI] [PubMed] [Google Scholar]
- Chandler P. M., Krishnapillai V. Isolation and properties of recombination-deficient mutants of Pseudomonas aeruginosa. Mutat Res. 1974 Apr;23(1):15–23. doi: 10.1016/0027-5107(74)90155-9. [DOI] [PubMed] [Google Scholar]
- Clark-Lewis I., Aebersold R., Ziltener H., Schrader J. W., Hood L. E., Kent S. B. Automated chemical synthesis of a protein growth factor for hemopoietic cells, interleukin-3. Science. 1986 Jan 10;231(4734):134–139. doi: 10.1126/science.3079915. [DOI] [PubMed] [Google Scholar]
- Clark D. J., Hill C. S., Martin S. R., Thomas J. O. Alpha-helix in the carboxy-terminal domains of histones H1 and H5. EMBO J. 1988 Jan;7(1):69–75. doi: 10.1002/j.1460-2075.1988.tb02784.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale R. M., McClure B. A., Houchins J. P. A rapid single-stranded cloning strategy for producing a sequential series of overlapping clones for use in DNA sequencing: application to sequencing the corn mitochondrial 18 S rDNA. Plasmid. 1985 Jan;13(1):31–40. doi: 10.1016/0147-619x(85)90053-8. [DOI] [PubMed] [Google Scholar]
- Darzins A., Chakrabarty A. M. Cloning of genes controlling alginate biosynthesis from a mucoid cystic fibrosis isolate of Pseudomonas aeruginosa. J Bacteriol. 1984 Jul;159(1):9–18. doi: 10.1128/jb.159.1.9-18.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V., Dikshit R., Konyecsni W. M., Chakrabarty A. M., Misra T. K. The algR gene, which regulates mucoidy in Pseudomonas aeruginosa, belongs to a class of environmentally responsive genes. J Bacteriol. 1989 Mar;171(3):1278–1283. doi: 10.1128/jb.171.3.1278-1283.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V., Gill J. F., Chakrabarty A. M. Gene algD coding for GDPmannose dehydrogenase is transcriptionally activated in mucoid Pseudomonas aeruginosa. J Bacteriol. 1987 Jan;169(1):351–358. doi: 10.1128/jb.169.1.351-358.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V., Gill J. F., Chakrabarty A. M. Pseudomonas aeruginosa infection in cystic fibrosis: nucleotide sequence and transcriptional regulation of the algD gene. Nucleic Acids Res. 1987 Jun 11;15(11):4567–4581. doi: 10.1093/nar/15.11.4567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V., Konyecsni W. M. Control of mucoidy in Pseudomonas aeruginosa: transcriptional regulation of algR and identification of the second regulatory gene, algQ. J Bacteriol. 1989 Jul;171(7):3680–3688. doi: 10.1128/jb.171.7.3680-3688.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drlica K., Rouviere-Yaniv J. Histonelike proteins of bacteria. Microbiol Rev. 1987 Sep;51(3):301–319. doi: 10.1128/mr.51.3.301-319.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ELLMAN G. L. A colorimetric method for determining low concentrations of mercaptans. Arch Biochem Biophys. 1958 Apr;74(2):443–450. doi: 10.1016/0003-9861(58)90014-6. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Finlay B. B., Falkow S. Common themes in microbial pathogenicity. Microbiol Rev. 1989 Jun;53(2):210–230. doi: 10.1128/mr.53.2.210-230.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn J. L., Ohman D. E. Use of a gene replacement cosmid vector for cloning alginate conversion genes from mucoid and nonmucoid Pseudomonas aeruginosa strains: algS controls expression of algT. J Bacteriol. 1988 Jul;170(7):3228–3236. doi: 10.1128/jb.170.7.3228-3236.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franssen H. J., Nap J. P., Gloudemans T., Stiekema W., Van Dam H., Govers F., Louwerse J., Van Kammen A., Bisseling T. Characterization of cDNA for nodulin-75 of soybean: A gene product involved in early stages of root nodule development. Proc Natl Acad Sci U S A. 1987 Jul;84(13):4495–4499. doi: 10.1073/pnas.84.13.4495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freytag J. W., Noelken M. E., Hudson B. G. Physical properties of collagen--sodium dodecyl sulfate complexes. Biochemistry. 1979 Oct 16;18(21):4761–4768. doi: 10.1021/bi00588a042. [DOI] [PubMed] [Google Scholar]
- Fyfe J. A., Govan J. R. Alginate synthesis in mucoid Pseudomonas aeruginosa: a chromosomal locus involved in control. J Gen Microbiol. 1980 Aug;119(2):443–450. doi: 10.1099/00221287-119-2-443. [DOI] [PubMed] [Google Scholar]
- Govan J. R., Fyfe J. A., Baker N. R. Heterogeneity and reduction in pulmonary clearance of mucoid Pseudomonas aeruginosa. Rev Infect Dis. 1983 Nov-Dec;5 (Suppl 5):S874–S879. doi: 10.1093/clinids/5.supplement_5.s874. [DOI] [PubMed] [Google Scholar]
- Govan J. R., Fyfe J. A., McMillan C. The instability of mucoid Pseudomonas aeruginosa: fluctuation test and improved stability of the mucoid form in shaken culture. J Gen Microbiol. 1979 Jan;110(1):229–232. doi: 10.1099/00221287-110-1-229. [DOI] [PubMed] [Google Scholar]
- Holck A., Kleppe K. Cloning and sequencing of the gene for the DNA-binding 17K protein of Escherichia coli. Gene. 1988 Jul 15;67(1):117–124. doi: 10.1016/0378-1119(88)90014-5. [DOI] [PubMed] [Google Scholar]
- Høiby N., Döring G., Schiøtz P. O. Pathogenic mechanisms of chronic Pseudomonas aeruginosa infections in cystic fibrosis patients. Antibiot Chemother (1971) 1987;39:60–76. [PubMed] [Google Scholar]
- Hübscher U., Lutz H., Kornberg A. Novel histone H2A-like protein of escherichia coli. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5097–5101. doi: 10.1073/pnas.77.9.5097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles J. A., Childs G. J. Comparison of the late H1 histone genes of the sea urchins Lytechinus pictus and Strongelocentrotus purpuratus. Nucleic Acids Res. 1986 Oct 24;14(20):8121–8133. doi: 10.1093/nar/14.20.8121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konyecsni W. M., Deretic V. Broad-host-range plasmid and M13 bacteriophage-derived vectors for promoter analysis in Escherichia coli and Pseudomonas aeruginosa. Gene. 1988 Dec 30;74(2):375–386. doi: 10.1016/0378-1119(88)90171-0. [DOI] [PubMed] [Google Scholar]
- Konyecsni W. M., Deretic V. DNA sequence and expression analysis of algP and algQ, components of the multigene system transcriptionally regulating mucoidy in Pseudomonas aeruginosa: algP contains multiple direct repeats. J Bacteriol. 1990 May;172(5):2511–2520. doi: 10.1128/jb.172.5.2511-2520.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levinson G., Gutman G. A. High frequencies of short frameshifts in poly-CA/TG tandem repeats borne by bacteriophage M13 in Escherichia coli K-12. Nucleic Acids Res. 1987 Jul 10;15(13):5323–5338. doi: 10.1093/nar/15.13.5323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levinson G., Gutman G. A. Slipped-strand mispairing: a major mechanism for DNA sequence evolution. Mol Biol Evol. 1987 May;4(3):203–221. doi: 10.1093/oxfordjournals.molbev.a040442. [DOI] [PubMed] [Google Scholar]
- Lipman D. J., Pearson W. R. Rapid and sensitive protein similarity searches. Science. 1985 Mar 22;227(4693):1435–1441. doi: 10.1126/science.2983426. [DOI] [PubMed] [Google Scholar]
- MacGeorge J., Korolik V., Morgan A. F., Asche V., Holloway B. W. Transfer of a chromosomal locus responsible for mucoid colony morphology in Pseudomonas aeruginosa isolated from cystic fibrosis patients to P. aeruginosa PAO. J Med Microbiol. 1986 Jun;21(4):331–336. doi: 10.1099/00222615-21-4-331. [DOI] [PubMed] [Google Scholar]
- Mermod N., O'Neill E. A., Kelly T. J., Tjian R. The proline-rich transcriptional activator of CTF/NF-I is distinct from the replication and DNA binding domain. Cell. 1989 Aug 25;58(4):741–753. doi: 10.1016/0092-8674(89)90108-6. [DOI] [PubMed] [Google Scholar]
- Mian F. A., Jarman T. R., Righelato R. C. Biosynthesis of exopolysaccharide by Pseudomonas aeruginosa. J Bacteriol. 1978 May;134(2):418–422. doi: 10.1128/jb.134.2.418-422.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. F., Mekalanos J. J., Falkow S. Coordinate regulation and sensory transduction in the control of bacterial virulence. Science. 1989 Feb 17;243(4893):916–922. doi: 10.1126/science.2537530. [DOI] [PubMed] [Google Scholar]
- Misra T. K. DNA sequencing: a new strategy to create ordered deletions, modified M13 vector, and improved reaction conditions for sequencing by dideoxy chain termination method. Methods Enzymol. 1987;155:119–139. doi: 10.1016/0076-6879(87)55012-1. [DOI] [PubMed] [Google Scholar]
- Mitchell P. J., Tjian R. Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins. Science. 1989 Jul 28;245(4916):371–378. doi: 10.1126/science.2667136. [DOI] [PubMed] [Google Scholar]
- Murphy G. L., Connell T. D., Barritt D. S., Koomey M., Cannon J. G. Phase variation of gonococcal protein II: regulation of gene expression by slipped-strand mispairing of a repetitive DNA sequence. Cell. 1989 Feb 24;56(4):539–547. doi: 10.1016/0092-8674(89)90577-1. [DOI] [PubMed] [Google Scholar]
- Ohman D. E., West M. A., Flynn J. L., Goldberg J. B. Method for gene replacement in Pseudomonas aeruginosa used in construction of recA mutants: recA-independent instability of alginate production. J Bacteriol. 1985 Jun;162(3):1068–1074. doi: 10.1128/jb.162.3.1068-1074.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng H. L., Novick R. P., Kreiswirth B., Kornblum J., Schlievert P. Cloning, characterization, and sequencing of an accessory gene regulator (agr) in Staphylococcus aureus. J Bacteriol. 1988 Sep;170(9):4365–4372. doi: 10.1128/jb.170.9.4365-4372.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pier G. B. Pulmonary disease associated with Pseudomonas aeruginosa in cystic fibrosis: current status of the host-bacterium interaction. J Infect Dis. 1985 Apr;151(4):575–580. doi: 10.1093/infdis/151.4.575. [DOI] [PubMed] [Google Scholar]
- Plasterk R. H., Simon M. I., Barbour A. G. Transposition of structural genes to an expression sequence on a linear plasmid causes antigenic variation in the bacterium Borrelia hermsii. Nature. 1985 Nov 21;318(6043):257–263. doi: 10.1038/318257a0. [DOI] [PubMed] [Google Scholar]
- Pon C. L., Calogero R. A., Gualerzi C. O. Identification, cloning, nucleotide sequence and chromosomal map location of hns, the structural gene for Escherichia coli DNA-binding protein H-NS. Mol Gen Genet. 1988 May;212(2):199–202. doi: 10.1007/BF00334684. [DOI] [PubMed] [Google Scholar]
- Pustell J. M. Interactive molecular biology computing. Nucleic Acids Res. 1988 Mar 11;16(5):1813–1820. doi: 10.1093/nar/16.5.1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronson C. W., Nixon B. T., Ausubel F. M. Conserved domains in bacterial regulatory proteins that respond to environmental stimuli. Cell. 1987 Jun 5;49(5):579–581. doi: 10.1016/0092-8674(87)90530-7. [DOI] [PubMed] [Google Scholar]
- Rouvière-Yaniv J., Gros F. Characterization of a novel, low-molecular-weight DNA-binding protein from Escherichia coli. Proc Natl Acad Sci U S A. 1975 Sep;72(9):3428–3432. doi: 10.1073/pnas.72.9.3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert H. S., So M. Genetic mechanisms of bacterial antigenic variation. Microbiol Rev. 1988 Sep;52(3):327–336. doi: 10.1128/mr.52.3.327-336.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman M., Simon M. Phase variation: genetic analysis of switching mutants. Cell. 1980 Apr;19(4):845–854. doi: 10.1016/0092-8674(80)90075-6. [DOI] [PubMed] [Google Scholar]
- Stern A., Brown M., Nickel P., Meyer T. F. Opacity genes in Neisseria gonorrhoeae: control of phase and antigenic variation. Cell. 1986 Oct 10;47(1):61–71. doi: 10.1016/0092-8674(86)90366-1. [DOI] [PubMed] [Google Scholar]
- Stibitz S., Aaronson W., Monack D., Falkow S. Phase variation in Bordetella pertussis by frameshift mutation in a gene for a novel two-component system. Nature. 1989 Mar 16;338(6212):266–269. doi: 10.1038/338266a0. [DOI] [PubMed] [Google Scholar]
- Suzuki M. SPKK, a new nucleic acid-binding unit of protein found in histone. EMBO J. 1989 Mar;8(3):797–804. doi: 10.1002/j.1460-2075.1989.tb03440.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka I., Appelt K., Dijk J., White S. W., Wilson K. S. 3-A resolution structure of a protein with histone-like properties in prokaryotes. Nature. 1984 Aug 2;310(5976):376–381. doi: 10.1038/310376a0. [DOI] [PubMed] [Google Scholar]
- Watanabe F. Cooperative interaction of histone H1 with DNA. Nucleic Acids Res. 1986 Apr 25;14(8):3573–3585. doi: 10.1093/nar/14.8.3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiser J. N., Love J. M., Moxon E. R. The molecular mechanism of phase variation of H. influenzae lipopolysaccharide. Cell. 1989 Nov 17;59(4):657–665. doi: 10.1016/0092-8674(89)90011-1. [DOI] [PubMed] [Google Scholar]
- West S. E., Iglewski B. H. Codon usage in Pseudomonas aeruginosa. Nucleic Acids Res. 1988 Oct 11;16(19):9323–9335. doi: 10.1093/nar/16.19.9323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winans S. C., Ebert P. R., Stachel S. E., Gordon M. P., Nester E. W. A gene essential for Agrobacterium virulence is homologous to a family of positive regulatory loci. Proc Natl Acad Sci U S A. 1986 Nov;83(21):8278–8282. doi: 10.1073/pnas.83.21.8278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods J. P., Spinola S. M., Strobel S. M., Cannon J. G. Conserved lipoprotein H.8 of pathogenic Neisseria consists entirely of pentapeptide repeats. Mol Microbiol. 1989 Jan;3(1):43–48. doi: 10.1111/j.1365-2958.1989.tb00102.x. [DOI] [PubMed] [Google Scholar]